Abstract
Genetic and epigenetic features of sessile serrated adenoma (SSA), a precursor lesion to colon cancer with microsatellite instability (MSI), were investigated. The aim of this study was to clarify whether there are location-dependent genetic and epigenetic features in SSA. Twenty-two patients with proximal SSAs and 8 with distal SSAs were recruited. Twenty-two patients with tubular adenoma (TA) and 66 with proximal colon cancer were studied for comparison. Genetic and epigenetic features were evaluated as BRAF and KRAS mutations, MSI, hMLH1 methylation and CpG island methylator phenotype (CIMP). BRAF mutation (p=0.007) and CIMP (p=0.012) were more frequently found in proximal than in distal SSAs. Furthermore, the KRAS mutation was found only in distal SSAs. In TAs, no location-related molecular features were observed. All SSAs, TAs and 42 colon cancer lesions were microsatellite stable (MSS). Twenty-four colon cancer lesions exhibited MSI and had more frequent BRAF mutations (p<0.001), hMLH1 methylation (p<0.001) and CIMP (p<0.001). BRAF mutation occurred in only 9.5% of MSS cancers (p=0.01). In MSI cancers with the BRAF mutation, a higher correlation with CIMP (p=0.032) was observed. We demonstrated the distinct genetic and epigenetic features between proximal and distal SSAs. Similar genetic and epigenetic features were shared between proximal SSAs and proximal MSI cancers harboring the BRAF mutation. By contrast, our results allow the possibility of carcinogenesis in SSAs leading to MSS cancer with the BRAF mutation.
Keywords: sessile serrated adenoma, serrated neoplastic pathway, BRAF mutation, CpG island methylator phenotype
Introduction
In colonic carcinogenesis, a serrated neoplastic pathway has been recommended as an alternative pathway distinct from the adenoma-carcinoma sequence. A hyperplastic polyp has been regarded as a benign lesion without malignant potential, whereas a tubular adenoma (TA) is a precursor of colorectal cancer in the adenoma-carcinoma sequence. Recently, polyps with serrated structures have been classified into subgroups, including hyperplastic polyps, traditional serrated adenomas and sessile serrated adenomas (SSAs) (1–3).
SSA is a new entity identified by Torlakovic et al in 1996 (4). Endoscopically, SSAs are flat, pale and large in size (5,6), which are different from the endoscopic features of hyperplastic polyps. The histological appearance of SSAs is distinguished from that of hyperplastic polyps on the basis of their abnormal architectural features, such as branching of crypts, dilatation of the base of the crypts, growth of crypts parallel to the muscularis mucosa and surface villosity or papillarity (6,7). Recent studies have shown that SSAs are more likely to occur in the proximal colon and show a high frequency of BRAF mutations, DNA methylation of the promoter region of the mismatch repair gene, such as hMLH1 and CpG island methylator phenotype (CIMP) (8–12). Since SSA has characteristics similar to colon cancer with microsatellite instability (MSI), it has attracted considerable attention as a possible precursor lesion to MSI cancers (13).
Clinicopathological findings suggest that MSI cancers and SSAs are frequently located in the proximal colon (14,15). Recent studies have shown distinct characteristics of MSI cancers emerging from the proximal and distal colon (16,17). Compared to distal MSI cancers, the proximal forms show female predominance, a more aggressive differentiation and a significantly higher frequency of hMLH1 methylations. The reason for the occurrence of these differences has not been elucidated. Since the location determines the characteristics of MSI cancers, precursors should also share location-associated features.
This study was designed to investigate the location-associated genetic and epigenetic features of SSA to evaluate its role as a precursor of MSI cancers. TA and colon cancer were also investigated for comparison.
Materials and methods
This study was approved by the Jichi Medical University Institutional Review Board prior to commencement. SSAs, TAs and colorectal cancer tissues were prospectively collected from patients at the Jichi Medical University Hospital and the Jichi Medical University Saitama Medical Center. SSAs and TAs were obtained endoscopically and classified into two categories according to their location, i.e., proximal or distal. Colon cancer tissues were obtained from patients who underwent surgical treatment. For all lesions, a part of the fresh tissue was immediately frozen for genetic analysis, and the remaining tissue was used for histological analysis. Proximal lesions were defined as those proximal to the splenic flexure, whereas distal lesions were defined as those distal to the splenic flexure. All colon cancer tissues were collected from the proximal colon.
SSA was diagnosed by five architectural features: basal crypt serration, basal dilatation of crypts, crypts that run horizontal to the basement membrane, crypt branching and surface villosity or papillarity (6,7). When the endoscopically resected polyp had two or more of the above-mentioned features, it was diagnosed as SSA. The lesions showing typical histological features of so called ‘traditional serrated adenoma’ (2) were excluded from analysis.
In total, 30 SSAs, 22 TAs and 66 colon cancer tissues were evaluated for their genetic and epigenetic features.
BRAF and KRAS mutation analysis
BRAF (T1799A) and KRAS mutations were studied by direct sequencing after polymerase chain reaction (PCR) amplification of exon 15 of the BRAF gene and codon 12 and 13 of the KRAS gene. For detection of the BRAF mutation, genomic DNA obtained from fresh frozen samples was amplified using the following primers: forward, TCATAATGCTTGCTCTGATAGGA and reverse, GGCCAAAAATTTAATCAGTGGA. For the detection of the KRAS mutation, the following primers were used: forward, CTGAAAATGACTGAATATAAACTTGT and reverse, ATA TGCATATTAAAACAAGATTTACC as previously described (18,19). PCR products were purified on a YM-30 Microcon column (Millipore) and sequenced using the BigDye terminator v3.1 cycle sequencing kit on ABI Prism 3100 (both from Applied Biosystems, Tokyo, Japan).
MSI analysis
Genomic DNA was extracted from fresh frozen samples using the EZ1 DNA tissue kit (Qiagen, Tokyo, Japan) and was amplified by PCR using the monomorphic markers BAT25 and BAT26 as previously described (20). PCR products were analyzed by Gene Scan using ABI Prism 3100, and the sample was scored showing MSI if there were additional peaks in the PCR products, or otherwise scored as microsatellite stable (MSS).
hMLH1 methylation and CIMP
Combined bisulfite restriction analysis was performed to assess gene methylation using primers that were designed to amplify the regions around the transcription sites of the target genes (21). Bisulfite modification was performed according to the manufacturer’s instructions using the Epitect Bisulfite kit (Qiagen). Genomic DNA (1 μg) was used for conversion with the bisulfite reagent. The mismatch repair gene hMLH1 and five MINT loci (MINT1, MINT2, MINT12, MINT25 and MINT31) were examined as previously described (22–24). The primer sequences, annealing temperatures and restriction enzymes utilized were identical to those previously described (24). After digestion, products were electrophoresed on 2% agarose gels and stained with ethidium bromide. Methylation density was confirmed using the image analysis program ImageJ, and positive methylation was defined when the methylation-sensitive restriction enzyme digested ≥10% of the DNA (24). All samples with methylation at three or more loci were designated as CIMP-positive.
Statistical analysis
Differences between the groups were evaluated using the Student’s t-test, Chi-square test or Fisher’s exact test. Statistical significance tests were two-tailed, and p<0.05 was considered statistically significant. Analysis was performed using SPSS software (version 17; SPSS Japan Inc., Tokyo, Japan).
Results
Sessile serrated adenoma
The location-associated clinical and molecular features of SSAs were evaluated (Table I). Patient age, gender and polyp size were not significantly different between the proximal and distal SSAs. The BRAF mutation was found in 86.4% of the proximal SSAs and 37.5% of the distal SSAs (p=0.007). However, the KRAS mutation was found only in the distal SSAs (p=0.015). BRAF and KRAS mutations were mutually exclusive. Epigenetic analysis demonstrated no hMLH1 methylation in both SSAs and, hence, both were MSS. However, CIMP was observed in 50% of the proximal SSAs, whereas no distal SSA showed CIMP.
Table I.
Clinical and molecular features of the proximal and distal sessile serrated adenoma (SSA) cases.
| Proximal SSA (n=22) | Distal SSA (n=8) | p-value | |
|---|---|---|---|
| Patient age (years) (mean ± SD) | 60.9±9.2 | 59.1±11.5 | 0.672 |
| Gender (male/female) | 14/8 | 4/4 | 0.678 |
| Size (mm) (mean ± SD) | 12.5±5.3 | 14.0±6.9 | 0.664 |
| BRAF (V600E) | 86.4% (19/22) | 37.5% (3/8) | 0.007 |
| KRAS (codon 12/13) | 0.0% (0/22) | 25.0% (2/8) | 0.015 |
| hMLH1 methylation | 0.0% (0/22) | 0.0% (0/8) | - |
| MSI | 0.0% (0/21) | 0.0% (0/8) | - |
| CIMP | 50.0% (11/22) | 0.0% (0/8) | 0.012 |
MSI, microsatellite instability; CIMP, CpG island methylator phenotype.
Ttubular adenoma
Location-associated features of TAs were investigated (Table II). Patient age, gender and polyp size were not significantly different between the two locations. The BRAF mutation, hMLH1 methylation, MSI and CIMP were not found in either the proximal or distal TAs. The KRAS mutation was found in 36.4% of the proximal TAs and in 40% of the distal TAs, showing no significant difference (p=0.864).
Table II.
Comparison of the clinical and molecular features between the proximal and distal tubular adenoma (TA) cases.
| Proximal TA (n=12) | Distal TA (n=10) | p-value | |
|---|---|---|---|
| Patient age (years) (mean ± SD) | 65.1±8.2 | 58.1±9.8 | 0.068 |
| Gender (male/female) | 8/4 | 5/5 | 0.666 |
| Size (mm) (mean ± SD) | 18.5±16.0 | 19.1±17.9 | 0.934 |
| BRAF (V600E) | 0.0% (0/12) | 0.0% (0/10) | - |
| KRAS (codon 12/13) | 36.4% (4/11) | 40.0% (4/10) | 0.864 |
| hMLH1 methylation | 0.0% (0/12) | 0.0% (0/10) | - |
| MSI | 0.0% (0/12) | 0.0% (0/10) | - |
| CIMP | 0.0% (0/12) | 0.0% (0/10) | - |
MSI, microsatellite instability; CIMP, CpG island methylator phenotype.
Proximal colon cancer
Proximal colon cancer was investigated for comparison to proximal SSA. The proximal colon cancer samples were divided according to the presence of MSI. In this study, 24 colon cancer tissues showed MSI and 42 were MSS. The clinical and molecular features of the MSI and MSS cancers were compared (Table III). Patient age and gender were not significantly different between both types of cancers. Mucinous or poorly differentiated cancer was more frequently noted in the MSI cancers (p=0.007). The BRAF mutation was found in 58.3% of the MSI cancers and in 9.5% of the MSS cancers (p<0.001). In contrast, the KRAS mutation was more frequently present in the MSS than in the MSI cancers (38.8 and 8.3%, respectively; p=0.01). Epigenetic analysis showed that hMLH1 methylation was frequently present in the MSI cancers (50%), and no methylation was found in the MSS cancers (p<0.001). Furthermore, CIMP was frequently found in the MSI cancers (66.7%), whereas it was relatively rare in the MSS cancers (11.9%) (p<0.001).
Table III.
Comparison of the clinical and molecular features between the proximal MSI and MSS cancers.
| MSI cancer (n=24) | MSS cancer (n=42) | p-value | |
|---|---|---|---|
| Patient age (years) (mean ± SD) | 68±8.8 | 66.9±13.9 | 0.704 |
| Gender (male/female) | 8/16 | 19/23 | 0.438 |
| Stage (-I/-II) | 5/19 | 8/31 | 0.861 |
| Mucinous/poorly differentiated | 37.5% (9/24) | 7.7% (3/39) | 0.007 |
| BRAF (V600E) | 58.3% (14/24) | 9.5% (4/42) | <0.001 |
| KRAS (codon 12/13) | 8.3% (2/24) | 38.1% (16/42) | 0.01 |
| hMLH1 methylation | 50.0% (12/24) | 0.0% (0/42) | <0.001 |
| CIMP | 66.7% (16/24) | 11.9% (5/42) | <0.001 |
MSI, microsatellite instability; MSS, microsatellite stability; CIMP, CpG island methylator phenotype.
Next, MSI cancers were divided into two groups based on the presence or absence of the BRAF mutation (Table IV). Patient age and gender were not significantly different between the MSI cancers with or without the BRAF mutation. Although the frequency of hMLH1 methylation was not statistically different, CIMP was more frequently observed in BRAF-mutated MSI cancers (85.7%) than in the BRAF wild-type MSI cancers (40%) (p=0.032). A high frequency of CIMP was found only in the proximal SSAs and the proximal MSI cancers harboring the BRAF mutation.
Table IV.
Comparison of MSI cancers with or without BRAF mutation.
| BRAF (+) (n=14) | BRAF (−) (n=10) | p-value | |
|---|---|---|---|
| Patient age (mean ± SD) | 62.4±14.6 | 71.9±11.6 | 0.070 |
| Female | 71.4% (10/14) | 60% (6/10) | 0.441 |
| KRAS | 0.0% (0/14) | 20% (2/10) | 0.163 |
| hMLH1 | 57.1% (8/14) | 40% (4/10) | 0.680 |
| CIMP | 85.7% (12/14) | 40% (4/10) | 0.032 |
CIMP, CpG island methylator phenotype; MSI, microsatellite instability.
Comparison of genetic and epigenetic features of the proximal SSAs, TAs, MSI cancers and MSS cancers
Genetic and epigenetic features and differences were compared between the proximal SSAs, TAs, MSI cancers and MSS cancers (Fig. 1). The BRAF mutation was more frequently found in the SSAs and MSI cancers than in the TAs and MSS cancers. On the other hand, the KRAS mutation was more frequently found in the TAs and MSS cancers than in the SSAs and MSI cancers. CIMP status was similar to that of the BRAF mutation, with the SSAs and MSI cancers being much more frequently CIMP-positive than the TAs or MSS cancers. The hMLH1 mutation was only observed in MSI cancers.
Figure 1.
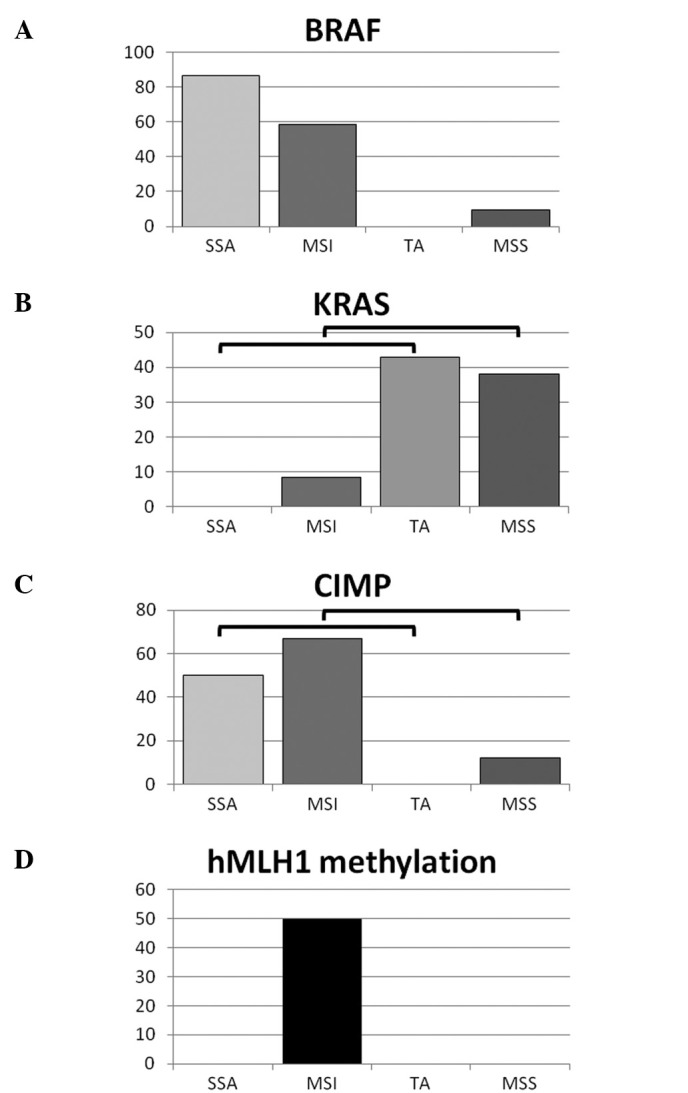
(A) The BRAF mutation is more frequent in SSAs and MSI cancers than in TAs (p<0.001) and MSS cancers (p<0.001), respectively. (B) The KRAS mutation is more frequent in TAs and MSS cancers than in SSAs (p=0.002) and MSI cancers (p=0.01), respectively. (C) hMLH1 methylation was only found in MSI cancers. (D) CIMP was more frequent in SSAs (50%) and MSI cancers than in TAs (0%) (p=0.001) and MSS cancers (p<0.001).
Discussion
In the present study, we demonstrated the distinct genetic and epigenetic features between proximal and distal SSA. Regarding genetic features, the BRAF mutation dominantly occurred in proximal SSAs rather than in distal SSAs. The KRAS mutation was more likely to be detected in distal than in proximal SSA. Epigenetic analysis exhibited no hMLH1 methylation in either proximal or distal SSAs, whereas proximal SSA was likely to possess CIMP (p=0.012). MSI, which is speculated to be caused by both genetic and epigenetic alterations, was detected in neither proximal nor distal SSA. Features, such as harboring the BRAF mutation and CIMP, characterized proximal SSA. On the other hand, TA did not show any difference in genetic and epigenetic features for either location.
Since MSI cancer is dominantly located in the proximal colon, we compared the features of proximal MSI cancers to those of proximal MSS cancers. Proximal MSI cancers showed a high frequency of BRAF mutations, a low frequency of KRAS mutations, and hMLH1 methylation in 50 and 66.7% were positive for CIMP. On the other hand, proximal MSS cancers exhibited less frequent BRAF mutations and more frequent KRAS mutations compared to proximal MSI cancers (p<0.001 and p=0.01, respectively). Epigenetic alterations, such as hMLH1 methylation and CIMP, were much less frequently observed in proximal MSS cancers.
SSA was characterized by a high frequency of BRAF mutations, and MSI cancers were distinguished by BRAF status. Therefore, the molecular features of MSI cancers with or without the BRAF mutation were further examined. Proximal MSI cancers harbored significantly more CIMP in the presence of the BRAF mutation than in its absence. High correlation was observed between positive CIMP and the BRAF mutation in proximal MSI cancers. These molecular features were also observed in proximal SSAs, but not in proximal TAs, suggesting that proximal SSA could be a precursor lesion of proximal MSI cancers harboring the BRAF mutation. Considering the distinct characteristics of MSI cancers of the proximal and distal colon (16,17), MSI cancers probably share their molecular features with SSA in a location-dependent manner. By contrast, MSS cancers shared their molecular features with TAs in a location-independent manner.
Several reports have indicated that SSAs are microsatellite stable (9,13,25), and our data showed the same result. Vaughn et al reported that hMLH1 methylation in proximal hyperplastic polyps is rare and most MSI cancers show methylation (26). Similarly, in our study, proximal SSAs did not show any hMLH1 methylation, whereas a considerable amount of MSI cancers exhibited hMLH1 methylation. Similar CIMP frequency was observed in proximal SSAs and proximal MSI cancers. These results suggest the possibility that DNA methylation initially occurs in a limited range of the genome sparing critical regions for cancer progression in proximal SSAs. Then the promoter region of the DNA mismatch repair gene can be methylated and silenced, resulting in MSI. This hypothesis is supported by previous reports which demonstrated that limited methylation in the hMLH1 promoter region did not inhibit hMLH1 protein expression (27,28).
Notably, a considerable number of proximal MSI cancers did not show the BRAF mutation; however, a small percentage of proximal MSS cancers did show the BRAF mutation. There are several reports with similar results for the frequency of the BRAF mutation in MSI and MSS cancers (29–31). Lubomierski et al (31) suggested that there were two genetically distinct entities of MSI cancer with and without the BRAF mutation, which is consistent with our data. We observed that 8.3% (2/24) of MSI cancers showed the KRAS mutation, one of which was CIMP-positive and another one exhibited hMLH1 methylation (data not shown). On the other hand, 9.5% (4/42) of MSS cancers exhibited the BRAF mutation. Considering the extremely high correlation between SSA and the BRAF mutation, we hypothesized that MSS cancers with the BRAF mutation could be derived from SSA followed by intricate genetic/epigenetic alterations in the adenoma-carcinoma pathway.
Another interesting finding was that more than 40% of MSI cancers did not show the BRAF mutations and were negative for CIMP, a characteristic of SSA. We found that 8.3% of MSI cancers showed the KRAS mutation, which was frequently found in TAs. From these results, we postulate that a certain number of MSI cancers emerge from TA. A minor population of proximal TAs may acquire MSI, which results in MSI cancers harboring KRAS and not the BRAF mutation. This putative mutual relationship for carcinogenesis of proximal colon cancer is shown in Fig. 2. We believe this concept is consistent with the emerging fusion pathway proposed by Jass et al (32) in which molecular alterations that are characteristic of the serrated and adenoma-carcinoma sequence co-occur in a minority of advanced colorectal polyps.
Figure 2.
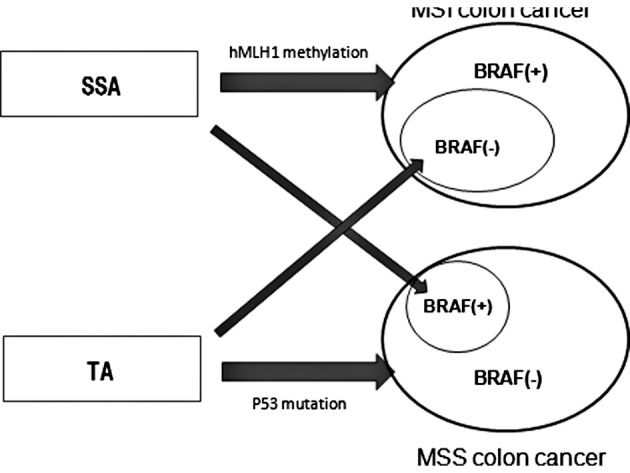
Putative mutual relationship for carcinogenesis between proximal colon cancer and precursor lesion. Whether colon cancer acquires MSI or not is highly influenced by its origin, SSA or TA, but it is not a definitive pathway for carcinogenesis.
In conclusion, similar genetic and epigenetic features between proximal SSAs and MSI cancers with the BRAF mutation suggest the strong possibility of proximal SSA as a precursor lesion for proximal MSI cancers with the BRAF mutation. The genetic and epigenetic features of proximal SSA and TA suggest the possibility that both polyps develop into either proximal MSI or MSS cancers.
Furthermore, the genetic and epigenetic features of proximal SSAs and TAs suggest that in a small percentage of cases, the former develop into proximal MSS cancers and the latter into proximal MSI cancers.
References
- 1.Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas. A distinct form of colorectal neoplasia. Am J Surg Pathol. 1990;14:524–537. doi: 10.1097/00000478-199006000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65–81. doi: 10.1097/00000478-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Torlakovic EE, Gomez JD, Driman DK, et al. Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA) Am J Surg Pathol. 2008;32:21–29. doi: 10.1097/PAS.0b013e318157f002. [DOI] [PubMed] [Google Scholar]
- 4.Torlakovic E, Snover DC. Serrated adenomatous polyposis in humans. Gastroenterology. 1996;110:748–755. doi: 10.1053/gast.1996.v110.pm8608884. [DOI] [PubMed] [Google Scholar]
- 5.Lee SK, Chang HJ, Kim TI, et al. Clinicopathologic findings of colorectal traditional and sessile serrated adenomas in Korea: a multicenter study. Digestion. 2008;77:178–183. doi: 10.1159/000142077. [DOI] [PubMed] [Google Scholar]
- 6.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi T, Jass JR. My approach to serrated polyps of the colorectum. J Clin Pathol. 2004;57:682–686. doi: 10.1136/jcp.2003.015230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandmeier D, Benhattar J, Martin P, Bouzourene H. Serrated polyps of the large intestine: a molecular study comparing sessile serrated adenomas and hyperplastic polyps. Histopathology. 2009;55:206–213. doi: 10.1111/j.1365-2559.2009.03356.x. [DOI] [PubMed] [Google Scholar]
- 10.Carr NJ, Mahajan H, Tan KL, Hawkins NJ, Ward RL. Serrated and non-serrated polyps of the colorectum: their prevalence in an unselected case series and correlation of BRAF mutation analysis with the diagnosis of sessile serrated adenoma. J Clin Pathol. 2009;62:516–518. doi: 10.1136/jcp.2008.061960. [DOI] [PubMed] [Google Scholar]
- 11.Kambara T, Simms LA, Whitehall VL, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lash RH, Genta RM, Schuler CM. Sessile serrated adenomas: prevalence of dysplasia and carcinoma in 2139 patients. J Clin Pathol. 2010;63:681–686. doi: 10.1136/jcp.2010.075507. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 14.Spring KJ, Zhao ZZ, Karamatic R, et al. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology. 2006;131:1400–1407. doi: 10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 15.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe T, Kobunai T, Toda E, et al. Distal colorectal cancers with microsatellite instability (MSI) display distinct gene expression profiles that are different from proximal MSI cancers. Cancer Res. 2006;66:9804–9808. doi: 10.1158/0008-5472.CAN-06-1163. [DOI] [PubMed] [Google Scholar]
- 17.Cho YK, Kim HC, Kim SH, et al. Location-related differences in sporadic microsatellite unstable colorectal cancer. Dig Liver Dis. 2010;42:611–615. doi: 10.1016/j.dld.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 19.Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878–4881. [PubMed] [Google Scholar]
- 20.Takemoto N, Konishi F, Yamashita K, et al. The correlation of microsatellite instability and tumor-infiltrating lymphocytes in hereditary non-polyposis colorectal cancer (HNPCC) and sporadic colorectal cancers: the significance of different types of lymphocyte infiltration. Jpn J Clin Oncol. 2004;34:90–98. doi: 10.1093/jjco/hyh018. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 23.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusano M, Toyota M, Suzuki H, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106:1467–1479. doi: 10.1002/cncr.21789. [DOI] [PubMed] [Google Scholar]
- 25.Kim YH, Kakar S, Cun L, Deng G, Kim YS. Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer. 2008;123:2587–2593. doi: 10.1002/ijc.23840. [DOI] [PubMed] [Google Scholar]
- 26.Vaughn CP, Wilson AR, Samowitz WS. Quantitative evaluation of CpG island methylation in hyperplastic polyps. Mod Pathol. 2010;23:151–156. doi: 10.1038/modpathol.2009.150. [DOI] [PubMed] [Google Scholar]
- 27.Miyakura Y, Sugano K, Konishi F, et al. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300–1309. doi: 10.1053/gast.2001.29616. [DOI] [PubMed] [Google Scholar]
- 28.Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 29.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zlobec I, Kovac M, Erzberger P, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. 2010;127:2569–2575. doi: 10.1002/ijc.25265. [DOI] [PubMed] [Google Scholar]
- 31.Lubomierski N, Plotz G, Wormek M, et al. BRAF mutations in colorectal carcinoma suggest two entities of microsatellite-unstable tumors. Cancer. 2005;104:952–961. doi: 10.1002/cncr.21266. [DOI] [PubMed] [Google Scholar]
- 32.Jass JR, Baker K, Zlobec I, et al. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a ‘fusion’ pathway to colorectal cancer. Histopathology. 2006;49:121–131. doi: 10.1111/j.1365-2559.2006.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


