Abstract
The hMSH2 gene, a member of the mismatch repair (MMR) pathway, plays a key role in the maintenance of genomic integrity. The common sequence variation in hMSH2, IVS12-6 T>C, has been implicated in cancer risk. However, the results of published studies on this polymorphism remain conflicting. Hence, we conducted a meta-analysis to clarify the role of the hMSH2 IVS12-6 T>C polymorphism in cancer. We performed a comprehensive literature search updated to March 2011 of studies on the associations between the hMSH2 IVS12-6 T>C polymorphism and cancer risk. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of the associations. Thirteen studies involving 7,527 patients and 8,762 control subjects were included in this meta-analysis. The overall results indicated no major influence of the polymorphism on cancer risk. However, stratified analysis by cancer types showed that the hMSH2 IVS12-6 polymorphism increased the risk for non-Hodgkin's lymphomas (heterozygote comparison: OR=1.62; 95% CI 1.06–2.47). When stratified by the source of controls, significant associations were observed in hospital-based populations (heterozygote comparison: OR=1.28; 95% CI 1.02–1.61). These results indicate that the polymorphism of hMSH2, IVS12-6, may cause a different effect in different types of cancers. To draw more comprehensive conclusions, further prospective studies with larger numbers of participants worldwide are required to examine the associations between this polymorphism and cancer risk.
Keywords: mismatch repair, hMSH2, polymorphism, meta-analysis
Introduction
During the last decade, a number of important genes responsible for the genesis of various types of cancers have been discovered, at the same time their mutations have been precisely established, and the pathway through which they act has been characterized (1). The mismatch repair (MMR) pathway, first described in bacteria, is involved in the maintenance of genomic integrity by repairing DNA replication errors (2), and MMR proteins correct base substitution mismatches and small insertion-deletion mismatches generated during DNA replication (3). The MMR system is well conserved from Escherichia coli to mammals, and the E. coli MMR system, where MutS, MutL and MutH complexes function, has been well analyzed. In mammalian cells, heterodimers of MutS homologues (MSH2-MSH6 and MSH2-MSH3) recognize replication errors, and the heterodimer of the MutL homologue (MLH1-PMS2) interacts with MutS homologues and recruits further repair proteins (4).
Seven MMR genes exist in humans: MLH1, MLH3, PMS1, PMS2, MSH2, MSH3 and MSH6 (5). The inactivation of these genes leads to increased genetic instability, which in turn results in an increased rate of mutation in ‘gatekeeper’ genes that regulate human cell proliferation and death (6). A role for hMSH2 in cancer has been firmly established in hereditary nonpolyposis colorectal cancer (HNPCC) (7–9). Several single nucleotide polymorphisms (SNPs), IVS12-6 T>C (rs2303428), G23A (rs4987188) and IVS10+12 A>G (rs3732183) were identified in the hMSH2 gene and among them, only the IVS12-6 T>C polymorphism was reported functional and therefore has been extensively studied in recent years (3,10–21).
The IVS12-6 T>C is a common polymorphism located at position −6 of the intronic splice acceptor site of exon 13 of hMSH2. Although the genetic function of this polymorphism has not been well determined, its variant type may result in alternative splicing and deficiency of hMSH2 protein (22,23).
To date, many studies have investigated the role of this polymorphism in the etiology of cancers of various organs including the lung, colon, rectum, ovary, and others (3,10–21). However, the results of these studies remain conflicting rather than conclusive. Considering the extensive role of hMSH2 in the carcinogenic process, we performed a meta-analysis on all eligible case-control studies to estimate the overall cancer risk of this polymorphism and to quantify the potential between-study heterogeneity.
Materials and methods
Identification and eligibility of relevant studies
We searched the electronic literature PubMed for all relevant reports (the last search update was March 22, 2011), using the key words ‘MSH2’ or ‘hMSH2’, ‘cancer’ and ‘polymorphism’. The search was limited to English language manuscripts. In addition, studies were identified by a manual search of the reference lists of reviews and retrieved studies. Studies were selected when there were available data for the hMSH2 IVS12-6 T>C polymorphism with cancer risk in a case-control design. Additional studies were identified by a hand search of the references of the original studies. We also used the PubMed option ‘Related Citations’ in each research article to search potentially relevant articles. In our meta-analysis, the studies had to meet the following criteria: i) was a study of the hMSH2 IVS12-6 T>C polymorphism and cancer risk, ii) used a case-control design and iii) contained available genotype frequency.
Data extraction
Two of the authors independently extracted data and reached a consensus on all of the items. For each study, the following information was sought: the first author's last name, year of publication, country of origin, ethnicity, source of control groups (population- or hospital-based controls), numbers of genotyped cases and controls, genotyping methods, and cancer type. Different ethnic descents were categorized as Caucasian, Asian and mixed (composed of an admixture of different ethnic groups). For studies including subjects of different ethnic groups, data were extracted separately for each ethnic group whenever possible.
Statistical analysis
For the control group of each study, the observed genotype frequencies of hMSH2 IVS12-6 T>C were assessed for Hardy-Weinberg equilibrium using the χ2 test. The strength of the association between hMSH2 IVS12-6 T>C and cancer risk was measured by odds ratios (ORs) with 95% confidence intervals (CIs). We first estimated the risks of the CC and CT genotypes on cancers, compared with the wild-type TT homozygote, and then evaluated the risks of (CC/CT) vs. TT and CC vs. (CT/TT) on cancers, assuming dominant and recessive effects of the variant C allele, respectively. In order to evaluate the ethnicity-specific effect, subgroup analyses were performed by ethnic group. In consideration of the possibility of heterogeneity across the studies, a statistical test for heterogeneity was performed based on the Q-test. When the P-value was >0.10 in the Q-test which indicates a lack of heterogeneity among studies, the summary OR estimate of each study was calculated by the fixed-effects model of Mantel-Haenszel (24). Otherwise, the random-effects model of DerSimonian and Laird (25) was used. An estimate of potential publication bias was carried out by the funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggested a possible publication bias. All statistical tests were performed with Stata software (version 10.0; Stata Corporation, College Station, TX, USA).
Results
Characteristics of the studies
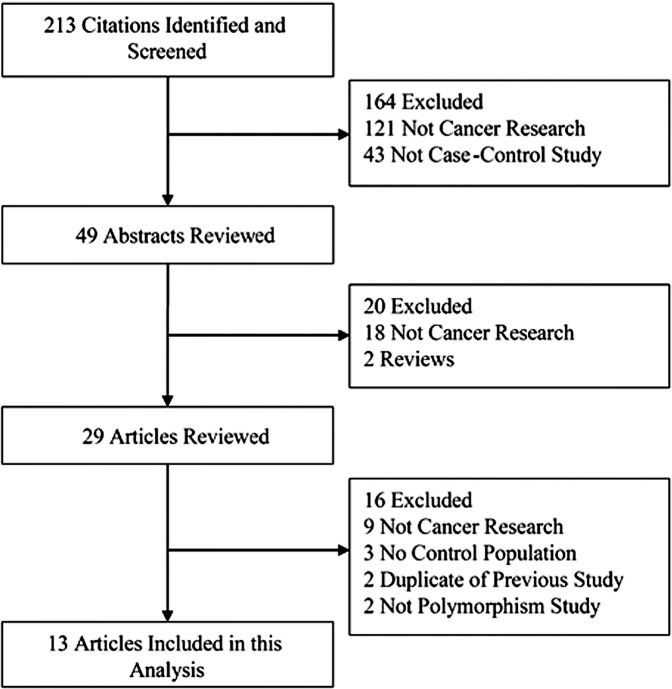
Thirteen eligible publications were identified on the association between the hMSH2 IVS12-6 T>C polymorphism and cancer risk including 7,527 cancer cases and 8,762 controls (3,10–21). The selected study characteristics are listed in Table I and the criteria for inclusion and exclusion are shown in Fig. 1. All studies were case-control studies. There were six studies in populations of Caucasian descent, four of Asian origin and three of mixed race. A classic polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay was conducted in four of the 13 studies. In addition, other methods were utilized to detect genotypes. The genotype distributions among the controls of all studies were in agreement with Hardy-Weinberg equilibrium for all except three studies (16,17,21).
Table I.
Characteristics of the eligible studies included in the meta-analysis.
| First author | Year | Country/Region | Ethnicity | Cancer type | Source of controls | Genotyping method | Cases | Controls | HWE |
|---|---|---|---|---|---|---|---|---|---|
| Goessl | 1997 | Germany | Caucasian | Colorectal cancer | PB | FLUPD | 106 | 125 | 0.540 |
| Paz-y-mino | 2002 | Ecuador | Mixed | Non-Hodgkin's lymphomas | HB | PCR-SSCP | 22 | 50 | 0.009 |
| Paz-y-mino | 2003 | Ecuador | Mixed | Lymphoma and leukemia | HB | PCR-SSCP | 181 | 50 | 0.009 |
| Hishida | 2003 | Japan | Asian | Non-Hodgkin's lymphomas | HB | PCR-RFLP | 103 | 487 | 0.942 |
| Kim | 2004 | Korea | Asian | Colorectal cancer | PB | TaqMan | 107 | 330 | 0.091 |
| Jung | 2006 | Korea | Asian | Lung cancer | PB | PCR-RFLP | 432 | 432 | 0.613 |
| Beiner | 2006 | Canada | Caucasian | Endometrial cancer | PB | MALDI-TOF | 665 | 654 | 0.000 |
| Song | 2006 | UK, Denmark, USA | Caucasian | Ovarian cancer | PB | TaqMan | 1325 | 2044 | 0.411 |
| Hsu | 2007 | Taiwan | Asian | Lung cancer | PB | PCR-RFLP | 156 | 235 | 0.493 |
| Raptisa | 2007 | Canada | Caucasian | Colorectal cancer | PB | TaqMan | 929 | 1098 | 0.444 |
| Raptisa | 2007 | Canada | Caucasian | Colorectal cancer | PB | TaqMan | 430 | 275 | 0.195 |
| Koessler | 2008 | UK | Caucasian | Colorectal cancer | PB | TaqMan | 2294 | 2279 | 0.752 |
| Tulupova | 2008 | Czech | Caucasian | Colorectal cancer | HB | TaqMan | 611 | 608 | 0.689 |
| Demokan | 2010 | Turkey | Mixed | Head and neck cancer | HB | PCR-RFLP | 166 | 95 | 0.054 |
Different populations existed in one study. HWE, Hardy-Weinberg equilibrium. PB, population-based; HB, hospital-based. FLUPD, fluorescent-labeled universal primer; PCR, polymerase chain reaction; SSCP, single-strand conformation polymorphism; RFLP, restriction fragment length polymorphism; MALDI-TOF, matrix assisted laser desorption/ionization time-of-flight.
Figure 1.
Studies identified with criteria for inclusion and exclusion.
Quantitative synthesis
There was a wide variation in the C allele frequency of the hMSH2 IVS12-6 T>C polymorphism among the controls across different ethnicities. For Asian populations, the IVS12-6 C allele frequency was 0.60 (95% CI 0.54–0.67), which was significantly higher than that in Caucasian populations (0.53; 95% CI 0.51–0.55; P=0.003) (Fig. 2).
Figure 2.
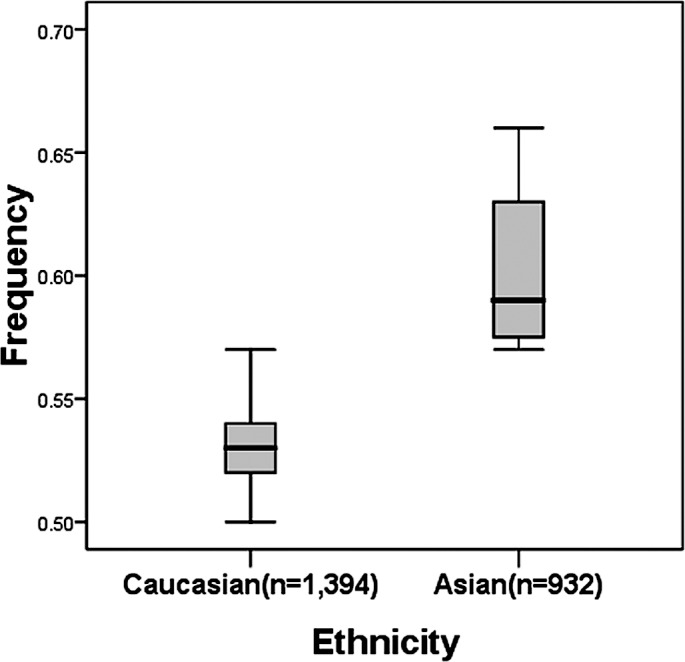
Frequencies of the variant alleles among controls stratified by ethnicity.
As shown in Table II, no significant association was found in any genetic model among studies of these cancers. Forest plot of heterozygote comparison was given (Fig. 3). In the stratified analysis by cancer type, significant increased risks were observed for non-Hodgkin's lymphoma patients (heterozygote comparison: OR=1.62; 95% CI 1.06–2.47) (Table II). According to the source of the controls, a significant effect was observed in hospital-based studies (heterozygote comparison: OR=1.28; 95% CI 1.02–1.61). Among studies of lung cancer, colorectal cancer and other cancers, no significant associations were found in any genetic model, in neither Asian nor Caucasian individuals (Table II).
Table II.
Stratified analyses of the hMSH2IVS12-6 T>C polymorphism on cancer risk.
| Variables | n | Cases/controls | CT vs. TT
|
CC vs. TT
|
CC/CT vs. TT (dominant)
|
CC vs. CT/TT (recessive)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | |||
| Total | 14 | 7527/8762 | 1.10 (0.94–1.28)b | 0.002 | 0.92 (0.74–1.15) | 0.881 | 1.07 (0.93–1.23)b | 0.003 | 0.89 (0.72–1.10) | 0.892 |
| Cancer types | ||||||||||
| Lung cancer | 2 | 588/667 | 1.15 (0.90–1.46) | 0.392 | 1.02 (0.70–1.46) | 0.818 | 1.11 (0.89–1.40) | 0.477 | 0.93 (0.66–1.30) | 0.878 |
| NHL | 2 | 125/537 | 1.62 (1.06–2.47) | 0.173 | 0.98 (0.43–2.27) | 0.948 | 1.50 (1.00–2.27) | 0.247 | 0.81 (0.36–1.81) | 0.951 |
| Colorectal cancer | 6 | 4477/4715 | 1.02 (0.82–1.28)b | 0.008 | 1.00 (0.68–1.45) | 0.896 | 1.03 (0.83–1.28)b | 0.008 | 1.04 (0.71–1.51) | 0.867 |
| Other cancers | 4 | 2337/2843 | 1.09 (0.77–1.55)b | 0.031 | 0.68 (0.42–1.09) | 0.261 | 1.03 (0.73–1.44)b | 0.027 | 0.67 (0.42–1.08) | 0.319 |
| Ethnicities | ||||||||||
| Asian | 4 | 798/1484 | 1.11 (0.92–1.34) | 0.114 | 1.04 (0.76–1.41) | 0.981 | 1.09 (0.91–1.31) | 0.243 | 0.96 (0.72–1.29) | 0.792 |
| Caucasian | 7 | 6360/7083 | 1.07 (0.88–1.28)b | 0.002 | 0.87 (0.63–1.20) | 0.809 | 1.06 (0.88–1.27)b | 0.002 | 0.87 (0.63–1.20) | 0.863 |
| Mixed | 3 | 369/195 | 1.78 (0.64–4.94)b | 0.080 | 0.25 (0.05–1.14) | 0.637 | 1.43 (0.57–3.64)b | 0.080 | 0.25 (0.05–1.12) | 0.694 |
| Source of controls | ||||||||||
| Hospital-based | 5 | 1083/1290 | 1.28 (1.02–1.61) | 0.214 | 0.78 (0.40–1.51) | 0.444 | 1.22 (0.98–1.52) | 0.223 | 0.70 (0.37–1.33) | 0.520 |
| Population-based | 9 | 6444/7472 | 1.04 (0.88–1.23)b | 0.003 | 0.94 (0.74–1.19) | 0.899 | 1.03 (0.88–1.20)b | 0.004 | 0.92 (0.73–1.15) | 0.900 |
P-value of the Q-test for heterogeneity test.
Random-effects model was used when the P-value for heterogeneity test was <0.10; otherwise, the fix-effects model was used.
Figure 3.
Forest plot of the cancer risk associated with hMSH2 IVS12-6 T>C (CT vs. TT). The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the study-specific weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Test of heterogeneity
There was significant heterogeneity for the heterozygote comparison (CT vs. TT, P=0.001) and dominant model comparison (CC/CT vs. TT, P=0.002). We then assessed the source of heterogeneity for the heterozygote comparison (CT vs. TT) by cancer type, ethnicity and source of controls. As a result, the source of controls (χ2=4.11, df=1, P=0.043), but not cancer type (χ2=6.21, df=3, P=0.102) or ethnicity (χ2=1.85, df=2, P=0.396) was found to contribute to the substantial heterogeneity.
Publication bias
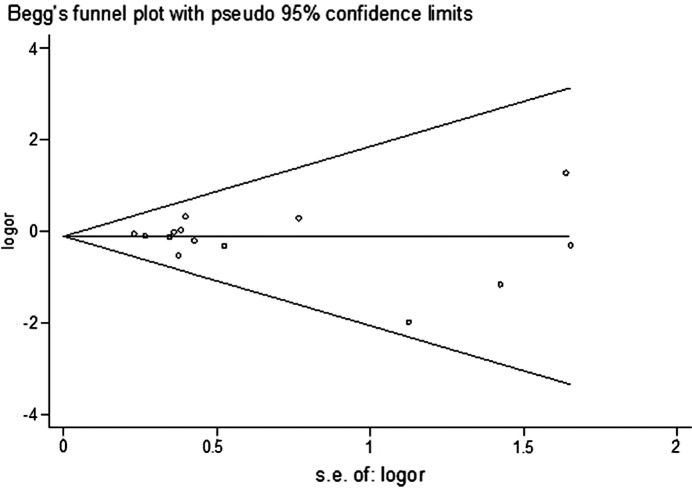
Begg's funnel plot and Egger's test were performed to assess the publication bias of the literature studies. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry. The Egger's test was subsequently used to provide statistical evidence of funnel plot symmetry. The results did not show any evidence of publication bias (t=0.83, P=0.423 for CC vs. TT) (Fig. 4).
Figure 4.
Begg's funnel plot for publication bias test (CC vs. TT). Each point represents a separate study for the indicated association. Log[or], natural logarithm of odds ratio. Horizontal line, mean effect size.
Discussion
The mismatch repair pathway plays an important role in the maintenance of genome integrity and acts conservatively in species. With its main function to repair mismatches during DNA duplication, the MMR pathway ensures the integrity and stability of the genome (3). Genome point mutations, microsatellite instability (MSI) and loss of heterozygosity (LOH) are all mutation phenotypes caused by impairment in any component of the MMR pathway (3,26). Studies have revealed the presence of MSI in more than 90% of hereditary non-polyposis colorectal cancer (HNPCC) and in 15% of non-family colorectal cancer cases (27), and mutations of MLH1, MSH2 and MSH6 germ cells in more than 95% of HNPCC cases with MSI (28). The presence of MSI has been confirmed in other primary cancers such as prostate (29), endometrial (30), pancreatic (31), gastric (32,33) and others (34). Meanwhile, a few SNPs were found to be involved in the formation and development of solid tumors through the alteration of the biological activities of DNA repairase (35–37); for example, MLH1 I219V and breast cancer (38), PMS2 rs7797466 and ovarian cancer (19), and MLH1 −93 G>A, MSH2 −118 T>C, MSH6 G39E and colorectal cancer (18,39). hMSH2 is the first MMR gene to be associated with HNPCC invasion. Its expression is varied not only in gastric cancer, lung cancer, endometrial cancer and others, but also is also associated with the prognosis of cancers. High expression indicates better repair and low expression is indicative of a worse repair ability (40). Although the genetic function of the hMSH2 IVS12-6 T>C polymorphism is not clear, its variant type may lead to alternative splicing and deficiency of hMSH2 protein (22,23).
In the present study, we performed a meta-analysis of published studies based on 13 case-control studies in order to reveal the association between the hMSH2 IVS12-6 T>C polymorphism and cancer risk. The results indicate that there is no significant association between this polymorphism and cancer risk. In the stratified analyses we found that the C allele was a risk factor for developing non-Hodgkin's lymphoma (NHL). This may be due to the limited number of studies analyzed and the small sample size. The ethnically mixed population in our meta-analysis consisted of only Ecuadorian and Trukese individuals from two continents representing a huge discrimination of human race. Our results also demonstrated that no significant associations were found in any genetic model among studies of lung cancer, colorectal cancer and others. A moderate association was observed in the hospital-based controls, but not in the population-based controls when stratifying the source of controls. This may have resulted from a differential effect of selection criteria in different cancers, which was dictated by the sample size in our meta-analysis, as well as the weight of each study. Other factors such as matched criteria may also have conferred an effect. The above differences may contribute to the inconsistent results. Therefore, it is very important to determine the unified selection criteria and to choose larger sample population studies.
We would like to note the differences in the genetic background and gene-environment interactions in the etiology. The IVS12-6 C allele frequency among the controls in Asian populations was significant higher than that in European populations, suggesting a possible ethnic difference (Fig. 2). Moreover, there is no reported study on African populations. Therefore, additional studies are needed to further validate ethnic differences on the effect of this SNP on cancer risk, particularly in Africans.
Identification of the source of heterogeneity is one of the most important goals of meta-analysis. In this analysis, we found that the source of heterogeneity was from the origin of the controls, suggesting that certain effects of the genetic polymorphism were population-specific.
Various limitations of this meta-analysis should be mentioned. First, the lack of original data of the reviewed studies limited our further evaluation of potential interactions, as interactions between gene-gene or gene-environment may modulate cancer risk. Second, our result was based on unadjusted estimates, while more precise analyses were needed to be performed had individual data been available, which would have allowed for an adjusted estimate by age or gender. However, our present meta-analysis also had advantages. First, a substantial number of cases and controls was pooled from different studies, which greatly increased the statistical power of the analysis. Second, the quality of case-control studies included in this meta-analysis was satisfactory according to our selection criteria.
In conclusion, our meta-analysis suggests that the hMSH2 IVS12-6 T>C polymorphism is not associated with cancer risk. A significant risk effect on cancer was observed in NHL patients while no statistical results were found for the other cancer types. Human race variation in the distribution of genotypes may have also affected the analysis. The role of this variant in other populations should be investigated by carrying out additional studies including a wider spectrum of subjects particularly African individuals. Further functional studies between the hMSH2 IVS12-6 T>C polymorphism and cancer risk should be conducted in order to reveal its mechanism. Additional well-designed extensive studies are warranted to validate the association between the hMSH2 IVS12-6 T>C polymorphism and the susceptibility of cancer.
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 3.Koessler T, Oestergaard MZ, Song H, et al. Common variants in mismatch repair genes and risk of colorectal cancer. Gut. 2008;57:1097–1101. doi: 10.1136/gut.2007.137265. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–4604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 6.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761, 763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 8.Kolodner RD. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 9.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 10.Demokan S, Suoglu Y, Ulusan M, Dalay N. Analysis of the hMSH2 gene variants in head and neck cancer. DNA Cell Biol. 2010;29:449–457. doi: 10.1089/dna.2009.1013. [DOI] [PubMed] [Google Scholar]
- 11.Goessl C, Plaschke J, Pistorius S, et al. An intronic germline transition in the HNPCC gene hMSH2 is associated with sporadic colorectal cancer. Eur J Cancer. 1997;33:1869–1874. doi: 10.1016/s0959-8049(97)00219-0. [DOI] [PubMed] [Google Scholar]
- 12.Hishida A, Matsuo K, Hamajima N, et al. Polymorphism in the hMSH2 gene (gIVS 12-6T-->C) and risk of non-Hodgkin lymphoma in a Japanese population. Cancer Genet Cytogenet. 2003;147:71–74. doi: 10.1016/s0165-4608(03)00185-7. [DOI] [PubMed] [Google Scholar]
- 13.Hsu HS, Lee IH, Hsu WH, Kao WT, Wang YC. Polymorphism in the hMSH2 gene (gISV12-6T > C) is a prognostic factor in non-small cell lung cancer. Lung Cancer. 2007;58:123–130. doi: 10.1016/j.lungcan.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Jung CY, Choi JE, Park JM, et al. Polymorphisms in the hMSH2 gene and the risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:762–768. doi: 10.1158/1055-9965.EPI-05-0834. [DOI] [PubMed] [Google Scholar]
- 15.Kim JC, Roh SA, Koo KH, et al. Genotyping possible polymorphic variants of human mismatch repair genes in healthy Korean individuals and sporadic colorectal cancer patients. Fam Cancer. 2004;3:129–137. doi: 10.1023/B:FAME.0000039919.66461.8f. [DOI] [PubMed] [Google Scholar]
- 16.Paz-y-Mino C, Fiallo BF, Morillo SA, et al. Analysis of the polymorphism [gIVS12-6T > C] in the hMSH2 gene in lymphoma and leukemia. Leuk Lymphoma. 2003;44:505–508. doi: 10.1080/1042819021000047038. [DOI] [PubMed] [Google Scholar]
- 17.Paz-y-Mino C, Perez JC, Fiallo BF, Leone PE. A polymorphism in the hMSH2 gene (gIVS12-6T>C) associated with non-Hodgkin lymphomas. Cancer Genet Cytogenet. 2002;133:29–33. doi: 10.1016/s0165-4608(01)00547-7. [DOI] [PubMed] [Google Scholar]
- 18.Raptis S, Mrkonjic M, Green RC, et al. MLH1 −93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J Natl Cancer Inst. 2007;99:463–474. doi: 10.1093/jnci/djk095. [DOI] [PubMed] [Google Scholar]
- 19.Song H, Ramus SJ, Quaye L, et al. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006;27:2235–2242. doi: 10.1093/carcin/bgl089. [DOI] [PubMed] [Google Scholar]
- 20.Tulupova E, Kumar R, Hanova M, et al. Do polymorphisms and haplotypes of mismatch repair genes modulate risk of sporadic colorectal cancer? Mutat Res. 2008;648:40–45. doi: 10.1016/j.mrfmmm.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Beiner ME, Rosen B, Fyles A, et al. Endometrial cancer risk is associated with variants of the mismatch repair genes MLH1 and MSH2. Cancer Epidemiol Biomarkers Prev. 2006;15:1636–1640. doi: 10.1158/1055-9965.EPI-06-0257. [DOI] [PubMed] [Google Scholar]
- 22.Bubb VJ, Curtis LJ, Cunningham C, et al. Microsatellite instability and the role of hMSH2 in sporadic colorectal cancer. Oncogene. 1996;12:2641–2649. [PubMed] [Google Scholar]
- 23.Xia L, Shen W, Ritacca F, et al. A truncated hMSH2 transcript occurs as a common variant in the population: implications for genetic diagnosis. Cancer Res. 1996;56:2289–2292. [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med. 2003;138:560–570. doi: 10.7326/0003-4819-138-7-200304010-00012. [DOI] [PubMed] [Google Scholar]
- 27.Rowley PT. Inherited susceptibility to colorectal cancer. Annu Rev Med. 2005;56:539–554. doi: 10.1146/annurev.med.56.061704.135235. [DOI] [PubMed] [Google Scholar]
- 28.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Wang J, Fraig MM, et al. Defects of DNA mismatch repair in human prostate cancer. Cancer Res. 2001;61:4112–4121. [PubMed] [Google Scholar]
- 30.Lax SF. Molecular genetic pathways in various types of endometrial carcinoma: from a phenotypical to a molecular-based classification. Virchows Arch. 2004;444:213–223. doi: 10.1007/s00428-003-0947-3. [DOI] [PubMed] [Google Scholar]
- 31.Wilentz RE, Goggins M, Redston M, et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: a newly described and characterized entity. Am J Pathol. 2000;156:1641–1651. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider BG, Bravo JC, Roa JC, et al. Microsatellite instability, prognosis and metastasis in gastric cancers from a low-risk population. Int J Cancer. 2000;89:444–452. doi: 10.1002/1097-0215(20000920)89:5<444::aid-ijc8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 33.Beghelli S, de Manzoni G, Barbi S, et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery. 2006;139:347–356. doi: 10.1016/j.surg.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Lawes DA, SenGupta S, Boulos PB. The clinical importance and prognostic implications of microsatellite instability in sporadic cancer. Eur J Surg Oncol. 2003;29:201–212. doi: 10.1053/ejso.2002.1399. [DOI] [PubMed] [Google Scholar]
- 35.Trojan J, Zeuzem S, Randolph A, et al. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122:211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 36.Hutter P, Couturier A, Rey-Berthod C. Two common forms of the human MLH1 gene may be associated with functional differences. J Med Genet. 2000;37:776–781. doi: 10.1136/jmg.37.10.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutter P, Wijnen J, Rey-Berthod C, et al. An MLH1 haplotype is over-represented on chromosomes carrying an HNPCC predisposing mutation in MLH1. J Med Genet. 2002;39:323–327. doi: 10.1136/jmg.39.5.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith TR, Levine EA, Freimanis RI, et al. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mrkonjic M, Raptis S, Green RC, et al. MSH2 118T>C and MSH6 159C>T promoter polymorphisms and the risk of colorectal cancer. Carcinogenesis. 2007;28:2575–2580. doi: 10.1093/carcin/bgm229. [DOI] [PubMed] [Google Scholar]
- 40.Lynch HT, Smyrk T, Lynch JF. Molecular genetics and clinical-pathology features of hereditary nonpolyposis colorectal carcinoma (Lynch syndrome): historical journey from pedigree anecdote to molecular genetic confirmation. Oncology. 1998;55:103–108. doi: 10.1159/000011843. [DOI] [PubMed] [Google Scholar]