Abstract
Aerobic granular sludge is attractive for high-rate biological wastewater treatment. Biomass wash-out conditions stimulate the formation of aerobic granules. Deteriorated performances in biomass settling and nutrient removal during start-up have however often been reported. The effect of wash-out dynamics was investigated on bacterial selection, biomass settling behavior, and metabolic activities during the formation of early-stage granules from activated sludge of two wastewater treatment plants (WWTP) over start-up periods of maximum 60 days. Five bubble-column sequencing batch reactors were operated with feast-famine regimes consisting of rapid pulse or slow anaerobic feeding followed by aerobic starvation. Slow-settling fluffy granules were formed when an insufficient superficial air velocity (SAV; 1.8 cm s−1) was applied, when the inoculation sludge was taken from a WWTP removing organic matter only, or when reactors were operated at 30°C. Fast-settling dense granules were obtained with 4.0 cm s−1 SAV, or when the inoculation sludge was taken from a WWTP removing all nutrients biologically. However, only carbon was aerobically removed during start-up. Fluffy granules and dense granules were displaying distinct predominant phylotypes, namely filamentous Burkholderiales affiliates and Zoogloea relatives, respectively. The latter were predominant in dense granules independently from the feeding regime. A combination of insufficient solid retention time and of leakage of acetate into the aeration phase during intensive biomass wash-out was the cause for the proliferation of Zoogloea spp. in dense granules, and for the deterioration of BNR performances. It is however not certain that Zoogloea-like organisms are essential in granule formation. Optimal operation conditions should be elucidated for maintaining a balance between organisms with granulation propensity and nutrient removing organisms in order to form granules with BNR activities in short start-up periods.
Keywords: biological wastewater treatment, aerobic granular sludge, granule formation, wash-out dynamics, bacterial selection, nutrient removal limitations
Introduction
Aerobic granular sludge (AGS) wastewater treatment processes are attractive for intensive and high-rate biological nutrient removal (BNR) and secondary clarification in single sequencing batch reactors (SBR; de Bruin et al., 2004; Giesen et al., 2012). Stable dense and fast-settling aerobic granules with tailored metabolic activities for the removal of carbon, nitrogen, and phosphorus are desired for the operation of robust AGS wastewater treatment plants (WWTP). For instance, the overgrowth of filamentous organisms must be avoided in order to prevent process disturbances by the deterioration of the settling properties of aerobic granules (van Loosdrecht et al., 2008).
The formation of aerobic granules has been stimulated by reactor start-up conditions leading to the wash-out of flocculent biomass and selecting for a fast-settling biomass, namely with the combination of short settling times of 3–5 min and short hydraulic retention times (HRT) of 6 h (Beun et al., 1999). Granulation can be impacted by additional operation parameters such as the influent feeding regime, the hydrodynamic shear force, and the concentration of dissolved oxygen (DO). For a review, refer to Lee et al. (2010). Granules have been successfully cultivated with feast-famine regimes involving pulse feeding (3–5 min) followed by prolonged aeration (3–4 h; Morgenroth et al., 1997; Beun et al., 1999; Tay et al., 2002), or anaerobic feeding (1 h) followed by aerobic starvation (2 h; de Kreuk et al., 2005). Granulation has only been observed with up-flow superficial air velocities (SAV) above 0.010 m s−1, typically between 0.025 and 0.045 m s−1. High up-flow aeration induce high shear and compaction forces at the surface of granules (Zima et al., 2007), and stimulate the production of exopolymeric substances (EPS) as well as hydrophobic adhesive interactions (Liu and Tay, 2002; Dulekgurgen et al., 2008).
Several studies have however reported on the deterioration of the settling properties of aerobic granules by overgrowth of filamentous microbial structures, called filamentous bulking. For a review, refer to Liu and Liu (2006). This phenomenon has been observed with volumetric organic loading rates (OLR) above 6 kgCODs d−1 m−3 (Shin et al., 1992; Moy et al., 2002), with high-energy carbon sources such as carbohydrates (Morgenroth et al., 1997; Weber et al., 2007), and at higher mesophilic temperatures of 30–35°C (Weber et al., 2007; Ebrahimi et al., 2010). Filamentous overgrowth has been limited with higher up-flow mixing or aeration velocities, and with the use of acetate as carbon source (Liu and Liu, 2006). Despite the reduction of filamentous bulking with this substrate, residual filamentous structures have still been observed, and have been presumed to act as backbones for the immobilization of microbial colonies (Martins et al., 2004). In studies investigating granulation in up-flow anaerobic sludge blanket reactors, it has been observed that the same Methanosaeta-affiliating phylotype was constantly dominating the bacterial community during the evolution of fluffy granules to compact granules under the progressive increase in shear forces in the reactor (Grotenhuis et al., 1992; Hulshoff Pol et al., 2004). In the case of aerobic granules, analysis of bacterial compositions of fluffy and dense granules is required to assess whether different granule structures exhibit the same dominant phylotypes or not.
From the nutrient removal point of view, AGS studies initially concentrated on the formation of aerobic granules, and on the removal of organic matter (Morgenroth et al., 1997; Beun et al., 1999; Tay et al., 2002). Emphasis has then been put on achieving nitrification (Tsuneda et al., 2003), denitrification (Beun et al., 2001; Mosquera-Corral et al., 2005), dephosphatation (Lin et al., 2003), and combined BNR (de Kreuk et al., 2005; Lemaire et al., 2008; Yilmaz et al., 2008) in AGS systems. However, it has been shown that 75–100 days have been required to obtain efficient nutrient removal activities in aerobic granules after reactor start-up with flocculent activated sludge (de Kreuk et al., 2005; Xavier et al., 2007; Ebrahimi et al., 2010; Gonzalez-Gil and Holliger, 2011). In these studies, only carbon has been removed during the start-up period. Some authors have achieved enhanced granulation with faster improvements in nutrient removal performances by seeding reactors with crushed pre-cultivated granules (Pijuan et al., 2011; Verawaty et al., 2012). However, the reason why phosphorus and nitrogen removal activities have been inhibited during the first 3 months of reactor start-up with flocculent inoculation sludge and wash-out conditions has not yet been further investigated. Different microbial ecology studies have mainly been conducted on mature granules (Adav et al., 2010; Gonzalez-Gil and Holliger, 2011), but only little information is available on the microbial composition of early-stage AGS.
The present study aimed to investigate the bacterial community dynamics during the formation of early-stage aerobic granules (0–60 days) in bubble-column SBRs operated under conditions selecting for a fast-settling biomass. The main objective was to assess the effect of wash-out dynamics and operation conditions on the underlying bacterial selection, the shape of aerobic granules, the biomass settling properties, and the nutrient removal performances. We first focused on the differences in predominant bacterial populations between compact and fluffy granules, and on a way to avoid filamentous bulking in AGS systems. We then carried out a detailed monitoring of the start-up of one reactor to detect correlations between operation conditions, bacterial community dynamics and nutrient removal performances. The knowledge gained at the microbial ecology level enabled to determine why nutrient removal deteriorated during the start-up of the granulation process in bubble-column SBRs operated under wash-out conditions.
Materials and Methods
Reactor infrastructure and sequencing batch operation
The design and the operation of the bubble-column SBRs were adapted from de Kreuk et al. (2005). The bubble-columns consisted of internal diameters of 52–62 mm, height-to-diameter ratios of 20–25, and working volumes of 2.1–3.1 L. The SBRs were operated in fixed cycles of 3 h comprising feeding of the influent wastewater through the settled sludge bed in pulse (6 min) or anaerobic regime (60 min), aeration (110 min), biomass settling (5 min or stepwise decrease from 15 to 3 min), and withdrawal of the treated effluent (remaining cycle time). The SBRs were inoculated with 2–3 gVSS L−1 of flocculent activated sludge originating from full-scale WWTPs. Biomass wash-out conditions were imposed with a short HRT of 6 h in order to stimulate granulation, according to Beun et al. (1999). A volume exchange ratio of 50% was applied to this end. The SBRs were operated under biomass dynamic conditions at undefined sludge retention time (SRT). The SRT was a function of the imposed settling time, of the height of the effluent withdrawal point, and of the intrinsic settling properties of the cultivated biomass. The composition of the synthetic cultivation media was similar to the one used by Ebrahimi et al. (2010), and is available in Table A1 in Appendix. Acetate was supplied as sole carbon and energy source at a constant concentration between 400 and 500 mgCODs L−1 in the influent wastewater. This resulted in a constant volumetric OLR of 200–250 mgCODs cycle−1 LR−1 (or 1.6–2.0 kgCODs d−1 mR−3 daily equivalents), and in an initial biomass specific OLR of 50–60 mgCODs cycle−1 gCODx−1 (or 0.4–0.6 kgCODs d−1 kgCODx−1 daily equivalents). The biomass specific OLR was a dynamic function of the residual biomass concentration evolving in the reactor. The phosphorous and nitrogenous nutrient ratios amounted to 4.8 gP-PO4 and 12.5 gN-NH4 per 100 gCODs, respectively. During aeration, air was supplied at the target flow-rate with mass flow controllers (Brooks Instrument, Netherlands), DO was not controlled and reached saturation (8–9 mgO2 L−1), and pH was regulated at 7.0 ± 0.2 by addition of 1 M HCl or NaOH with a proportional-integral controller.
Granulation experiments
In the first part, the granulation process was studied in five reactors (R1–R5) where the bacterial community compositions of early-stage AGS were analyzed in relation with the different combinations of operation parameters, as summarized in Table 1.
Table 1.
Operation parameters applied during the granulation start-up experiments.
| Reactor1 | Inoculation sludge2 | Temperature (°C) | Feeding regime4 | Up-flow SAV (cm s−1) | Settling time6 (min) |
|---|---|---|---|---|---|
| R1 | OMR | 23 ± 23 | Pulse | 1.8/4.05 | 5 |
| R2 | OMR | 23 ± 23 | Anaerobic | 1.8/4.05 | 5 |
| R3 | BNR | 20 | Anaerobic | 1.8 | 15 to 3 |
| R4 | BNR | 20 | Anaerobic | 2.0 | 15 to 3 |
| R5 | BNR | 30 | Anaerobic | 1.8 | 15 to 3 |
| R6 | BNR | 23 ± 23 | Anaerobic | 2.5 | 15 to 3 |
1R1–R5 were run in the first part of the study investigating differences in bacterial community compositions of early-stage aerobic granules. R6 was operated for detailed analysis of process conditions governing bacterial selection during granulation.
2The reactors were inoculated with flocculent activated sludge originating either from a WWTP designed for organic matter removal (OMR) only, or from a WWTP operated for full biological nutrient removal (BNR).
3R1, R2, and R6 were operated at ambient temperature without temperature control.
4The synthetic influent wastewater was supplied either in 6 min with a pulse-feeding regime, or in 60 min with an anaerobic-feeding plug-flow regime.
5R1 and R2 were operated first with a low up-flow superficial air velocity (SAV) of 1.8 cm s−1. This parameter was doubled after 4 weeks for remediating filamentous overgrowth.
6Two different biomass settling patterns were tested with a constant settling time of 5 min, or with stepwise decrease in the settling time from 15 to 3 min.
The first two reactors R1 and R2 were inoculated with a flocculent activated sludge originating from a WWTP designed for organic matter removal (OMR) only (ERM Morges, Switzerland). R1 and R2 were operated during at most 50 days at ambient temperature (23 ± 2°C), with pulse (6 min) or anaerobic plug-flow (60 min) feeding regimes, with an initially low up-flow SAV of 1.8 cm s−1 during aeration, and with a constant settling time of 5 min. After having observed proliferation of fluffy granules (30 days), the up-flow SAV was increased to 4.0 cm s−1 in order to obtain dense granules.
The reactors R3, R4, and R5 were inoculated with a flocculent activated sludge originating for a WWTP designed for full BNR along an anaerobic-anoxic-aerobic process (ARA Thunersee, Switzerland). These reactors were operated at either low (20°C, R3 and R4) or high (30°C, R5) mesophilic temperature, with anaerobic plug-flow feeding (60 min), with a low up-flow SAV of 1.8–2.0 cm s−1 during aeration, and with a stepwise decrease in the settling time from 15 to 3 min in 10–15 days. The operation of R4 and R5 has previously been described in detail by Ebrahimi et al. (2010), and lasted over 40 days. R3 was run on a shorter period of 15 days, but microbial ecology data were collected at higher frequency during the transition from flocculent to granular sludge.
In the second part, reactor R6 was operated with a combination of conditions selecting for the formation of dense fast-settling granules and a detailed monitoring of operation conditions, bacterial community compositions, and nutrient removal performances was carried out. R6 was inoculated with flocculent activated sludge taken from the BNR-WWTP, and was operated during 60 days at 23 ± 2°C and pH 7.0 ± 0.2, with anaerobic plug-flow feeding (60 min), a moderate up-flow SAV of 2.5 cm s−1, and a stepwise decrease in the settling time from 15 to 3 min. Temperature, pH, DO, and electrical conductivity signals were collected on-line. Concentrations of biomass present in the reactor and in the treated effluent, and microbial ecology data were collected on a daily basis. Liquid phase samples were taken every 3–5 days for physicochemical analyses of soluble compounds in the influent wastewater, in the reactor at the end of the anaerobic phase, and in the treated effluent.
Characterization of metabolic activities of the inoculation sludge taken from the BNR-WWTP
The nutrient removal capacities of the inoculation sludge taken from the BNR-WWTP were tested in anaerobic, aerobic, and anoxic batch tests, and compared to the operation data of the BNR-WWTP. The tests were run at 20°C in 2-L stirred tank reactors with a biomass concentration of 3–4 gVSS L−1 and with similar starting nutrient concentrations as in R6.
Analyses of soluble compounds and biomass
Acetate concentration was determined with a high performance liquid chromatograph equipped with an organic acids ion exclusion column ORH-801 (Transgenomics, UK) and a refraction index detector (HPLC Jasco Co-2060 Plus, Omnilab, Germany). The concentration of anions was measured with an ICS-90 ion exchange chromatograph equipped with an IonPacAS14A column and an electrical conductivity detector (Dionex, Switzerland). The concentration of cations was measured with an ICS-3000A ion exchange chromatograph equipped with an IonPacCS16 column and an electrical conductivity detector (Dionex, Switzerland).
The particulate concentrations of total (TSS), volatile (VSS), and inorganic suspended solids (ISS) were measured according to de Kreuk et al. (2005). Granules were observed by light microscopy.
Molecular analyses of bacterial community compositions
The compositions and dynamics of the bacterial communities were characterized by terminal-restriction fragment length polymorphism (T-RFLP) analysis targeting the v1–v3 hypervariable region of the Eubacteria 16S rRNA gene pool. The T-RFLP method was adapted from Rossi et al. (2009) and Ebrahimi et al. (2010), and contained the following modifications. DNA was extracted from 100 mg of homogenized biomass samples using the Maxwell 16 Tissue DNA Purification System (Promega, Switzerland). Gene fragments of 500 bp were amplified by PCR using universal eubacterial primers: a FAM-labeled 8-F forward primer (FAM-5′-GAGTTTGATCMTGGCTCAG-3′) and an unlabeled 518-R reversed primer (5′-ATTACCGCGGCTGCTGG-3′). The PCR program was run in a T3000 Thermocycler (Biometra GmbH, Germany) in 30 cycles comprising a longer denaturation time than the one used by the authors, for optimal amplification of Accumulibacter-related polyphosphate-accumulating organisms (PAO): 10 min initial denaturation (95°C), 1 min denaturation (95°C), 45 s primer annealing (56°C), 2 min elongation (72°C), 10 min final elongation (72°C). The amplicons were purified and concentrated using Invisorb MSB Spin PCRapace purification kits (Invitek Stratec Molecular GmbH, Germany). Amounts of 200 ng of purified PCR products were digested at 37°C for 3 h with 0.5 units of the HaeIII endonuclease (Promega, Switzerland). Volumes of 1 μL of digestion products were mixed with 8.5 μL of HiDi formamid and 0.5 μL of GeneScan 600-LIZ internal size standard (Applied Biosystems, USA), and were denaturated for 2 min at 95°C. The terminal-restriction fragments (T-RFs) were separated and analyzed by capillary gel electrophoresis in an ABI Prism 3100-Avant Genetic Analyzer using a fluid POP-6 gel matrix and fluorescence laser detection (Applied Biosystems, USA). The T-RFLP profiles were aligned using the Treeflap crosstab macro (Rees et al., 2004). The bacterial community structures were expressed as relative contributions of all operational taxonomic units (OTU) contributing to the total measured fluorescence. Predominant OTUs with relative abundances above 2% were presented in stacked bar plots for simplified visual observation. Three single biomass samples from the whole set of samples of the study were analyzed in triplicates to determine the overall relative standard deviation related to the T-RFLP method (6%). For reactor R6, biomass equivalents of target OTUs were expressed by multiplying their relative abundances by the mass of VSS present in the reactor.
Analysis of the richness and diversity of the bacterial community evolving in reactor R6
Richness and Shannon’s H’ diversity indices were computed with the R software version 2.14.1 equipped with the Vegan package (R-Development-Core-Team, 2008; Oksanen et al., 2009) based on the full T-RFLP profiles collected during experiment R6. Mathematical geometric evolution models were fitted to the measured richness and diversity profiles with the Berkeley Madonna software (Macey et al., 2000) in order to simulate the evolution of both indices in the reactor. Standard deviation intervals on model predictions were computed from 1000 Monte Carlo simulations on underlying parameters.
Phylogenetic affiliation of operational taxonomic units
Predominant OTUs detected in R1–R5 were affiliated to closest bacterial relatives by using the cloning-sequencing databank developed by Ebrahimi et al. (2010) and complemented in the present study. Two DNA extracts from biomass samples collected in R6 at day 2 (flocculent sludge) and day 59 (granular sludge) were sent to Research and Testing Laboratory (Lubbock, TX, USA) for 454 Tag-encoded FLX amplicon pyrosequencing with a Genome Sequencing FLX System (Roche, Switzerland) using the procedure developed by Sun et al. (2011), and the same primers (8-F and 518-R) as the ones used for T-RFLP analysis. The pyrosequencing datasets were denoised and processed with the PyroTRF-ID bioinformatics procedure developed by Weissbrodt and Shani, et al. (paper submitted), which includes sequence annotation with the Greengenes database (McDonald et al., 2012), digital T-RFLP profiling, comparison of digital and experimental T-RFLP profiles, and phylogenetic affiliation of OTUs. QIIME algorithms were used in the denoising process (Caporaso et al., 2010).
Bacteriome analysis
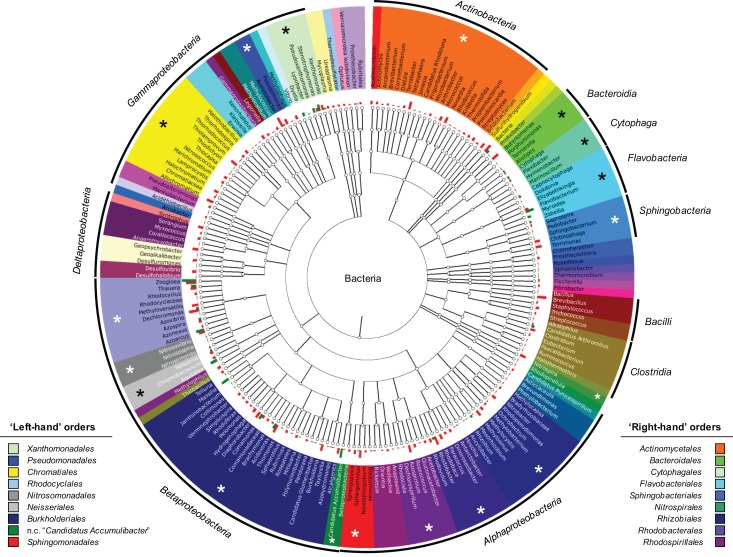
The pyrosequencing datasets of the two biomass samples collected in R6 were analyzed by the metagenomics RAST server (MG-RAST; Meyer et al., 2008) for annotation and comparative analysis. The Ribosomal Database Project (RDP; Cole et al., 2009) was used as sequence annotation source. A minimum identity cutoff of 97% was applied in order to retain only the closest bacterial affiliations. A circular phylogenetic tree was constructed with the pyrosequencing datasets of the two samples. The tree was complemented with two bar plots representing the relative abundances of the bacterial genera in both samples. Richness and Shannon’s H’ diversity indices were also computed from these datasets.
Results
Composition and activity of early-stage granules cultivated from flocculent OMR-sludge
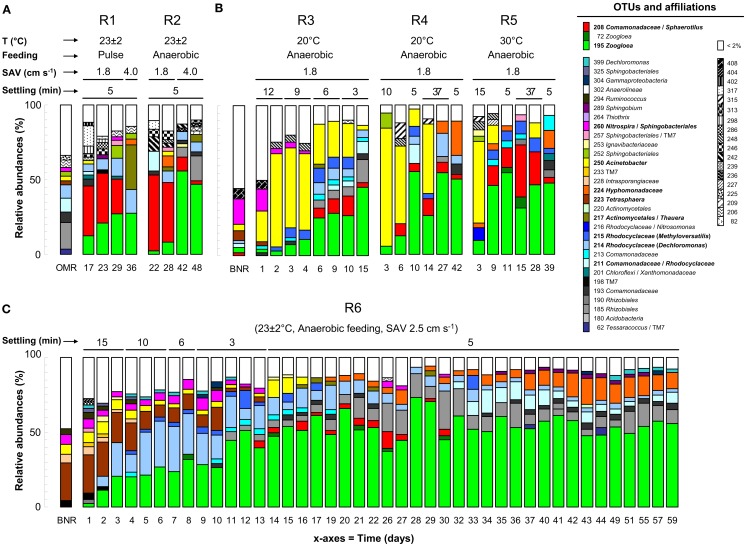
Reactors R1 and R2 were inoculated with flocculent activated sludge taken from an aeration tank of a WWTP designed for OMR only (OMR-sludge). Operation at ambient temperature (23 ± 2°C), with acetate as carbon source, a low up-flow SAV of 1.8 cm s−1, and a fixed settling time of 5 min resulted in the proliferation of slow-settling fluffy early-stage granules (Figure 1A). Segmented chain filamentous bacterial structures were detected by light microscopy (Figure 1B). The underlying bacterial community compositions are presented in Figure 2A. Closest bacterial affiliations of specific OTUs detected in these reactors are given in Table A2 in Appendix. The fluffy granules obtained in R1 with pulse feeding (6 min) were predominantly composed of Burkholderiales affiliates related to the Sphaerotilus-Leptothrix group (OTU-208, 23–33%), and by Zoogloea spp. belonging to Rhodocyclales (OTU-195, 12–27%). In R2 operated with anaerobic feeding (60 min), Sphaerotilus-Leptothrix affiliates dominated (39–50%) over Zoogloea spp. (2–8%) in fluffy granules.
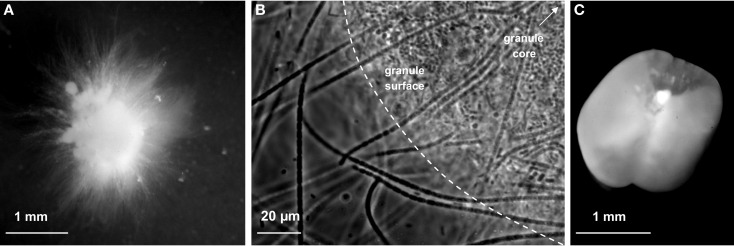
Figure 1.
Example of early-stage aerobic granule structures observed with light microscopy. Fluffy slow-settling granule obtained after 30 days in reactor R2 with OMR-sludge and low up-flow SAV of 1.8 cm s−1, and exhibiting filamentous outer structures (A). Filamentous segmented chain bacterial structures interspersing across the granular biofilm observed on a sample collected on day 22 in R2 (B). Dense fast-settling granule present after 50 days in R6 with BNR-sludge and moderate up-flow SA of 0.025 m s−1, and displaying a tulip-like folded structure around a more opaque internal core (C).
Figure 2.
Dynamics of predominant bacterial OTUs analyzed with T-RFLP during the six granulation experiments. Reactors R1 and R2 were inoculated with activated sludge from the OMR-WWTP (A). R3, R4, and R5 were inoculated with activated sludge from the BNR-WWTP (B). High resolution bacterial ecology data were collected from R6 to assess the effect of wash-out dynamics on bacterial selection during granulation (C). Main operation conditions are indicated at the top of each graph. Closest bacterial affiliations of target OTUs presented in Table 2 are given on the right.
The application of a higher up-flow SAV of 4.0 cm s−1 after 30 days resulted in the recovery of smooth and dense fast-settling granules. The predominant organisms shifted from Sphaerotilus-Leptothrix affiliates to Zoogloea spp. In the granules of R1, Zoogloea spp. (27%) and Thaurea spp. (OTU-217, 30%) outcompeted Burkholderiales (<1%). The dense granules of R2 were highly dominated by Zoogloea spp. (47–57%). Burkholderiales decreased to 2% after day 48. OTU-185 affiliating with Gammaproteobacteria and with Rhizobiales from Alphaproteobacteria was present up to 17%.
At physical reactor boundaries, filamentous bulking led to deteriorated sludge settling. Both reactors displayed poor nutrient removal performances. After the recovery of fast-settling granules, nutrient removal did not improve. After pulse feeding in R1, acetate was fully removed within 40 min during the aeration phase. In R2 where slow plug-flow anaerobic feeding was applied, more than 90% of the acetate leaked into the aeration phase during which it was fully consumed. Ammonium was not nitrified and biological dephosphatation did not occur. Only partial nitrogen (20%) and phosphorus removal (10%) was detected in both reactors which was probably due to anabolic requirements.
Composition and activities of early-stage granules cultivated from flocculent BNR-sludge
The reactors R3, R4, and R5 were operated with an inoculation sludge originating from a WWTP designed for full BNR, under anaerobic feeding, and with a stepwise decrease in the settling time from 15–3 min. The three reactors resulted in the formation of smooth and dense fast-settling granules after 9–10 days (Figure 1C). The underlying bacterial community dynamics are presented in Figure 2B. The inoculation sludge from the BNR-WWTP was dominated by Nitrospira and Sphingobacteriales (OTU-260, 17%), Tetrasphaera spp. (OTU-223, 7%), and an unidentified OTU-408 (7%). Zoogloea spp. (OTU-195), Burkholderiales (OTU-207), OTU-210 affiliating with Acidobacteriales and Firmicutes, and OTU-214 affiliating with Rhodocyclales-related organisms such as Dechloromonas and Methyloversatilis spp. were present in lower abundances (2–3%). In all three reactors, the sludge was still in the flocculent state over the first 6 days. The predominant organisms of the inoculum were replaced within 2–3 days by Acinetobacter spp. (OTU-250, 21–79%). Granulation correlated with the proliferation of Zoogloea spp. (28–55%). Dechloromonas (5–16%), Methyloversatilis (3–10%), and Rhizobiales (4–16%) were detected as flanking populations. Hyphomonadaceae affiliates were abundantly present after 42 days in R4 (23%), and after 39 days in R5 (12%). In contrast to the operation at 20°C in R3 and R4 where dense fast-settling granules were constantly present, the operation at 30°C in R5 led to the proliferation of organisms affiliated to the Sphaerotilus-Leptothrix group (35%) and resulted in a mixture of dense and fluffy granules. Even though a BNR-sludge was used as inoculum, nitrification and dephosphatation activities were not detected in the three AGS systems. Acetate was only consumed to a small extent during the anaerobic feeding phases (18–25%) and fully removed during the aeration phases.
Dynamics of bacterial communities and process performance under wash-out conditions
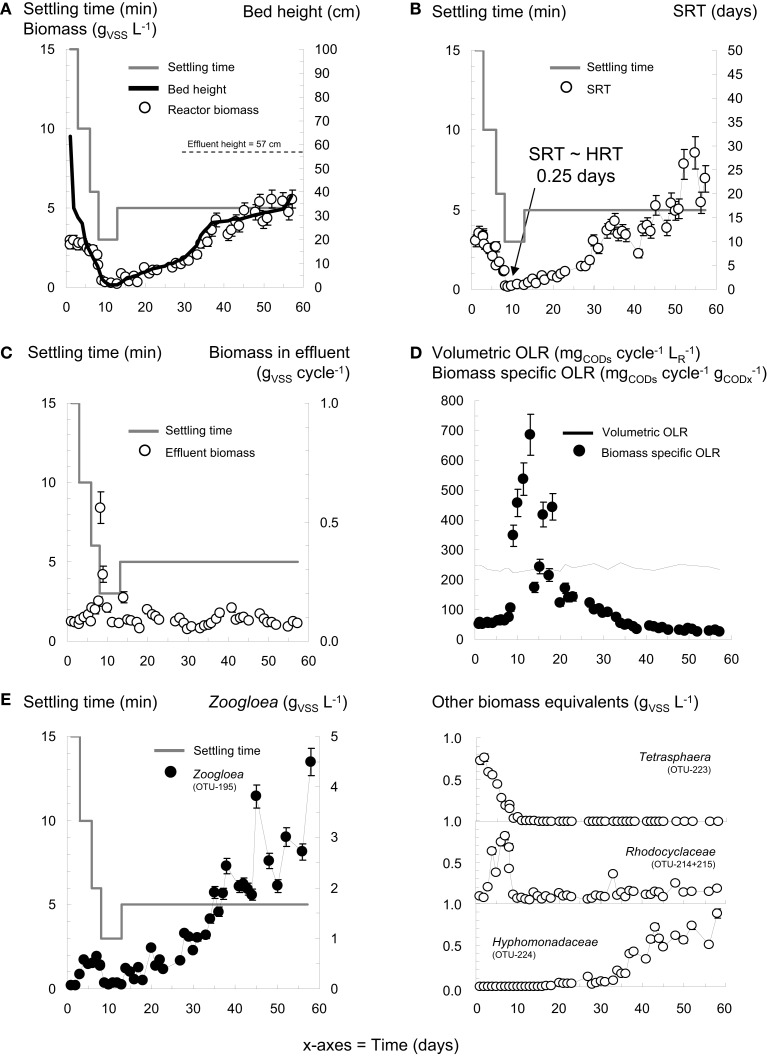
For reactor R6, high frequency of data collection allowed to detect correlations between operation conditions, bacterial dynamics, and BNR performances during early-stage granulation under wash-out conditions. Changes in biomass properties are presented in Figures 3A,B in function of the settling time. With initial settling times of 15 and then 10 min during the first 5 days, the activated sludge remained in the flocculent state and a biomass concentration of 2.45–2.95 gVSS L−1 was maintained in the reactor, forming a settled sludge blanket of 15–30 cm, and the SRT amounted to 12 days. The decrease in the settling time from 6 to 3 min at day 8 resulted in extensive biomass wash-out (Figure 3C). An extremely low residual biomass concentration of 0.2 gVSS L−1 was remaining in the system, and formed a settled sludge blanket of only 1 cm. The SRT dropped to 0.5 day, and approached the HRT of 0.25 day. First granule nuclei were observed after 10 days. At day 12, the settling time was increased to 5 min as safety measure to keep the granules in the system. The granular biomass increased to 4.0 gVSS L−1 at day 37, progressively stabilized at 5.3 ± 0.4 gVSS L−1 after 52 days, and formed a settled sludge blanket of 32–40 cm. The fraction of ISS amounted to 38% in the inoculation sludge, and to 12% in the early-stage AGS. An example of a dense fast-settling granule present in R6 after 50 days is presented in Figure 1C.
Figure 3.
Detailed evolution of biomass parameters during granulation in reactor R6 under wash-out dynamics, in function of the imposed settling time. The evolution of the biomass concentration and of the height of the settled sludge blanket displayed similar profiles as soon as the settling time was decreased to 3 min (A). The application of wash-out conditions resulted in the re-coupling of the SRT to the HRT at day 8 (B). A high amount of biomass was withdrawn with the effluent wastewater during wash-out from day 8 on (C). Under reactor operation with a constant volumetric OLR, the biomass specific OLR exhibited a drastic increase during the period of extensive wash-out between day 8 and day 20 (D). Zoogloea, Tetrasphaera, Rhodocyclaceae, and Hyphomonadaceae affiliates displayed distinct biomass evolutions (E). Early-stage granules nuclei formed after 10 days.
The reactor was operated with a constant volumetric OLR of 250 mgCODs cycle−1 LR−1. The biomass specific OLR however evolved with the residual biomass concentration from initially 51 to 685 mgCODs cycle−1 gCODx−1 between day 8 and day 20 after the intensive biomass wash-out (Figure 3D). As the AGS biomass grew in the system, the biomass specific OLR progressively decreased to 20 mgCODs cycle−1 gCODx−1.
Tetrasphaera spp. (OTU-223) dominated in the inoculation sludge (26%), and were progressively replaced in the flocculent sludge after 5 days by Zoogloea spp. (OTU-195, 21%) and by OTU-214 (29%; Figure 2C). During this initial phase, mainly Dechloromonas and Comamonadaceae relatives contributed to OTU-214, whereas Accumulibacter accounted for only 1% of this OTU (Table 2). When expressed as biomass concentration equivalents, OTU-195 and OTU-214 increased during this period up to 0.6 and 0.8 gVSS L−1, respectively (Figure 3E). The extensive biomass wash-out at day 8 resulted in the rapid decrease in the masses of all bacterial populations below 0.1 gVSS L−1. Zoogloea spp. then rapidly proliferated up to a relative abundance of 54 ± 8% in the early-stage AGS from day 15 to day 60. Other Rhodocyclales affiliates (OTUs 214 and 215) declined below 5% at day 26. The concentration of Zoogloea spp. amounted to 3.0 gVSS L−1 after 52 days. The concentration of other Rhodocyclales affiliates remained low, but exhibited a slight increase from 0.06 to 0.19 gVSS L−1 from day 10 to day 60.
Table 2.
Closest phylogenetic bacterial affiliations of specific OTUs detected in reactor R6, that were obtained after pyrosequencing, mapping, and digital T-RFLP profiling with the PyroTRF-ID bioinformatics procedure (Weissbrodt and Shani et al., paper submitted).
| T-RF1(bp) | Sample2 | Counts3 (−) | Fraction of T-RF4 (%) | Affiliation5 |
Accession number5 | Closest relative and original microbiota6 | Smith-Waterman mapping score (−)7 |
||
|---|---|---|---|---|---|---|---|---|---|
| Phylum → Order | Family → Genus | Absolute | Normalized | ||||||
| 62 | Day 2 | 22 | 67 | O: Actinomycetales | G: Tessaracoccus | GQ097568 | Uncultured bacterium clone nbw397a07c1 from human skin microbiome | 380 | 0.977 |
| 10 | 30 | P: candidate phylum TM7 | EU104134 | Uncultured bacterium clone M0509_49 from activated sludge | 373 | 0.912 | |||
| 72 | Day 2 | 18 | 40 | O: Rhodocyclales | G: Zoogloea | AJ011506 | Zoogloea resiniphilia strain DhA-35T | 376 | 0.962 |
| 13 | 35 | O: Rhodocyclales | G: Thauera | AY945909 | Uncultured bacterium clone DR-48 from denitrifying bioreactor | 348 | 0.926 | ||
| Day 59 | 11 | 100 | O: Rhodocyclales | G: Zoogloea | AJ011506 | Zoogloea resiniphilia strain DhA-35T | 373 | 0.942 | |
| 180 | Day 2 | 4 | 100 | P: Acidobacteria | GQ396818 | Uncultured bacterium clone AK1AB1_04A from recently deglaciated soils | 323 | 0.807 | |
| 185 | Day 2 | 8 | 89 | O: Rhizobiales | FJ719048 | Uncultured bacterium clone p03_D09 from aquifer sediments | 296 | 0.757 | |
| 1 | 11 | O: Rhodobacterales | FJ719099 | Uncultured bacterium clone p04_H05 from aquifer sediments | 318 | 0.941 | |||
| Day 59 | 14 | 100 | O: Rhizobiales | AF502218 | Uncultured bacterium clone HP1B02 from EBPR sludge | 409 | 0.969 | ||
| 190 | Day 2 | 20 | 49 | O: Rhizobiales | CU918969 | Uncultured Alphaproteobacterium clone QEEA1DD02 from anaerobic digester | 404 | 0.962 | |
| 18 | 44 | O: Burkholderiales | G: Acidovorax | EU539550 | Uncultured bacterium clone nbt241e12 from human skin microbiota | 356 | 0.944 | ||
| 193 | Day 2 | 22 | 30 | O: Burkholderiales | G: Acidovorax | EU375646 | Acidovorax sp. u41 from bacterial community degrading organic pollutants | 363 | 0.936 |
| 21 | 28 | O: Burkholderiales | G: Simplicispira | AJ505861 | Comamonadaceae bacterium strain PIV-16-2 from denitrifying bacterial community | 377 | 0.969 | ||
| 13 | 17 | O: Burkholderiales | G: Delftia | EF515241 | Uncultured bacterium clone 21g10 from electricigen enrichment in a MFC | 355 | 0.920 | ||
| Day 59 | 72 | 94 | O: Burkholderiales | G: Acidovorax | AJ864847 | Acidovorax sp. strain J33 from high mountain lake habitats | 383 | 0.923 | |
| 5 | 6 | O: Xanthomonadales | EU662583 | Uncultured bacterium clone MC1_16S_13 from sulfidic karst system | 343 | 0.868 | |||
| 195 | Day 2 | 231 | 84 | O: Rhodocyclales | G: Zoogloea | AF234684 | Uncultured sludge bacterium H7 from nitrifying-denitrifying industrial WWTP | 385 | 0.990 |
| 18 | 7 | O: Pseudomonadales | G: Acinetobacter | GQ073520 | Uncultured bacterium clone nbw209f04c1 from human skin microbiome | 337 | 0.988 | ||
| Day 59 | 4793 | 100 | O: Rhodocyclales | G: Zoogloea | EU639144 | Uncultured bacterium clone LPB19 from EBPR sludge | 402 | 0.854 | |
| 198 | Day 2 | 65 | 94 | P: candidate phylum TM7 | DQ640696 | Uncultured TM7 bacterium clone Skagenf80 from EBPR-WWTP | 367 | 0.870 | |
| 201 | Day 2 | 10 | 48 | P: Chloroflexi | CU927307 | Uncultured Chloroflexi bacterium clone EDN7BB07 from anaerobic digester | 443 | 1.000 | |
| 8 | 38 | O: Xanthomonadales | F: Xanthomonadaceae | FJ612198 | Uncultured bacterium clone DP3.5.36 from lake ecosystem | 360 | 0.923 | ||
| 208 | Day 2 | 6 | 30 | O: Burkholderiales | G: Rhodoferax | AB154311 | Uncultured bacterium clone S9F-53 from eutrophic lake | 351 | 0.931 |
| 1 | 10 | O: Burkholderiales | G: Sphaerotilus | AB087568 | Sphaerotilus sp. L19 from filamentous activated sludge bulking process | 374 | 0.979 | ||
| 210 | Day 2 | 6 | 42 | O: Acidobacteriales | FJ230900 | Uncultured bacterium clone F25 from river water receiving antibiotics-rich effluents | 403 | 0.988 | |
| 5 | 35 | P: Firmicutes | G: Trichococcus | EU234209 | Uncultured bacterium clone B14 from river water receiving antibiotics-rich effluents | 295 | 1.000 | ||
| 211 | Day 59 | 7 | 54 | O: Burkholderiales | F: Comamonadaceae | EF540425 | Uncultured soil bacterium clone MK4a from semi-coke | 345 | 0.925 |
| 6 | 46 | O: Rhodocyclales | F: Rhodocyclales | DQ088735 | Uncultured bacterium clone BE16FW031601GDW_hole1-9 from gold mine groundwater | 343 | 0.724 | ||
| 213 | Day 2 | 17 | 73 | O: Burkholderiales | F: Comamonadaceae | AY662010 | Uncultured bacterium clone 300A-D08 from groundwater contaminated with nitric acid | 365 | 0.979 |
| 214 | Day 2 | 136 | 48 | O: Rhodocyclales | G: Dechloromonas | AY064177 | Uncultured Betaproteobacterium clone UCT N123 from EBPR-WWTP | 382 | 0.977 |
| 45 | 16 | O: Burkholderiales | G: Rhodoferax | AB452981 | Betaproteobacterium clone HIBAF001 from freshwater bacterioplankton | 366 | 0.948 | ||
| 29 | 10 | O: Rhodocyclales | G: Methyloversatilis | AY436796 | Methyloversatilis universalis strain EHg5 isolated from sediments | 364 | 0.958 | ||
| 12 | 4 | O: Rhodocyclales | G: Zoogloea | DQ413172 | Zoogloea sp. EMB 357 isolated from anaerobic-aerobic SBR | 344 | 0.901 | ||
| 11 | 4 | O: Burkholderiales | G: Aquamonas | DQ521469 | Uncultured bacterium clone ANTLV1_A07 from Antarctica lake ice cover microbiota | 366 | 0.951 | ||
| 4 | 1 | O: Rhodocyclales | G: Rhodocyclus | EF565151 | Uncultured bacterium clone VIR_D5 from EBPR sludge rich in Accumulibacter | 368 | 0.981 | ||
| Day 59 | 14 | 88 | O: Rhodocyclales | G: Dechloromonas | DQ413103 | Uncultured bacterium clone 44 from anaerobic-aerobic SBR | 381 | 0.890 | |
| 2 | 12 | O: Rhodocyclales | G: Rhodocyclus | AF502224 | Uncultured bacterium clone HP1A13 from EBPR sludge | 372 | 0.923 | ||
| 215 | Day 2 | 14 | 44 | O: Rhodocyclales | G: Methyloversatilis | DQ066958 | Uncultured bacterium clone pLW-7 from sediment consortium metabolizing C1 compounds | 349 | 0.928 |
| 9 | 28 | P: Chloroflexi | G: Caldilinea | CU917747 | Uncultured Chloroflexi bacterium clone QEEB2DA06 from anaerobic digester | 279 | 0.730 | ||
| 2 | 6 | O: Rhodocyclales | G: Rhodocyclus | FJ719063 | Uncultured bacterium clone p04_A04 from aquifer sediments | 320 | 0.938 | ||
| 1 | 3 | O: Rhodocyclales | G: Dechloromonas | AY062126 | Uncultured Betaproteobacterium clone UCT N141 from EBPR-WWTP | 306 | 0.820 | ||
| 216 | Day 2 | 5 | 35 | O: Rhodocyclales | F: Rhodocyclales | NR029035 | Quatrionicoccus australiensis strain Ben 117 from activated sludge | 311 | 0.881 |
| 3 | 21 | O: Burkholderiales | F: Comamonadaceae | EU180529 | Betaproteobacterium BAC49 from granular activated carbon filters | 273 | 0.853 | ||
| 1 | 7 | O: Nitrosomonadales | G: Nitrosomonas | EU937892 | Uncultured bacterium clone 3BR-6DD from an iron oxidizing freshwater habitat | 348 | 0.909 | ||
| 217 | Day 2 | 15 | 46 | O: Actinomycetales | AF513101 | Uncultured bacterium clone 7 from foaming activated sludge | 385 | 0.955 | |
| 12 | 36 | O: Rhodocyclales | G: Thauera | AM084110 | Thauera sp. R-28312 from denitrifying sludge | 386 | 1.000 | ||
| 223 | Day 2 | 545 | 99 | O: Actinomycetales | G: Tetrasphaera | AF255629 | Uncultured bacterium clone Ebpr19 from EBPR-WWTP | 374 | 0.944 |
| Day 59 | 23 | 100 | O: Actinomycetales | G: Tetrasphaera | AF527583 | Uncultured bacterium clone LPB21 from EBPR sludge | 371 | 0.949 | |
| 224 | Day 59 | 135 | 96 | O: Rhodobacterales | F: Hyphomonadaceae | AF236001 | Alphaproteobacterium A0904 | 285 | 0.625 |
| 228 | Day 2 | 50 | 88 | O: Actinomycetales | F: Intrasporangiaceae | AF513091 | Uncultured bacterium clone 17 from faming activated sludge | 382 | 0.946 |
| 3 | 5 | C: Actinobacteria | F: Microthrixaceae | CU917839 | Uncultured Actinobacterium clone QEEB1AC11 from anaerobic digester | 388 | 0.965 | ||
| 233 | Day 2 | 26 | 87 | P: candidate phylum TM7 | FJ534960 | Uncultured bacterium clone 14 from anaerobic fermentation of waste activated sludge | 271 | 0.666 | |
| 2 | 7 | O: Phycisphaerales | FJ612210 | Uncultured bacterium clone DP7.3.10 from lake ecosystem | 283 | 0.625 | |||
| 250 | Day 2 | 35 | 92 | O: Pseudomonadales | G: Acinetobacter | EU467673 | Uncultured bacterium clone molerat_2g12_1 from gut microbiota | 415 | 0.883 |
| 252 | Day 2 | 4 | 80 | O: Sphingobacteriales | FJ793188 | Uncultured bacterium clone TDB87 from a hot spring dam | 295 | 0.905 | |
| 253 | Day 2 | 14 | 88 | O: Ignavibacteriales | F: Ignavibacteriaceae | AB186808 | Uncultured bacterium from polychlorinated-dioxin-dechlorinating microbial community | 462 | 0.977 |
| 257 | Day 2 | 7 | 58 | O: Sphingobacteriales | EF562554 | Uncultured bacterium clone CC_3 from consortium degrading complex organic matter | 380 | 0.997 | |
| 5 | 42 | P: candidate phylum TM7 | AB200304 | Uncultured bacterium clone UTFS-OF09-d22-29 from EBPR-WWTP | 283 | 0.663 | |||
| 260 | Day 2 | 16 | 76 | O: Nitrospirales | G: Nitrospira | AF314422 | Uncultured bacterium PHOS-HE34 from an aerobic EBPR ecosystem | 366 | 0.924 |
| 4 | 19 | O: Sphingobacteriales | FJ660602 | Uncultured bacterium clone A194 from full-scale WWTP | 334 | 0.859 | |||
| Day 59 | 3 | 100 | O: Sphingobacteriales | AY302128 | Uncultured bacterium clone DSBR-B082 from denitrifying community | 354 | 0.878 | ||
| 264 | Day 59 | 3 | 100 | O: Thiotrichales | G: Thiothrix | L79963 | Thiothrix fructosivorans strain I, a filamentous sulfur bacterium from WWTP | 334 | 0.933 |
| 289 | Day 2 | 4 | 57 | O: Sphingomonadales | G: Sphingobium | AB040739 | Sphingobium cloacae a non-ylphenol-degrading bacterium isolated from WWTP | 277 | 0.785 |
| 3 | 43 | O: Rhodospirillales | F: Rhodospirillaceae | EU864465 | Uncultured bacterium clone E52 from river water receiving antibiotics-rich effluents | 350 | 0.967 | ||
| 294 | Day 2 | 1 | 100 | O: Clostridiales | G: Ruminococcus | DQ796981 | Uncultured bacterium clone RL386_aao85c11 from human gut microbiome | 289 | 0.906 |
| 302 | Day 2 | 6 | 75 | C: Anaerolineae | EU332818 | Uncultured organism clone OTUI177 from aerobic EBPR-SBR | 310 | 0.831 | |
| 304 | Day 2 | 28 | 93 | C: Gammaproteobacteria | FJ356049 | Uncultured bacterium clone G5 from lab-scale EBPR system | 383 | 0.844 | |
| 325 | Day 2 | 4 | 100 | O: Sphingobacteriales | GQ396974 | Uncultured bacterium clone AK1DE1_04E from recently deglaciated soils | 299 | 0.779 | |
| 399 | Day 59 | 12 | 100 | O: Rhodocyclales | G: Dechloromonas | EF632559 | Dechloromonas sp. A34, a bacterium from phosphate mining overburden respiring selenate | 378 | 0.922 |
1Size of target terminal-restriction fragments (T-RF) obtained with HaeIII digestion and forming operational taxonomic units (OTU).
2Original biomass sample selected for pyrosequencing analysis.
3Number of sequences from the pyrosequencing dataset that were related to the particular reference organism.
4Different bacterial populations can contribute to the same T-RF. The percentage of contribution of each population to the target T-RF is given in this column.
5Closest bacterial affiliations and GenBank accession numbers obtained after mapping against the Greengenes reference sequences of 16S rRNA encoding gene (McDonald et al., 2012). Legend: P, phylum; C, class; O, order; F, family; G, genus.
6Description of closest relatives and original microbiota from which the reference clones were isolated were obtained after submitting the accession numbers into the GenBank public database (Benson et al., 2011).
7The Smith–Waterman (SW) score was used as mapping similarity measure. SW scores consider nucleotide positions and gaps in the sequence structures. The highest absolute SW score that can be obtained is the length of the sequence itself. Each absolute SW score was normalized to the length of the related denoised centroid sequence in order to allow comparison between sequences of various lengths. After mapping in MG-RAST (Meyer et al., 2008), the affiliations obtained for the two denoised pyrosequencing datasets were related to traditional sequence identity scores of 99.7 ± 0.5%.
After granulation, additional bacterial populations evolved in the AGS. The relative abundance of Rhizobiales (OTU-185) increased from 6% at day 11 to 26% at day 39, and stabilized subsequently at 10 ± 4% over the next 20 days. Hyphomonadaceae (OTU-224) were detected above 1% from day 17 on, and were present at 13 ± 3% after day 37. Comamonadaceae (OTU-211) increased up to 16% at day 34, and remained at 5 ± 2% until the end of the experiment. Acinetobacter spp. (OTU-250) were only detected during the first 17 days in relative abundances of 3–12%. OTU-260 affiliating with Sphingobacteriales (and Nitrospira probably to a less extent) was present up to 7% at day 10, but was only present at low relative abundances of <1–4% in the early-stage granules. Nitrifiers were not detected above the detection limit of the T-RFLP method.
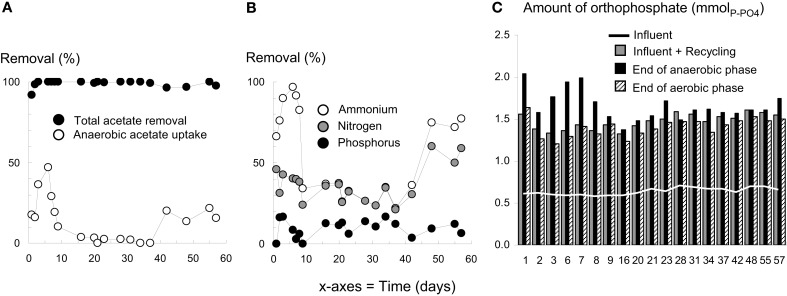
Wastewater treatment plants operation data and metabolic batch tests indicated that the inoculation sludge was efficiently removing organic matter (95%), nitrogen (97%), and phosphorus (92%). BNR activities were detected in R6 during initial operation with a high settling time (Figures 4A,B). After 6 days, 48% of the acetate load was consumed during anaerobic feeding, ammonium was efficiently nitrified to nitrate (97%), and 40% of nitrogen was removed. Two millimoles of orthophosphate were cycled in alternating anaerobic-aerobic conditions (Figure 4C), but only 9% of phosphorus was effectively removed. After intensive biomass wash-out at day 8, BNR activities were lost except carbon removal. Between day 10 and day 40, less than 4% of acetate was taken up during the anaerobic feeding phase, and only 31 ± 6% of ammonium was removed, presumably by assimilation into biomass. The orthophosphate cycling activity was lost, and phosphorus removal remained at 11 ± 4% until the end of the experiment. After day 40, ammonium and nitrogen removal recovered to 77 and 60%, respectively. However, nitrite instead of nitrate accumulated in the system. A slight increase in the anaerobic acetate uptake (up to 22%) was detected.
Figure 4.
Detailed evolution of the nutrient removal performances in reactor R6. The application of wash-out dynamics resulted in the transient loss of anaerobic acetate uptake (A), nitrification, nitrogen removal, and phosphorus removal performances (B) from day 6 to day 40. Orthophosphate cycling activities in alternating anaerobic-aerobic conditions were not detected during the same period (C).
Bacteriome analysis of the flocculent and granular sludge in R6
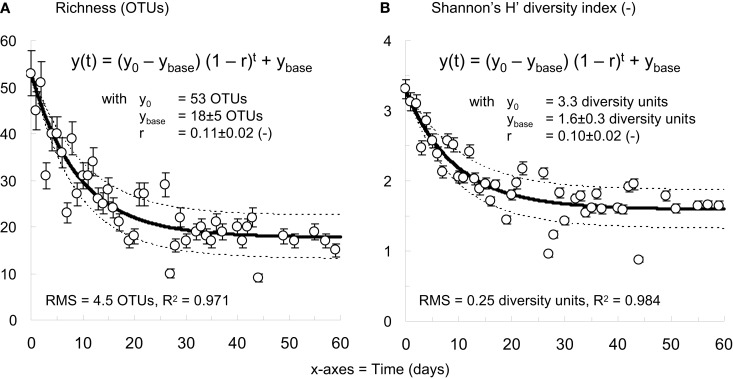
Based on the T-RFLP data collected from R6, the bacterial community was displaying a strong decrease of 66% in richness and 52% in diversity during the start-up of the reactor (Figure 5). The bacterial community of the activated sludge taken from the full-scale BNR was associated with a richness of 53 OTUs and a diversity index of 3.3. The bacterial community of the early-stage granules (day 30–60) was composed of 18 ± 5 OTUs, and showed a diversity index of 1.6 ± 0.3. The best fits of the mathematical geometric evolution models to the evolution of richness and diversity indices (R2 = 0.97 and 0.98, respectively) were obtained with finite rates of decrease of 11 and 10%, respectively. According to the models, the decrease in richness and diversity before extensive biomass wash-out (day 0–8) amounted to 38 and 30%, i.e., apparent decrease rates of about 2.5 OTUs and 0.13 diversity units per day. During the formation of early-stage granules occurring after biomass wash-out (day 8–27), the richness and diversity decreased by another 27 and 21% (0.8 OTUs and 0.04 diversity units per day).
Figure 5.
The bacterial community present in R6 exhibited a strong decrease of about 66% in richness (A) and of about 52% in Shannon’s H’ diversity (B) from inoculation with flocculent activated sludge from a full-scale BNR-WWTP fed with real wastewater to formation of early-stage granules fed with an acetate-based synthetic influent. Mathematical negative exponential growth models successfully explained the evolution of both indices during reactor start-up (R2 = 0.97–0.98). The model trends are given with standard deviation intervals computed from 1000 Monte Carlo simulations. Legend: y0: initial richness or diversity value, ybase: average final richness or diversity value after 60 days, r: negative growth rate, RMS: root mean square error.
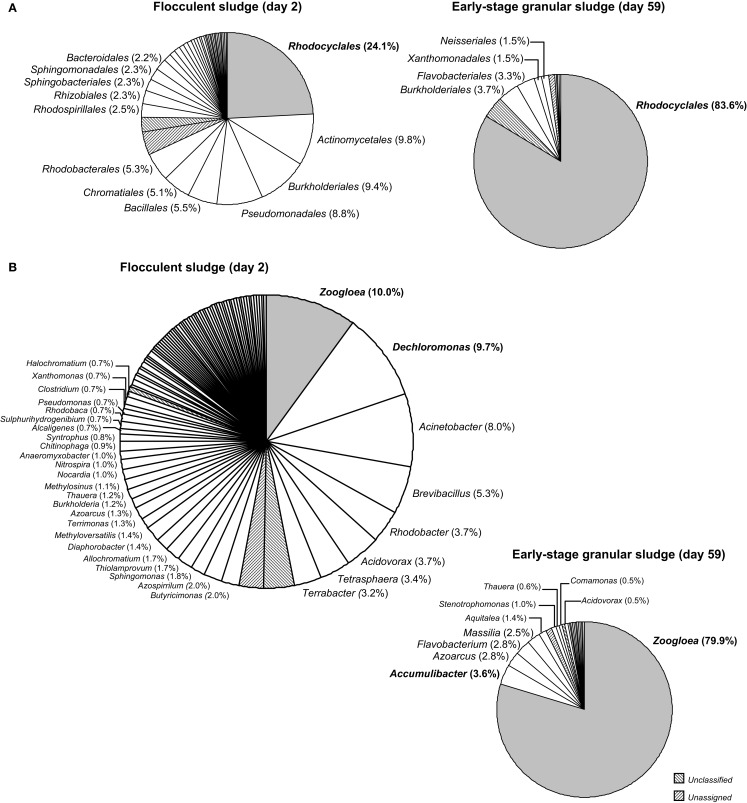
The pyrosequencing analyses of biomass samples collected on day 2 and day 59 confirmed that the early-stage AGS displayed a strongly reduced richness and diversity compared to the initial flocculent sludge (Table 3). The bacteriome of the flocculent sludge on day 2 was composed of 50 orders and 170 genera that were evenly distributed (3.9 diversity units). The bacteriome of the early-stage AGS was composed of only 20 orders and 57 genera that were unevenly distributed (1.1 diversity units). This analysis also showed that the T-RFLP method was covering at least 85% of the diversity obtained by pyrosequencing. Within the Rhodocyclales order, Zoogloea affiliates became very predominant in the early-stage AGS, and Dechloromonas-related organisms that were abundant in the flocculent sludge at day 2 were replaced by Accumulibacter and Azoarcus relatives in the early-stage AGS at day 59.
Table 3.
Summary of the main bacterial orders and genera identified by pyrosequencing analysis of the flocculent sludge and early-stage AGS samples taken from R6.
| Main bacterial orders | %1 | Main bacterial genera and relative abundances |
|---|---|---|
| FLOCCULENT SLUDGE (DAY 2) | ||
| Rhodocyclales | 24 | Zoogloea (10%), Dechloromonas (9.7%), Methyloversatilis (1.4%), Azoarcus (1.3%), Thauera (1.2%) |
| Actinomycetales | 10 | Tetrasphaera (3.4%), Terrabacter (3.2%), Nocardia (1.0%), Micrococcus (0.4%), Streptomyces (0.3%) |
| Burkholderiales | 9 | Acidovorax (3.7%), Diaphorobacter (1.4%), Burkholderia (1.2%), Alcaligenes (0.7%), Hydrogenophaga (0.4%) |
| Pseudomonadales | 9 | Acinetobacter (8.0%), Pseudomonas (0.7%) |
| Bacillales | 6 | Brevibacillus (5.3%), Trichococcus (0.3%) |
| Chromatiales | 5 | Thiolamprovum (1.7%), Allochromatium (1.7%), Halochromatium (0.7%) |
| Rhodobacterales | 5 | Azospirillum (2.0%) |
| Rhodospirillales | 3 | Rhodobacter (3.7%), Rhodobaca (0.7%) |
| Rhizobiales | 2 | Methylosinus (1.1%), Rhodopseudomonas (0.5%), Methylocystis (0.3%), Bradyrhizobiaceae (0.1%) |
| Sphingobacteriales | 2 | Terrimonas (1.3%), Chitinophaga (0.9%) |
| Sphingomonadales | 2 | Sphingomonas (1.8%) |
| Bacteroidales | 2 | Butyricimonas (2.0%) |
| 38 residual orders (<2%) | 21 | 137 residual genera |
| EARLY-STAGE AGS (DAY 59) | ||
| Rhodocyclales | 84 | Zoogloea (80%), Accumulibacter (3.6%), Azoarcus (2.8%), Thauera (0.6%) |
| Burkholderiales | 4 | Massilia (2.5%), Comamonas (0.5%), Acidovorax (0.5%) |
| Flavobacteriales | 3 | Flavobacterium (2.8%) |
| Xanthomonadales | 2 | Stenotrophomonas (1.0%), Pseudoxanthomonas (0.3%), Dyella (0.2%) |
| Neisseriales | 2 | Aquitalea (1.4%) |
| 15 residual orders (<2%) | 5 | 45 residual genera |
At the level of the nitrifiers, the pyrosequencing analysis enabled detection of ammonium- (AOB) and nitrite-oxidizing bacteria (NOB). AOB were only detected at relative abundances below 0.5% in the flocculent sludge, namely Nitrosococcus (0.24%), Nitrosomonas (0.12%), and Nitrosovibrio spp. (0.06%), and represented a biomass concentration of 0.012 gVSS −1. The NOB-related Nitrospira spp. were detected in higher abundance (1.02%) than Nitrobacter spp. (0.06%). The two genera together accounted for a biomass concentration of 0.032 gVSS L−1. The AOB and NOB present in the flocculent sludge were not detected in the early-stage granules. Only the AOB Nitrosospira spp. were detected at 0.03%, and accounted for 0.002 gVSS L−1 on this particular day.
Discussion
Fluffy and dense fast-settling granules harbored different predominant phylotypes
Unfavorable filamentous bulking occurring during early-stage granulation was related to the application of an insufficient SAV (1.8 cm s−1) in the case of an inoculum taken from OMR-WWTP, or when operation was conducted at higher mesophilic temperature (30°C). The bacterial community of slow-settling fluffy granules was dominated by filamentous Sphaerotilus and Leptothrix bacterial genera. These organisms are known to cause severe filamentous bulking in conventional WWTPs (Richard et al., 1985). During the formation of compact flocs and granular biofilms, the proliferation of filamentous organisms toward the outside of microbial aggregates is enhanced by substrate gradients generated by diffusion limitations across the biofilm matrices (Martins et al., 2004; Liu and Liu, 2006). The ecology data showed that filamentous bulking can also occur with acetate as carbon source, and not only with carbohydrates that have been proposed as main bulking vectors (Liu and Liu, 2006).
The application of a more intensive SAV (4.0 cm s−1) was successful for the recovery of smooth and dense fast-settling granules. In the study of McSwain et al. (2004), filamentous overgrowth was counteracted by high shear forces. In analogy to chlorine addition in conventional WWTPs, high shear forces helped to break the superficial filamentous structures. Specific remedial actions that suppress the cause of filamentous proliferation are however preferred for sustainable reactor operation (van Loosdrecht et al., 2008). The inoculation sludge taken from the BNR-WWTP was beneficial for the production of compact granules at 20°C with a low SAV. At full-scale level, this corresponds to definite energetic advantages. With the BNR-sludge, fluffy granules were only observed at 30°C. The growth kinetics of filamentous bacteria are enhanced at such temperature (Richard et al., 1985). In BNR-WWTPs, the successive anaerobic, anoxic, and aerobic zones are clearly separated. The readily biodegradable substrates are fully removed by PAO under anaerobic conditions, and are not available in the aerobic zone for fast-growing heterotrophs such as filamentous bacteria (van Loosdrecht et al., 2008). Thus, BNR-sludge exhibits a lower filamentous bulking potential than OMR-sludge, and can be advantageous for the granulation process. Ensuring full anaerobic acetate uptake in AGS-SBRs might also favorably suppress filamentous overgrowth.
Dense fast-settling early-stage aerobic granules were dominated by Zoogloea relatives. In contrast to fluffy and dense anaerobic granules that have been both dominated by Methanosaeta spp. (Grotenhuis et al., 1992; Hulshoff Pol et al., 2004), fluffy and dense aerobic granules were composed of different predominant phylotypes. Zoogloea spp. have also previously been detected in other granulation studies involving wash-out conditions (Etterer, 2006; Li et al., 2008; Ebrahimi et al., 2010; Gonzalez-Gil and Holliger, 2011).
The possible role of Rhodocyclales-related organisms in granulation
The T-RFLP and metagenomics analyses revealed that Rhodocyclales-affiliated Zoogloea, Dechloromonas, Thauera, and Rhodocyclus spp. were abundant in the communities of fast-settling early-stage granules. Acinetobacter spp. were present during the transition from flocs to granules with anaerobic feeding. The Rhodocyclales-affiliated organisms share some physiological properties in BNR-WWTPs (Hesselsoe et al., 2009). They produce EPS and store poly-β-hydroxyalcanoates (PHA) when high organic loads are present under aerobic conditions, hold an arsenal of surface adhesins, and form flocs and biofilms (Sich and Van Rijn, 1997; Allen et al., 2004; Dugan et al., 2006; Oshiki et al., 2008; Nielsen et al., 2010; Seviour et al., 2012). Acetate was abundantly present under aerobic conditions due to pulse feeding, or to incomplete anaerobic uptake. Feast-famine regimes and high shear stress also stimulate EPS production during granulation (Liu and Tay, 2002; Dulekgurgen et al., 2008; Seviour et al., 2010). In contrast to flocculent sludge settling that can suffer from Zoogloea-mediated viscous bulking (Norberg and Enfors, 1982; van Niekerk et al., 1987; Lajoie et al., 2000), AGS settling was not hampered by the proliferation of Zoogloea relatives. High shear stress and compaction forces generated by up-flow aeration (Zima et al., 2007) were likely to counteract viscous bulking. In addition, storage compounds such as PHA confer higher density and settling velocity to bacterial cells (Mas et al., 1985; Schuler et al., 2001). PHA storage was confirmed by confocal laser scanning microscopy analysis with Nile Red staining of cross-sectioned granules dominated by Zoogloea spp. (data not shown). Hence, the physiology of Zoogloea-like and other Rhodocyclales-affiliated organisms might be relevant for the cohesion of granular biofilms. However, microbial aggregation is probably not restricted to single organisms, and specific process conditions could select for other organisms with similar functions (Bossier and Verstraete, 1996; Beun et al., 1999; Wang et al., 2009).
Wash-out conditions as drastic bacterial selection pressure during aerobic granulation
Even though inoculation with BNR-sludge and anaerobic feeding were combined, active PAO and nitrifiers were outcompeted by Zoogloea spp. during start-up. Two tentative explanations of this specific bacterial selection were formulated from the results of R3–R5 based on wash-out dynamics. Firstly, the wash-out dynamics resulted in an insufficient SRT that did not enable bacterial populations with lower growth rates such as PAO and nitrifiers to maintain themselves in the system. Secondly, during anaerobic feeding, the influent wastewater was not long enough in contact with the low residual biomass after wash-out. With a constant volumetric OLR and a fixed anaerobic plug-flow feeding phase, a large acetate fraction was present during aeration and selected for fast-growing Zoogloea spp. over PAO. The data collected with R6 were used to confirm these explanations, and are discussed hereafter.
Tetrasphaera spp. and other Rhodocyclales-affiliated organisms such as Dechloromonas, Methyloversatilis spp., and Rhodocyclus spp. to a lower extent, were able to compete with Zoogloea spp. for the carbon source when 2.45 gVSS L−1 and 30 cm of settled flocculent biomass was initially present in the system. By considering a bed porosity of 0.5 and an influent flow-rate of 21 mL min−1, each volume fraction of the influent wastewater was in contact with the settled biomass during 15 min on average. With this contact time, 50% of acetate was removed under anaerobic conditions with concomitant release of orthophosphate showing that PAO activity was still present. Accumulibacter was only present in low abundance in the flocculent sludge (0.1–0.5%). However, additional organisms could have contributed to the detected PAO activity. Tetrasphaera spp. have been described as putative PAO in full-scale BNR-WWTP, but their underlying dephosphatating metabolism has not yet been deciphered (Nielsen et al., 2012). Dechloromonas spp. have been described as an accompanying guild of Accumulibacter, and have been proposed as putative PAO as well (Kong et al., 2007; Oehmen et al., 2010). The genus Methyloversatilis that affiliates to Rhodocyclales has only recently been discovered, and has been shown to metabolize nitrogen (Kalyuzhnaya et al., 2006; Baytshtok et al., 2008; Kittichotirat et al., 2011). However, more research is required on its metabolism under alternating anaerobic-aerobic conditions.
The combination of a low settling time (3 min) and a low HRT (6 h) resulted in intensive biomass wash-out. The SRT dropped to a value close to the HRT, and the reactor system was governed by the hydraulic properties. Aerobic heterotrophic organisms such as Zoogloea, Dechloromonas, Acinetobacter, and filamentous Burkholderiales affiliates that are related to maximum growth rates of 0.229–0.690 h−1 (Lau et al., 1984; van Niekerk et al., 1987; Logan et al., 2001; Kim and Pagilla, 2003) that are above 1/HRT, were able to proliferate over slower-growing PAO (0.042 h−1, Henze et al., 1999) and nitrifiers (0.017–0.046 h−1, Xavier et al., 2007). During reactor start-up, strong decreases in richness and diversity were observed. The apparent decrease rates were about 3 times higher before than after wash-out, indicating that the use of a synthetic wastewater with acetate as sole carbon source significantly contributed to the change in the bacterial community structure before wash-out. Gonzalez-Gil and Holliger (2011) have also reported that early-stage AGS cultivated with acetate or propionate as sole carbon and energy sources displayed half of the richness of the inoculation sludge. Winkler et al. (2011) have reported that, although distant, denaturing gradient gel electrophoresis profiles of bacterial communities of a conventional WWTP and of a pilot AGS reactor fed with the same urban wastewater exhibited similar richness and eveness.
After wash-out, only 0.3 gVSS L−1 of biomass was remaining in the system and the biomass specific OLR increased by a factor of 13 from 51 to 685 mgCODs cycle−1 gCODx−1. Granulation started with a biomass specific OLR above 2.7 kgCODs d−1 kgCODx−1 equivalents, which is in agreement with the bottom value of 1.3 kgCODs d−1 kgCODx−1 considered by Morgenroth et al. (1997) to enable sludge granulation. However, with a settled biomass height of only 1 cm, the contact time with the influent wastewater was extremely short (30 s). The fixed anaerobic plug-flow feeding phase thus resulted in the leakage of more than 90% of the acetate load into the aeration phase where it was available for fast-growing aerobic heterotrophs. This also explains why Zoogloea spp. outcompeted Accumulibacter, and why phosphorus was not removed. Deteriorated dephosphatation has also been correlated in flocculent sludge SBRs with Zoogloea proliferation over PAO caused by the concomitant presence of acetate as electron donor and oxygen or nitrate as terminal electron acceptors (Fang et al., 2002; Montoya et al., 2008). Proper anaerobic selector operation has been recommended to suppress this zoogloeal overgrowth (van Loosdrecht et al., 2008).
In conclusion, the detailed microbial ecology investigation involving T-RFLP, pyrosequencing, and PyroTRF-ID analyses conducted in this study in combination with a bioprocess engineering approach showed that slow-settling fluffy granules and dense fast-settling early-stage granules cultivated under wash-out dynamics were displaying distinct predominant phylotypes, namely filamentous Burkholderiales affiliates and Zoogloea relatives, respectively. Filamentous bulking could be remediated by the application of intensive up-flow aeration, or by the use of an inoculation sludge taken from a BNR-WWTP. A combination of insufficient SRT and of leakage of acetate into the aeration phase was the cause for the proliferation of Zoogloea spp. in dense fast-settling granules, and for the deterioration of BNR performances which has been commonly observed by different authors during granulation start-ups. It is however not certain that Zoogloea-like organisms are essential in granule formation. Additional research is needed to determine if they are required to stimulate early-stage granulation in BNR systems, or if granules can be cultivated without their involvement. Furthermore, optimal operation conditions should be elucidated for maintaining a balance between organisms with granulation propensity and nutrient removing organisms in order to form granules with BNR activities in short start-up periods.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financed by the Swiss National Science Foundation, Grant No. 205321-120536. David Weissbrodt obtained a grant from the EPFL FEE Foundation. We are grateful to ERM Morges and ARA Thunersee for providing the inoculation sludge. We recognize the excellent technical assistance of Sébastien Gabus, Jean-Pierre Kradolfer and Marc Deront in reactor operation, and of Corinne Weis, Yoan Rappaz and Emmanuelle Rohrbach in molecular ecology analyses. Scot E. Dowd, Yan Sun and Lars Koenig from the Research and Testing Laboratory (Lubbock, TX, USA) are acknowledged for pyrosequencing analyses and advice.
Appendix
Composition of the cultivation media (Table A1). Cloning-sequencing databank constructed for OTUs detected in reactors R1–R5 (Table A2). Full bacteriome phylogenetic tree constructed with the two pyrosequencing datasets (Figure A1). Sector graph representation of the bacteriome composition of the two bacteriomes of flocculent sludge and granular sludge (Figure A2).
Table A1.
Composition of the cultivation media, adapted from de Kreuk et al. (2005) and Ebrahimi et al. (2010).
| Compound | CAS no. | Molecular formula | Molecular weight (g mol−1) | Amount per 20 L of medium1 | Concentration in medium (mmol L−1) |
|---|---|---|---|---|---|
| Carbon source medium2 | |||||
| Sodium acetate | 127-09-3 | C2H3O2Na ·3H2O | 136.09 | 170.11/212.64 g | 362.5/78.1 |
| Magnesium sulfate | 7487-88-9 | MgSO4·7H2O | 246.51 | 17.75 g | 3.6 |
| Potassium chloride | 7447-40-7 | KCl | 74.55 | 7.16 g | 4.8 |
| Nitrogen and phosphorus sources medium2 | |||||
| Ammonium chloride | 12125-02-9 | NH4Cl | 53.49 | 37.87 g | 35.4 |
| Dipotassium hydrogen phosphate | 7758-11-4 | K2HPO4 | 174.18 | 14.62 g | 4.2 |
| Potassium dihydrogen phosphate | 7778-77-0 | KH2PO4 | 136.09 | 5.72 g | 2.1 |
| Trace element solution | – | – | – | 100 mL | – |
| Compound | CAS no. | Molecular formula | Molecular weight (g mol−1) | Amount per 5 L of stock solution1 (g) | Concentration in stock solution (mmol L−1) |
| Trace element solution | |||||
| Disodium ethylenediaminetetraacetate | 139-33-3 | C10H14N2O8Na2·2H2O | 372.25 | 637.0 | 342.2 |
| Zinc sulfate | 7733-02-0 | ZnSO4·7H2O | 287.59 | 22.0 | 15.3 |
| Calcium chloride | 10043-52-4 | CaCl2·2H2O | 147.02 | 81.8 | 111.3 |
| Manganese chloride | 7773-01-5 | MnCl2·4H2O | 197.92 | 50.6 | 51.1 |
| Iron(II) sulfate | 7720-78-7 | FeSO4·7H2O | 278.05 | 49.9 | 35.9 |
| Ammonium heptamolybdate | 12027-67-7 | (NH4)6Mo7O24·4H2O | 1’235.88 | 16.4 | 2.7 |
| Copper(II) sulfate | 7758-98-7 | CuSO4·5H2O | 249.71 | 15.7 | 12.6 |
| Cobalt(II) chloride | 7646-79-9 | CoCl2·6H2O | 237.96 | 16.1 | 13.5 |
1The cultivation media and the trace element solution were prepared in 20 and 5 L of demineralized water.
2During the feeding phase of each SBR cycle, the synthetic influent wastewater was composed of 10% (v/v) of carbon source medium, of 10% (v/v) of nitrogen and phosphorus source medium, and of 80% (v/v) of tap water.
3The reactors R1–R5 and R6 were operated with 400 and 500 mgCOD L−1 of acetate in the influent wastewater, respectively. The chemical oxygen demand (COD) conversion factor of acetate is 64 gCOD molAc−1.
Table A2.
Phylogenetic affiliation of predominant clones isolated from biomass samples of reactors R1–R5.
| T-RF1 (bp) | Reactor2 | Affiliation3 |
Accession number3 | Closest relative and original microbiota4 | Sequence identity5 (%) | |
|---|---|---|---|---|---|---|
| Phylum → Family | Genus | |||||
| 71 | R3 | O: Rhodocyclales | G: Zoogloea | AJ505853 | Zoogloea resiniphila strain PIV-3C2w from denitrifying bacterial community | 99 |
| 185 | R4 | C: Gammaproteobacteria | AM180057 | Uncultured bacterium clone A1-41 from a N- and P-removing system | 97 | |
| O: Rhizobiales | G: Ochrobactrum | CP000759 | Ochrobactrum anthropi ATCC 49188 | 100 | ||
| 193 | R4, R5 | O: Rhodocyclales | G: Dechloromonas | AF314420 | Uncultured bacterium PHOS-HE23 from aerobic EBPR system | 99 |
| 195 | R1, R3 | O: Rhodocyclales | G: Zoogloea | DQ413172 | Zoogloea sp. EMB 357 from anaerobic-aerobic SBR | 100 |
| R4, R5 | O: Rhodocyclales | G: Zoogloea | AF527582 | Uncultured bacterium clone LPB19 from EBPR system | 99 | |
| O: Enterobacteriales | G: Escherichia/Shigella | AY838362 | Bacterium HPC775 from effluent treatment plant of chemical and dye industry | 98 | ||
| 208 | R1, R3 | O: Burkholderiales | G: Sphaerotilus | AB087568 | Sphaerotilus sp. L19 involved in filamentous sludge bulking process | 100 |
| O: Burkholderiales | G: Leptothrix | FM175128 | Uncultured Leptothrix sp. from tufa core sample | 98 | ||
| R4, R5 | O: Burkholderiales | AB087576 | Leptothrix sp. L18 involved in filamentous sludge bulking process | 98 | ||
| 213 | R5 | O: Burkholderiales | F: Comamonadaceae | AY491594 | Uncultured bacterium clone oc53 from oligotrophic microbial fuel cells | 99 |
| 214 | R4, R5 | O: Rhodocyclales | F: Rhodocyclales | EF565147 | Uncultured bacterium clone UWMH_4 in Accumulibacter enrichment | 99 |
| 215 | R3 | O: Rhodocyclales | G: Dechloromonas | EF632559 | Dechloromonas sp. A34 from phosphate mining overburden | 98 |
| R4, R5 | O: Rhodocyclales | G: Dechloromonas | AY823964 | Uncultured Betaproteobacterium clone DFAW-071 from acetate-fed denitrifying community | 100 | |
| 216 | R5 | O: Nitrosomonadales | G: Nitrosomonas | AJ621027 | Ammonium-oxidizing Nitrosomonas sp. Is32 | 97 |
| 217 | R3 | O: Rhodocyclales | G: Zoogloea | DQ413157 | Zoogloea sp. EMB 108 from anaerobic-aerobic SBR | 98 |
| R1 | O: Rhodocyclales | G: Thauera | AM084110 | Thauera sp. R-28312 from denitrifying community | 95 | |
| 224 | R4, R5 | O: Sphingomonadales | G: Novosphingobium | AJ746094 | Novosphingobium sp. MG37 from haemodialysis water and fluid | 100 |
| O: Rhodobacterales | F: Hyphomonadaceae | AF502221 | Uncultured bacterium clone HP1B26 from EBPR sludge | 99 | ||
| 239 | R4, R5 | C: Gammaproteobacteria | AY651306 | Uncultured bacterium clone Cont82 from EBPR ecosystem | 99 | |
| 250 | R3 | O: Pseudomonadales | G: Acinetobacter | GQ383923 | Acinetobacter sp. XJ127 from sewage water | 99 |
| R4, R5 | O: Pseudomonadales | G: Acinetobacter | DQ205305 | Kartchner Caverns bacterium HI-O4 | 100 | |
| O: Flavobacteriales | G: Chryseobacterium | AF502204 | Uncultured bacterium clone HP1B06 from EBPR sludge | 99 | ||
| 289 | R4, R5 | O: Sphingomonadales | F: Sphingomonadaceae | U37341 | Sphingomonas paucimobilis isolate EPA 505 from PAH-degrading soil consortia | 100 |
The initial cloning-sequencing database of Ebrahimi et al. (2010) was complemented in the present study.
1Size of target terminal-restriction fragments (T-RF) obtained with HaeIII digestion and forming operational taxonomic units (OTU).
2Reactor biomass system from which the clones were isolated. A number of 1–7 clones were sequenced per T-RF.
3Closest bacterial affiliations and GenBank accession numbers obtain after mapping in the Ribosomal Database Project (RDP; Cole et al., 2009).
Legend: P, phylum; C, class; O, order; F, family; G, genus.
4Description of closest relatives and original microbiota from which the reference clones were isolated obtained from GenBank (Benson et al., 2011).
5Sequence identity scores obtained after mapping in RDP.
Figure A1.
Bacteriome phylogenetic tree constructed in MG-RAST (Meyer et al., 2008) with the pyrosequencing datasets of the two biomass samples collected on day 2 (flocculent sludge, red bar plot) and day 59 (early-stage AGS, green bar plot) in the R6 reactor. Each bar plot is related to the number of pyrosequencing reads detected per bacterial affiliation. The tree is presented with classes (outer black circle segments) and orders subdivisions (colored slices), and bacterial genera names. The identity of target orders marked with an asterisk is given for each left-hand and right-hand half of the circular tree. The RDP database (Cole et al., 2009) was used as annotation source, and a minimum identity cutoff of 97% was applied.
Figure A2.
Differences in the bacteriome composition of the flocculent sludge present in R6 before wash-out, and of the early-stage granular sludge present at day 59, at the order level (A) and at the genus level (B).
References
- Adav S. S., Lee D. J., Lai J. Y. (2010). Microbial community of acetate utilizing denitrifiers in aerobic granules. Appl. Microbiol. Biotechnol. 85, 753–762 10.1007/s00253-009-2317-9 [DOI] [PubMed] [Google Scholar]
- Allen M. S., Welch K. T., Prebyl B. S., Baker D. C., Meyers A. J., Sayler G. S. (2004). Analysis and glycosyl composition of the exopolysaccharide isolated from the floc-forming wastewater bacterium Thauera sp. MZ1T. Environ. Microbiol. 6, 780–790 10.1111/j.1462-2920.2004.00615.x [DOI] [PubMed] [Google Scholar]
- Baytshtok V., Kim S., Yu R., Park H., Chandran K. (2008). Molecular and biokinetic characterization of methylotrophic denitrification using nitrate and nitrite as terminal electron acceptors. Water Sci. Technol. 58, 359–365 10.2166/wst.2008.391 [DOI] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. (2011). GenBank. Nucleic Acids Res. 39, D32–D37 10.1093/nar/gkq1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beun J. J., Heijnen J. J., van Loosdrecht M. C. M. (2001). N-removal in a granular sludge sequencing batch airlift reactor. Biotechnol. Bioeng. 75, 82–92 10.1002/bit.1167 [DOI] [PubMed] [Google Scholar]
- Beun J. J., Hendriks A., van Loosdrecht M. C. M., Morgenroth E., Wilderer P. A., Heijnen J. J. (1999). Aerobic granulation in a sequencing batch reactor. Water Res. 33, 2283–2290 10.1016/S0043-1354(98)00463-1 [DOI] [PubMed] [Google Scholar]
- Bossier P., Verstraete W. (1996). Triggers for microbial aggregation in activated sludge? Appl. Microbiol. Biotechnol. 45, 1–6 10.1007/s002530050640 [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., Kulam-Syed-Mohideen A. S., McGarrell D. M., Marsh T., Garrity G. M., Tiedje J. M. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 10.1093/nar/gkp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin L. M. M., de Kreuk M. K., van der Roest H. F. R., Uijterlinde C., van Loosdrecht M. C. M. (2004). Aerobic granular sludge technology: an alternative to activated sludge? Water Sci. Technol. 49, 1–7 [PubMed] [Google Scholar]
- de Kreuk M. K., Heijnen J. J., van Loosdrecht M. C. M. (2005). Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnol. Bioeng. 90, 761–769 10.1002/bit.20470 [DOI] [PubMed] [Google Scholar]
- Dugan P., Stoner D., Pickrum H. (2006). “The genus Zoogloea,” in The Prokaryotes, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (New York: Springer; ), 960–970 [Google Scholar]
- Dulekgurgen E., Artan N., Orhon D., Wilderer P. A. (2008). How does shear affect aggregation in granular sludge sequencing batch reactors? Relations between shear, hydrophobicity, and extracellular polymeric substances. Water Sci. Technol. 58, 267–276 10.2166/wst.2008.382 [DOI] [PubMed] [Google Scholar]
- Ebrahimi S., Gabus S., Rohrbach-Brandt E., Hosseini M., Rossi P., Maillard J., Holliger C. (2010). Performance and microbial community composition dynamics of aerobic granular sludge from sequencing batch bubble column reactors operated at 20°C, 30°C, and 35°C. Appl. Microbiol. Biotechnol. 87, 1555–1568 10.1007/s00253-010-2621-4 [DOI] [PubMed] [Google Scholar]
- Etterer T. J. (2006). Formation, Structure and Function of Aerobic Granular Sludge. Ph.D. thesis, Technische Universität München, Munich [Google Scholar]
- Fang H. H. P., Zhang T., Liu Y. (2002). Characterization of an acetate-degrading sludge without intracellular accumulation of polyphosphate and glycogen. Water Res. 36, 3211–3218 10.1016/S0043-1354(02)00027-1 [DOI] [PubMed] [Google Scholar]
- Giesen A., Niermans R., van Loosdrecht M. C. M. (2012). Aerobic granular biomass: the new standard for domestic and industrial wastewater treatment? Water21 4, 28–30 10.3390/w4010028 [DOI] [Google Scholar]
- Gonzalez-Gil G., Holliger C. (2011). Dynamics of microbial community structure and enhanced biological phosphorus removal of propionate- and acetate-cultivated aerobic granules. Appl. Environ. Microbiol. 77, 8041–8051 10.1128/AEM.05738-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotenhuis J. T. C., Plugge C. M., Stams A. J. M., Zehnder A. J. B. (1992). Hydrophobicities and electrophoretic mobilities of anaerobic bacterial isolates from methanogenic granular sludge. Appl. Environ. Microbiol. 58, 1054–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze M., Gujer W., Mino T., Matsuo T., Wentzel M. C., Marais G. V. R., Van Loosdrecht M. C. M. (1999). Activated sludge model No.2d, ASM2d. Water Sci. Technol. 39, 165–182 10.1016/S0273-1223(99)00332-7 [DOI] [Google Scholar]
- Hesselsoe M., Fureder S., Schloter M., Bodrossy L., Iversen N., Roslev P., Nielsen P. H., Wagner M., Loy A. (2009). Isotope array analysis of Rhodocyclales uncovers functional redundancy and versatility in an activated sludge. ISME J. 3, 1349–1364 10.1038/ismej.2009.78 [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol L. W., De Castro Lopes S. I., Lettinga G., Lens P. N. L. (2004). Anaerobic sludge granulation. Water Res. 38, 1376–1389 10.1016/j.watres.2003.12.002 [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G., De Marco P., Bowerman S., Pacheco C. C., Lara J. C., Lidstrom M. E., Chistoserdova L. (2006). Methyloversatilis universalis gen. nov., sp. nov., a novel taxon within the Betaproteobacteria represented by three methylotrophic isolates. Int. J. Syst. Evol. Microbiol. 56, 2517–2522 10.1099/ijs.0.64191-0 [DOI] [PubMed] [Google Scholar]
- Kim H., Pagilla K. R. (2003). Competitive growth of Gordonia and Acinetobacter in continuous flow aerobic and anaerobic/aerobic reactors. J. Biosci. Bioeng. 95, 577–582 10.1016/S1389-1723(03)80028-2 [DOI] [PubMed] [Google Scholar]
- Kittichotirat W., Good N. M., Hall R., Bringel F., Lajus A., Médigue C., Smalley N. E., Beck D., Bumgarner R., Vuilleumier S., Kalyuzhnaya M. G. (2011). Genome sequence of Methyloversatilis universalis FAM5 T, a methylotrophic representative of the order Rhodocyclales. J. Bacteriol. 193, 4541–4542 10.1128/JB.05331-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Xia Y., Nielsen J. L., Nielsen P. H. (2007). Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology 153, 4061–4073 10.1099/mic.0.2007/007245-0 [DOI] [PubMed] [Google Scholar]
- Lajoie C. A., Layton A. C., Gregory I. R., Sayler G. S., Don E. T., Meyers A. J. (2000). Zoogleal clusters and sludge dewatering potential in an industrial activated-sludge wastewater treatment plant. Water Environ. Res. 72, 56–64 10.2175/106143000X137112 [DOI] [Google Scholar]
- Lau A. O., Strom P. F., Jenkins D. (1984). Growth kinetics of Sphaerotilus natans and a floc former in pure and dual continuous culture. J. Water Pollut. Control Fed. 56, 41–51 [Google Scholar]
- Lee D.-J., Chen Y.-Y., Show K.-Y., Whiteley C. G., Tay J.-H. (2010). Advances in aerobic granule formation and granule stability in the course of storage and reactor operation. Biotechnol. Adv. 28, 919–934 10.1016/j.biotechadv.2010.08.007 [DOI] [PubMed] [Google Scholar]
- Lemaire R., Yuan Z., Blackall L. L., Crocetti G. R. (2008). Microbial distribution of Accumulibacter spp. and Competibacter spp. in aerobic granules from a lab-scale biological nutrient removal system. Environ. Microbiol. 10, 354–363 10.1111/j.1462-2920.2007.01456.x [DOI] [PubMed] [Google Scholar]
- Li A. J., Yang S. F., Li X. Y., Gu J. D. (2008). Microbial population dynamics during aerobic sludge granulation at different organic loading rates. Water Res. 42, 3552–3560 10.1016/j.watres.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Lin Y. M., Liu Y., Tay J. H. (2003). Development and characteristics of phosphorus-accumulating microbial granules in sequencing batch reactors. Appl. Microbiol. Biotechnol. 62, 430–435 10.1007/s00253-003-1359-7 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu Q.-S. (2006). Causes and control of filamentous growth in aerobic granular sludge sequencing batch reactors. Biotechnol. Adv. 24, 115–127 10.1016/j.biotechadv.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Liu Y., Tay J. H. (2002). The essential role of hydrodynamic shear force in the formation of biofilm and granular sludge. Water Res. 36, 1653–1665 10.1016/S0043-1354(02)00022-2 [DOI] [PubMed] [Google Scholar]
- Logan B. E., Zhang H., Mulvaney P., Milner M. G., Head I. M., Unz R. F. (2001). Kinetics of perchlorate- and chlorate-respiring bacteria. Appl. Environ. Microbiol. 67, 2499–2506 10.1128/AEM.67.6.2499-2506.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey R., Oster G., Zahnley T. (2000). Berkeley Madonna User’s Guide. Berkeley, CA: University of California [Google Scholar]
- Martins A. M. P., Picioreanu C., Heijnen J. J., van Loosdrecht M. C. M. (2004). Three-dimensional dual-morphotype species modeling of activated sludge flocs. Environ. Sci. Technol. 38, 5632–5641 10.1021/es049659l [DOI] [PubMed] [Google Scholar]
- Mas J., Pedros-Alio C., Guerrero R. (1985). Mathematical model for determining the effects of intracytoplasmic inclusions on volume and density of microorganisms. J. Bacteriol. 164, 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., Hugenholtz P. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwain B. S., Irvine R. L., Wilderer P. A. (2004). The influence of settling time on the formation of aerobic granules. Water Sci. Technol. 50, 195–202 [PubMed] [Google Scholar]
- Meyer F., Paarmann D., D’Souza M., Olson R., Glass E., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya T., Borrás L., Aguado D., Ferrer J., Seco A. (2008). Detection and prevention of enhanced biological phosphorus removal deterioration caused by Zoogloea overabundance. Environ. Technol. 29, 35–42 10.1080/09593330802008560 [DOI] [PubMed] [Google Scholar]
- Morgenroth E., Sherden T., van Loosdrecht M. C. M., Heijnen J. J., Wilderer P. A. (1997). Aerobic granular sludge in a sequencing batch reactor. Water Res. 31, 3191–3194 10.1016/S0043-1354(97)00216-9 [DOI] [Google Scholar]
- Mosquera-Corral A., de Kreuk M. K., Heijnen J. J., van Loosdrecht M. C. M. (2005). Effects of oxygen concentration on N-removal in an aerobic granular sludge reactor. Water Res. 39, 2676–2686 10.1016/j.watres.2005.04.065 [DOI] [PubMed] [Google Scholar]
- Moy B. Y. P., Tay J. H., Toh S. K., Liu Y., Tay S. T. L. (2002). High organic loading influences the physical characteristics of aerobic sludge granules. Lett. Appl. Microbiol. 34, 407–412 10.1046/j.1472-765X.2002.01108.x [DOI] [PubMed] [Google Scholar]
- Nielsen P. H., Mielczarek A. T., Kragelund C., Nielsen J. L., Saunders A. M., Kong Y., Hansen A. A., Vollertsen J. (2010). A conceptual ecosystem model of microbial communities in enhanced biological phosphorus removal plants. Water Res. 44, 5070–5088 10.1016/j.watres.2010.09.011 [DOI] [PubMed] [Google Scholar]
- Nielsen P. H., Saunders A. M., Hansen A. A., Larsen P., Nielsen J. L. (2012). Microbial communities involved in enhanced biological phosphorus removal from wastewater – a model system in environmental biotechnology. Curr. Opin. Biotechnol. 23, 452–459 10.1016/j.copbio.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Norberg A. B., Enfors S. O. (1982). Production of extracellular polysaccharide by Zoogloea ramigera. Appl. Environ. Microbiol. 44, 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehmen A., Carvalho G., Lopez-Vazquez C. M., van Loosdrecht M. C. M., Reis M. A. M. (2010). Incorporating microbial ecology into the metabolic modelling of polyphosphate accumulating organisms and glycogen accumulating organisms. Water Res. 44, 4992–5004 10.1016/j.watres.2010.06.071 [DOI] [PubMed] [Google Scholar]
- Oksanen J., Kindt R., Legendre P., O’Hara B., Simpson G. L., Solymos P., Stevens M. H. H., Wagner H. (2009). Vegan: Community Ecology Package. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Oshiki M., Onuki M., Satoh H., Mino T. (2008). PHA-accumulating microorganisms in full-scale wastewater treatment plants. Water Sci. Technol. 58, 13–20 10.2166/wst.2008.652 [DOI] [PubMed] [Google Scholar]
- Pijuan M., Werner U., Yuan Z. (2011). Reducing the startup time of aerobic granular sludge reactors through seeding floccular sludge with crushed aerobic granules. Water Res. 45, 5075–5083 10.1016/j.watres.2011.07.009 [DOI] [PubMed] [Google Scholar]
- R-Development-Core-Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Rees G., Baldwin D., Watson G., Perryman S., Nielsen D. (2004). Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86, 339–347 10.1007/s10482-004-0498-x [DOI] [PubMed] [Google Scholar]
- Richard M., Hao O., Jenkins D. (1985). Growth kinetics of Sphaerotilus species and their significance in activated sludge bulking. J. Water Pollut. Control Fed. 57, 68–81 [Google Scholar]
- Rossi P., Gillet F., Rohrbach E., Diaby N., Holliger C. (2009). Statistical assessment of variability of terminal restriction fragment length polymorphism analysis applied to complex microbial communities. Appl. Environ. Microbiol. 75, 7268–7270 10.1128/AEM.00135-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler A. J., Jenkins D., Ronen P. (2001). Microbial storage products, biomass density, and setting properties of enhanced biological phosphorus removal activated sludge. Water Sci. Technol. 43, 173–180 [PubMed] [Google Scholar]
- Seviour T., Donose B. C., Pijuan M., Yuan Z. (2010). Purification and conformational analysis of a key exopolysaccharide component of mixed culture aerobic sludge granules. Environ. Sci. Technol. 44, 4729–4734 10.1021/es102658s [DOI] [PubMed] [Google Scholar]
- Seviour T., Yuan Z., van Loosdrecht M. C. M., Lin Y. (2012). Aerobic sludge granulation: a tale of two polysaccharides? Water Res. 46, 4803–4813 10.1016/j.watres.2012.06.018 [DOI] [PubMed] [Google Scholar]
- Shin H. S., Lim K. H., Park H. S. (1992). Effect of shear stress on granulation in oxygen aerobic upflow sludge bed reactors. Water Sci. Technol. 26, 601–605 [Google Scholar]
- Sich H., Van Rijn J. (1997). Scanning electron microscopy of biofilm formation in denitrifying, fluidised bed reactors. Water Res. 31, 733–742 10.1016/S0043-1354(97)80988-8 [DOI] [Google Scholar]
- Sun Y., Wolcott R. D., Dowd S. E. (2011). Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol. Biol. 733, 129–141 10.1007/978-1-61779-089-8_9 [DOI] [PubMed] [Google Scholar]
- Tay J. H., Liu Q. S., Liu Y. (2002). Aerobic granulation in sequential sludge blanket reactor. Water Sci. Technol. 46, 13–18 [PubMed] [Google Scholar]
- Tsuneda S., Nagano T., Hoshino T., Ejiri Y., Noda N., Hirata A. (2003). Characterization of nitrifying granules produced in an aerobic upflow fluidized bed reactor. Water Res. 37, 4965–4973 10.1016/j.watres.2003.08.017 [DOI] [PubMed] [Google Scholar]
- van Loosdrecht M. C. M., Martins A. M., Ekama G. A. (2008). “Bulking sludge,” in Biological Wastewater Treatment: Principles, Modelling and Design, eds Henze M., Van Loosdrecht M. C. M., Ekama G. A., Brdjanovic D. (London: IWA Publishing; ), 291–308 [Google Scholar]
- van Niekerk A. M., Jenkins D., Richard M. G. (1987). The competitive growth of Zoogloea ramigera and type 021N in activated sludge and pure culture – a model for low F:M bulking. J. Water Pollut. Control Fed. 59, 262–273 [Google Scholar]
- Verawaty M., Pijuan M., Yuan Z., Bond P. L. (2012). Determining the mechanisms for aerobic granulation from mixed seed of floccular and crushed granules in activated sludge wastewater treatment. Water Res. 46, 761–771 10.1016/j.watres.2011.11.054 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang Z., Wu W. (2009). Research advances in aerobic granular sludge. Acta Scientiae Circumstantiae 29, 449–473 [Google Scholar]
- Weber S. D., Ludwig W., Schleifer K. H., Fried J. (2007). Microbial composition and structure of aerobic granular sewage biofilms. Appl. Environ. Microbiol. 73, 6233–6240 10.1128/AEM.01002-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M. K. H., Kleerebezem R., de Bruin L. M. M., Habermacher J., Abbas B., van Loosdrecht M. C. M. (2011). “Microbial diversity differences in aerobic granular sludge in comparison to conventional treatment plant,” in IWA Biofilm Specialist Conference, ed. Qi Z. (London: IWA), 23–26 [Google Scholar]
- Xavier J. B., de Kreuk M. K., Picioreanu C., van Loosdrecht M. C. M. (2007). Multi-scale individual-based model of microbial and byconversion dynamics in aerobic granular sludge. Environ. Sci. Technol. 41, 6410–6417 10.1021/es070264m [DOI] [PubMed] [Google Scholar]
- Yilmaz G., Lemaire R., Keller J., Yuan Z. (2008). Simultaneous nitrification, denitrification, and phosphorus removal from nutrient-rich industrial wastewater using granular sludge. Biotechnol. Bioeng. 100, 529–541 10.1002/bit.21774 [DOI] [PubMed] [Google Scholar]
- Zima B. E., Diez L., Kowalczyk W., Delgado A. (2007). Sequencing batch reactor (SBR) as optimal method for production of granular activated sludge (GAS) – fluid dynamic investigations. Water Sci. Technol. 55, 151–158 10.2166/wst.2007.223 [DOI] [PubMed] [Google Scholar]