Abstract
The actin binding protein α-actinin is a major component of focal adhesions found in vertebrate cells and of the focal adhesion-like structures found the body wall muscle of the nematode Caenorhabditis elegans. To study its in vivo function in this genetic model system we isolated a strain carrying a deletion of the single C. elegans α-actinin gene. We assessed the cytological organization of other C. elegans focal adhesion proteins, and the ultrastructure of the mutant. The mutant does not have normal dense bodies by EM but these dense body-like structures still contain the focal adhesion proteins integrin, talin and vinculin by immunofluorescence microscopy. Actin is found in normal-appearing I-bands, but with abnormal accumulations near muscle cell membranes. Although swimming in water appeared grossly normal, use of automated methods for tracking locomotion of individual worms revealed a defect in bending. We propose that the reduced motility of the α-actinin null is due to abnormal dense bodies that are less able to transmit the forces generated by actin/myosin interactions.
Keywords: α-actinin, focal adhesion, muscle, Caenorhabditis elegans
INTRODUCTION
The actin binding protein α-actinin is found at both cell-cell and cell-ECM adherens junctions where, in part, it functions to link the actin cytoskeleton to the junctional complex 1; 2. It is known to bind to multiple proteins at adherens junctions, and so multiple models have been proposed to describe its organization at such junctions. At focal adhesions α- actinin may link actin to integrin via a direct interaction 3; 4 or via an indirect interaction mediated by the proteins vinculin and talin 5; 6; 7; 8; 9. As an actin binding protein, α-actinin is the best characterized of all the proteins at adherens junctions, but other adherens junction proteins like vinculin and talin can also bind to actin 10; 11; 12; 13; 14; 15.
Understanding the complex interplay between these various actin-binding proteins in the establishment and maintenance of cytoskeletal-adherens junction linkage is a significant challenge. Though vertebrates have multiple isoforms of α-actinin, all isoforms have a subunit molecular weight of about 100 kDa 2. For all isoforms the actin-binding domain, which is similar to that found in spectrin and dystrophin, falls within approximately the first 250 amino acids. The actin-binding domain is followed by four 120 residue spectrin-like repeats that allow for assembly into antiparallel homodimers (Figure 1b). The spectrin repeats are critical for the function of the protein 16; 17; 18. Both the muscle and non-muscle isoforms of the protein contain two EF-hand calcium-binding motifs at the C-terminus. The EF hand of the muscle isoform, however, is slightly divergent and calcium does not affect its actin binding properties.
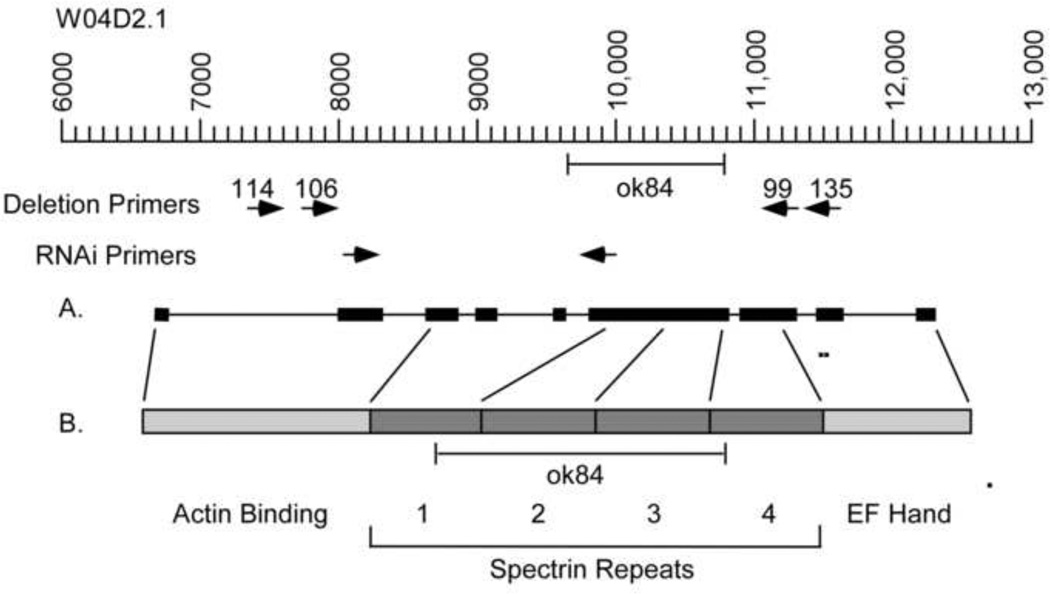
Figure 1. The ok84 allele is an 1136 base pair internal deletion.
The sequence coordinates are taken from cosmid W04D2 that encodes the α-actinin gene W04D2.1 (Barstead et al., 1991; Waterston and Sulston, 1995). (A). The coding sequence starts at position 6654 and ends at position 12182. The oligonucleotide primers used for PCR reactions were located as shown. The outer primer set generated a 3478 base pair PCR fragment that extended between base pairs 7681 and 11159. The inner primer set generated a 3250 base pair PCR fragment that extended between base pairs 7980 and 11130. The 1136 base pair ok84 deletion extends from position 9656 to position 10792. (B). The domains of the 920 amino acid α-actinin polypeptide are shown. The deletion not only eliminates a portion of the gene but also likely disrupts splicing so that the protein would be truncated at amino acid 250. If the mutant protein were stable in vivo, it would contain the actin binding site, and a portion of the first spectrin repeat.
Several genetic and pseudo-genetic studies point to a significant function for α-actinin in the assembly or maintenance of various actin structures in vivo. Either the over or under expression of α-actinin in vertebrate cells affects cell motility and can lead to tumorigenic cell lines 19. Pavalko and Burridge showed that the injection of proteolytic fragments of α- actinin into living cells leads to the disassembly of stress fibers 20. Drosophila mutants that lack α-actinin are not viable, though the protein is not necessary for the initial assembly of muscle Z-disks but rather for the maintenance of their integrity under the stress of contraction 21; 22; 23. Finally, two groups have shown that the absence of α-actinin-3 is associated with certain forms of congenital muscular dystrophy 24; 25. Although mutation of α-actinin has not yet been associated with nemaline myopathy, this disorder is characterized by the presence of rod-shaped structures called nemaline bodies which consist of skeletal α-actin and Z-disk proteins, including α-actinin26. Recently, several missense mutations in α-actinin-2 have been found to cause hypertrophic cardiomyopathy27. Finally, point mutations in α-actinin-4 in humans result in one type of glomerulosclerosis28; 29.
In the nematode Caenorhabditis elegans α-actinin is found at adherens junctions in the gut and at focal adhesion-like structures, called dense bodies, in the body wall muscle 30; 31 (Figure 2). Other proteins found with α-actinin at dense bodies include integrin 32, talin 33, and vinculin 34 (Figure 2d). The body wall muscle dense bodies, therefore, are good general models for the attachment of actin to membranes in vertebrate cells. The proper assembly of these adherens junctions is critical to the viability of the nematode, as mutations in vinculin or integrin that interfere with assembly lead to complete paralysis of the muscle, incomplete elongation, and a characteristic embryonic arrest, the so called PAT phenotype 35; 36; 37. Based on this, and on the information from the study of vertebrate adherens junctions, we expected that α-actinin would be the major actin binding protein in the dense body and, therefore, that it would be as critical to the function of the dense body as is vinculin and integrin. To examine the function of α-actinin in C. elegans we devised a genetic strategy to eliminate it from the dense body and then to determine the consequences for dense body assembly, actin filament organization, and the behavior of the mutant animal. We were surprised to discover that mutations eliminating C. elegans α-actinin had remarkably mild effects. Such mutants not only did not show a PAT phenotype, like that caused by mutations in vinculin and integrin, but rather were viable as homozygotes, showed nearly normal looking muscle as assayed by polarized light microscopy, and nearly normal dense body arrays as assayed by immunofluorescence microscopy using antibodies to integrin, talin and vinculin. The mutants, however, showed abnormal accumulations of actin at the ends of the muscle cells and, as assayed by electron microscopy, had dense body analogues that were shorter and broader at the base. Further, although casual observation of worm locomotion or the use of a standard liquid motility assay did not show abnormality, quantitative analysis of the locomotion of individual worms revealed a defect in body bending. We conclude that α-actinin has a role in the final assembly of dense bodies, and that a fully assembled dense body is required for efficient transmission of force.
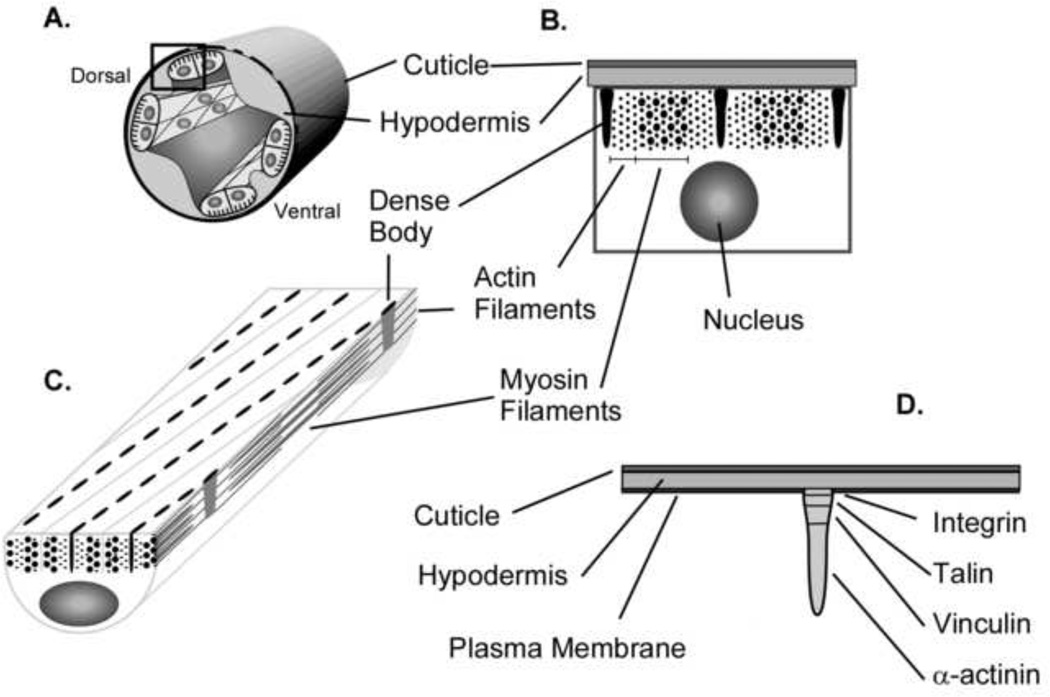
Figure 2. Dense bodies are focal adhesion-like adherens junctions.
(A). The diagram shows a cross section through the C. elegans body wall including the cuticle on the surface, an epidermal cell layer adjacent to the cuticle known as the hypodermis, and four quadrants of muscle cells around the circumference. The muscle quadrants run the length of the animal. In adult C. elegans each quadrant has twenty-three to twenty-four mononucleate, striated muscle cells. (B). In a cross section the dense bodies are seen adjacent to thin filaments that emanate from other close-by dense bodies. A zone of myosin filaments is seen to overlap with a zone of actin filaments. (C). A section taken from diagram A showing 1) the arrangement of dense bodies in the body wall muscle, 2) the projection of the wedge shaped dense bodies from the membrane into the cell, and 3) the relationship of the actin and myosin filament system to the dense bodies. (D). Four of the known protein components of dense bodies are shown. These proteins include integrin, talin, vinculin and α-actinin. Their relative positions within the dense body are not known with certainty, but can be inferred from other work (Francis and Waterston, 1985; Barstead and Waterston, 1989; Gettner et al., 1995b; Moulder et al., 1996a).
MATERIALS AND METHODS
C. elegans Strains
General methods for the growth and maintenance of nematode strains were as described in Brenner 38. The N2 strain was used for the analysis of wild type α-actinin organization. The C. elegans α-actinin mutant atn-1(ok84) was isolated as described below. For transformation rescue experiments we used a strain carrying a temperature sensitive allele of the pha-1 gene pha-1(e2123). For nematode behavior assays, we used N2 as wildtype, atn-1 (ok84), and the atn-1 (ok84); sfIs2 rescued line.
Isolation of an α-actinin Mutant
Synchronous L4 larvae were mixed with 15 ml of M9 buffer containing 4, 5', 8- trimethylpsoralen (Sigma, St. Louis) at 30 mg/ml. After 15 minutes at room temperature, the mixture was transferred to a sterile 15-ml petri plate. The suspension was irradiated with 360 nm UV light for 90 seconds at 340 mW/cm with gentle shaking (Model UVL-21 Blak-Ray Lamp, longwave UV-365 nm, Fisher Scientific). The mutagenized animals were cultured for twenty-four hours. They were then treated with basic hypochlorite to collect their eggs. After the eggs hatched the resulting animals were arrayed in 1152 cultures of 500 animals. After five days we harvested a portion of each culture, prepared and strategically pooled the DNA according to our standard procedures (Barstead and Waterston, 1991), and performed nested PCR with primers flanking the α-actinin gene. The primer sequences were as follows:
Outer Primers:
#114 - AGATGCCATTGACACCTTCC
#135 - TATTCTGTCTGTACCGGACG
Inner Primers:
#106 - ATTCACAGCCTGGTGCAACT
#99 - ATGGAATCGCTTCGTGTCGG
Putative deletion fragments were identified by the presence of a PCR fragment that was shorter than the wild type fragment. We identified one such DNA pool. The reserved sibling population that corresponded to the PCR address was harvested, divided into smaller sub populations, allowed to reproduce, and the process was repeated until we captured a single animal carrying the deletion. After recovering the α-actinin mutant, the strain was backcrossed four times to wild type animals to reduce the possibility that our analysis would be affected by unrelated mutations that were unintentional by products of the mutagenesis. The allele name for this isolate is ok84.
Transformation Rescue
We constructed a strain with the following genotype: atn-1(ok84); pha-1(e2123). We transformed this strain with a cloned wild type copy of the pha-1 gene, as the selectable marker, and the C. elegans cosmid W04D2.1 that carried a wild type copy of the C. elegans α-actinin. Rescue was determined by examining the organization of the muscle. To construct atn-1 (ok84); sfIs2, first we prepared transgenic worms of atn-1 (ok84) harboring an extrachromosomal array containing a 9 kb genomic PCR fragment representing the wild type atn-1 gene (generated by primers GAGTGTTCTTCATCACCAATTGAATATTTTTCGAGTGTTG and CTCAGGATATTATCCATTTCGCAGCTAAAAAAACTCAAAC) and the transformation marker sur-5::GFP 39. Second, this extrachromosomal array was integrated into the genome by UV irradiation 40 (P. Barrett, personal communication)).
RNAi
We did RNAi according to protocols described by Ahringer and colleagues 41. The fragment designated for RNAi was obtained by polymerase chain reaction (PCR) from an α-actinin cDNA. The following primer pair used for PCR amplification: 5'- GTG TCA AGT TGG TCT CCA TTG G -3' and 5'- CGG GGT ACC TGA TAT CCT TGA TAA GAT -3'. Their location relative to the gene are shown in Figure 1. These primers were designed such that the resulting PCR fragment would contain Asp718 and SacI restriction sites at its ends. The RNAi fragment was cloned into the corresponding restriction sites in the vector pPD129.36 such that the insert was flanked by T7 polymerase initiation sites. The resulting plasmid, pRB100, was transformed into the E. coli strain HT115 [F-, mcrA, mcrB, IN(rrnD-rrnE)1, rnc14::Tn10(DE3 lysogen: lacUV5 promoter -T7 polymerase) (IPTG-inducible T7 polymerase) (RNAse III minus)]. In the presence of IPTG, we expected that this transformed E. coli strain would produce double stranded RNA matching the corresponding α-actinin cDNA sequence from pRB100. To introduce this double stranded RNA, we fed this E. coli strain either to wild type C. elegans or to a C. elegans strain carrying the ok84 deletion in the α-actinin gene. We first placed 10 first larval stage worms on culture plates seeded with E. coli HT115 as a control, or with HT115 carrying pRB100. Carbenicillin (25mg/ml) was used to select for the presence of pRB100. Both the control and experimental cultures contained 1 mM IPTG, which induces expression of double stranded RNA from pRB50. After five days, we harvested the progeny from the original worm cultures. The harvested worms were fixed and stained with AlexaFluor-546-phalloidin (Molecular Probes, Eugene, Oregon) as described below.
Immunocytochemistry
Antibodies to α-actinin (monoclonals MH35 and MH40.3), integrin (monoclonal MH25), tight junctions (monoclonal MH27), and vinculin (monoclonal MH24) were generated in the laboratory of Robert Waterston (Francis and Waterston, 1985; Francis and Waterston, 1991). We used commercial antibodies to actin (monoclonal C4) (ICN, Costa Mesa, California). For staining with antibodies to integrin, nematodes were fixed with a variation of the freeze-crack method of Albertson 42; 43. Slides with freeze-cracked nematodes were incubated in 3% formaldehyde (EM grade, Ted Pella, Inc, Redding, CA) in 0.1M-phosphate buffer for 20 minutes at room temperature. For staining with antibodies to actin, α-actinin, talin, or vinculin, nematodes were fixed with either the freeze-crack method or with a variation of the whole nematode fixation method of Nonet 44. Slides with freeze-cracked nematodes were fixed by incubation for 1 hour in a modified Bouin's fixative (50% methanol, 35% saturated picric acid, 12% formalin, 2% acetic acid, 1% ß-mercaptoethanol). For whole nematode fixation, rinsed nematodes were placed in a 1.5 ml tube containing modified Bouin’s fixative and shaken at room temperature for 30 minutes. The tube was then placed in liquid nitrogen for one minute (to crack open the cuticle), defrosted under warm water, then shaken for an additional 30 minutes at room temperature. Nematodes were rinsed three times with a borate buffer with ß-mercaptoethanol (20 mM H3BO3, 10 mM NaOH, 0.5% Triton X-100, 2% ß-mercaptoethanol), and then shaken for 1 hour at room temperature. The rinses and incubation were repeated two more times, followed by thorough rinsing in borate buffer without b-mercaptoethanol, and rinses in antibody buffer (0.5% Triton X-100, 1 mM EDTA, 0.1% BSA in PBS with 0.05% sodium azide). After either type of fixation, slides or nematodes were rinsed in PBS, followed by incubation in 10% donkey serum in antibody buffer for one hour. Primary antibody incubations (1:100–1:2, 000 in antibody buffer) were done overnight. After thorough rinsing with antibody buffer, slides were incubated in secondary antibody for four hours. Unlabeled and Cy3-labeled secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA); Oregon Green 488 was coupled to secondary antibodies using the Oregon Green labeling kit from Molecular Probes (Eugene, OR). After rinsing, slides were mounted in anti-bleaching medium containing DAPI to label nuclei 45.
Staining with rhodamine-phalloidin or AlexaFluor-546-phalloidin was done on animals fixed in the following way: Worms were washed from culture plates with M9 buffer. They were pelleted in a centrifuge and the M9 was removed. We used formaldehyde made from paraformadehyde to fix the worms. The worms were suspended in a solution of 3% formaldehyde in 0.2 M NaPO4, pH 7.0, at room temperature. We removed the fix after thirty minutes and added a solution of 3% formaldehyde in 0.2 M NaPO4, pH 8.0. The worms were then incubated an additional thirty minutes at room temperature. To remove the fix we washed the worms five times in ten volumes of PBS, pH 7.0. The worms were then treated with EM grade acetone for two minutes at –20°. To remove the acetone we washed the worms five times with PBS, pH 7.0. The worms were incubated with 50-µM rhodamine-phalloidin for one hour. We removed the unbound phalloidin by washing five times with PBS, pH 7.0. Images were collected with either a Leica TCS or a Zeiss LSM510 confocal microscope
Electron microscopy
We harvested L2 to L4 larval staged worms from standard cultures by washing the culture plates with M9 buffer. The worms were allowed to settle in a 15-ml centrifuge tube and withdrew all but 0.5 ml of the M9 buffer. Approximately 200 µl of the worm suspension were added to Millipore filters (Type HA, 0.45 µm) cut to 7 mm × 10 mm dimensions. A house vacuum was used to gently pull liquid through the filter. Being careful to insure that the worms did not dry, we placed the glass filter on wet Whatman filter paper (~9 mm × 11 mm). The worms were placed on a Styrofoam disk and frozen ultra-rapidly by slamming the disk onto a copper block cooled with liquid nitrogen (LN2) (Reichert MM80E slam freezer). We removed the frozen filters, with attached worms, from the Styrofoam disk under LN2 and placed them into a nylon mesh basket 46; 47 in −80º C anhydrous acetone containing 1% tannic acid and 1% anhydrous glutaraldehyde. Freeze substitution was done in an ultralow freezer at −80º C for 72 hr with occasional agitation. The freeze substituted worms were rinsed three times in 50 ml of anhydrous acetone at −80º C, transferred to acetone with 1% OsO4 at −80º C and then warmed slowly to −20º C over 8 h and then to 4º C over 16 h. After three rinses with acetone, we pipetted the worms from the mesh basket to Eppendorf microfuge tubes. The fixed worms were infiltrated with Spurr's resin 48, which was then polymerized at 65º C for 18 hr. We cut silver sections with a diamond knife using a Reichert Ultracut E Ultramicrotome. The samples were stained with the double lead staining method of Daddow 49 using a modified Sato's lead stain. We viewed the samples using a JEOL 1200EX scanning transmission electron microscope at an accelerating voltage of 100 kV.
Nematode Behavior Assays
A standard motility assay, which counts the number of times a worm moves back and forth in a drop of M9 liquid was performed as described in Mercer et al. 50. For a more detailed analysis of worm locomotion, we performed the following procedure: All strains were grown using standard protocol 38 on NGM plates with OP50 (cultivation plates). Well-fed young adults were used for the assays. Animals were assayed on 2.1 % NGM plates without bacterial food (assay plates). Assay plates were stored at 4 °C and warmed to room temperature 2–3 hours before the assay. For the assay, young adult worms were picked individually from cultivation plate to an assay NGM plate and allowed to crawl for 3 minutes to remove residual bacteria and adjust to the new plate condition. After 3 minutes, the worms were touched on the head under a dissection microscope (20× magnification on a Zeiss Stemi SV 11 Dissecting Scope) with a platinum pick to induce reversals, and their behaviors were recorded by videomicroscopy. Taps were administered at least three minutes apart, and three or four taps were given in 10–15 minutes. Behavior was recorded at 8 fps using a CCD camera (Infinity 2-1 CCD, Lumenera Corporation). The movies were analyzed using a custom written LabView program. For each frame, the program thresholded the image to identify the worm, created a mid-body spline, and calculated a length of the worm from the spline 51. Amplitude in the body bend can also be calculated for each frame (Figure 10). We define the reversal amplitude to be the largest amplitude during a reversal after being tapped on the head and before an omega turn or changing directions52. To account for small length differences in worms, the ratio of amplitude to length was reported. The Mann Whitney Test was used to assess statistical significance.
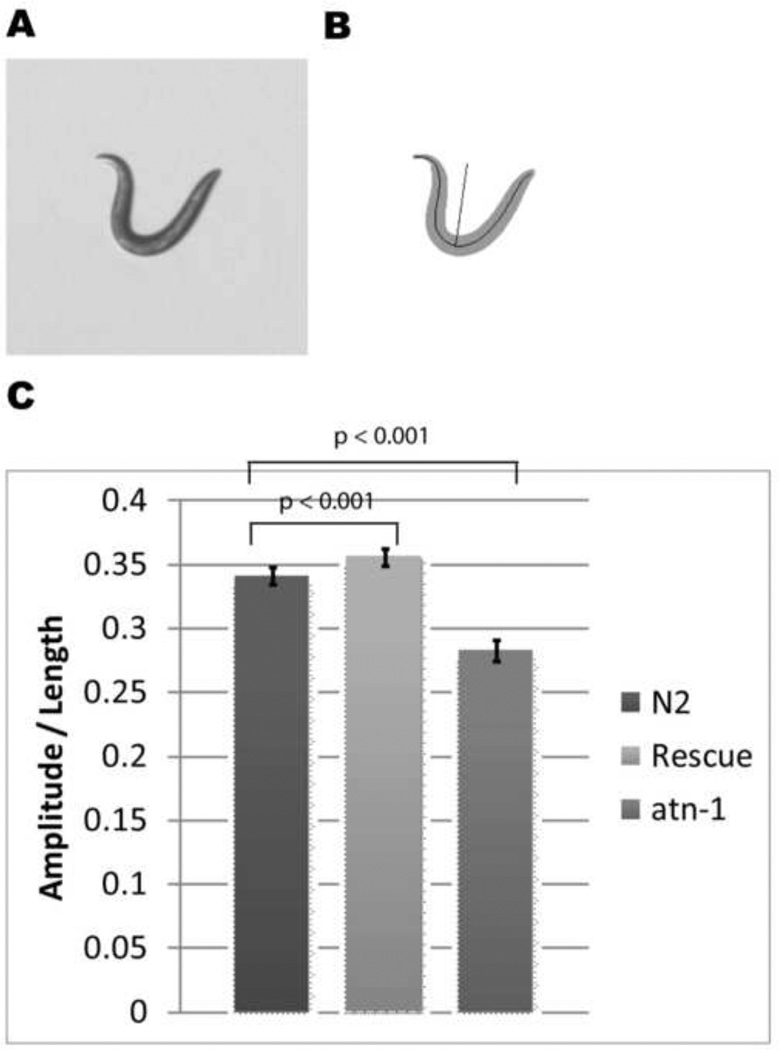
Figure 10. Behavior assay based on reversal amplitude reveals a defect in atn-1(ok84) mutant animals, which can be fully rescued.
(a) Bright field image of a worm during a reversal with a deep ventral bend. (b) Thresholded image of (a); a spline can be fit to the worm, and an amplitude can be measured. (c) Normalized amplitude to body length for wild type N2 (n=14), rescued strain (atn-1(ok84); sfIs2[atn-1; sur-5::GFP])(n=12), and atn-1(ok84) (n=19). Error bars represent SEM.
Tandem Mass Spectrometry
50–100 micrograms of total protein extract from the mutant strain atn-1(ok84) were subjected to SDS-PAGE. The separated proteins were stained with Coomassie blue. The portion of the gel containing proteins running 25–35 kDa was excised and the proteins in the gel slice were subjected to in-gel digestion. The resulting peptides were analyzed by reverse-phase liquid chromatography coupled with tandem mass spectrometry53 using an LTQ-Orbitrap mass spectrometer (Thermo Finnigan, San Jose, CA). We used a reverse database strategy to evaluate false discovery rate; the matched peptides were filtered according to matching scores to remove all false matches from the reverse database54. Finally, we only accepted proteins that were matched by at least two peptides to further improve the confidence of identification; i.e., where modified peptide forms are considered to be different peptides (e.g. ABC* and ABC are considered to be two peptides).
RESULTS
The mutant allele ok84 eliminates seventy percent of the C-terminus of α-actinin
The solitary C. elegans α-actinin gene is organized as shown in Figure 1a. We used PCR with primers flanking the gene to isolate a strain carrying a deletion within the gene (Figure 1a; see Materials and Methods). The allele was given the designation ok84. We purified the deleted PCR fragment and determined its DNA sequence. The sequence showed that the 1136 base pair deletion extended from base pair 9656 to 10792. The deletion creates a fusion between sequences in the fifth intron and the sixth exon. The remaining splice sites nearest the deletion endpoints cannot be spliced to create an in-frame protein fusion. If the mutant protein were stable in vivo, it would be truncated at amino acid 250, with only the actin binding domain remaining.
Though the putative truncated protein would likely be non-functional, we took two steps to confirm that it was not present at substantial levels in the atn-1 mutant. First, the truncated protein could not be detected with either of two monoclonal antibodies on Western blots (data not shown). Second, the predicted truncated protein of ~29 kDa from atn-1(ok84) was not detected when we performed mass spectrometry analysis of proteins in the range of 25–35 kDa, although we identified 547 proteins in that gel slice (data not shown).
To clean up the genetic background of the original mutant isolate we crossed the strain four times to wild type worms. We then did two tests to establish the validity of the strain for phenotypic analysis. First, we tested for the presence of α-actinin in situ in the mutant using two monoclonal antibodies (Figure 3). We determined that the antigens recognized by these two antibodies were completely absent from the mutant. Second, after we found that actin was disorganized in the α-actinin mutant (see below), we showed that a wild type transgenic copy of the α-actinin gene could restore normal actin organization (Figure 4).
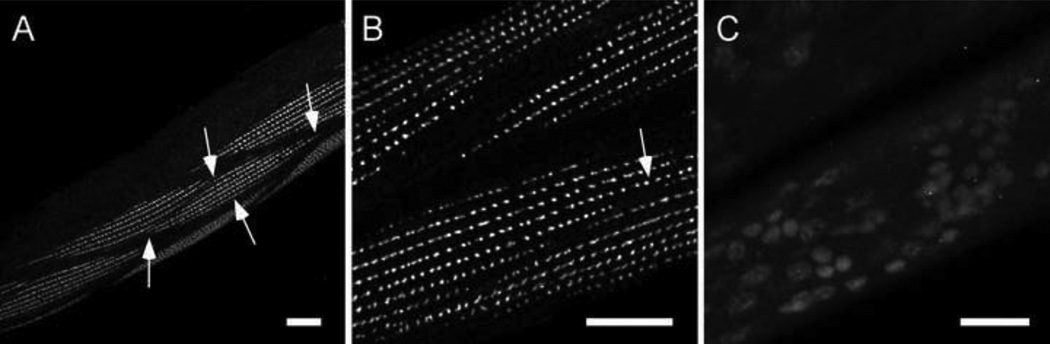
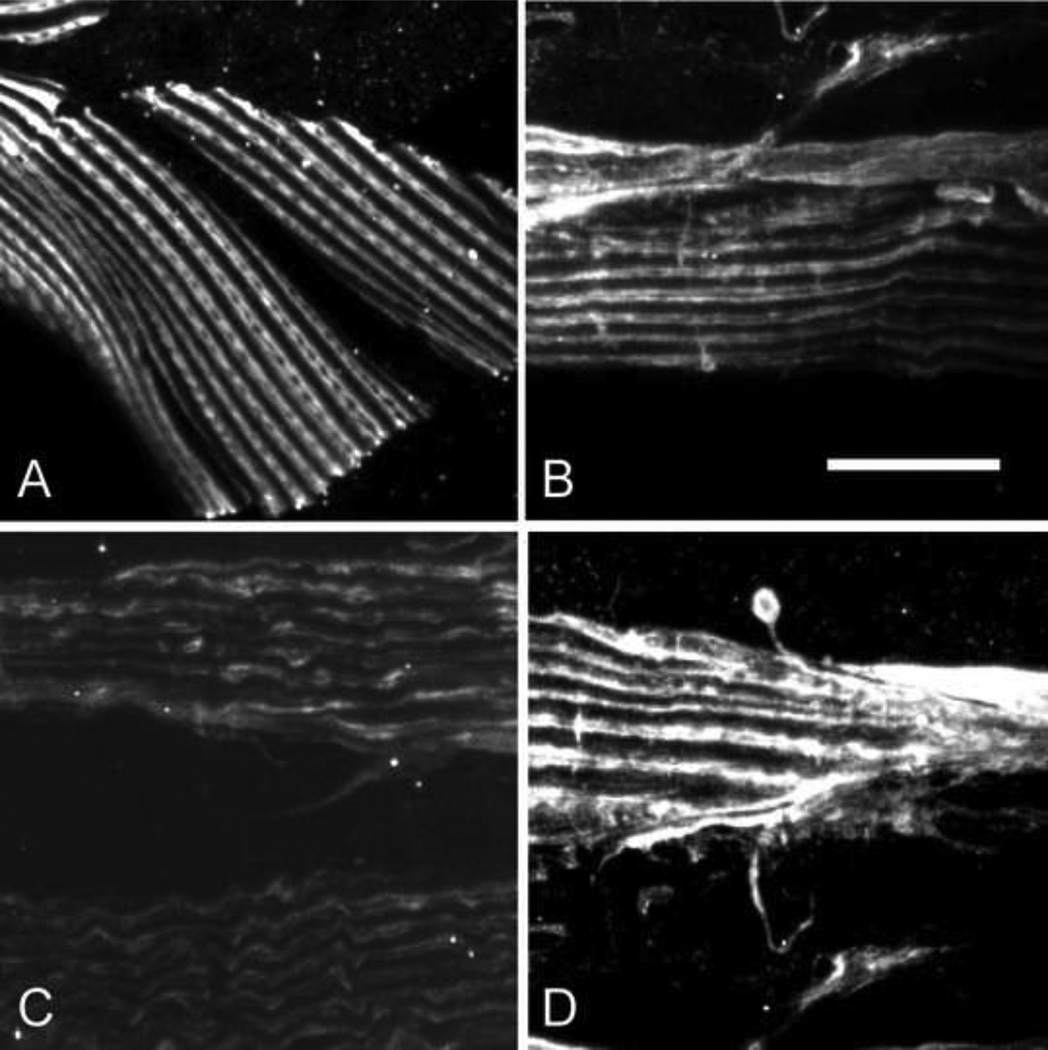
Figure 3. α-actinin is absent from the mutant in situ.
We used two monoclonal antibodies to C. elegans α-actinin (Francis and Waterston, 1985) to look for the protein in the mutant. The results from one of these antibodies, MH35, are shown. The antibody MH40 gives the same results. Panels A and B show a normal dense body array in wild type muscle. The arrows in panel A demark the boundaries of a single muscle cell. The arrow in panel B points to a dense body. Panel C shows that the epitope recognized by MH35 is missing in the mutant. Bleed through of DAPI stained nuclei are visible, however. Bar = 10 microns.
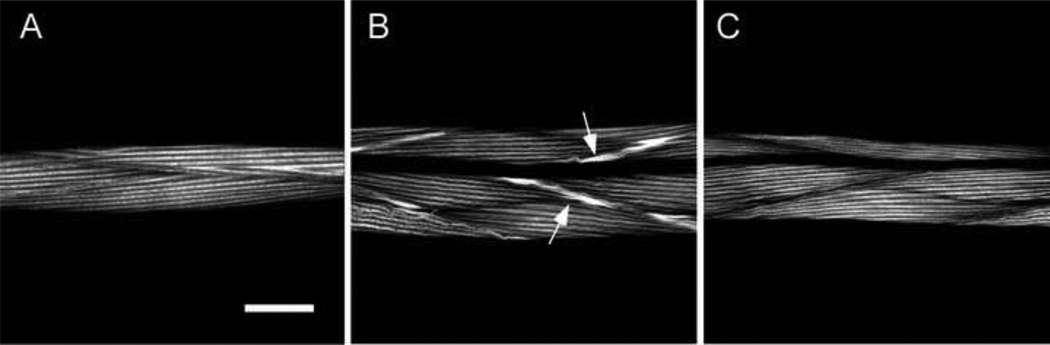
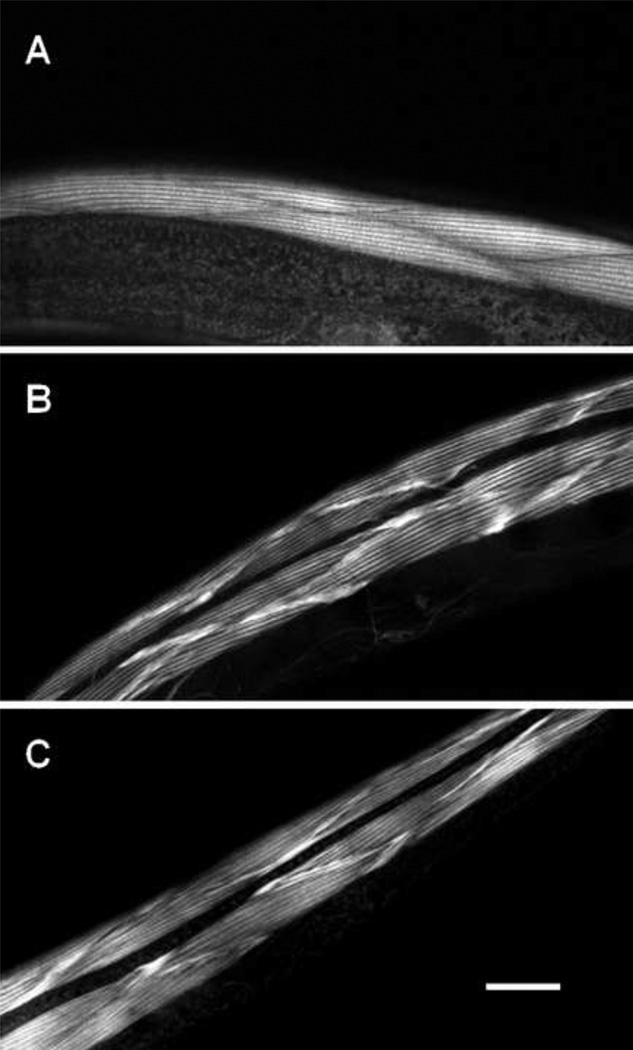
Figure 4. A transgenic copy of the wild type gene rescues the mutant phenotype.
We used rhodamine-phalloidin to examine the organization of actin in wild type animals (A), in animals with the ok84 deletion (B), and in animals with the ok84 deletion that carry a transgenic copy of the wild type α-actinin gene (C). In both A and C one sees normal actin filament striations. In B one sees relatively well-ordered striations, with abnormal accumulations of actin at the cell boundaries (arrows). Bar = 20 microns.
Actin is variably disorganized in the body wall muscle of the mutant
We wanted to determine whether the elimination of α-actinin from the body wall muscle would affect the organization of muscle actin. First, as assayed using rhodamine phalloidin, which binds to filamentous actin, actin in the body wall muscle was for the most part well organized. The actin striations in the mutant, however, were less uniform. The mutant muscle also showed abnormal bundles of actin at the cell boundaries (Figure 4). Second, as assayed using a monoclonal antibody to actin we found a range of disorganization in the mutant (Figure 5). In most cells, the actin was organized normally in well-ordered striations. In some cells, or regions within cells, however, the actin accumulated into large bundles. Finally, in some cases actin was missing from its normal location.
Figure 5. Actin is variably disorganized in the mutant.
We used a monoclonal antibody to actin to examine the range of organization in the α-actinin mutant. One sees normal actin striations in wild type muscle (A). In the mutant one can see some cells with nearly normal actin organization (B), with weakly staining, poorly defined striations (C), or with well staining, but ragged striations with abnormal actin bundles (D). Bar = 10 microns.
Other adherens junction proteins are organized relatively normally
In models of adherens junctions α-actinin is placed as the most distal component relative to the plasma membrane. An examination of the organization of other known adherens junction proteins in cells lacking α-actinin, therefore, bears on the mechanism by which the structure is assembled. We, therefore, examined the organization of integrin and vinculin in α-actinin mutant animals (Figure 6). We found that both of these proteins were in the correct cellular domain, that is, at the basal side of the cell along with all of the other components of the contractile apparatus. Further, they were all relatively well ordered at sites that likely corresponded to partially formed adherens junctions that lacked α-actinin. Similar results were seen for talin (not shown). These membrane proximal structures, however, appeared broader than normal dense bodies, and the edges were poorly delineated.
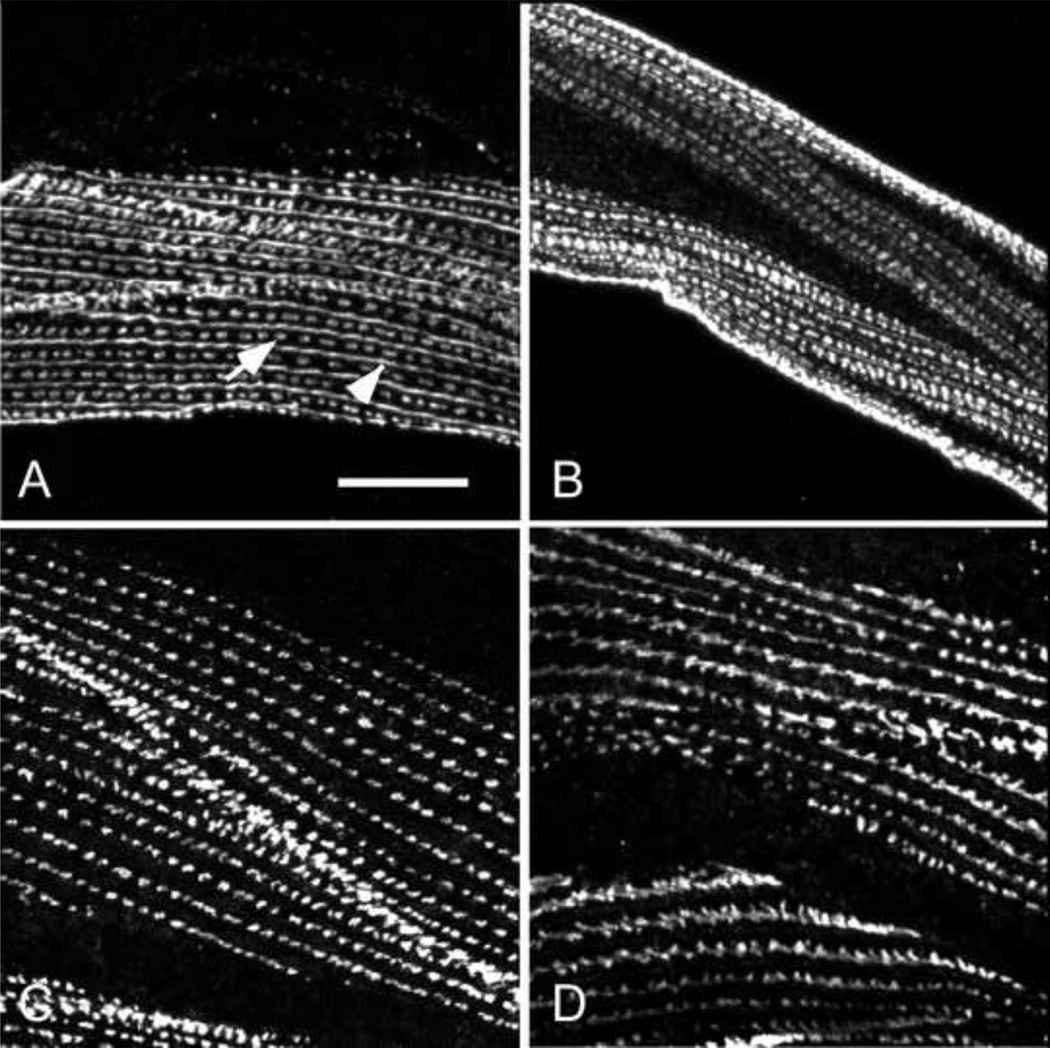
Figure 6. Integrin and vinculin are mildly disorganized in the mutant.
We used the monoclonal antibodies MH24 and MH25 that recognize C. elegans vinculin30; 34 and ßintegrin30; 37, respectively, to examine the organization of these proteins in wild type (A and C) and mutant (B and D) body wall muscle. As is normally seen in wild type C. elegans muscle, integrin is found at each of the dense body adherens junctions (arrow) and at the base of M-lines (arrow head) (A). Integrin is found at both of these sites in the mutant (B). In wild type muscle vinculin is found at dense bodies (C). Vinculin is more-or-less normally arrayed in the mutant at membrane-associated sites. Both integrin and vinculin, however, appear more broadly in structures that are less distinct than in wild type muscle. Bar = 10 microns.
The dense bodies are abnormal but the acto-myosin filament arrays are relatively well ordered
As we determined by light microscopy, the actin and other adherens junction proteins were surprisingly well ordered in the mutant, and so to detect subtle defects we examined the structure in more detail using electron microscopy (Figure 7). As seen in cross sections through the muscle filament array, in most cases the actin filaments are organized normally around the myosin filaments. In the mutant, however, where we should see finger-like dense bodies at the membrane that are about 150 nm wide at the base and that project up to 1 mm into the cell, we instead see dense structures with a spread of about 500 nm and a height of about 20 nm. Just above this dense plaque, we see an accumulation of actin filaments in that encompass an area of about 500 nm wide and with a height between 0.15 to 0.25 nm. The entire contractile filament array, including thick and thin filaments, however, extended up to 1 mm away from the membrane, nearly the depth seen in wild type muscle.
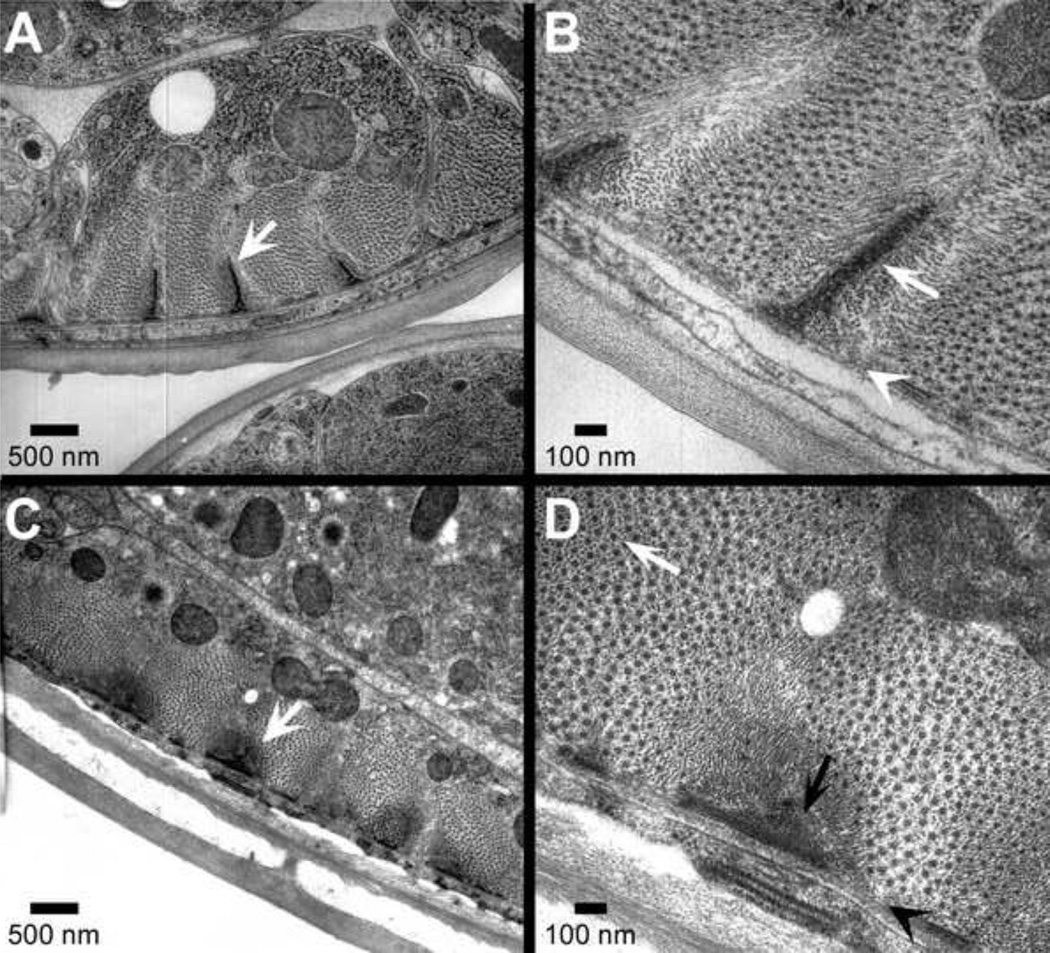
Figure 7. By EM, the mutant does not contain normal dense body structures.
Panels A and B show EM cross-sections through wild type muscle. The finger-like dense bodies (arrows) project approximately 1 micron into the cell from the plasma membrane and are approximately 150 nm at their base. Such structures are absent in the mutant, panels C and D. Still, actin filaments are organized around myosin filaments, however (panel D white arrow). Rather than terminating at normal dense bodies, the actin filaments seem to terminate at dense structures that are approximately 150 nm deep and 500 nm at their base (panel C white arrow). The filament array, however, extends up to 1 micron into the cell.
Epigenetic Inactivation via RNAi confirms the null phenotype
Double stranded RNA interference (RNAi) is a simple and rapid method to specifically inactivate gene function in the nematode Caenorhabditis elegans. As the genetic deletion (ok84) did not remove the entire gene, (and therefore strains carrying this mutation may not represent the null phenotype), we used RNAi to buttress our genetic analysis. As described in above, we placed a portion of the α-actinin coding sequence in a cloning vector that is designed for the production of double stranded RNA in an appropriate E. coli strain. Feeding is a standard way to introduce double stranded RNA for RNAi experiments in C. elegans, so the resulting E. coli strain was fed both to wild type and to worms carrying the ok84 deletion. As assayed using antibodies, animals where the α-actinin gene was inactivated via RNAi did not produce detectable α-actinin protein. The phenotype of animals where the α-actinin gene was inactivated via RNAi was identical to that seen for the genetic deletion mutant (Figure 8). Further, ok84 animals showed no further muscle degradation when tested with RNAi.
Figure 8. The RNAi phenotype is same as that for the genetic mutant.
All panels show C. elegans body wall muscle stained with AlexaFluor-546-phalloidin to visualize filamentous actin. (A). Actin organization in control animals that were fed a control E. coli strain that lacked the plasmid pRB100. (B). Wild type animals fed the RNAi bacterial strain carrying pRB100 with a cloned copy of the C. elegans α-actinin gene. The phenotype is identical to that seen for animals carrying a mutation in the α-actinin gene atn-1(ok84) (Figure 4b). (C). An α-actinin genetic mutant fed the same bacteria as those animals shown in panel B. The phenotype of such animals is no worse than either the RNAi or genetic phenotypes alone. Bar =20 µm.
Loss of Function of α-actinin Results in Nematodes Less Able to Bend
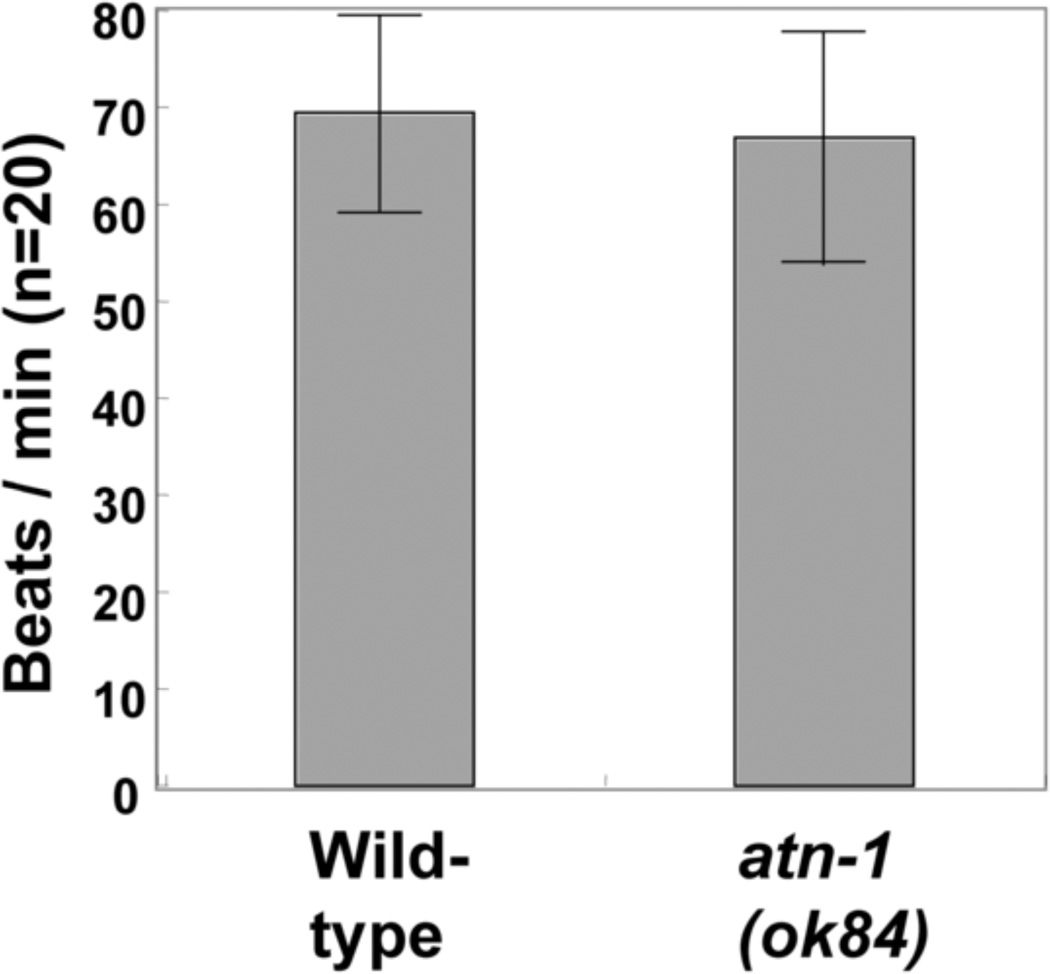
Because the α-actinin mutant displays shorter and broader dense bodies and variably disorganized actin, we wanted to determine what effect these abnormalities would have on muscle function. As the activity of body wall muscle is required for the worm to move, one way to assess the activity of this muscle is to assay the locomotion of the whole worm. By casual observation, ok84 and wild type animals seem to move with the same speed and general sinusoidal motion. A typical liquid motility assay, counting the number of times an animal moves back and forth in buffer, also showed no significant difference between wild type and ok84 (Figure 9). Using videomicroscopy and image processing software which track, records and analyzes the movement of individual worms, we compared the backward movements of wild type and ok84 animals. The amplitude of body bending, normalized for body length, was calculated. As shown in Figure 9, the amplitude of reversals was significantly smaller for ok84 than that of wild type. To be sure that this difference was not due to closely linked mutations that had not been eliminated by outcrossing, we performed the same assay on atn-1(ok84) animals that were rescued for the muscle structural defect by carrying an integrated array of the wild type atn-1 gene (Supplemental Figure 1). As shown in Figure 10, these rescued animals show a restoration of body bending in reversals to wild type levels. Therefore, the loss of α-actinin in body wall muscles results in nematodes that are less able to bend, perhaps because they transmit the force of myosin/actin interaction less efficiently.
Figure 9. A standard swimming assay reveals no difference in motility between wild type and atn-1(ok84) mutant adult worms.
The number of times an adult worm moved back and forth in liquid during 1 minute was counted as described in Mercer et al. 2003. The means and standard deviations are shown. For each genotype, n=20.
DISCUSSION
To test the function of α-actinin in the body wall muscle of the nematode Caenorhabditis elegans we isolated a strain carrying a deletion within the single known α-actinin gene. The deletion extended from within the first of the spectrin repeats through to the fourth and last repeat. Further, the coding sequence distal to the deletion likely cannot be spliced in the normal reading frame. Such a deletion likely prevents the formation of normal anti-parallel α-actinin homodimers 16; 17; 18. Moreover, we did not detect any α-actinin in situ in the mutant using two monoclonal antibodies that recognize the protein. Further, the predicted truncated protein of ~29 kDa from ok84 was not detected when we performed mass spectrometry analysis of proteins in the range of 25–35 kDa, although we identified 547 proteins in that gel slice. The mutant, therefore, likely is a molecular null. The RNAi results add to the evidence that reported phenotype is a result of a complete loss of function in the α-actinin gene. The phenotype of wild type animals where α-actinin was eliminated by RNAi was the same as that seen for the genetic mutant. Further, if the genetic mutant were not null, we would expect that the application of double stranded α-actinin RNA would lead to further reductions in the amount of functional α-actinin and a worse phenotype. That the phenotype was not worse supports our contention that the genetic mutant represents the true null phenotype. Finally, the similarity between the phenotypes of the genetic mutant and that seen with RNAi strongly argues against the possibility that the genetic mutant contains an opportunistic suppressor mutation linked to the atn-1 gene.
Given others and our past results showing that several other dense body proteins (vinculin, integrin, PAT-4 (ILK), PAT-6 (actopaxin) and UNC-97) are essential genes 55 we were surprised to find that α-actinin was not an essential gene. Animals homozygous for the deletion were viable, with mild effects on actin organization. Some cells showed abnormal accumulations of actin at both their anterior and posterior boundaries. We do not know whether such accumulations result from the misassembly of actin, or as a consequence of a structural failure in the contractile apparatus that leads to actin disorganization. Electron micrographs of the mutant muscle showed areas where actin and myosin filaments were organized normally, with six actin filaments surrounding a single myosin filament. Other areas, however, were clearly abnormal, with large accumulations of actin. The dense bodies were shorter and broader at the base, suggesting that the cells accommodate to the absence of α-actinin by changing the organization of other dense body proteins, perhaps by increasing the level of other actin binding proteins with an alternative architecture that keeps them closer to the plasma membrane. Nevertheless, the overall filament number and depth of the filament array in the mutant is close to that found in wild type muscle. Perhaps the main function of α-actinin is not primarily structural, but rather it serves as a mechanosensitive scaffold56. Given the architecture of the mutant structures and their proximity to the membrane, the actin filaments likely arc up from the membrane to achieve their necessary position within the filament array. Based on our results, it is seems clear that other proteins within the dense body can partially compensate when α-actinin is absent. Both talin and vinculin are known to bind to actin, and are present at appropriate sites in the mutant, and so either of these proteins might substitute for the actin binding function of α-actinin. Perhaps the over expression of either of these candidates might lead to an improvement in the phenotype of the α-actinin mutant.
Nevertheless, α-actinin mutant muscle does not function normally. Quantitative analysis of backward locomotion reveals that these animals have reduced ability to bend. This is significant since the transmission of force by body wall muscle myofibrils via dense bodies and M-lines through the muscle cell membrane, basement membrane, hypodermis and cuticle bends the worm and this allows the worm to move. Thus, our interpretation is that in the α-actinin mutant with dense bodies of abnormal structure, the force is less efficiently transmitted. Interestingly, we were able to restore the normal bending ability in ok84 only when every body wall muscle cell expressed wild type atn-1 and had normal actin organization with an integrated transgene; use of an extrachromosomal array, in which some cells lost the array, as detected by the typical abnormal actin organization of ok84, did not show rescue of the bending phenotype (data not shown). Moreover, according to the promoter analysis results available on WormBase (http://wormbase.org/), atn-1 is expressed in body wall muscle, the pharynx (probably pharyngeal muscle), and a number of unidentified cells in the tail. Therefore, the behavioral phenotype that we have found is most likely due to its body wall muscle expression.
The fact that we had to use a sophisticated assay of locomotion to detect a defect in ok84, is compatible with the idea that this muscle is partially compensated for the loss of α-actinin. There is a growing list of components of nematode muscle focal adhesions for which the intragenic deletions (consistent with loss of function) display either mild or no obvious defects in myofibril organization or motility (e.g. UIG-1 57, LIM-8 and LIM-9 58, SCPL-1 59, and ZYX-1 60. One explanation is that these proteins have non-crucial functions or are redundant with other proteins. An alternative explanation is that our ability to discern a phenotype depends on the assay used. We can hypothesize that under the usual lab growth conditions and motility assays, nematode muscle operates far below its maximal capacity. It will be interesting in the future to determine if more quantitative assays of motility, such as the one described here, will reveal phenotypes for these many other mutants.
Supplementary Material
Rhodamine-phalloidin was used to visualize actin thin filaments. In this strain, every body wall muscle cell expresses the array (as determined by nuclear localized SUR-5::GFP) and shows normal thin filament organization.
ACKNOWLEDGMENTS
This work was supported by a grant from the Muscular Dystrophy Association of America to RJB, and grant AR052133 from the National Institutes of Health to GMB. The mass spectrometric analyses were performed by the Emory University Proteomics Core Service Center. We appreciate the help of the Core’s director, Dr. Junmin Peng. HL is a Sloan Foundation fellow in neuroscience.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Burridge K, Kelly T, Connell L. Proteins involved in the attachment of actin to the plasma membrane. Philos Trans R Soc Lond B Biol Sci. 1982;299:291–299. doi: 10.1098/rstb.1982.0133. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard A, Ohanian V, Critchley D. The structure and function of alpha-actinin. J Muscle Res Cell Motil. 1989;10:280–289. doi: 10.1007/BF01758424. [DOI] [PubMed] [Google Scholar]
- 3.Otey CA, Pavalko FM, Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J Cell Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavalko FM, Otey CA, Simon KO, Burridge K. Alpha-actinin: a direct link between actin and integrins. Biochem Soc Trans. 1991;19:1065–1069. doi: 10.1042/bst0191065. [DOI] [PubMed] [Google Scholar]
- 5.Otto JJ. Detection of vinculin-binding proteins with an 125I-vinculin gel overlay technique. J Cell Biol. 1983;97:1283–1287. doi: 10.1083/jcb.97.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 7.Belkin AM, Koteliansky VE. Interaction of iodinated vinculin, metavinculin and alpha-actinin with cytoskeletal proteins. FEBS Lett. 1987;220:291–294. doi: 10.1016/0014-5793(87)80832-3. [DOI] [PubMed] [Google Scholar]
- 8.Wachsstock DH, Wilkins JA, Lin S. Specific interaction of vinculin with alpha-actinin. Biochem Biophys Res Commun. 1987;146:554–560. doi: 10.1016/0006-291x(87)90564-x. [DOI] [PubMed] [Google Scholar]
- 9.Lee SW, Wulfkuhle JD, Otto JJ. Vinculin binding site mapped on talin with an anti-idiotypic antibody. J Biol Chem. 1992;267:16355–16358. [PubMed] [Google Scholar]
- 10.Gilmore AP, Jackson P, Waites GT, Critchley DR. Further characterisation of the talin-binding site in the cytoskeletal protein vinculin. J Cell Sci. 1992;103(Pt 3):719–731. doi: 10.1242/jcs.103.3.719. [DOI] [PubMed] [Google Scholar]
- 11.Gilmore AP, Wood C, Ohanian V, Jackson P, Patel B, Rees DJ, Hynes RO, Critchley DR. The cytoskeletal protein talin contains at least two distinct vinculin binding domains. J Cell Biol. 1993;122:337–347. doi: 10.1083/jcb.122.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–12619. [PubMed] [Google Scholar]
- 13.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem Biophys Res Commun. 1995;210:159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- 15.Hemmings L, Rees DJ, Ohanian V, Bolton SJ, Gilmore AP, Patel B, Priddle H, Trevithick JE, Hynes RO, Critchley DR. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci. 1996;109(Pt 11):2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- 16.Flood G, Kahana E, Gilmore AP, Rowe AJ, Gratzer WB, Critchley DR. Association of structural repeats in the alpha-actinin rod domain. Alignment of inter-subunit interactions. J Mol Biol. 1995;252:227–234. doi: 10.1006/jmbi.1995.0490. [DOI] [PubMed] [Google Scholar]
- 17.Flood G, Rowe AJ, Critchley DR, Gratzer WB. Further analysis of the role of spectrin repeat motifs in alpha-actinin dimer formation. Eur Biophys J. 1997;25:431–435. doi: 10.1007/s002490050057. [DOI] [PubMed] [Google Scholar]
- 18.Flood GJ, Gratzer WB, Kahana E, Rowe AJ, Critchley DR. Association of structural repeats in alpha-actinin. Biochem Soc Trans. 1995;23:399S. doi: 10.1042/bst023399s. [DOI] [PubMed] [Google Scholar]
- 19.Gluck U, Ben-Ze'ev A. Modulation of alpha-actinin levels affects cell motility and confers tumorigenicity on 3T3 cells. J Cell Sci. 1994;107(Pt 7):1773–1782. doi: 10.1242/jcs.107.7.1773. [DOI] [PubMed] [Google Scholar]
- 20.Pavalko FM, Burridge K. Disruption of the actin cytoskeleton after microinjection of proteolytic fragments of alpha-actinin. J Cell Biol. 1991;114:481–491. doi: 10.1083/jcb.114.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fyrberg E, Kelly M, Ball E, Fyrberg C, Reedy MC. Molecular genetics of Drosophila alpha-actinin: mutant alleles disrupt Z disc integrity and muscle insertions. J Cell Biol. 1990;110:1999–2011. doi: 10.1083/jcb.110.6.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fyrberg C, Ketchum A, Ball E, Fyrberg E. Characterization of lethal Drosophila melanogaster alpha-actinin mutants. Biochem Genet. 1998;36:299–310. doi: 10.1023/a:1018789227987. [DOI] [PubMed] [Google Scholar]
- 23.Roulier EM, Fyrberg C, Fyrberg E. Perturbations of Drosophila alpha-actinin cause muscle paralysis, weakness, and atrophy but do not confer obvious nonmuscle phenotypes. J Cell Biol. 1992;116:911–922. doi: 10.1083/jcb.116.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.North KN, Beggs AH. Deficiency of a skeletal muscle isoform of alpha-actinin (alpha-actinin-3) in merosin-positive congenital muscular dystrophy. Neuromuscul Disord. 1996;6:229–235. doi: 10.1016/0960-8966(96)00361-6. [DOI] [PubMed] [Google Scholar]
- 25.Vainzof M, Costa CS, Marie SK, Moreira ES, Reed U, Passos-Bueno MR, Beggs AH, Zatz M. Deficiency of alpha-actinin-3 (ACTN3) occurs in different forms of muscular dystrophy. Neuropediatrics. 1997;28:223–228. doi: 10.1055/s-2007-973704. [DOI] [PubMed] [Google Scholar]
- 26.Ochala J. Thin filament proteins mutations associated with skeletal myopathies: defective regulation of muscle contraction. J Mol Med. 2008;86:1197–1204. doi: 10.1007/s00109-008-0380-9. [DOI] [PubMed] [Google Scholar]
- 27.Chiu C, Bagnall RD, Ingles J, Yeates L, Kennerson M, Donald JA, Jormakka M, Lind JM, Semsarian C. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 55:1127–1135. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, Pollak MR. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 29.Weins A, Kenlan P, Herbert S, Le TC, Villegas I, Kaplan BS, Appel GB, Pollak MR. Mutational and Biological Analysis of alpha-actinin-4 in focal segmental glomerulosclerosis. J Am Soc Nephrol. 2005;16:3694–3701. doi: 10.1681/ASN.2005070706. [DOI] [PubMed] [Google Scholar]
- 30.Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barstead RJ, Kleiman L, Waterston RH. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil Cytoskeleton. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- 32.Hess D, Isenberg G. A new fluorescence-based, hydrophobic photolabeling technique for analyzing membrane-associated proteins. FEBS Lett. 1999;445:279–282. doi: 10.1016/s0014-5793(99)00155-6. [DOI] [PubMed] [Google Scholar]
- 33.Moulder GL, Huang MM, Waterston RH, Barstead RJ. Talin requires beta-integrin, but not vinculin, for its assembly into focal adhesion-like structures in the nematode Caenorhabditis elegans. Mol Biol Cell. 1996;7:1181–1193. doi: 10.1091/mbc.7.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- 35.Barstead RJ, Waterston RH. Vinculin is essential for muscle function in the nematode. J Cell Biol. 1991;114:715–724. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gettner SN, Kenyon C, Reichardt LF. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitani S. Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Development Growth & Differentiation. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- 41.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 42.Chuang PT, Albertson DG, Meyer BJ. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 43.Duerr JS, Frisby DL, Gaskin J, Duke A, Asermely K, Huddleston D, Eiden LE, Rand JB. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 46.Fields SD, Conrad MN, Clarke M. The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J Cell Sci. 1998;111(Pt 12):1717–1727. doi: 10.1242/jcs.111.12.1717. [DOI] [PubMed] [Google Scholar]
- 47.Fields SD, Strout GW, Russell SD. Spray-freezing freeze substitution (SFFS) of cell suspensions for improved preservation of ultrastructure. Microsc Res Tech. 1997;38:315–328. doi: 10.1002/(SICI)1097-0029(19970801)38:3<315::AID-JEMT12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 48.Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 49.Daddow LY. An abbreviated method of the double lead stain technique. J Submicrosc Cytol. 1986;18:221–224. [PubMed] [Google Scholar]
- 50.Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 54.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 55.Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 57.Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem Biophys Res Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- 58.Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM. A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (Obscurin) in Caenorhabditis elegans. Mol Biol Cell. 2008;19:2424–2432. doi: 10.1091/mbc.E08-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lecroisey C, Martin E, Mariol MC, Granger L, Schwab Y, Labouesse M, Segalat L, Gieseler K. DYC-1, a protein functionally linked to dystrophin in Caenorhabditis elegans is associated with the dense body, where it interacts with the muscle LIM domain protein ZYX-1. Mol Biol Cell. 2008;19:785–796. doi: 10.1091/mbc.E07-05-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rhodamine-phalloidin was used to visualize actin thin filaments. In this strain, every body wall muscle cell expresses the array (as determined by nuclear localized SUR-5::GFP) and shows normal thin filament organization.