Abstract
Although metastasis or relapse is a leading cause of death for patients with gastric cancer, the hematogenous spread of cancer cells remains undetected at the time of initial therapy. The development of novel diagnostic molecular marker(s) to detect circulating gastric cancer cells is an issue of great clinical importance. We obtained peripheral blood samples from 10 patients with gastric cancer who underwent laparotomy and 4 healthy volunteers. Microarray analysis consisting of 30,000 genes or ESTs was carried out using eight gastric cancer tissues and normal gastric mucosae. We selected 53 genes up-regulated in gastric cancer compared to normal gastric mucosae from our microarray data set, and, among these, identified five candidate marker genes (TSPAN8, EPCAM, MMP12, MMP7 and REG3A) which were not expressed in peripheral blood mononuclear cells (PBMCs) from 4 healthy volunteers. We further carried out semi-quantitative nested reverse transcription-polymerase chain reaction (RT-PCR) for HRH1, EGFR, CK20 and CEA in addition to the five newly identified genes using PBMCs of patients with gastric cancer, and found that expression of one or more genes out of the nine was detected in 80% of the patients with gastric cancer. Moreover, the numbers of genes expressed in PBMCs were ≤2 and ≥2 in all vascular invasion-negative cases and in 5 of 6 positive cases, respectively, showing significant differences between the two groups (P=0.041). Nested RT-PCR analysis for the set of nine marker genes using PBMCs may provide the potential for detection of circulating gastric cancer cells prior to metastasis formation in other organs.
Keywords: microarray, gastric cancer, molecular marker, nested RT-PCR, peripheral blood
Introduction
Gastric cancer causes approximately 800,000 deaths worldwide per year and is still one of the leading causes of cancer-related death in the world (1). Most gastric cancers at an early stage can be cured by surgical resection; however, patients with advanced gastric cancers have worse prognosis than those with early stage disease (2). Although metastasis or relapse is the main cause of death for patients with gastric cancer (3), the hematogenous spread of malignant cells remains undetected at the time of initial therapy. During the development of cancer, tumor cells may detach from the primary tumor and disseminate into the lymph system and/or blood circulation, and grow in the bone marrow, liver, kidney and other organs, which is called micrometastasis (4). Micrometastasis is barely detected by routine biochemical and histopathological assays or graphical methods, such as X-ray, CT and MRI (3). Detection of circulating tumor cells at the mRNA level [reverse transcription-polymerase chain reaction (RT-PCR)] in blood samples of patients with cancer could serve as a unique and easy diagnostic tool to predict cancer recurrence and to monitor treatment effectiveness (5–7). However, molecular marker(s) that detect circulating gastric cancer cells for routine clinical use have not yet been identified. Hence, the development of novel diagnostic molecular marker(s) to detect circulating gastric cancer cells is an issue of great clinical importance.
Carcinoembryonic antigen (CEA) is a well-known tumor marker and has been used to detect small amounts of adenocarcinoma cells in the blood, peritoneal wash or other body fluids (8–12). However, the expression of CEA mRNA is not specific to cancer cells and often produces false-positive results (13). Profiling of gene expression patterns on genome-wide microarrays enables investigators to perform comprehensive characterization of molecular activities in cancer cells (14–17). Systematic analysis of expression levels for thousands of genes is also a useful approach for identifying molecular markers to detect small amounts of circulating cancer cells (18). In this study, we identified genes whose expression had been altered during gastric carcinogenesis using genome-wide information obtained from 8 cases on a microarray consisting of 30,000 transcribed elements. Based on the results of the microarray assay, we identified five candidate genes for the specific detection of circulating gastric cancer cells at the mRNA level. We suggest that such information may lead ultimately to improve the prognosis of patients with gastric cancers.
Materials and methods
Blood and tissue samples
Blood samples were obtained from 10 patients with gastric cancer who underwent laparotomy and 4 healthy volunteers after obtaining informed consent. Heparinized blood samples (5 ml) from the 10 patients with gastric cancer were obtained from a peripheral artery through a catheter used for monitoring arterial blood pressure during surgical operation. Peripheral venous blood was obtained from 4 healthy volunteers for control after discarding the initial 10 ml of blood to protect the mixture from epithelial cells. Clinicopathological characteristics of the 10 patients are shown in Table I. Clinical stage of each patient was judged according to the Union for International Cancer Control (UICC) TNM classification. Among the 10 patients with gastric cancer, 8 primary gastric cancer tissues and corresponding non-cancerous gastric mucosae from surgically resected tissues were obtained at Sapporo Medical University and Douto Hospital after each patient had provided informed consent. The samples that had been confirmed histologically as gastric adenocarcinoma were used for microarray study. These samples were immediately frozen and stored at −80°C. All cancer tissues were obtained from the margin of the tumor mass, while non-cancerous tissues were obtained from corresponding normal mucosae of the same stomach. This study was approved by the Ethics Committee of Sapporo Medical University, School of Medicine, Hokkaido, Japan.
Table I.
Characteristics of patients included in the nested RT-PCR analysis of PBMCs.
| Parameters | No. of patients |
|---|---|
| Gender (male:female) | 5:5 |
| Age range (average), in years | 41–82 (61.9) |
| Depth of tumor invasion (T1:T2:T3:T4) | 1:6:2:1 |
| Lymph node metastasis (N0:N1:N2:N3) | 4:3:2:1 |
| Distant metastasis (M0:M1) | 10:0 |
| Liver metastasis (H0:H1) | 10:0 |
| Peritoneal metastasis (P0:P1) | 9:1 |
| Peritoneal lavage cytology (CY0:CY1) | 9:1 |
| Stage (I:II:III:IV) | 4:1:2:3 |
| Lymphatic invasion (ly0:ly1–3) | 2:8 |
| Vessel invasion (v0:v1–3) | 4:6 |
RNA extraction of blood samples
We prepared peripheral blood mononuclear cells (PBMCs) using Ficoll (Amersham Biosciences, Buckinghamshire, UK) and extracted total RNA using TRIzol (Invitrogen, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. Before the synthesis of cDNA, deoxyribonuclease I (DNase I) (Nippon Gene, Japan) was added to each sample of total RNA according to the manufacturer’s instructions.
Analysis of microarray
Total RNA was extracted from each gastric tissue using TRIzol according to the manufacturer’s instructions. To guarantee the quality of RNAs, total RNA extracted from the residual tissue of each case was electrophoresed on a denaturing agarose gel, and the quality was confirmed by the presence of rRNA bands. After treatment with DNase I, T7-based RNA amplification was carried out as described previously with a few modifications (19). Using 5 μg of total RNA from each tissue sample as starting material, one round of amplification was performed; the amount of each amplified RNA (aRNA) was measured by a spectrophotometer. A mixture of normal gastric mucosae from 8 patients was prepared as a universal control and was amplified in the same manner; 2.5 μg of aRNAs from each cancerous tissue and from the control was reversely transcribed in the presence of Cy5-dCTP and Cy3-dCTP, respectively (15). AceGene 30K-1 Chip Version (Hitachi Software Engineering Co., Japan) was used for microarray analysis. The procedures for hybridization, washing, photometric quantification of signal intensities of each spot and normalization of data were according to the manufacturer’s instructions. To normalize the amount of mRNA between tumors and controls, the fluorescence intensities of Cy5 (gastric cancer) and Cy3 (control) for each target spot were adjusted so that the mean Cy5/Cy3 ratio of 30,000 genes equaled 1. Genes were categorized into three groups according to the cancer/normal ratio of their mean signal intensity: up-regulated (expression ratio >5.0), down-regulated (expression ratio <0.2) and unchanged expression (expression ratio between 0.2 and 5.0).
Semi-quantitative RT-PCR
To validate the result of the microarray analysis, we examined the expression levels of the genes up-regulated in gastric cancer by means of semi-quantitative RT-PCR analysis. Total RNAs (3 μg) extracted from each cancerous tissue and normal gastric mucosa were reversely transcribed for single-stranded cDNAs using oligo(dT)12–18 primer with Superscript II reverse transcriptase (Life Technologies, Inc.). Each single-stranded cDNA was diluted for subsequent PCR amplification. A housekeeping gene, GAPDH, served as the internal control. The PCR reaction was conducted at 95°C for 5 min, and then for 30 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min followed by 72°C for 10 min, in the Gene Amp PCR System 9700 (Perkin-Elmer Applied Biosystems, Foster City, CA, USA).
Nested RT-PCR using blood samples
We performed nested RT-PCR using total RNAs extracted from PBMCs to accurately examine mRNA levels of the candidate marker genes. Initially, RT-PCR was carried out as described above. In nested RT-PCR, 1 ml of the initial PCR product, 4 ml of 10X PCR buffer, 200 mmol/l dNTP mixture, 0.2 mmol/l primers and 1 unit Taq DNA polymerase (Takara) were added to a 40-ml aliquot of the reaction mixture. The PCR reaction was conducted at 95°C for 5 min, and then 30 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min followed by 72°C for 10 min, in the Gene Amp PCR System 9700. The RT-PCR products were detected using 2% agarose gel electrophoresis. The primer sequences are summarized in Table II.
Table II.
Primer sequences for semi-quantitative nested RT-PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| TSPAN8 | 5′-TCAACTTCTTGTTCTGGCTATGT-3′ | 5′-TATAGCTTTGGCATGGTCTCTGC-3′ |
| EPCAM | 5′-TGATCCTGACTGCGATGAGAGC-3′ | 5′-CAGCTTTCAATCACAAATCAGT-3′ |
| MMP12 | 5′-AACCAGCTCTCTGTGACCCCA-3′ | 5′-TCCAAGGATGTTAGGAAGCAAC-3′ |
| MMP7 | 5′-TCTCTGGACGGCAGCTATGCG-3′ | 5′-AATAAGACACAGTCACACCATAA-3′ |
| REG3A | 5′-GTATCTTGGATGCTGCTTTCCTG-3′ | 5′-GTATGACAAAATGAAGAGACTGA-3′ |
| HRH1 | 5′-TACAAGGCCGTACGACAACACT-3′ | 5′-TCTGCTGTTCTTCTATGGTGCCT-3′ |
| EGFR | 5′-ATGTCCCCACGGTACTTACTCCC-3′ | 5′-TCTTAACAATGCTGTAGGGGCTC-3′ |
| CK20 | 5′-TGGATTTCAGTCGCAGA-3′ | 5′-ATGTAGGGTTAGGTCATCAAAG-3′ |
| CEA | 5′-TTCTCCTGGTCTCTCAGCTGGG-3′ | 5′-AATGCTTTAAGGAAGAAGCAA-3′ |
Results
Identification of up- or down-regulated genes in the gastric cancers
We extracted RNAs from eight primary gastric cancer tissues and corresponding normal gastric mucosae as control, and carried out gene expression analysis using a microarray consisting of 30,000 genes or ESTs. We then selected genes from our data set according to the criterion that the cancer/ normal ratio of the mean signal intensity of a given gene was >5.0 or <0.2, and 53 genes were identified as up-regulated and 123 genes as down-regulated in the gastric cancer tissues compared to the normal gastric mucosa (Tables III and IV). The up-regulated genes represented a variety of functions, including genes associated with signal-transduction pathways (SFRP4 and TSPAN8), genes encoding transcription factors (TRIM33), genes involved in various metabolic pathways (ADH4, USP33, RNF128, MAN2A1, UBD and GCNT3), apoptosis (SPP1 and RIPK2), chemokines (CCL20), DNA replication and recombination (SNRPA1), cell adhesion and cytoskeleton (LAMB3, EPCAM, MMP7 and COL1A1), cell-cell signaling (CEACAM6 and CXCL9), cell cycle (CDC2, BUB1 and CCNB2), cell proliferation (REG1B and REG3A), or other functions (SPINK4, TMC5, LGALS2, KYNU, DDX58, LY96, UMPS and RNF157).
Table III.
Genes up-regulated in advanced gastric cancer.
| No. | Accession no. | Gene symbol | Description |
|---|---|---|---|
| 1 | NM_006507 | REG1B | Regenerating islet-derived 1 β (pancreatic stone protein, pancreatic thread protein) |
| 2 | NM_004577 | PSPH | Phosphoserine phosphatase |
| 3 | NM_138938 | REG3A | Regenerating islet-derived 3 α |
| 4 | NM_014471 | SPINK4 | Serine peptidase inhibitor, Kazal type 4 |
| 5 | NM_001105249 | TMC5 | Transmembrane channel-like 5 |
| 6 | NM_002426 | MMP12 | Matrix metallopeptidase 12 (macrophage elastase) |
| 7 | NM_002423 | MMP7 | Matrix metallopeptidase 7 (matrilysin, uterine) |
| 8 | NM_002483 | CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| 9 | NM_001786 | CDC2 | Cell division cycle 2, G1 to S and G2 to M |
| 10 | NM_000582 | SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) |
| 11 | NM_004751 | GCNT3 | Glucosaminyl (N-acetyl) transferase 3, mucin type |
| 12 | NM_000574 | CD55 | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| 13 | NM_002443 | MSMB | Microseminoprotein, β |
| 14 | NM_004336 | BUB1 | UB1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| 15 | NM_015017 | USP33 | Ubiquitin-specific peptidase 33 |
| 16 | NM_004591 | CCL20 | Chemokine (C-C motif) ligand 20 |
| 17 | NM_017633 | FAM46A | Family with sequence similarity 46, member A |
| 18 | NM_000088 | COL1A1 | Collagen, type I, α 1 |
| 19 | XR_017717 | ADAMTSL3 | ADAMTS-like 3 |
| 20 | NM_138938 | REG3A | Regenerating islet-derived 3 α |
| 21 | NM_017934 | PHIP | Pleckstrin homology domain interacting protein |
| 22 | XR_016124 | Similar to p21-activated kinase 2 | |
| 23 | NM_006398 | UBD | Ubiquitin D |
| 24 | NM_002358 | MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) |
| 25 | NM_002483 | CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross-reacting antigen) |
| 26 | NM_173164 | IPO9 | Importin 9 |
| 27 | NM_003014 | SFRP4 | Secreted frizzled-related protein 4 |
| 28 | NM_004616 | TSPAN8 | Tetraspanin 8 |
| 29 | NM_002354 | EPCAM | Epithelial cell adhesion molecule |
| 30 | NM_006498 | LGALS2 | Lectin, galactoside-binding, soluble, 2 |
| 31 | NM_002372 | MAN2A1 | Mannosidase, α, class 2A, member 1 |
| 32 | NM_003937 | KYNU | Kynureninase (L-kynurenine hydrolase) |
| 33 | NM_003821 | RIPK2 | Receptor-interacting serine-threonine kinase 2 |
| 34 | NM_00108039 | ITGA7 | Integrin, α 7 |
| 35 | NM_000670 | ADH4 | Alcohol dehydrogenase 4 (class II), π polypeptide |
| 36 | NM_014314 | DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| 37 | NM_006418 | OLFM4 | Olfactomedin 4 |
| 38 | NM_198187.3 | ASTN2 | Astrotactin 2 |
| 39 | NM_015364 | LY96 | Lymphocyte antigen 96 |
| 40 | NM_000574 | CD55 | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| 41 | NM_018964 | SLC37A1 | Solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| 42 | NM_018455 | CENPN | Centromere protein N |
| 43 | NM_001710 | CFB | Complement factor B |
| 44 | NM_033020 | TRIM33 | Tripartite motif-containing 33 |
| 45 | NM_003090 | SNRPA1 | Small nuclear ribonucleoprotein polypeptide A' |
| 46 | NM_000373 | UMPS | Uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5′-decarboxylase) |
| 47 | NM_144584 | C1orf59 | Chromosome 1 open reading frame 59 |
| 48 | NM_052916.2 | RNF157 | Ring finger protein 157 |
| 49 | NM_006332 | IFI30 | Interferon, γ-inducible protein 30 |
| 50 | NM_002416 | CXCL9 | Chemokine (C-X-C motif) ligand 9 |
| 51 | NM_001017402 | LAMB3 | Laminin, β 3 |
| 52 | NM_004701 | CCNB2 | Cyclin B2 |
| 53 | NM_194463 | RNF128 | Ring finger protein 128 |
Table IV.
Genes down-regulated in advanced gastric cancer.
| No. | Accession no. | Gene symbol | Description |
|---|---|---|---|
| 1 | NM_004190 | LIPF | Lipase, gastric |
| 2 | NM_020143 | PNO1 | Partner of NOB1 homolog (S. cerevisiae) |
| 3 | NM_000257 | MYH7 | Myosin, heavy chain 7, cardiac muscle, β |
| 4 | NM_015173 | TBC1D1 | TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| 5 | NM_005408 | CCL13 | Chemokine (C-C motif) ligand 13 |
| 6 | NM_174929 | ZMIZ2 | Zinc finger, MIZ-type containing 2 |
| 7 | NM_004747 | DLG5 | Discs, large homolog 5 (Drosophila) |
| 8 | NM_024872.2 | DOK3 | Docking protein 3 |
| 9 | NM_201653 | CHIA | Chitinase, acidic |
| 10 | NM_003893 | LDB1 | LIM domain binding 1 |
| 11 | NM_012455.2 | PSD4 | Pleckstrin and Sec7 domain containing 4 |
| 12 | NM_005213 | CSTA | Cystatin A (stefin A) |
| 13 | NM_005416 | SPRR3 | Small proline-rich protein 3 |
| 14 | NM_014989 | RIMS1 | Regulating synaptic membrane exocytosis 1 |
| 15 | NM_001018005 | TPM1 | Tropomyosin 1 (α) |
| 16 | NM_213589 | RAPH1 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| 17 | NM_004898 | CLOCK | Clock homolog (mouse) |
| 18 | NM_013292 | Fast skeletal myosin light chain 2 | |
| 19 | NM_020321 | ACCN3 | Amiloride-sensitive cation channel 3 |
| 20 | NM_002754 | MAPK13 | Mitogen-activated protein kinase 13 |
| 21 | NM_013443 | ST6GALNAC6 | ST6 (α-N-acetyl-neuraminyl-2,3-β-galactosyl-1,3)-N-acetylgalactosaminide α-2,6-sialyltransferase 6 |
| 22 | NM_001042453 | Serine/threonine protein kinase MST4 | |
| 23 | NM_032646 | TTYH2 | Tweety homolog 2 (Drosophila) |
| 24 | NM_015089 | p53-associated parkin-like cytoplasmic protein | |
| 25 | NM_003609 | HIRIP3 | HIRA interacting protein 3 |
| 26 | NR_002219 | BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) |
| 27 | NM_000068 | CACNA1A | Calcium channel, voltage-dependent, P/Q type, α 1A subunit |
| 28 | NM_203377 | MB | Myoglobin |
| 29 | NM_003768 | PEA15 | Phosphoprotein enriched in astrocytes 15 |
| 30 | NM_053013 | ENO3 | Enolase 3 (β, muscle) |
| 31 | XR_018802 | PI4K2A | Phosphatidylinositol 4-kinase type 2 α |
| 32 | NM_003725 | HSD17B6 | Hydroxysteroid (17-β) dehydrogenase 6 homolog (mouse) |
| 33 | NM_006063 | KBTBD10 | Kelch repeat and BTB (POZ) domain containing 10 |
| 34 | NM_012288 | TRAM2 | Translocation associated membrane protein 2 |
| 35 | NM_000730 | CCKAR | Cholecystokinin A receptor |
| 36 | NM_000290 | PGAM2 | Phosphoglycerate mutase 2 (muscle) |
| 37 | NM_199354 | PRB1 | Proline-rich protein BstNI subfamily 1 |
| 38 | XR_019039 | ACTB | Actin, β |
| 39 | NM_006478 | GAS2L1 | Growth arrest-specific 2 like 1 |
| 40 | NM_024674 | LIN28 | Lin-28 homolog (C. elegans) |
| 41 | NM_001070 | TUBG1 | Tubulin, γ 1 |
| 42 | NM_015654 | NAT9 | N-acetyltransferase 9 |
| 43 | NM_003643 | GCM1 | Glial cells missing homolog 1 (Drosophila) |
| 44 | NM_006901.2 | MYO9A | Myosin IXA |
| 45 | NM_017785 | CCDC99 | Coiled-coil domain containing 99 |
| 46 | NM_025135 | FHOD3 | Formin homology 2 domain containing 3 |
| 47 | NM_022566 | MESDC1 | Mesoderm development candidate 1 |
| 48 | NM_198255 | TERT | Telomerase reverse transcriptase |
| 49 | NM_018231 | Amino acid transporter | |
| 50 | NM_002458 | MUC5B | Mucin 5B, oligomeric mucus/gel-forming |
| 51 | NM_001001522 | TAGLN | Transgelin |
| 52 | NM_002631 | PGD | Phosphogluconate dehydrogenase |
| 53 | NM_006984 | CLDN10 | Claudin 10 |
| 54 | NM_004359 | CDC34 | Cell division cycle 34 homolog (S. cerevisiae) |
| 55 | NM_001824 | CKM | Creatine kinase, muscle |
| 56 | NM_002274 | KRT13 | Keratin 13 |
| 57 | XR_019039 | ACTB | Actin, β |
| 58 | NM_000477 | ALB | Albumin |
| 59 | NM_001519.2 | BRF1 | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae) |
| 60 | NM_006790 | MYOT | Myotilin |
| 61 | NM_021948 | BCAN | Brevican |
| 62 | NM_001142404.1 | CD164 | CD164 molecule, sialomucin |
| 63 | BC050364.1 | C7orf13 | Chromosome 7 open reading frame 13 |
| 64 | NM_005177 | ATP6V0A1 | ATPase, H+ transporting, lysosomal V0 subunit a1 |
| 65 | NM_020393 | PGLYRP4 | Peptidoglycan recognition protein 4 |
| 66 | XM_937007 | FRMPD3 | FERM and PDZ domain containing 3 |
| 67 | NM_024754 | PTCD2 | Pentatricopeptide repeat domain 2 |
| 68 | NM_001098511 | KIF2A | Kinesin heavy chain member 2A |
| 69 | NM_025058 | TRIM46 | Tripartite motif-containing 46 |
| 70 | AK126458.1 | MYO15B | Myosin XVB pseudogene |
| 71 | NM_018659 | CYTL1 | Cytokine-like 1 |
| 72 | NM_002965 | S100A9 | S100 calcium binding protein A9 |
| 73 | NM_032566 | SPINK7 | Serine peptidase inhibitor, Kazal type 7 (putative) |
| 74 | NM_001669 | ARSD | Arylsulfatase D |
| 75 | NM_206820 | MYBPC1 | Myosin binding protein C, slow type |
| 76 | NM_003200 | TCF3 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) |
| 77 | NM_031413 | CECR2 | Cat eye syndrome chromosome region, candidate 2 |
| 78 | NM_017539 | DNAH3 | Dynein, axonemal, heavy chain 3 |
| 79 | NM_017426 | NUP54 | Nucleoporin 54 kDa |
| 80 | NM_002020 | FLT4 | Fms-related tyrosine kinase 4 |
| 81 | NM_007320 | RANBP3 | RAN binding protein 3 |
| 82 | NM_005286 | NPBWR2 | Neuropeptides B/W receptor 2 |
| 83 | NM_006428 | MRPL28 | Mitochondrial ribosomal protein L28 |
| 84 | NM_014280.2 | DNAJC8 | DnaJ (Hsp40) homolog, subfamily C, member 8 |
| 85 | NM_020679 | MIF4GD | MIF4G domain containing |
| 86 | NM_001823 | CKB | Creatine kinase, brain |
| 87 | NM_000477 | ALB | Albumin |
| 88 | NM_001927 | DES | Desmin |
| 89 | NM_005416 | SPRR3 | Small proline-rich protein 3 |
| 90 | NM_022468 | MMP25 | Matrix metallopeptidase 25 |
| 91 | NM_016599 | MYOZ2 | Myozenin 2 |
| 92 | NM_000243 | MEFV | Mediterranean fever |
| 93 | NM_002272 | KRT4 | Keratin 4 |
| 94 | NM_003279 | TNNC2 | Troponin C type 2 (fast) |
| 95 | NM_006685 | SMR3B | Submaxillary gland androgen regulated protein 3 homolog B (mouse) |
| 96 | NM_014760 | TATDN2 | TatD DNase domain containing 2 |
| 97 | NM_006928 | SILV | Silver homolog (mouse) |
| 98 | NM_016522 | Neurotrimin | |
| 99 | NM_000760 | CSF3R | Colony stimulating factor 3 receptor (granulocyte) |
| 100 | NM_003167 | SULT2A1 | Sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| 101 | NM_183360 | DTNB | Dystrobrevin, β |
| 102 | NM_001711 | BGN | Biglycan |
| 103 | NM_023077 | C1orf163 | Chromosome 1 open reading frame 163 |
| 104 | NM_015926.4 | TEX264 | Testis expressed 264 |
| 105 | NM_006757 | TNNT3 | Troponin T type 3 (skeletal, fast) |
| 106 | NM_002675 | PML | Promyelocytic leukemia |
| 107 | XR_018113 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| 108 | NM_021245 | MYOZ1 | Myozenin 1 |
| 109 | NM_000383 | AIRE | Autoimmune regulator |
| 110 | NM_006846 | SPINK5 | Serine peptidase inhibitor, Kazal type 5 |
| 111 | XM_939725 | AP1S2 | Adaptor-related protein complex 1, sigma 2 subunit |
| 112 | NM_024505 | NOX5 | NADPH oxidase, EF-hand calcium binding domain 5 |
| 113 | NM_020145 | SH3GLB2 | SH3-domain GRB2-like endophilin B2 |
| 114 | NM_016192 | TMEFF2 | Transmembrane protein with EGF-like and two follistatin-like domains 2 |
| 115 | NM_006472 | TXNIP | Thioredoxin interacting protein |
| 116 | NM_031866 | FZD8 | Frizzled homolog 8 (Drosophila) |
| 117 | NM_003808 | TNFSF13 | Tumor necrosis factor (ligand) superfamily, member 13 |
| 118 | NM_015503 | SH2B1 | SH2B adaptor protein 1 |
| 119 | NM_014047 | C19orf53 | Chromosome 19 open reading frame 53 |
| 120 | NM_022754 | SFXN1 | Sideroflexin 1 |
| 121 | NM_003061 | SLIT1 | Slit homolog 1 (Drosophila) |
| 122 | NM_003047 | SLC9A1 | Solute carrier family 9 (sodium/hydrogen exchanger), member 1 (antiporter, Na+/H+, amiloride sensitive) |
| 123 | NM_021991 | JUP | Junction plakoglobin |
On the other hand, the down-regulated genes included those associated with various metabolic pathways (CKM, ARSD and BGN), small molecule transport (ACCN3, ATP6V0A1 and SFXN1), signal transduction (FLT4, NPBWR2, CSF3R and FZD8), cell cycle regulation (TBC1D1, DLG5, GAS2L1, CDC34 and SH2B1), cell adhesion (RAPH1, CLDN10, BCAN, CD164 and JUP), transcription factors (LDB1, CLOCK, GCM1, BRF1, TCF3, PML and AIRE), cell-cell signaling (PGD, S100A9, CCL13 and RIMS1) or other functions.
Identification of candidate genes as molecular markers for the detection of circulating gastric cancer cells in human peripheral blood
Of the 53 genes that were up-regulated in the gastric cancer compared to the normal gastric tissues, we identified five candidate marker genes [tetraspanin 8 (TSPAN8), epithelial cell adhesion molecule (EPCAM), matrix metallopeptidase 12 (MMP12), matrix metallopeptidase 7 (MMP7) and regenerating islet-derived 3 α (REG3A)] for the detection of circulating gastric cancer cells in peripheral blood in accordance with the following criteria: i) no or weak expression in human normal tissues in the published database (20), ii) no expression in PBMCs from 4 healthy volunteers by nested RT-PCR. In addition to the above five newly identified genes, we analyzed histamine receptor H1 (HRH1) since a previous study reported that this gene was overexpressed in gastric cancer cells, and the expression of this gene satisfied the above criteria (21). Moreover, three candidate marker genes [keratin 20 (CK20), epidermal growth factor receptor (EGFR) and carcinoembryonic antigen (CEA)], which have been reported to be promising markers for the detection of cancer cells, were further analyzed (11,13,22,23).
Association of the expression of the nine marker genes for the detection of circulating gastric cancer cells with clinicopathological parameters by nested RT-PCR
We carried out semi-quantitative nested RT-PCR analysis of the nine candidate marker genes for the detection of circulating cancer cells using PBMCs of patients with gastric cancer. Of the nine candidate genes, the expression of MMP12 and CEA mRNAs was positive in 40% of the patients with gastric cancer. However, the expression of the other seven genes was positive in ≤30% of the patients, respectively (Table V). We then investigated a combined effect of the expression of the nine genes on the detection of circulating cancer cells. Expression of one or more genes out of the nine was detected in 80% of the patients with gastric cancer by nested RT-PCR (Table VI).
Table V.
Positive ratio of the nine marker genes for detection of circulating gastric cancer cells.
| Marker genes | TSPAN8 | EPCAM | HRH1 | CK20 | MMP12 | MMP7 | EGFR | REG3A | CEA |
|---|---|---|---|---|---|---|---|---|---|
| Positive cases (%) | 20 | 30 | 30 | 20 | 40 | 10 | 10 | 20 | 40 |
Table VI.
Number of positive genes in 10 cases by nested RT-PCR.
| Cases | GC-1 | GC-2 | GC-3 | GC-4 | GC-5 | GC-6 | GC-7 | GC-8 | GC-9 | GC-10 |
|---|---|---|---|---|---|---|---|---|---|---|
| No. of positive genes | 1 | 0 | 2 | 3 | 5 | 6 | 1 | 2 | 0 | 2 |
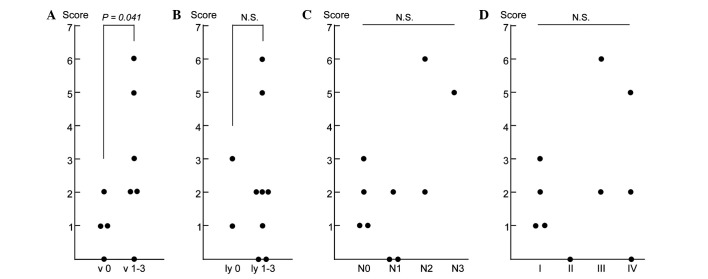
We further investigated the association of the expression of the nine candidate marker genes with clinicopathological parameters of the 10 cases. We focused on four parameters: vascular invasion (v factor), lymphatic invasion (ly factor), lymph node metastasis (N factor) and pathological stage I–IV, and investigated the association of these parameters with the total number of positive genes in the PBMCs of each patient (Fig. 1). Of the four parameters, the numbers of genes expressed in the PBMCs were ≤2 in all of the vascular invasion-negative cases (v 0), while the numbers of genes were ≥2 in 5 of 6 positive cases (v 1–3), exhibiting a significant difference between the two groups (P=0.041; Fig. 1A). However, no significant association was observed for the other three parameters (Fig. 1B–D), suggesting that the combined expression analysis of the nine marker genes using PBMCs detected micrometastasis through vascular invasion in the primary gastric cancer tissues.
Figure 1.
Relationships between clinicopathological parameters and the expression of the set of nine marker genes in PBMCs of gastric cancer patients. Associations of the total number of marker genes which were expressed in the PBMCs with (A) vascular invasion (v factor), (B) lymphatic invasion (ly factor), (C) lymph node metastasis (N factor) and (D) pathological stage are shown. The score indicates the number of marker genes expressed in PBMCs of each patient. N.S., not significant (P>0.05). Student’s t-test was used for A and B, and Cochran-Armitage test was used for C and D.
Discussion
Microarrays, at present, are widely used to analyze expression of thousands of genes simultaneously in cancer tissues. In the present study, we identified five genes (TSPAN8, EPCAM, MMP12, MMP7 and REG3A) as potential markers for the detection of circulating cancer cells in the peripheral blood of patients with gastric cancer through genome-wide gene expression profiling in combination with nested RT-PCR. Some of these genes have previously been reported to be up-regulated in gastric cancer cells; however, they have not previously been designated for the detection of circulating gastric cancer cells by nested RT-PCR. Furthermore, the combined expression analysis of the five genes and four previously reported marker genes, HRH1, EGFR, CK20 and CEA, revealed that one or more mRNAs among the nine genes could be detected in 80% of the patients with gastric cancer by nested RT-PCR, suggesting that a set of nine marker genes is more sensitive than a single marker gene for detection of circulating gastric cancer cells. In this study, we did not investigate the association of distant metastasis with expression of the nine marker genes since no patients had distant metastasis among the 10 studied patients. Although we could not exclude false-positive cases due to non-malignant epithelial cells which may have contaminated the blood samples during collection and which may have expressed the targeted transcripts (18), pathological v factor showed a significant association with the total number of marker genes expressed in the PBMCs of the patients. Hence, the set of nine marker genes may be promising for the detection of minimal amounts of circulating gastric cancer cells prior to the metastatic growth of gastric cancer cells in organ(s).
Among the five marker genes which were newly identified in the microarray analysis, we identified epithelial cell adhesion molecule (EPCAM) which is a member of a family of type I membrane proteins and pan-epithelial differentiation antigen expressed in many types of carcinomas (24–28). Magnetic beads or structures coated with EPCAM monoclonal antibodies have been recently used for circulating cancer cell separation (29–31). Although we did not compare the accuracy of the detection of gastric cancer cells by these methods to that of nested RT-PCR since we did not conduct the former assays, 30% of patients with gastric cancer exhibited EPCAM-positivity in PBMCs by nested RT-PCR. Further clinical study investigating the relationship between the clinical outcome of patients and EPCAM expression in PBMCs by nested RT-PCR may clarify whether this method could be clinically applied for the detection of circulating gastric cancer cells. Two matrix metalloproteinases, MMP7 and MMP12, were among the five marker genes which were newly identified in this study. MMPs are a family of zinc-dependent proteolytic enzymes capable of cleaving extracellular matrix proteins, and the expression of MMPs in cancer tissue has been reported to be associated with the risk of metastasis (32–38). These two MMPs may play important roles in tumor invasion and the formation of metastasis in gastric cancer.
In conclusion, five novel marker genes were designated for the detection of circulating gastric cancer cells. The nested RT-PCR analysis for the set of nine marker genes, TSPAN8, EPCAM, MMP12, MMP7, REG3A, HRH1, EGFR, CK20 and CEA, using PBMCs of patients with gastric cancer may provide the potential for the detection of circulating gastric cancer cells prior to the formation of metastasis in other organs. Our data suggest that early detection and personalized therapy for gastric cancers, by prescribing the appropriate treatment to patients with a high risk of metastasis, may be achievable by utilizing specific sets of marker genes according to the approach shown here.
Acknowledgments
We thank Tomohisa Furuhata, Yasutoshi Kimura, Chikashi Kihara, Kenji Okita and Noriko Nishikawa for the helpful discussions.
References
- 1.Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM. The global burden of cancer: priorities for prevention. Carcinogenesis. 2009;31:100–110. doi: 10.1093/carcin/bgp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott K, Lordick F, Blank S, Buchler M. Gastric cancer: surgery in 2011. Langenbecks Arch Surg. 2011 Jan. doi: 10.1007/s00423-010-0738-7. (E-pub ahead of print). [DOI] [PubMed] [Google Scholar]
- 3.Chen XM, Chen GY, Wang ZR, Zhu FS, Wang XL, Zhang X. Detection of micrometastasis of gastric carcinoma in peripheral blood circulation. World J Gastroenterol. 2004;10:804–808. doi: 10.3748/wjg.v10.i6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vardakis N, Messaritakis I, Papadaki C, et al. Prognostic significance of the detection of peripheral blood CEACAM5 mRNA-positive cells by real-time polymerase chain reaction in operable colorectal cancer. Clin Cancer Res. 2011;17:165–173. doi: 10.1158/1078-0432.CCR-10-0565. [DOI] [PubMed] [Google Scholar]
- 6.Miyazono F, Natsugoe S, Takao S, et al. Surgical maneuvers enhance molecular detection of circulating tumor cells during gastric cancer surgery. Ann Surg. 2001;233:189–194. doi: 10.1097/00000658-200102000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 8.Qiu MZ, Li ZH, Zhou ZW, et al. Detection of carcinoembryonic antigen messenger RNA in blood using quantitative real-time reverse transcriptase-polymerase chain reaction to predict recurrence of gastric adenocarcinoma. J Transl Med. 2010;8:107. doi: 10.1186/1479-5876-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guadagni F, Kantor J, Aloe S, et al. Detection of blood-borne cells in colorectal cancer patients by nested reverse transcription-polymerase chain reaction for carcinoembryonic antigen messenger RNA: longitudinal analyses and demonstration of its potential importance as an adjunct to multiple serum markers. Cancer Res. 2001;61:2523–2532. [PubMed] [Google Scholar]
- 10.Ikeguchi M, Kaibara N. Detection of circulating cancer cells after a gastrectomy for gastric cancer. Surg Today. 2005;35:436–441. doi: 10.1007/s00595-004-2978-z. [DOI] [PubMed] [Google Scholar]
- 11.Dardaei L, Shahsavani R, Ghavamzadeh A, et al. The detection of disseminated tumor cells in bone marrow and peripheral blood of gastric cancer patients by multimarker (CEA, CK20, TFF1 and MUC2) quantitative real-time PCR. Clin Biochem. 2011;44:325–330. doi: 10.1016/j.clinbiochem.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Katsuragi K, Yashiro M, Sawada T, Osaka H, Ohira M, Hirakawa K. Prognostic impact of PCR-based identification of isolated tumour cells in the peritoneal lavage fluid of gastric cancer patients who underwent a curative R0 resection. Br J Cancer. 2007;97:550–556. doi: 10.1038/sj.bjc.6603909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori K, Aoyagi K, Ueda T, et al. Highly specific marker genes for detecting minimal gastric cancer cells in cytology negative peritoneal washings. Biochem Biophys Res Commun. 2004;313:931–937. doi: 10.1016/j.bbrc.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Zembutsu H, Ohnishi Y, Daigo Y, et al. Gene-expression profiles of human tumor xenografts in nude mice treated orally with the EGFR tyrosine kinase inhibitor ZD1839. Int J Oncol. 2003;23:29–39. [PubMed] [Google Scholar]
- 15.Zembutsu H, Ohnishi Y, Tsunoda T, et al. Genome-wide cDNA microarray screening to correlate gene expression profiles with sensitivity of 85 human cancer xenografts to anticancer drugs. Cancer Res. 2002;62:518–527. [PubMed] [Google Scholar]
- 16.Zembutsu H, Suzuki Y, Sasaki A, et al. Predicting response to docetaxel neoadjuvant chemotherapy for advanced breast cancers through genome-wide gene expression profiling. Int J Oncol. 2009;34:361–370. [PubMed] [Google Scholar]
- 17.Zembutsu H, Yanada M, Hishida A, et al. Prediction of risk of disease recurrence by genome-wide cDNA microarray analysis in patients with Philadelphia chromosome-positive acute lymphoblastic leukemia treated with imatinib-combined chemotherapy. Int J Oncol. 2007;31:313–322. [PubMed] [Google Scholar]
- 18.Obermayr E, Sanchez-Cabo F, Tea MK, et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC. 2010;10:666. doi: 10.1186/1471-2407-10-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo L, Salunga RC, Guo H, et al. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 20.Saito-Hisaminato A, Katagiri T, Kakiuchi S, Nakamura T, Tsunoda T, Nakamura Y. Genome-wide profiling of gene expression in 29 normal human tissues with a cDNA microarray. DNA Res. 2002;9:35–45. doi: 10.1093/dnares/9.2.35. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa S, Furukawa Y, Li M, et al. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012–7017. [PubMed] [Google Scholar]
- 22.Amin AT, Shiraishi N, Ninomiya S, Tajima M, Inomata M, Kitano S. Increased mRNA expression of epidermal growth factor receptor, human epidermal receptor, and survivin in human gastric cancer after the surgical stress of laparotomy versus carbon dioxide pneumoperitoneum in a murine model. Surg Endosc. 2010;24:1427–1433. doi: 10.1007/s00464-009-0793-8. [DOI] [PubMed] [Google Scholar]
- 23.Mori M, Mimori K, Inoue H, et al. Detection of cancer micrometastases in lymph nodes by reverse transcriptase-polymerase chain reaction. Cancer Res. 1995;55:3417–3420. [PubMed] [Google Scholar]
- 24.Trebak M, Begg GE, Chong JM, Kanazireva EV, Herlyn D, Speicher DW. Oligomeric state of the colon carcinoma-associated glycoprotein GA733-2 (Ep-CAM/EGP40) and its role in GA733-mediated homotypic cell-cell adhesion. J Biol Chem. 2001;276:2299–2309. doi: 10.1074/jbc.M004770200. [DOI] [PubMed] [Google Scholar]
- 25.Maghzal N, Vogt E, Reintsch W, Fraser JS, Fagotto F. The tumor-associated EpCAM regulates morphogenetic movements through intracellular signaling. J Cell Biol. 2010;191:645–659. doi: 10.1083/jcb.201004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du W, Ji H, Cao S, et al. EpCAM: a potential antimetastatic target for gastric cancer. Dig Dis Sci. 2009;55:2165–2171. doi: 10.1007/s10620-009-1033-8. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn S, Koch M, Nubel T, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee S, Richardson AM, Rodriguez-Canales J, et al. Identification of EpCAM as a molecular target of prostate cancer stroma. Am J Pathol. 2009;175:2277–2287. doi: 10.2353/ajpath.2009.090013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sha MY, Xu H, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J Am Chem Soc. 2008;130:17214–17215. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myung JH, Launiere CA, Eddington DT, Hong S. Enhanced tumor cell isolation by a biomimetic combination of E-selectin and anti-EpCAM: implications for the effective separation of circulating tumor cells (CTCs) Langmuir. 2010;26:8589–8596. doi: 10.1021/la904678p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosokawa M, Hayata T, Fukuda Y, et al. Size-selective microcavity array for rapid and efficient detection of circulating tumor cells. Anal Chem. 2010;82:6629–6635. doi: 10.1021/ac101222x. [DOI] [PubMed] [Google Scholar]
- 32.Shi WD, Meng ZQ, Chen Z, Lin JH, Zhou ZH, Liu LM. Identification of liver metastasis-related genes in a novel human pancreatic carcinoma cell model by microarray analysis. Cancer Lett. 2009;283:84–91. doi: 10.1016/j.canlet.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 33.Szarvas T, Becker M, vom Dorp F, et al. Matrix metallo-proteinase-7 as a marker of metastasis and predictor of poor survival in bladder cancer. Cancer Sci. 2010;101:1300–1308. doi: 10.1111/j.1349-7006.2010.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshima T, Akaike M, Yoshihara K, et al. Clinicopathological significance of the gene expression of matrix metalloproteinase-7, insulin-like growth factor-1, insulin-like growth factor-2 and insulin-like growth factor-1 receptor in patients with colorectal cancer: insulin-like growth factor-1 receptor gene expression is a useful predictor of liver metastasis from colorectal cancer. Oncol Rep. 2008;20:359–364. [PubMed] [Google Scholar]
- 35.Fang YJ, Lu ZH, Wang GQ, et al. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis. 2009;24:875–884. doi: 10.1007/s00384-009-0725-z. [DOI] [PubMed] [Google Scholar]
- 36.Kerkela E, Ala-aho R, Klemi P, et al. Metalloelastase (MMP-12) expression by tumour cells in squamous cell carcinoma of the vulva correlates with invasiveness, while that by macrophages predicts better outcome. J Pathol. 2002;198:258–269. doi: 10.1002/path.1198. [DOI] [PubMed] [Google Scholar]
- 37.Balaz P, Friess H, Kondo Y, Zhu Z, Zimmermann A, Buchler MW. Human macrophage metalloelastase worsens the prognosis of pancreatic cancer. Ann Surg. 2002;235:519–527. doi: 10.1097/00000658-200204000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofmann HS, Hansen G, Richter G, et al. Matrix metalloproteinase-12 expression correlates with local recurrence and metastatic disease in non-small cell lung cancer patients. Clin Cancer Res. 2005;11:1086–1092. [PubMed] [Google Scholar]