Abstract
Proteins released into the culture medium by Staphylococcus aureus (S. aureus) strain 6 were determined at the end of the exponential growth phase (4.5 h). Eleven proteins were identified by liquid chromatography coupled with mass spectrometry. Three proteins were predicted to have signal peptides indicating their extracellular localization. The other proteins were presumably located in the cytoplasm of the bacteria. Five out of the 11 proteins were involved in carbohydrate metabolism. Other intracellular proteins of S. aureus were not detected in the culture medium. This indicates that the release of these 11 proteins was specific and that unspecific protein release due to damaged or dying bacteria did not play a role. It is suggested that enzymes associated with carbohydrate metabolism may provide the energy necessary for the transition of bacteria from a resting to a proliferative state. Another enzyme released by S. aureus, superoxide dismutase, may catalyze redox reactions in this context. The production of other proteolytic enzymes and toxins may take place at later stages of bacterial growth. A cocktail of these 11 proteins was used for the immunization of mice. Indeed, vaccination with these proteins prolonged the survival times of mice upon infection with S. aureus strain 6. Therefore, these proteins may have implications for the development of novel strategies for the prevention and therapy of S. aureus infections.
Keywords: antioxidant enzymes, carbohydrate metabolism, liquid chromatography coupled with mass spectrometry, proteomics
Introduction
It is well known that microorganisms release primary and secondary metabolites as well as peptides and proteins into their environment (in either the host organism or the nutrition medium). The function of releasing these products is mainly unknown. Recently, we demonstrated that Klebsiella pneumoniae (K. pneumoniae) and Streptococcus pneumoniae release proteins with a molecular weight of 30–50 kDa into the nutritional medium during the exponential growth phase (1). The use of this protein fraction for vaccination conferred complete protection of mice from K. pneumoniae infection. It was proven that this protection was not due to protease, one of the key invasion factors of K. pneumoniae, nor by antigens of the bacterial cell wall. Notably, optimal protection was observed with proteins obtained in the exponential growth phase of the bacteria, i.e., proteins involved in the transition of bacteria from a resting to a proliferative state. This result may be of significant practical value for the onset of the infection process. However, the absence of the amino acid sequences of these proteins did not allow identification of these proteins.
Recently, the genomes of 9 strains of Staphylococcus aureus (S. aureus) have been sequenced and annotated [http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi; http://www.tigr.org] (14). A more detailed database of S. aureus is now available with both genomic and proteomic information (2). During the past decade, several investigations have addressed the issue of secreted proteins from S. aureus (3,4). It can be hypothesized that the cellular and molecular mechanisms responsible for transition from a resting to a proliferative state are common among microorganisms. Nutritional substances taken up by bacteria are proposed primer signals to start growth. Due to the energy required to boost metabolism, subsets of proteins rather than the total spectrum of enzymes are induced. Therefore, the aim of the present investigation was to determine the protein spectrum synthesized and released by S. aureus at the early stage of growth.
Materials and methods
Animal experimentation
The experiments were carried out using 2- to 3-month-old Balb/c male mice. The animals were obtained from Stolbovaya Company (Moscow, Russia). The mice received standard laboratory feed and tap water ad libitum. All experiments were carried out in accordance with the legal regulations for animal experimentation in Russia and with official permission of the Institute of Experimental Diagnosis and Therapy of Tumors of the N.N. Blokhin Russian Cancer Research Center and the Mechnikov Research Institute for Vaccine and Sera, Russian Academy of Medical Science, Moscow, Russia (registration code 076). Mice were sacrificed prior to death during the experiments.
Bacterial strains and growth conditions
S. aureus strain 6 was obtained from the collection of the L.A. Tarasevich State Scientific Research Institute for Standardization and Quality Control of Biologicals, Moscow, Russia. It was previously shown that this strain is the most virulent for conventional mice among all S. aureus strains (5). Liquid nutritional medium based on casein hydrolyzed from pancreas supplemented with yeast extract and glucose was used for cultivation (6). Five to seven colonies isolated from agar culture were transferred into a flask with 200 ml nutritional medium. After 12–14 h of incubation, the growing culture was transferred into a flask with 2 l of medium at a 1:10 proportion of culture to medium, and incubated at 37°C at 90 rmp/min on a Multi-Shaker Multi PSU-20 (BioSan Ltd., Riga, Latvia). The bacterial growth was monitored using a spectrophotometer (Genesys 10 UV; Thermo Spectronic, Rochester, NY, USA) by measuring the optical density at 565 nm. The number of colonies was determined, calculated 24 h after incubation on agar plates and inoculated with serial dilutions of the bacterial culture. The efficacy of the cultivation process was evaluated every hour by the density of the biomass calculated as the number of colony forming units (CFUs).
Extraction of S. aureus extracellular proteins
Bacterial cells were separated from the culture media by sterilization filtration using 0.22-μM pore filters (Millipore, MA, USA). For subsequent protein extraction, 80% ammonium sulphate precipitation was used. The precipitate was centrifuged (1,7). The pellet was dissolved in 0.01 M Tris-buffer (pH 7.4) with 0.01% sodium aside and transferred into 50 mM ammonium carbonate using a PD-10 column (GE Health Care, USA).
One-dimensional SDS-PAGE
Proteins were separated by electrophoresis in 12.5% polyacrylamide gel (8). The samples were dissolved in buffer consisting of 63 mM Tris/HCL, pH 6.8, 10% (v/v) glycerol, 2% (m/v sodium dodecyl sulphate (SDS) and 30 μM bromophenol blue. Samples were applied in quantities of 20 μg protein per lane.
Liquid chromatography coupled with mass spectrometry (LC-MS) analysis
After SDS-PAGE electrophoresis (12.5% gel), gels were stained with Coomasse Brilliant Blue R-250, and protein strips were dissected. Pieces, 1x1 mm, were cut out from the protein strip. These gel pieces were washed for 5 min in 70 μl 50% 200 mM NH4HCO3 – 50% acetonitryl (v/v) and transferred to 70 μl acetonitryl for 20 min. Acetonitryl was removed, and the gels were dried using a SpeedVac for 20 min. Dried gel pieces were transferred to a solution containing 3 μl trypsin (15 ng/μl) in 50 mM NH4HCO3, incubated on ice for 30 min, and then kept overnight at 37°C.
LC-MS analysis was performed on a 1200 Series HPLC-Chip (Agilent Technology, Inc., Santa Clara, CA, USA) in combination with an Agilent 6520 Accurate-Mass Q-TOF LC/MS system (Agilent Technology, Inc.).
In the present investigation, an ID chip with a 150×0.075 mm analytic column and 40 nl concentrating column were used. The conditions of analysis were as follows. A 5-μl peptide solution sample was obtained from the gel after trypsin digestion. The flow speed was 300 nl/min for the pumps of the analytic and 3 μl/min for the pump of the concentrating columns. The composition of liquid phase A was 5% acetonitryl and 0.1% formic acid. Liquid phase B consisted of 90% acetonitryl and 0.1% formic acid. Gradients: 0% B to 50% during 50 min, then 80% B during 60 min up to 65 min, then 0% B at 65.1 min; stopping the separation. The samples were applied on the chip in 100% of buffer A.
Bioinformatic methods
Data analysis was performed by comparing MS/MS spectra against the European Bioinformatics Institute non-redundant proteome set of Swiss-Prot entries through the Spectrum Mill MS Proteomics Workbench (Agilent), with no static or dynamic modifications. The other settings of the Spectrum Mill MS Proteomics Workbench were: species subset database, human; instrument, Agilent Q-TOF; precursor mass tolerance, 5 ppm; digest specificity, no enzyme; and search mode, identity. Results were autovalidated after each search using the default parameters for autovalidation.
Protein database searches were performed with Spectrum Mill MS Proteomic Workbench Rev A.03.03.084. SR4 (licensed to Kurchatov Institute, Moscow, Russia) using the MSDB database of protein sequences (http://www.proteomics.leeds.ac.uk/bioninf/msdb.html; restoration August 31, 2006; Proteomics Department at the Hammersmith Campus of Imperial College, London, UK), with no static or dynamic modifications. The other settings of the Spectrum Mill MS Proteomics Workbench were: species subset database, S. aureus; instrument, Agilent Q-TOF; precursor mass tolerance, 5 ppm; digest specificity, trypsin; maximum number of missed cleavages, 1; and search mode, identity. Results were autovalidated after each search using the default parameters for autovalidation.
Protein measurement
The amount of secreted proteins in the immune doses was measured by the Bradford method (9).
Statistics
Analysis of variance with the Bonferroni post hoc test was used for statistical analyses. Analyses were performed with Prism (version 5.0b) software. Values were considered significant at a P-value <0.05.
Results
Bacterial growth and SDS-PAGE
We first analyzed the growth kinetics under cultivation conditions described in Materials and methods. As shown in Fig. 1, the exponential growth phase of S. aureus strain 6 ended after 5 h. At the end of the exponential growth phase (after 4.5 h of cultivation), the number of CFUs was counted. A linear correlation between the number of seeded cells and the efficacy of the cultivation process was observed as measured by biomass density (Fig. 2). All further experiments were performed with a seeding number of 1×108 CFUs and a 4.5-h cultivation time.
Figure 1.
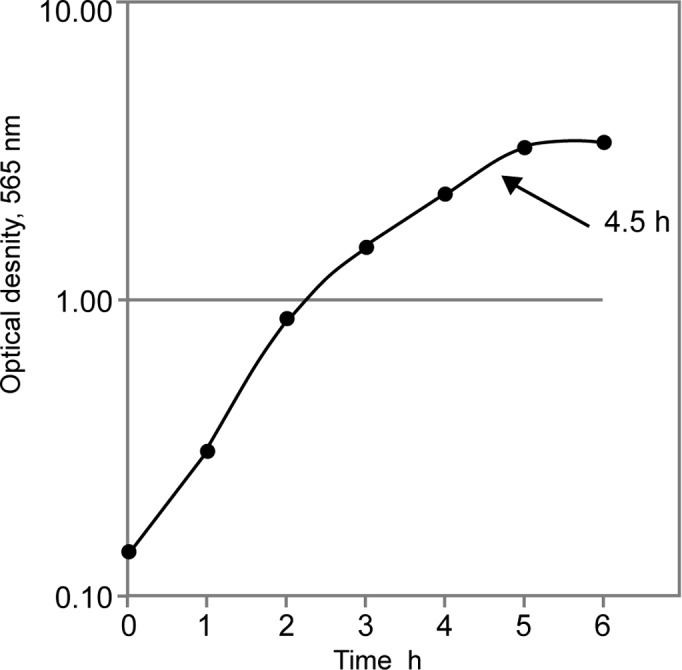
Growth kinetics of S. aureus strain 6. The arrow shows the time of harvesting of the culture for subsequent experiments.
Figure 2.
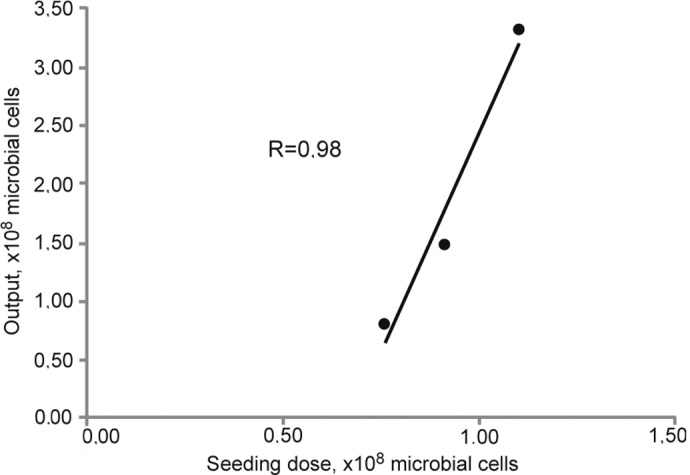
Determination of biomass density using different seeding numbers of S. aureus strain 6.
The bacterial cells were separated from the culture medium which was further processed as previously described (1,7). A filtrate of the culture medium was subjected to SDS-PAGE (Fig. 3). Electrophoretic separation showed two faint bands in the region of 100 kDa and four intense and several weak bands between 30 and 50 kDa. Below 30 kDa, no distinct bands were observed, although silver staining revealed the presence of undefined biological material. This may be indicative of polysaccharides and proteoglycans as previously identified in K. pneumoniae (1).
Figure 3.
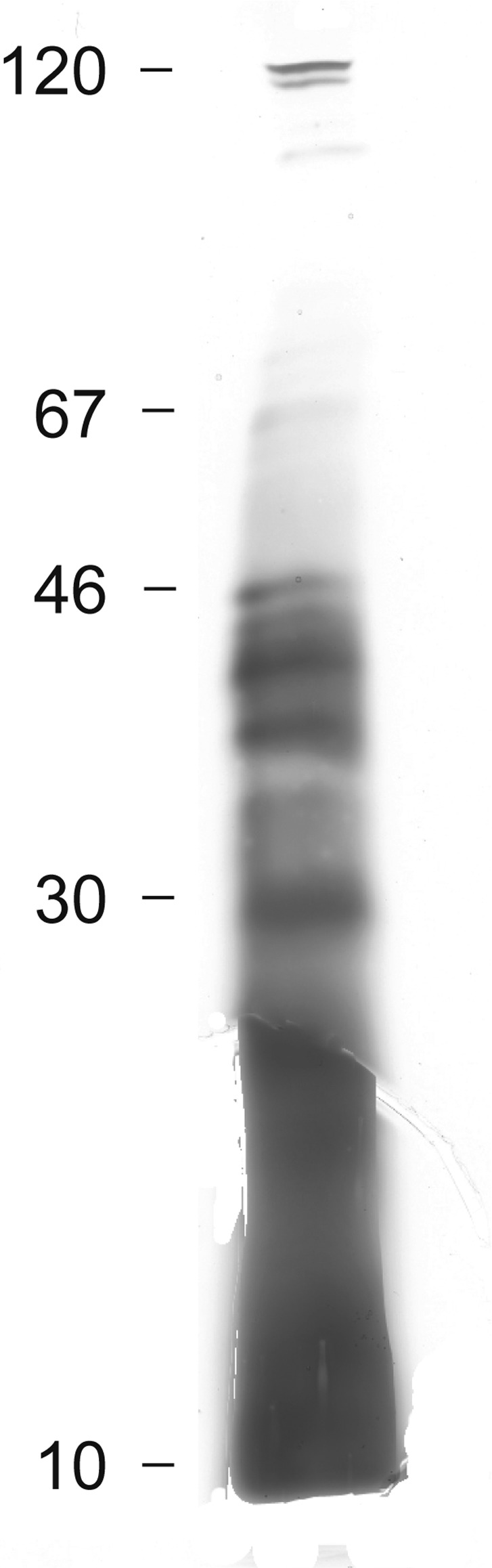
One-dimensional electrophoresis in polyacrylamide gel with sodium dodecyl sulphate (SDS-PAGE) of the extracellular proteins, extracted from the culture medium after 4.5 h of S. aureus growth. On the left side of the figure, molecular weights (kDa) are indicated; arrows on the right side indicate the protein traces mentioned in the text.
LC-MS analysis
The protein bands between 30 and 50 kDa were cut out and digested by trypsin, and subjected to LC-MS and MSDB analyses. A total of 208 peptides were identified. One hundred and fourteen of them were unique, and the average number of the unique peptides for one protein was 10.4 (median seven peptides/protein). As a result, 11 proteins were determined. Using PSOTb software (10), the isoelectric points (pI) of the proteins were calculated. A cytosolic localization was determined for 3 proteins (CP-N3, 5 and 11). One protein appeared to be localized in the cell membrane (CM-N4-lipoprotein) and one protein was extracellular (EC-N9-superoxide dismutase). The localization of the other 6 proteins could not be determined using PSORTb (Table I). These 6 proteins were further analyzed using the Uniprot Knowledgebase and the database of the European Institute for Bioinformatics (11). This analysis revealed that 4 of these 6 proteins were cytoplasmatically located and played a role in cellular metabolism: carbohydrate metabolism (N8 – protein SAS0528, hexulose-6-phosphate synthase), glycolysis (N1 – fructose bisphosphate aldolase class 1 and N10 – transaldolase) and glycine cleavage (N2). The supposed glycerophosphoryl-diester phosphodiesterase (N7) and protein SA0295 (N6) contained N-terminal signal peptides, indicating that they were extracellular localized or non-covalently linked to the bacterial cell wall. SA0295 (N6) was highly similar to the amino acid sequence of S. aureus acid phosphatase. Hence, by using two approaches, 7 out of 11 proteins were identified as having a cytoplasmic origin and 3 out of 11 were secreted (3,4).
Table I.
Extracellular proteins identified by MALDI-TOF mass-spectrometry in the exponential growth phase of S. aureus strain 6.
| N | Accession no. | Gene symbol | Ordered locus name | Protein identification | MW (Da) | pI | Protein score | Coincident peptides | The most similar strains | Localization according to PSORTb
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP | CM | CW | EC | Total | ||||||||||
| 1 | Q2YWF3 | fba | SAB2479 | Fructose bisphosphate aldolase class 1 | 32907 | 4.69 | 788 | 25 | RF122, MRSA252 | 2.50 | 2.50 | 2.50 | 2.50 | U |
| 2 | B89855 | gcvH | SA0760 | Glycine cleavage system H protein | 14072 | 3.70 | 140 | 2 | N315, RF122, NCTC 8325, MW2 | 2.50 | 2.50 | 2.50 | 2.50 | U |
| 3 | A89896 | fabD | SA1073 | Malonyl CoA-acyl carrier protein transacylase | 33628 | 4.59 | 97 | 3 | N315, RF122, MRSA252, NCTC 8325, MRSA476 | 9.67 | 0.01 | 0.15 | 0.17 | CP |
| 4 | H89832 | SA0587 | SA0587 | Lipoprotein, Streptococcal adhesin PsaA homologue | 35049 | 9.16 | 120 | 2 | N315 | 0.00 | 9.68 | 0.17 | 0.16 | CMa |
| 5 | F89850 | pgk | SA0728 | Phosphoglycerate kinase | 42575 | 4.96 | 373 | 14 | N315, RF122 | 10.00 | 0.00 | 0.00 | 0.00 | CP |
| 6 | D89795 | SA0295 | SA0295 | Similar to outer membrane protein precursor | 33331 | 10.08 | 353 | 20 | N315, MRSA252 | 0.00 | 3.33 | 3.33 | 3.33 | Ua |
| 7 | H89862 | glpQ | SA0820 | Putative glycerophosphoryl diester phosphodiesterase | 35289 | 9.14 | 87 | 2 | N315, MRSA252 | 0.00 | 0.15 | 4.55 | 5.15 | Ua |
| 8 | D89825 | SAS0528 | SAS0528 | Putative hexulose-6-phosphate synthase | 22422 | 4.32 | 244 | 3 | N315, RF122, MW2, USA300 | 2.50 | 2.50 | 2.50 | 2.50 | U |
| 9 | H89935 | sodA | SA1382 | Superoxide dismutase | 22697 | 4.86 | 387 | 17 | N315 | 0.01 | 0.09 | 0.18 | 9.72 | EC |
| 10 | F89963 | SA1599 | SA1599 | Similar to transaldolase | 25742 | 4.51 | 260 | 7 | N315, RF122, JH1 | 2.50 | 2.50 | 2.50 | 2.50 | U |
| 11 | G89850 | tpiA | SA0729 | Triosephosphate isomerase | 27245 | 4.51 | 539 | 19 | N315, RF122, NCTC 8325 | 9.67 | 0.01 | 0.15 | 0.17 | CP |
The Mascot search results displayed above [accession no., molecular weight (MW), protein score, coincident peptides and the most similar strains) are from the best protein match to published S. aureus genomes. Gene designations and ordered locus names are according to UniProt (http://www.uniprot.org) for S. aureusN315. pI values were determined using Isoelectric Point Calculator (http://isoelectric.ovh.org) and EMBOSS (http://emboss.sourceforge.net) pK values. Localization was predicted using PSORTb 3.0 (http://psort.org/psortb/index.html). CP, cytoplasmic; CM, cytoplasmic membrane; CW, cell wall-associated; EC, extracellular. Protein localization was based upon which category scored the highest value as determined by PSORTb. If localization could not be determined by PSORTb, the protein was listed as unknown (U).
Presence of the consensus amino acid sequence for N-terminal signal peptide.
Protective activity of the secreted proteins
The protein bands between 30 and 50 kDa were used for immunization of mice. S. aureus strain 6 (4×108 cells) was then injected into the animals. The protective activity of this vaccination has been estimated by the survival of animals after infection. Two different doses of secreted proteins (2 and 30 μg) were injected into mice, both of which revealed protective activity. As shown in Table II, all of the animals of the control group died. Vaccination of mice with the secreted proteins led to survival of half of the animals (P<0.05). In a second experiment, the LD50 values were determined in vaccinated and non-vaccinated mice by injection of varying numbers of injected S. aureus strain 6 cells. Inoculates with six different cell densities of S. aureus strain 6 ranging from 1×107 to 6×108 cells were injected into the peritoneal cavity of mice. In the immunized mice, the LD50 was 4.3×108 microbial cells and in control mice 1.8×108 cells.
Table II.
Protective activity of the secreted proteins of S. aureus strain 6.
| Immunization dose (μg/mice) | No. of mice per group | No. of surviving mice | Statistical significance | |
|---|---|---|---|---|
| Control | - | 10 | 0 | |
| 1st vaccinated group | 30 | 10 | 5 | P<0.05 |
| 2nd vaccinated group | 2 | 10 | 6 | P<0.05 |
Mice were immunized twice subcutaneously with two doses of secreted proteins at a 2-week interval. Staphylococci were used to infect anesthetized mice 14 days after the last immunization. Each animal was injected i.p. with 4×108 microbial cells. Statistical significance was calculated using the Bonferroni test.
Discussion
In the present investigation, we found 11 proteins in the culture medium of exponentially growing S. aureus strain 6. Additional compounds identified by gel electrophoresis presumably were polysaccharides and proteoglycans as recently observed in the culture medium of K. pneumonie (1). A significant finding was that we did not identify proteins which are known to be secreted by bacteria, but also membrane-bound and cytoplasmic proteins. To minimize the contamination of the nutritional medium with dying cells, which non-specifically release their intracellular constituents, we only used cells in the exponential growth phase. Thereby, a massive destruction of cells releasing intracellular proteins was prevented. For this reason, we conclude that these proteins were secreted from the bacteria into the culture medium by specific transport processes. Another reason for the specificity of this observation is that only this small profile of 11 proteins was present in the medium and not the broad spectrum of hundreds of intracellular proteins as could be expected from cell lysis. Notably, similar profiles of secreted proteins as determined in the present investigation for S. aureus, were also described for K. pneumoniae and Streptococcus pneumoniae (1).
If we assume that these proteins were specifically released by the exponentially growing cells via yet unknown secretory pathways, the question arises as to the function of these proteins. The determination of the isoelectric points revealed that two proteins were in the alkaline diapason (pI for protein N4 is 9.16 and for protein N6 is 10.08), whereas all others had their isoelectric point at acidic pH values. N6 was assigned as a protein transporter by database mining, while the function of N4 is unknown. According to the general physicochemical properties of proteins, it is highly probable that they form complexes with acidic proteins. These complexes would then have a neutral charge. A neutral charge of protein complexes is one of the conditions sufficient for molecular transport through cellular membranes. Hence, it can be hypothesized that complex formation of alkaline and acidic proteins may explain the secretion of the 11 proteins identified by us.
It has been shown that decreased isoelectric points are associated with weaker immunogenicity of proteins (12). Hence, it can be expected that the 9 acidic proteins in our study may exert weak immunogenicity, whereas the two alkaline proteins provoke strong immune reactions. The explanation for a causative association between molecular charge and immunogenicity is that cell membranes normally have a negative charge. Therefore, acidic molecules (with negative charge) are rejected by membranes of immune competent cells and alkaline molecules (with a positive charge) are attracted.
Determination of the proposal functions of the identified proteins
Superoxide dismutase (N9) [sodA (SA1382)] is well known as a component of the antioxidant stress response. This enzyme is predominantly expressed at the end of the exponential growth phase of S. aureus (13) and may take part in carbohydrate metabolism-related redox reactions, i.e., in energy transport. Two other extracellular proteins – glycerophosphoryl diester phosphodiesterase (N7) [glpQ (SA0820)] (14) and protein N6 (SA0295), with similarity to acid phosphatase S. aureus (15), probably hydrolyze biological macromolecules assimilated by bacterial cells, e.g., glycerine, fat acids and phosphatases.
As predicted by the PSORTb software, protein N4 [lipoprotein (SA0587)] is localized in the cell membrane, as this protein contains a transmembrane internal spiral as a signal peptide. There is a high similarity of this protein with ATP-binding cassette substrate-linking lipoprotein SA0587, which transports metal ions into cells. Furthermore, it was reported that this protein, which has a positive charge, may transport negatively charged enzymes (16). It is conceivable, therefore, that basic residues in this protein may be involved in electrostatic interactions with negatively charged substrates
Of the 7 cytoplasmic proteins, 5 are involved in carbohydrate metabolism: N1 – fructose bisphosphate aldolase class 1 [fba (SAB2479)], N11 – triosephosphate isomerase [tpiA (SA0729)] and N5 – phosphoglycerate kinase [pgk (SA0728)] – are glycolys enzymes. N10 – transaldolase (SA1599) links glycolys with the pentose phosphate shunt. N8 – proposed hexulose-6-phosphate synthase [sgaH (SAS0528)] catalyses the reversible reaction of assimilated formaldehyde by ribuloso-5-phosphate to arabinoso-6-phosphate. The reversed reaction is also a part of the de novo pyrimidine biosynthesis. The other two enzymes, N3 – transacylase malonyl CoA-acyl carrier protein [fabD (SA1073)] and N2 – protein of the H system cleavage glycine [gcvH (SA0760)] are involved in the biosynthesis of fatty acids and decarboxylation of glycine, respectively.
The exponential growth phase is very important for the development of bacteria, since resting bacterial cells have only limited energy reserves. Therefore, the start of growth is causatively linked with the activation of energy metabolism.
The result of the present investigation that S. aureus releases only a limited number rather than thousands of proteins was confirmed by comparable investigations of other authors. Ziebandt et al identified 43 extracellular S. aureus proteins, including those controlled by the additional regulator-gene (agr) and/or alternative factor σB (sigB) (17). Nakano et al identified 29 extracellular proteins produced by methicillin-resistant strains of S. aureus (14) using two-dimensional electrophoresis linked with the N-terminal sequence of amino acids. At the time point of the transition from the resting to the proliferative state, bacterial cells do not have enough energy and nutritional resources to synthesize the entire spectrum of proteins. Therefore, only a limited number of proteins with crucial functional importance is synthesized by the microorganisms. We speculate that the host immune response may inhibit bacterial growth by attracting extracellular bacterial proteins leading to energy depletion rather than acting on the bacterial cell itself.
Our hypothesis that enzymes of the carbohydrate metabolism act as signals for the development of infections is supported by the fact that patients with diabetes mellitus more frequently suffer from severe bacterial complications than non-diabetic subjects. There are several hints from the literature supporting this point of view. Diabetic patients are prone to develop all types of infections. Pneumococcal infections are a common cause of morbidity and mortality among individuals affected with diabetes (18). Furthermore, pyomyositis is a relatively infrequent, subacute primary bacterial muscle infection. Due to its non-specific clinical symptoms, it is unlikely to be diagnosed early particularly in diabetic patients. This delay in diagnosis may be fatal (19).
In a large nationwide survey of diabetic patients undergoing a variety of non-cardiac surgical procedures, glucose control in the first 24 h after surgery was poor, and mean serum glucose concentrations of ≥150 mg/dl during this time period were associated with increased rates of postoperative infectious complications (20).
These data are in accord with results showing that various growth factors, which regulate cell growth also influence glucose metabolism (21). In this case, increased concentrations of carbohydrates may be a signal for microorganisms ‘to wake up’ and grow. Such a growth signal may also be too strong to kill microorganisms by chemotherapy, as they have enough energy to survive a challenge by drugs.
In conclusion, the identification of carbohydrate metabolism-related enzymes and transporter proteins in the extracellular medium of S. aureus may have implications for the development of new strategies for prevention and therapy of S. aureus infections. Preliminary data concerning the protective activity of the immunization of animals by these secreted proteins confirms our assumption of the importance of these proteins for the viability of microorganisms.
References
- 1.Trishin AV, Donenko FV, Kurbatova EA, Voyushin KE, Kiselevsky MV, Egorova NB, Gruber IM, Semenova IB, Semenov BF. Protective activity of secreted proteins of Streptococcus pneumoniae and Klebsiella pneumoniae. Zh Microbiol. 2008;4:46–50. [PubMed] [Google Scholar]
- 2.Jones RC, Deck J, Edmondson RD, Hart ME. Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using 1D-PAGE and nanocapillary LC coupled with tandem MS. J Bacteriol. 2008;190:5265–5278. doi: 10.1128/JB.00383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burlak C, Hammer CH, Robinson MA, Whitney AR, McGavin MJ, Kreiswirth BN, Deleo FR. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sibbald MJ, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJ, Raangs GC, Stokroos I, Arends JP, Dubois JY, van Dijl JM. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol Mol Biol Rev. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egorova NB, Korzaya LI. The effect of repeated administration of staphylococcal immune preparations on the development of local staphylococcal infection in mice. J Hyg Epidemiol Microbiol Immunol. 1983;27:203–210. [PubMed] [Google Scholar]
- 6.Kuzmenko OM, Zlygostev SA, Mikhaylova NA, Gruber IM, Akhmatova NK, Kurbatova EA, Cherkasova LS. Characteristics of antigenic complexes of Staphylococcus aureus vaccine strains obtained in different cultivation conditions. Zh Microbiol. 2010;2:51–54. [PubMed] [Google Scholar]
- 7.Donenko FV, Sitdikova SM, Syrtsev AV, Gradyushko AT, Kiselevsky MV, Serebryakova MV, Efferth T. Hemoglobin-associated proteins isolated from blood serum of Ehrlich carcinoma-bearing mice. Int J Oncol. 2008;32:885–893. doi: 10.3892/ijo.32.4.885. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli UK. Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FSL. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 11.The UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattori M, Miyakawa S, Ohama Y, Kawamura H, Yoshida T, To-o K, Kuriki T, Takahashi K. Reduced immunogenicity of beta-lactoglobulin by conjugation with acidic oligosaccharides. J Agric Food Chem. 2004;52:4546–4553. doi: 10.1021/jf0353887. [DOI] [PubMed] [Google Scholar]
- 13.Clements MO, Watson SP, Foster SJ. Characterisation of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano M, Kawano Y, Kawagish M, Hasegawa T, Iinuma Y, Oht M. Two-dimensional analysis of exoproteins of methicillin-resistant Staphylococcus aureus (MRSA) for possible epidemiological applications. Microbiol Immunol. 2002;46:11–22. doi: 10.1111/j.1348-0421.2002.tb02671.x. [DOI] [PubMed] [Google Scholar]
- 15.Du Plessis EM, Theron J, Joubert L, Lotter T, Watson TG. Characterization of a phosphatase secreted by Staphylococcus aureus strain 154, a new member of the bacterial class C family of nonspecific acid phosphatases. Syst Appl Microbiol. 2002;25:21–30. doi: 10.1078/0723-2020-00098. [DOI] [PubMed] [Google Scholar]
- 16.Alexeyev MF, Winkler HH. Complete replacement of basic amino acid residues with cysteines in Rickettsia prowazekii ATP/ADP translocase. Biochim Biophys Acta. 2002;1565:136. doi: 10.1016/s0005-2736(02)00544-8. [DOI] [PubMed] [Google Scholar]
- 17.Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, Engelmann S. The influence of agr and sigmaB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics. 2004;4:3034–3047. doi: 10.1002/pmic.200400937. [DOI] [PubMed] [Google Scholar]
- 18.Mohan V, Unnikrishnan R, Thomas N, Bhansali A, Wangnoo SK, Thomas K. Pneumococcal infections and immunization in diabetic patients. J Postgrad Med. 2011;57:78–81. doi: 10.4103/0022-3859.74299. [DOI] [PubMed] [Google Scholar]
- 19.Abdullah ZS, Khan MU, Kodali SK, Javaid A. Pyomyositis mimicking osteomyelitis detected by SPET/CT. Hell J Nucl Med. 2010;13:277–279. [PubMed] [Google Scholar]
- 20.King JT, Goulet JL, Perkal MF, Rosenthal RA. Glycemic control and infections in patients with diabetes undergoing noncardiac surgery. Ann Surg. 2011;253:158–165. doi: 10.1097/SLA.0b013e3181f9bb3a. [DOI] [PubMed] [Google Scholar]
- 21.Heiden MGV, Plas DR, Rathmell JC, Harris MH, Thompson CB. Growth factors can influence cell growth and survival though effects on glucose metabolism. Mol Cell Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


