Abstract
Drug resistance of cancer stem/initiating cells has been considered to be one of the main reasons for tumor relapse. However, knowledge concerning the changes in stem/ initiating cells during chemotherapy is limited. In the present study, the breast cancer cell line MDA-MB-468 was cultured with 5-fluorouracil and serially passaged. Six cell generations were collected. Semi-quantitative RT-PCR and flow cytometric techniques were used to evaluate the protein and mRNA expression of stem/initiating factors (CD44+/CD24−, Oct 3/4, SOX2 and β-catenin), drug-resistance genes (BCRP and MRP1) and an anti-apoptosis gene (survivin). The clone formation rate was also examined in every generation of cells. The results showed that, under conditions of persistent chemotherapy, the factors representing the quantity of stem/initiating cells (β-catenin, Oct 3/4 and SOX2) followed a fluctuating trend of decrease-increase-further increase-decrease-increase-decrease, and factors representing the proportion of stem/initiating cells (proportion of CD44+/CD24− and the clone formation rate) demonstrated a fluctuating trend of increase-further increase-further increase-decrease. The drug-resistance genes (BCRP and MRP1) and the anti-apoptosis gene (survivin) demonstrated a wave of increase-further increase-further increase-decrease-increase (MRP1 decrease)-decrease. β-catenin, Oct 3/4 and SOX2 showed a positive correlation (r=1, p<0.01). Our study confirmed that the drug resistance of cancer cells is mainly due to tumor stem/initiating cells, and that under conditions of persistent chemotherapy, the quantity or function of breast cancer stem/initiating cells increases and decreases alternately.
Keywords: breast cancer, stem cell, 5-fluorouracil, drug resistance, anti-apoptosis
Introduction
Currently, comprehensive therapy, including surgery, chemotherapy and radiotherapy, is the main method for treating breast cancer, but the curative ratio is low due to frequent relapse. The resistance of cancer cells to chemotherapeutic drugs is thought to be the principal cause for relapse. Numerous theories have attempted to explain this phenomenon, and recently the cancer stem/initiating cell (CSC) theory has attracted much interest.
In 2006, to evaluate the chemosensitivity of glioblastoma multiforme (GBM) stem cells, Eramo et al (1) treated stem cell clones derived from different GBM patients with eight chemotherapeutic agents and assessed the rate of cell death in comparison to chemosensitive Jurkat leukemic cells and primary immature erythroblast. They observed a marked resistance of GBM stem cells to all the compounds used, whereas both Jurkat cells and erythroblasts displayed high rates of cell death. After treatment with chemotherapeutic agents, GBM stem cells were able to recover and proliferate. In a study by Phillips et al, CD24(−/low)/CD44(+) cancer-initiating cells were isolated from MCF-7 and MDA-MB-231 breast cancer cell lines, and their response to radiation was investigated. The results showed that the cancer-initiating cells were more resistant to radiation than the unisolated cells (2). In another study which included primary breast cancer patients (n=108) treated with neoadjuvant chemotherapy consisting of sequential paclitaxel and epirubicin-based chemotherapy, breast cancer stem cells were identified by immunohistochemical staining of aldehyde dehydrogenase 1 (ALDH1). ALDH1-positive tumors were significantly associated with a low rate of complete response (3). Similar results steadily emerged, urging researchers to relate drug-resistance to cancer stem/initiating cells (4).
CSCs which are not killed by chemotherapy, may become the cause of relapse. However, knowledge concerning the changes manifested in CSCs which are resistent to chemotherapy is limited. Answers to questions concerned with the effects of traditional chemotherapy drugs on CSC, the changes in CSCs treated by chemotherapy, the relationship between CSCs and drug-resistance proteins, and the relationship between CSCs and anti-apoptosis genes may help to understand the mechanism of CSC drug-resistance and to identify more effective methods of therapy which eventually kill CSCs.
In the present study, the MDA-MB-468 breast cancer cell line was cultured and different passages were treated continually with the traditional chemotherapy drug 5-fluorouracil (5-FU). For every cell generation, certain markers of CSCs, drug-resistance proteins and the anti-apoptosis gene were examined and compared to find a clue with which to resolve the abovementioned questions. The CSC factors included β-catenin, Octamer binding factor 3/4 (Oct 3/4), SRY-box 2 (SOX2) and CD44+/CD24− cells. The markers involved with drug-resistance proteins and anti-apoptosis genes were multidrug resistance protein 1 (MRP1), breast cancer resistance protein (BCRP) and survivin, respectively. Our results suggest that the drug-resistance of cancer cells is mainly due to CSCs, and chemotherapy leads to an alteration in the quantity or function of breast CSCs demonstrating an alternate increase and decrease.
Materials and methods
Cell culture and chemotherapy
The MDA-MB-468 breast cancer cell line was a gift from the Chinese-United Kingdom Medical Laboratory. The cells were cultured at 37°C with 5% CO2 in DMEM (HyClone, USA) and supplemented with 10% fetal calf serum (HyClone), 100 U/ml penicillin G and 100 μg/ml streptomycin (Gibco, USA). The cells were cultured in 75 cm2 flasks, and a 0.1 μg/ml (concentration data were obtained from a preliminary experiment) terminal concentration of 5-FU was added to the nutrient fluid when cells grew to a 60–70% base area. Cell cultures were harvested using 0.05% trypsin/EDTA (Gibco) in PBS and subcultured until achieving a 80–90% base area. The second generation was cultured, treated with chemotherapy and passaged in the same manner. Thus, six generations of cells were cultured. The cells in every generation were harvested and used in corresponding procedures in the subsequent experiments. Three independent experiments were performed. The control group was not treated with chemotherapy.
RNA extraction and semi-quantitative RT-PCR assay
Cells were harvested and stored at −70°C in TRIzol® (Invitrogen, USA). Total RNA was isolated by extracting the TRIzol samples with chloroform followed by separation and purification according to the manufacturer’s instructions. RNase inhibitor mix (1 μl) was added per 50 μl total RNA prior to treatment with DNase (Promega, USA) for 20 min. The RNA concentration and purity were quality controlled by analysis using a spectrophotometer (U-3010, Japan). First-strand cDNA was synthesized from the total RNA (3 μg) using the First-Strand Synthesis system (Promega) and used as a template for PCR amplification with Taq polymerase (Qiagen, Germany). Specific primer sequences are listed in Table I. PCR conditions included an initial denaturation at 94°C for 5 min, and 35 cycles at 94°C for 30 sec, annealing (Table I) for 45 sec, 72°C for 45 sec and a final extension step at 72°C for 5 min. RT-PCR products were separated by 1.2% agarose gel electrophoresis in a 0.5% Tris-acetate buffer and stained with ethidium bromide. The bands were photographed using a digital camera (Canon, Japan), and the absorbance of every band was calculated using Software Quantity One (Bio-Rad, USA). The absorbances of the inspected bands were divided by the absorbance of β-actin in the same sample to obtain the relative value of the marker in the sample. At least six independent experiments were performed.
Table I.
Information regarding the PCR primers.
| Primer | NCBI ID | Primer sequence | Product size (bp) | Melting temperature (°C) |
|---|---|---|---|---|
| β-catenin | NM_001098209.1 | F: CCCACTAATGTCCAGCGTTT | 382 | 54.0 |
| R: AACCAAGCATTTTCACCAGG | ||||
| Oct 3/4 | NM_002701.4 | F: TTCAGCCAAACGACCATC | 484 | 57.4 |
| R: GGAAAGGGACCGAGGAGTA | ||||
| SOX2 | NM_003106.2 | F: ACACCAATCCCATCCACACT | 223 | 61.0 |
| R: CAAACTTCCTGCAAAGCTCC | ||||
| MRP1 | NM_019898.2 | F: CTGAGTTCCTGCGTACCTAT | 212 | 56.2 |
| R: TTCTGCGGTGCTGTTGT | ||||
| BCRP | NM_004827.2 | F: GCCATTCTCCCAGTCA | 494 | 49.7 |
| R: GGGCGTCTATACACCAT | ||||
| Survivin | NM_001012271.1 | F: CTTGCCAGAGCCACGAA | 632 | 55.4 |
| R: GGAACCTCACCCATAGCC | ||||
| β-actin | NM_001101.3 | F: GCCCTGAGGCACTCTTC | 330 | 54.9 |
| R: GGCCGGACTCGTCATAC |
F, forward; R, reverse.
Flow cytometry
To identify the CD44+/CD24− cell proportion in every generation, cells were harvested with 0.05% trypsin/ EDTA (Gibco) and then suspended (2×106 cells/100 μl) in Stain Buffer containing 1% FBS (EBioscience, USA). Phycoerythrin (PE)-conjugated mouse against human CD44 monoclonal antibody (EBioscience) and fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD24 monoclonal antibody (EBioscience) were added to the cell suspension at the concentrations recommended by the manufacturer, and incubation was carried out at 4°C in the dark for 60 min. Proper isotype controls were used for each cell labeling experiment. The labeled cells were fixed in 100% methanol on ice for 5 min. Flow cytometric analysis was performed in triplicate using a flow cytometer (Partec, Germany). Four generations were assessed.
Colony formation experiment
Single cells were prepared in each generation and seeded in culture dishes (60 mm in diameter); each dish initially had 10×103 cells. The cells were cultured in the same culture medium as the former (but without 5-FU) for 2–3 weeks and fixed with methanol when colonies were visible by the naked eye. The colonies were stained for 2–3 min with haematoxylin and those that consisted of ≥50 cells were counted under a microscope. Four generations were assessed. At least six independent experiments were performed.
Statistical analysis
Data are expressed as the means ± standard deviation (SD). The statistical significance of differences between groups was analyzed by one-way analysis of variance, and the relationship between variables was calculated by two-tailed Pearson correlation analysis using the Statistical Product and Service Solutions version 13.0 program for Windows (SPSS, USA). Differences with p-values <0.05 were considered significant.
Results
mRNA expression of CSC factors, drug-resistance genes and an anti-apoptosis gene
In the experimental (treated with 5-FU) and control groups (without 5-FU), the mRNA expression of the markers, β-catenin, Oct 3/4, SOX2, MRP1, BCRP, survivin and actin, in six cell generations was investigated.
In the control group, each marker exhibited no differences among the cell generations (p>0.05) (Fig. 1). In the experimental group, the markers fluctuated in the different cell generations (p<0.05) (Figs. 2 and 3). All markers selected in the present study showed a similar wave of increases and decreases (Fig. 3) The factors related to CSCs (β-catenin, Oct 3/4 and SOX2) exhibited a fluctuating trend of increase-increase-further increase-decrease-increase-decrease. The drug-resistance genes (BCRP and MRP) and the anti-apoptosis gene (survivin) exhibited waves of increase-further increase-further increase-decrease-increase (MRP1 decrease)-decrease.
Figure 1.
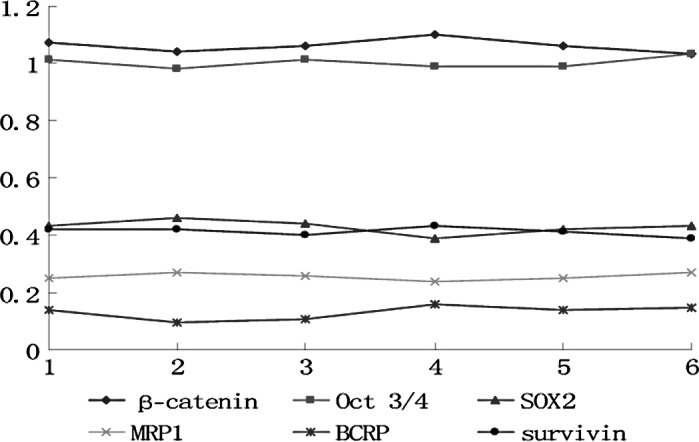
Trend of mRNA expression of the relative markers in each cell generation in the control group. The x-axis indicates cell generation, whereas the y-axis indicates the relative value of mRNA expression.
Figure 2.
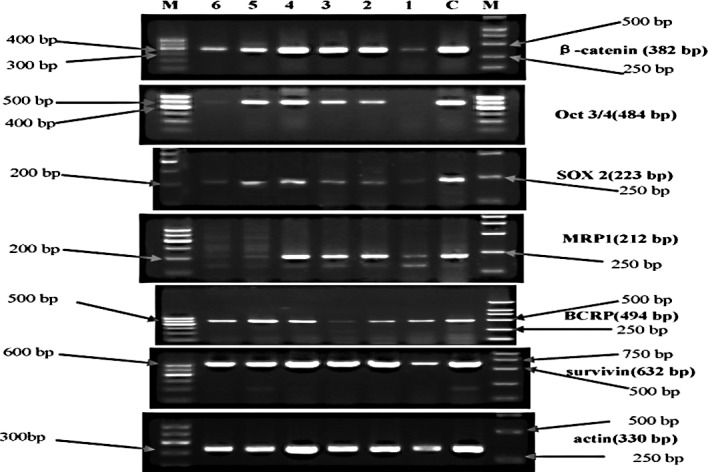
Electrophorogram of mRNA expression of the relative markers in each cell generation in the experimental group. M, molecular weight marker; the left marker is the 100-bp ladder, the right marker is DL2000 (apart from Oct 3/4, whose marker was the 100-bp ladder). C, control group. Lanes 1, 2, 3, 4, 5 and 6 represent the first, second, third, fourth, fifth and sixth cell generation, respectively.
Figure 3.
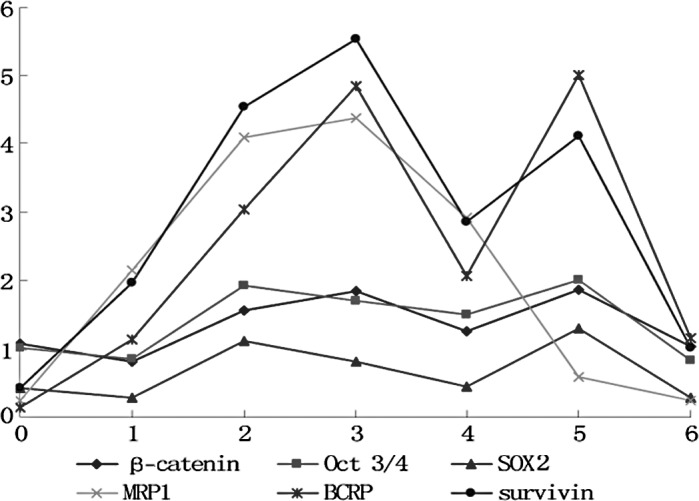
Trend of mRNA expression of the relative markers in each cell generation in the experimental group. The x-axis indicates the cell generation, whereas the y-axis indicates the relative value of mRNA expression.
The correlationship among the markers was also analyzed and most exhibited a positive correlation (Table II).
Table II.
Correlation among the stem/initiating-related factors, chemoresistance and anti-apoptotic factors.
| Factors | β-catenin | Oct 3/4 | SOX2 | MRP1 | BCRP | Survivin | |
|---|---|---|---|---|---|---|---|
| β-catenin | r-value | 1 | 0.922 | 0.882 | 0.389 | 0.936 | 0.859 |
| p-value | 0.003 | 0.009 | 0.389 | 0.002 | 0.013 | ||
| Oct 3/4 | r-value | 0.922 | 1 | 0.933 | 0.486 | 0.861 | 0.860 |
| p-value | 0.003 | 0.002 | 0.269 | 0.013 | 0.013 | ||
| SOX2 | r-value | 0.882 | 0.933 | 1 | 0.279 | 0.824 | 0.759 |
| p-value | 0.009 | 0.002 | 0.544 | 0.023 | 0.048 | ||
| MRP1 | r-value | 0.389 | 0.486 | 0.279 | 1 | 0.436 | 0.753 |
| p-value | 0.389 | 0.269 | 0.544 | 0.329 | 0.051 | ||
| BCRP | r-value | 0.936 | 0.861 | 0.824 | 0.436 | 1 | 0.917 |
| p-value | 0.002 | 0.013 | 0.023 | 0.329 | 0.004 | ||
| Survivin | r-value | 0.859 | 0.860 | 0.759 | 0.753 | 0.917 | 1 |
| p-value | 0.013 | 0.013 | 0.048 | 0.051 | 0.004 |
Proportion of CD44+/CD24− cells
The proportion of CD44+/ CD24− cells in the experimental group also exhibited a trend of decrease and increase in each cell generation (p<0.05) (Fig. 4). The proportion of CD44+/CD24− cells in each cell generation in the control group showed no difference among the different cell generations (p>0.05; data not shown).
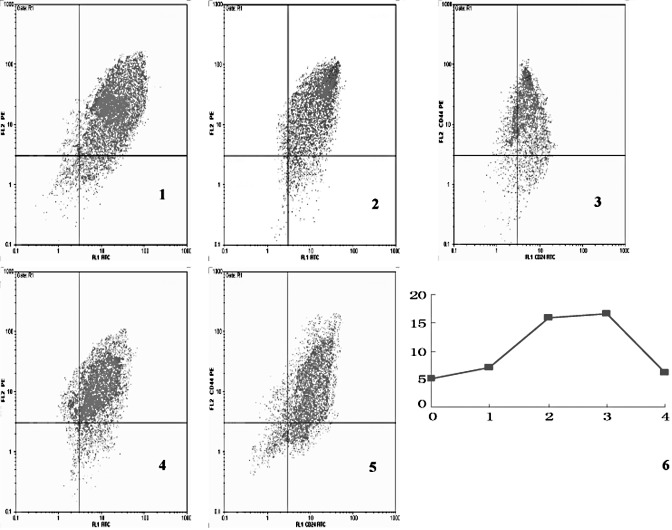
Figure 4.
Proportion of CD44+/CD24− cells in the experimental group. Images 1, 2, 3, 4 and 5 are the proportion of CD44+/CD24− cells in the non-treated group, the first, second, third and fourth generation of cells, respectively. The CD44+/CD24− cells were located in the upper left quadrant. Image 6 is a line graph showing the fluctuation in the proportion of CD44+/ CD24− cells in different cell generations. The x-axis indicates the cell generation, whereas the y-axis denotes the percentage.
The proportion of CD44+/CD24− cells in the experimental group showed a positive correlation with other markers (Table III).
Table III.
Correlation between the proportion of CD44+/CD24− cells and the other factors.
| β-catenin | Oct 3/4 | SOX2 | MRP1 | BCRP | Survivin | |
|---|---|---|---|---|---|---|
| Proportion of CD44+/CD24− | ||||||
| r-value | 0.857 | 0.804 | 0.889 | 0.846 | 0.891 | 0.921 |
| p-value | 0.063 | 0.101 | 0.043 | 0.071 | 0.042 | 0.026 |
Colony formation
In the different cell generations, the colonies formed per 104 seeded cells varied (p<0.05) (Fig. 5). The decrease and increase fluctuation was very similar to that shown in Fig. 4. The number of colonies formed in the experimental group exhibited a positive correlation with several of the other markers (Table IV).
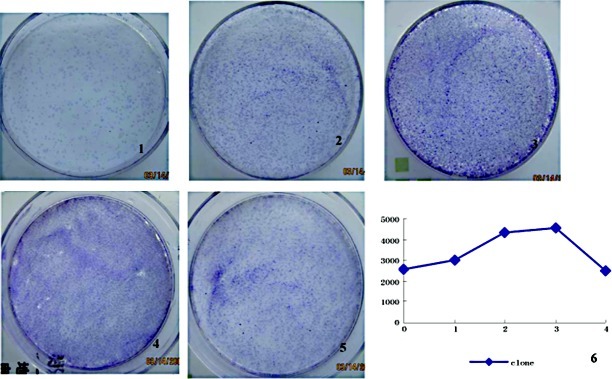
Figure 5.
Colony formation in the experimental group. Images 1, 2, 3, 4 and 5 show the colony formation in the non-treated group, and first, second, third, fourth generation cells, respectively. Image 6 is a line graph showing fluctuations in the colony forming rate (colonies/104 seeding cells) in the different cell generations. The x-axis indicates the cell generation, whereas the y-xis denotes the quantity of colonies.
Table IV.
Correlation between the rate of clone formation and the other stem cell-related factors.
| β-catenin | Oct 3/4 | SOX2 | MRP1 | BCRP | Survivin | CD44+/CD24− | |
|---|---|---|---|---|---|---|---|
| Clone formation | |||||||
| r-value | 0.811 | 0.722 | 0.850 | 0.792 | 0.858 | 0.880 | 0.992 |
| p-value | 0.096 | 0.168 | 0.068 | 0.110 | 0.063 | 0.049 | 0.001 |
Discussion
In the present study, three factors were selected to represent the existence or quantity/functional change in CSCs: mRNA expression of β-catenin, Oct 3/4 and SOX2. β-catenin is the key component in the Wnt/β-catenin signaling pathway. The Wnt/β-catenin signaling pathway plays an important role in the differentiation and proliferation of stem cells (5). Experiments have confirmed that activation of the Wnt/β-catenin signaling pathway maintains stem cell renewal capacity and restrains stem cell differentiation in normal tissue (6–9), whereas it prompts tumor stem cell amplification in tumor tissue (9). Oct 3/4 is an embryo-specific transcription factor, which maintains totipotency (10) of the embryo stem cells and participates in the renewal of stem cells (11). Oct 3/4 is thought to be one of the markers of totipotency or stem cells. Its expression was found to be extremely low or absent in differentiated cells, but was increased in normal stem and cancer stem cells (12,13). SOX2 was found to be expressed in totipotency cells and early multipotential cells in the embryo, regulating the self-renewal process (14,15) of embryo stem cells. SOX2 has been associated with the maintainence of cell pluripotency (16,17) and is also considered as a marker of stem cells (18). In light of the above findings, the mRNA expression of β-catenin, Oct 3/4 and SOX2 may be considered to represent the quantity of activated stem/initiating cells.
The other two markers of stem/initiating cells are CD44+/ CD24− cells and the rate of colony formation. According to a study by Al-Hajj et al (19) as well as other previous studies (20–23), CD44+/CD24− cells in breast cancer tissue exhibit certain stem cell-like properties, such as self-renewal and multi-directional differentiation, therefore they are considered to be breast cancer stem/initiating cells. The capacity to form colonies reflects renewal ability, so under equivalent seeding conditions, the rate of colony formation reflects the proportion of stem/initiating cells in the primitive seeded cells. In the present study, these two markers represented the proportion of stem/initiating cells.
In the experimental group, the mRNA expression of the above markers in the six generations exhibited an evident fluctuation with statistical significance, whereas the mRNA expression in the control group did not fluctuate. The results indicate that chemotherapy surely affects the quantity or function of CSCs.
The mRNA expression of three markers representing the quantity of CSCs (β-catenin, Oct 3/4 and SOX2) exhibited the following trend of fluctuation: decrease-increase-further increase-decrease-increase-decrease (1–6 generation). The two markers representing the proportion of CSCs exhibited the following fluctuating trend: increase-further increase-further increase-decrease (1–4 generation). In the first cell generation, the quantity of CSCs decreased slightly since some of them were killed by 5-FU (even though they had drug-resistance properties). More non-stem/initiating cells were killed compared to CSCs, therefore the proportion of CSCs increased in the first generation cells. CSCs are activated and proliferate through stimulation of 5-FU, a decrease in cell number, and factors released from dead cells. Thus, the quantity and proportion of CSCs increased significantly in the second and third generations. In the fourth generation, most of the CSCs differentiated to relatively mature tumor cells, leading to the quantity and proportion of CSCs to decrease. The differentiated tumor cells had no or low drug-resistance, so a large portion of them were killed and the CSCs were again activated and proliferated, leading to an increase in the quantity and proportion of CSCs in the fifth generation cells. The situation in the sixth generation was similar to the fourth generation, reflecting the alteration resulting from CSC differentiation.
The results indicate that the quantity and proportion of CSCs increase and decrease in turn in different cell generations under a condition of persistent chemotherapy. This finding has not been previously reported.
The fact that the mRNA expression of β-catenin, Oct 3/4 and SOX2 exhibits a nearly perfect positive correlation (p<0.01) suggests that they come from the same population of cells, proving their reliability to represent CSCs. The percentage of CD44+/CD24− cells exhibited nearly a perfect positive correlation (r=0.99, p<0.01) with the rate of colony formation, confirming their reliability as indicators of the proportion of CSCs.
Another key point in the present study is the drug-resistance of CSC. The MRP1 gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. This full transporter is a member of the MRP subfamily which is involved in multi-drug resistance (24–27). MRP proteins markedly increase in side population (SP) cells (28–30), in which CSCs are rich. The BCRP gene is also included in the superfamily of ABC transporters. BCRP has been proven to be involved in the multi-drug resistance of many types of cancers (particularly breast cancer) (31–33) and in CSCs (24–27).
mRNA expression of BCRP fluctuated along with an increase and decrease in the mRNA expression of β-catenin, Oct 3/4 and SOX2 in the second, third, fourth, fifth and sixth generation, showing a positive correlation between them. The results suggest that BCRP expression is a property of CSCs, and there is low BCRP expression in differentiated cells. This finding is consistent with that of Doyle et al (32), who argued that the number of SP cells (abundant in CSCs) in MCF-7/AdrVp (a multidrug-resistant human breast cancer subline) was higher than in parental MCF-7 cells, and BCRP was overexpressed in the former. A similar study (34) showed that CD133-positive cells (assumed markers of glioblastoma cancer stem cells) isolated from human glioblastoma expressed higher levels of BCRP1 mRNA. MRP1 did not demonstrate a positive correlation with CSC-related factors; thus, MRP1 may be expressed in both CSCs and differentiated cells.
Survivin is a member of the inhibitor of apoptosis gene family, which encodes negative regulatory proteins that prevent apoptotic cell death (35,36). In the present study, the expression of survivin mRNA was positivlye correlated with factors of CSCs, suggesting that the expression of survivin is associated with the drug-resistance of CSCs. In fact, overexpression of anti-apoptosis genes in cancer stem cells has been previously reported (34,37,38). ‘Survivor cells’ surviving chemotherapy in acute myelocytic leukemia, for example, were found to be enriched in CD34+ stem cells and accompanied by overexpression of many anti-apoptosis genes. BCRP may cooperate with survivin to maintain CSC survival.
In summary, the overexpression of drug-resistance genes and anti-apoptosis genes in CSCs is an important cause of traditional chemotherapy failure and cancer relapse. To cure cancer, effective ways to eliminate CSCs must be developed.
References
- 1.Eramo A, Ricci-Vitiani L, Zeuner A, et al. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13:1238–1241. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 2.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 3.Tanei T, Morimoto K, Shimazu K, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 4.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 5.Mimeault M, Hauke R, Mehta PP, Batra SK. Recent advances in cancer stem/progenitor cell research: therapeutic implications for overcoming resistance to the most aggressive cancers. J Cell Mol Med. 2007;11:981–1011. doi: 10.1111/j.1582-4934.2007.00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reguart N, He B, Taron M, You L, Jablons DM, Rosell R. The role of Wnt signaling in cancer and stem cells. Future Oncol. 2005;1:787–797. doi: 10.2217/14796694.1.6.787. [DOI] [PubMed] [Google Scholar]
- 7.Mishra L, Shetty K, Tang Y, Stuart A, Byers SW. The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene. 2005;24:5775–5789. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- 8.He B, Barg RN, You L, et al. Wnt signaling in stem cells and non-small-cell lung cancer. Clin Lung Cancer. 2005;7:54–60. doi: 10.3816/CLC.2005.n.022. [DOI] [PubMed] [Google Scholar]
- 9.Kruger JA, Kaplan CD, Luo Y, et al. Characterization of stem cell-like cancer cells in immune-competent mice. Blood. 2006;108:3906–3912. doi: 10.1182/blood-2006-05-024687. [DOI] [PubMed] [Google Scholar]
- 10.Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 12.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 13.Hubner K, Fuhrmann G, Christenson LK, et al. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- 14.Chew JL, Loh YH, Zhang W, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 16.Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J Biol Chem. 2005;280:24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
- 17.Masui S, Nakatake Y, Toyooka Y, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 18.Ginis I, Luo Y, Miura T, et al. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 21.Balic M, Lin H, Young L, et al. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12:5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 22.Mylona E, Giannopoulou I, Fasomytakis E, et al. The clinicopathologic and prognostic significance of CD44+/CD24(-/low) and CD44−/CD24+ tumor cells in invasive breast carcinomas. Hum Pathol. 2008;39:1096–1102. doi: 10.1016/j.humpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stride BD, Valdimarsson G, Gerlach JH, Wilson GM, Cole SP, Deeley RG. Structure and expression of the messenger RNA encoding the murine multidrug resistance protein, an ATP-binding cassette transporter. Mol Pharmacol. 1996;49:962–971. [PubMed] [Google Scholar]
- 25.D’Hondt V, Caruso M, Bank A. Retrovirus-mediated gene transfer of the multidrug resistance-associated protein (MRP) cDNA protects cells from chemotherapeutic agents. Hum Gene Ther. 1997;8:1745–1751. doi: 10.1089/hum.1997.8.15-1745. [DOI] [PubMed] [Google Scholar]
- 26.Lautier D, Canitrot Y, Deeley RG, Cole SP. Multidrug resistance mediated by the multidrug resistance protein (MRP) gene. Biochem Pharmacol. 1996;52:967–977. doi: 10.1016/0006-2952(96)00450-9. [DOI] [PubMed] [Google Scholar]
- 27.Abaan OD, Mutlu PK, Baran Y, Atalay C, Gunduz U. Multidrug resistance mediated by MRP1 gene overexpression in breast cancer patients. Cancer Invest. 2009;27:201–205. doi: 10.1080/07357900802173562. [DOI] [PubMed] [Google Scholar]
- 28.Dean M. ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia. 2009;14:3–9. doi: 10.1007/s10911-009-9109-9. [DOI] [PubMed] [Google Scholar]
- 29.Chuthapisith S, Eremin J, El-Sheemey M, Eremin O. Breast cancer chemoresistance: emerging importance of cancer stem cells. Surg Oncol. 2010;19:27–32. doi: 10.1016/j.suronc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Dave B, Chang J. Treatment resistance in stem cells and breast cancer. J Mammary Gland Biol Neoplasia. 2009;14:79–82. doi: 10.1007/s10911-009-9117-9. [DOI] [PubMed] [Google Scholar]
- 31.Allikmets R, Schriml LM, Hutchinson A, Romano-Spica V, Dean M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998;58:5337–5339. [PubMed] [Google Scholar]
- 32.Doyle LA, Yang W, Abruzzo LV, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross DD, Yang W, Abruzzo LV, et al. Atypical multidrug resistance: breast cancer resistance protein messenger RNA expression in mitoxantrone-selected cell lines. J Natl Cancer Inst. 1999;91:429–433. doi: 10.1093/jnci/91.5.429. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan S, Aspe JR, Asumen MG, et al. Extracellular, cell-permeable survivin inhibits apoptosis while promoting proliferative and metastatic potential. Br J Cancer. 2009;100:1073–1086. doi: 10.1038/sj.bjc.6604978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- 37.Wang J, Guo LP, Chen LZ, Zeng YX, Lu SH. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;67:3716–3724. doi: 10.1158/0008-5472.CAN-06-4343. [DOI] [PubMed] [Google Scholar]
- 38.Kornblau SM, Qiu YH, Bekele BN, et al. Studying the right cell in acute myelogenous leukemia: dynamic changes of apoptosis and signal transduction pathway protein expression in chemotherapy resistant ex-vivo selected ‘survivor cells’. Cell Cycle. 2006;5:2769–2777. doi: 10.4161/cc.5.23.3507. [DOI] [PubMed] [Google Scholar]