Abstract
The amino acid sequence of the canine adenosine A1 receptor and the atomic coordinates of a structurally related protein, bacteriorhodopsin, were combined to generate a three-dimensional model for the adenosine A1 receptor. This model consists of seven amphipathic alpha-helices, forming a pore that has a rather distinct partition between hydrophobic and hydrophilic regions. Subsequently, a highly potent and selective ligand, N6-cyclopentyladenosine, was docked into this cavity. A binding site is proposed that takes into account the conformational characteristics of the ligand, obtained from indirect modeling studies by the ‘active analog approach’. Moreover, it involves two histidine residues that were shown to be important for ligand coordination from chemical modification studies. Finally, the deduced binding site was used to model other potent ligands that could all be accommodated consistent with earlier biochemical and pharmacological findings.
Keywords: Molecular modeling; adenosine receptor (s); cyclopentyladenosine (CPA); 1,3-dipropyl-8-cyclopentylxanthine (DPCPX); bacteriorhodopsin; histidine residues
INTRODUCTION
Adenosine receptors belong to the large family of G protein-coupled receptors. There are at least two subtypes of adenosine receptors, A1 and A2 (ref. 1). Activation of A1 receptors can lead to an inhibition of the enzyme adenylate cyclase through their interaction with (subtypes of) Gi, the inhibitory guanine nucleotide binding protein. Activation of A2 receptors (of which two further subtypes may exist, A2a and A2b) leads to a stimulation of adenylate cyclase via the activation of Gs, the stimulatory guanine nucleotide binding protein, with a concomitant increase in the intracellular cAMP concentration.2 Recently, both A1 and A2a receptors have been cloned,3–6 yielding the amino acid sequences of both macromolecules (see also Figure 1). A comparison with other known primary structures of G protein-coupled receptors indicated that adenosine receptors also show a typical pattern of seven predominantly hydrophobic stretches of 20–25 amino acids that could span the cell membrane as alpha-helices (see also Figure 2).3
FIGURE 1.
A comparison of the amino acid sequences of the canine A1 and A2 receptors with the human β2-adrenergic receptor, the rat ml acetylcholine receptor, and several neuropeptidc receptors. Alignment was performed with the MACAW multiple sequence alignment program.27 Putative transmembrane sections (indicated by horizontal bars and roman numbers) were treated as blocks without allowing gaps. Nucleotide sequences were retrieved from the GenBank data base.
FIGURE 2.
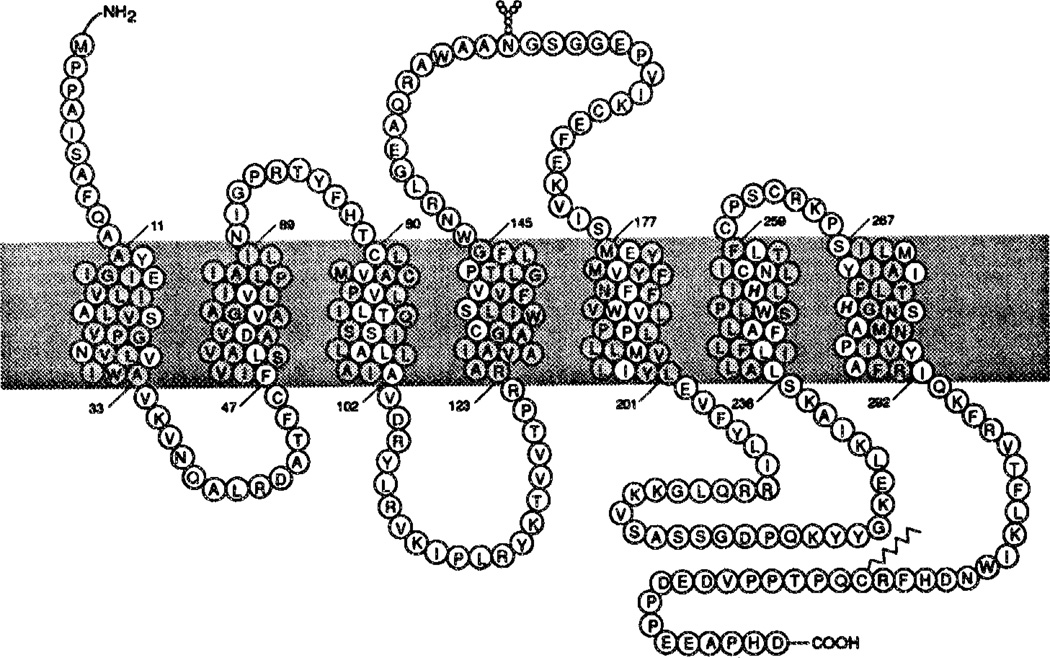
Two-dimensional representation of the canine A1 receptor (see also ref. 10). The N-terminus is located on the extracellular side, and the C-terminus on the cytosolic side. Histidines on transmembrane helices that are putatively involved in ligand binding as shown in italics. Amino acid residues in the transmembrane regions that are proposed to be surrounding the central cavity are highlighted. Leu65, Ile69, Ala124, and Ala127 are also aligned in the direction of the cavity, but are not highlighted since the analogous regions in the structure of bacteriorhodopsin are protruding out of the area of the central cavity.
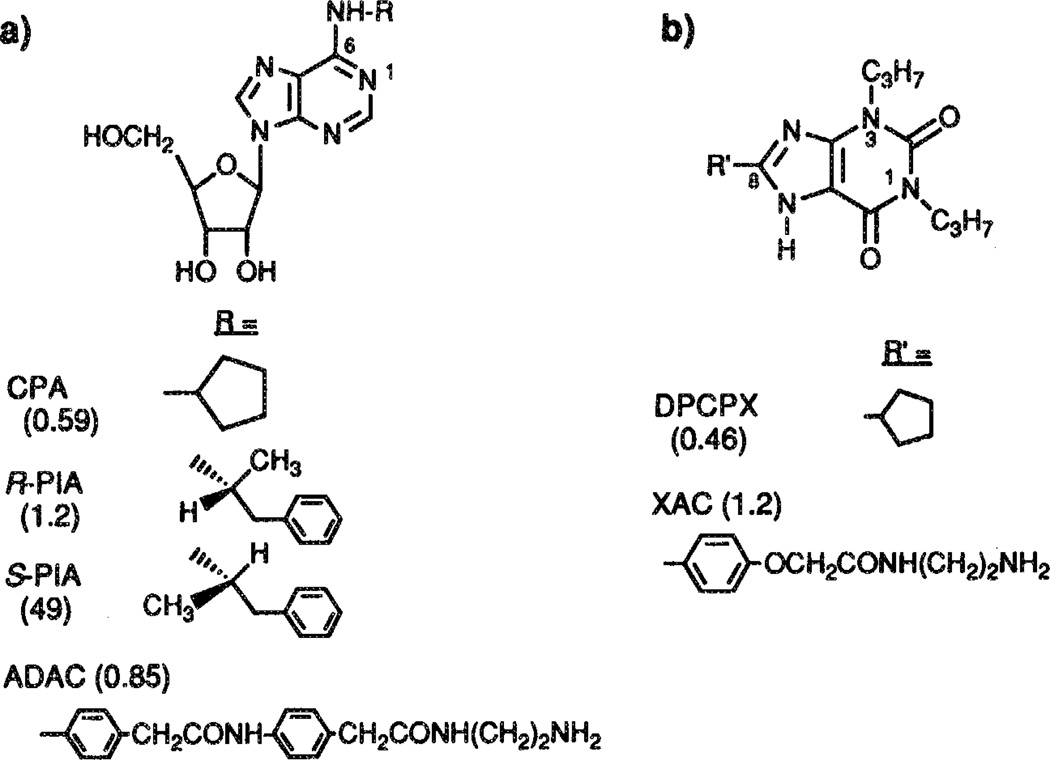
Adenosine, the endogenous ligand for these receptors, has many potent although transient effects, e.g. on the cardiovascular and nervous systems, since adenosine is rapidly degraded in the body.7 In the last two decades many metabolically more stable analogs of adenosine have been synthesized, enabling a much more detailed characterization of the receptors.8 The discovery that xanthines, such as theophylline and caffeine, are moderately active adenosine receptor antagonists,9 likewise stimulated synthetic efforts in this area. As a result, highly potent and selective agonists and antagonists are known for the A1 receptor.8 Selective and potent agonists are available for the A2a receptor, but truly selective antagonists are lacking.10 Examples of adenosine receptor ligands used in the present study are depicted in Figure 3, together with their affinities for rat A1 receptors.
FIGURE 3.
Structures of various adenosine agonists (a) and antagonists (b) having high affinity at A1 receptors. Their Ki values (in nM), determined on rat A1 receptors (refs. 8 and 10), are between parentheses.
It would appear that there are now enough ligands and data available to generate a three-dimensional model of the adenosine receptor–ligand interaction. With this goal we examined specific conformational (3D) information with respect to both ligand and receptor.
With the aid of molecular modeling techniques the region to which the N6-substituents of adenosine receptor agonists bind, has been explored.11 The ‘active analog approach’12 has been used to define the conformational space of this N6-region on the receptor in a detailed and quantitative way. The C8-substituents in xanthine derivatives have been analyzed similarly.13 These two and other studies also generated ideas how agonists and antagonists might overlap in the receptor binding site.11, 13–16 Three models could thus be evaluated, in which the (i) the adenine ring overlaps with the xanthine ring by coincidence of the four nitrogen atoms (‘normal’), (ii) the adenine ring overlaps with the xanthine ring upside down (‘flipped’, as in the relative orientation shown in Figure 3), and (iii) the N6- and C8-substituents are largely superimposed with lesser overlap in the adenine and xanthine ring systems (‘N6/C8’). The first model (‘normal’), although intuitively appealing, is probably incorrect, since the molecular electrostatic potentials of the two ring systems differ greatly. The latter two models have been used to design novel (and potent) antagonists,16, 17 and might both have validity. From theoretical and NMR studies on adenosine, theophylline and xanthine-7-ribosides it became apparent that adenosine binds to the receptor in the anti conformation (as depicted in Figure 3), i.e. with the ribose turned away from the adenine ring system,18 corroborating earlier suggestions.19 Thus, in summary, the conformational characteristics upon interaction with the receptor of adenosine, its analogs, and of receptor antagonists, such as the xanthines, are well defined.
With respect to the atomic coordinates of the receptor protein there is much more uncertainty. Crystallization of membrane proteins with subsequent structure determination by X-ray spectroscopy is not feasible here. The limited quantities available of receptor proteins still preclude the use of NMR methodology. Hence, at present we have to rely on the three-dimensional structural analogy with a model membrane protein, bacteriorhodopsin.20 This macromolecule is of similar size and shares other characteristics with the mammalian G protein-coupled receptors. It displays the typical seven transmembrane alpha-helical architecture that was mentioned before, and, hence, it could serve as a template for receptor modeling.21 From chemical modification studies of adenosine receptors22–24 it appeared that at least one, and probably two histidine residues are involved in ligand binding. Within the transmembrane regions (see also Figures 1 and 2) there are indeed two such residues present, one in helix VI, the other in helix VII.10
In the present study we propose a model for the ligand binding site that takes into account conformational characteristics of the ligands, the involvement of the histidine residues mentioned above, as well as the bacteriorhodopsin-like integrity of the A1 receptor protein. It should be emphasized that the model is not meant to represent the definitive structural characterization of the receptor, but rather serves, hopefully, as a conceptual framework for further efforts in the medicinal chemistry of adenosine receptors.
COMPUTATIONAL METHODS
All model building, docking, energy minimization, and molecular dynamics studies were carried out using the software package BIOGRAF version 2.2 (Molecular Simulations, Sunnyvale, CA, USA). All manipulations were performed on a Silicon Graphics 4D/25GT workstation.
Homology Model Building
The nucleotide sequences for the canine adenosine A1 and A2 receptors, other G protein-coupled receptors, and bacteriorhodopsin were accessed from the GenBank data base. The implied amino acid sequences were generated and further sequence analysis was performed with the aid of the Sequence Analysis Software Package,25 running on a Convex C-240 computer. The prediction of the transmembrane sections was based on the Kyte-Doolittle method,26 with a 9-residue window. Multiple sequence alignment was performed with MACAW,27 with no gaps within the transmembrane domains allowed. The rationale behind this limitation is that these residues which are conserved throughout a family, and certainly between closely related receptors like both adenosine receptor subtypes, are likely to serve similar functions. The introduction of only a one-residue gap would already cause a major spatial reorientation (a 100° shift) of the residues to follow.
A three-dimensional model structure of the seven transmembrane domains of the canine A1 receptor was constructed by using the atomic coordinates of the bacterial protein bacteriorhodopsin (Brookhaven Protein Data Bank, code 1BRD).20 The amino acid residues of the seven transmembrane alpha-helices were ‘mutated’ on the computer screen to provide an initial structure of the A1 receptor. It should be mentioned here that there is virtually no amino acid homology between bacteriorhodopsin and G protein-coupled receptors (see also Discussion for further remarks). However, all helices in both proteins are of similar size, and we used this feature for the alignment of helical residues in the A1 receptor with those present in 1BRD as follows (bacteriorhodopsin/A1 receptor): helix I: 10–32/11–33; helix II: 39–61/47–69; helix III: 79–100/80–101; helix IV: 107–127/124–144; helix V: 137–157/179–199; helix VI: 168–191/236–259; helix VII: 202–225/267–290. Although other, slightly different alignments are conceivable, the further manipulations with the alpha-helices of the A1 receptor (rotations, energy minimizations and molecular dynamics) allow the amino acid residues to ‘scan’ ample space in the receptor architecture. Hence, the initial, rather arbitrary, alignment appeared of little importance for the eventual model.
Subsequently, the helices of the A1 receptor were rotated in such a way that most conserved, most hydrophilic (in particular the histidine residues in helices VI and VII), and all charged residues were located at the ‘inside’ of the receptor. Rotations were constrained about the helical axes as defined by the 1BRD structure. Translational motions were not allowed. The orientation of the helices relative to each other remained virtually unchanged during this process. The rationale here was that hydrophobic residues, now mainly located at the outside of the receptor, will interact with the lipid environment.
Energy Minimizations, Docking and Molecular Dynamics
Throughout all the calculations default values for the various parameters in the BIOGRAF molecular mechanics (Dreiding28) force field and molecular dynamics were used. The Dreiding force field employs a distance-dependent dielectric constant (ε). The default value for ε was 1.0. Hydrogen atoms bound to carbon atoms were not represented explicitly, in order to save computer time. Conjugate gradient energy minimizations were continued until the rms energy gradient was less than 0.1 kcal/mol·Å. First, the initial A1 receptor structure as described above was energy minimized in this way, thereby eliminating unfavorable nonbonded contacts from the side chains of the amino acid residues, and allowing the A1 receptor proline residues to adapt an appropriate configuration.
Reference ligand structures were an A1-selective agonist, CPA, and a competitive, A1-selective antagonist, DPCPX. The initial conformations of these two compounds were those that were derived from previous molecular modeling studies with ligands only.11,13 In these studies the semi-empirical molecular orbital program MOPAC had been used to calculate the minimum energy conformations of the compounds, starting from the crystal structures of adenosine and theophylline, as retrieved from the Cambridge Structural Database.29 By a careful analysis of a whole series of structural analogs it appeared possible to delineate in a rather detailed way the receptor bound conformations of CPA and DPCPX, without knowing the receptor structure itself. The other compounds used in the present study were built and energy minimized in BIOGRAF, starting from the two reference structures
CPA was docked into the pore, formed by the seven alpha-helices, in several orientations, all characterized by the proximity of the two histidine residues in helices VI and VII. The N6-cyclopentyl substituent could thus point in several directions, i.e. piercing between either helices V and VI or VII and I, or three orientations with CPA entirely within the pore. These three orientations comprised the following: (i) CPA in the center of the cavity with the cyclopentyl substituent pointing to the extracellular side, being close to the helices IV, V and VI; (ii) CPA in the center of the cavity with the cyclopentyl substituents more or less equatorial, close to helices VII, I and II, and (iii) CPA somewhat closer to the extracellular side with the cyclopentyl substituent pointing to the extracellular side, close to helices VII, I and II. Each receptor-CPA complex was energy minimized and then relaxed by a 15 ps molecular dynamics run. A constant temperature of 300 K was maintained during the runs by velocity scaling. All atoms were allowed to move, A time step of 0.001 ps was used, with updates of the nonbonded pair list after every 50 time steps. The eventual complexes after the molecular dynamics runs were again energy minimized, and the (gain in) interaction energy between the receptor and CPA was calculated for each of the possibilities described above. Energy values were calculated as follows: ΔEinteraction = (Ereceptor + ECPA) − Ecomplex. As an example, data for the preferred complex were: 78 kcal/mol = (257 + 73) kcal/mol − 252 kcal/mol. The energy values do not correspond to the ‘real’ energetic values in an absolute way. They can only be compared to each other in terms of ‘more’ or ‘less’ favorable states. The energy values cannot be used to calculate exact values of affinities between molecules out of interaction energies, since changes in entropy and solvation effects are not taken into account. The most favorable orientation was used for further docking procedures with all compounds in the bacteriorhodopsin-like A1 receptor template.
RESULTS
A1 Receptor Topology
The alignment of the adenosine A1 and A2 receptor subtypes and some other G protein-coupled receptors is shown in Figure 1. Unfortunately, bacteriorhodopsin does not have enough sequence homology to align it with the G protein-coupled receptors. The seven helical domains are indicated by horizontal bars (H I–H VII). Three extracellular (E I–E III) and three cytoplasmic (C 1–C III) loops connect the transmembrane domains. The N-terminus is located on the extracellular side, whereas the C-terminus is in the intracellular compartment. Shaded amino acids indicate structural homology with (some of) the other receptors. The aspartic acid residue in helix III, common to all receptors for biogenic amines, such as β2 and ml (Figure 1), is absent in adenosine and peptide (substance P and endothelin) receptors. In these macromolecules a histidine residue in helix VI is conserved.
A two-dimensional representation of the adenosine A1 receptor is given in Figure 2. A more detailed analysis of structure-function relationships for the adenosine receptors, e.g. sites for glycosylation, phosphorylation and G protein interaction, will be described elsewhere.10
For the construction of a three-dimensional model of the A1 receptor we used the atomic coordinates of bacteriorhodopsin.20 Since no data for the non-membrane parts of this protein (i.e. loops and tails) are available, we refrained from modeling equivalent parts of the adenosine A1 receptor. There are more prolines in the A1 receptor transmembrane domains than in the corresponding regions in bacteriorhodopsin (8 and 5, respectively) and on different positions, which renders the topology of the Al receptor slightly different from bacteriorhodopsin, since prolines act as ‘helix benders’. Due to the procedure followed (‘mutation’ with subsequent minimization), however, the A1 receptor retains the overall appearance of bacteriorhodopsin, with only slight changes in helical architecture.
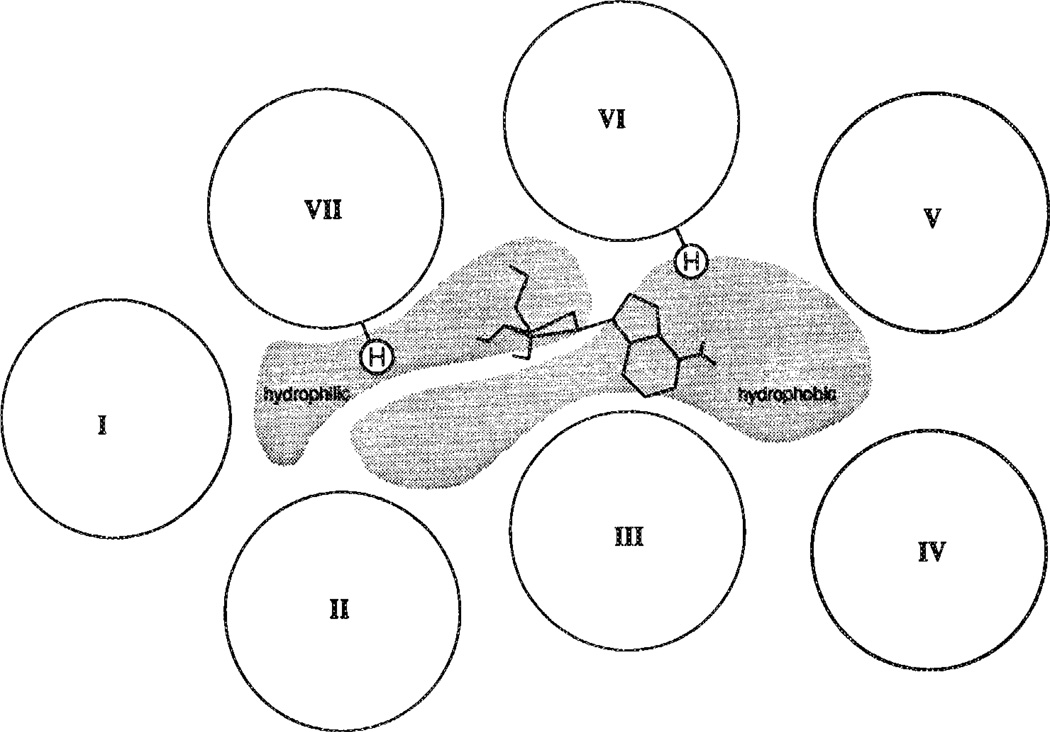
Upon analysis of the characteristics of the bacteriorhodopsin-like pore a rather distinct partition between hydrophobic and hvdrophilic regions in the upper part of the A1 receptor cavity emerged (Figure 4; see also ref. 10). No such explicit partition is observed in bacteriorhodopsin (data not shown).
FIGURE 4.
A generalized map of the upper half of each transmembrane helix of the A1 receptor (modified from ref. 10), showing separate domains of hydrophobic and hydrophilic environment. The proposed orientation of adenosine in relation to the histidines in transmembrane helices that are putatively involved in ligand binding is shown.
Docking of N6-cyclopentyladenosine (CPA) and Other Ligands
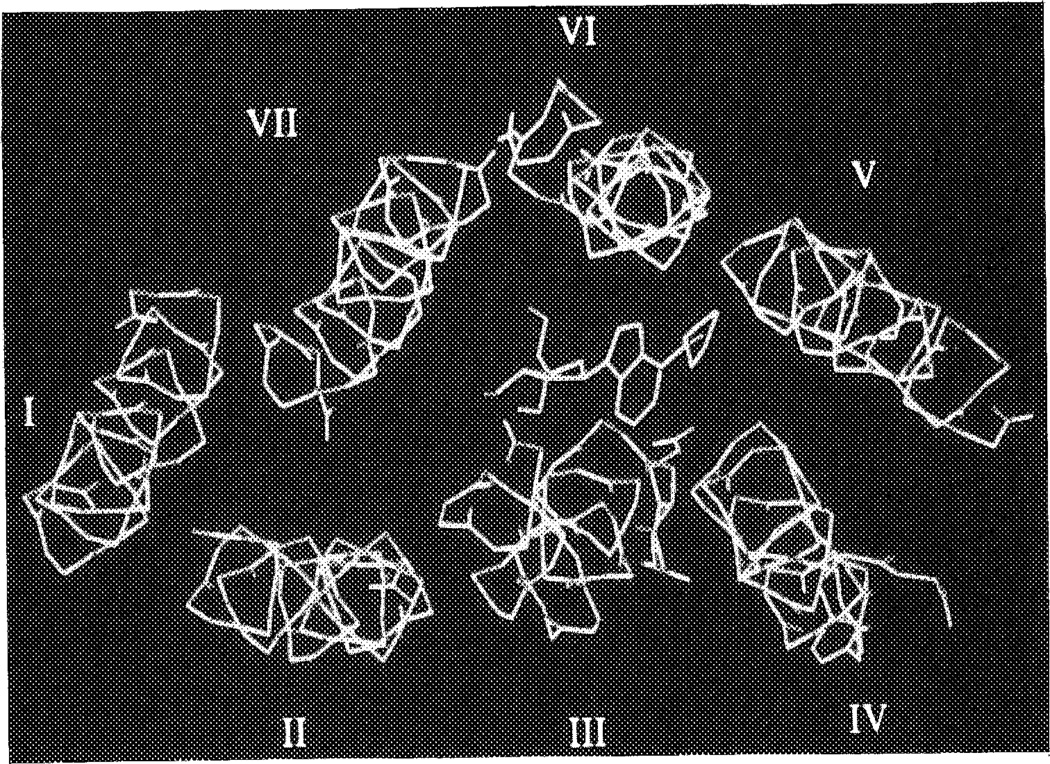
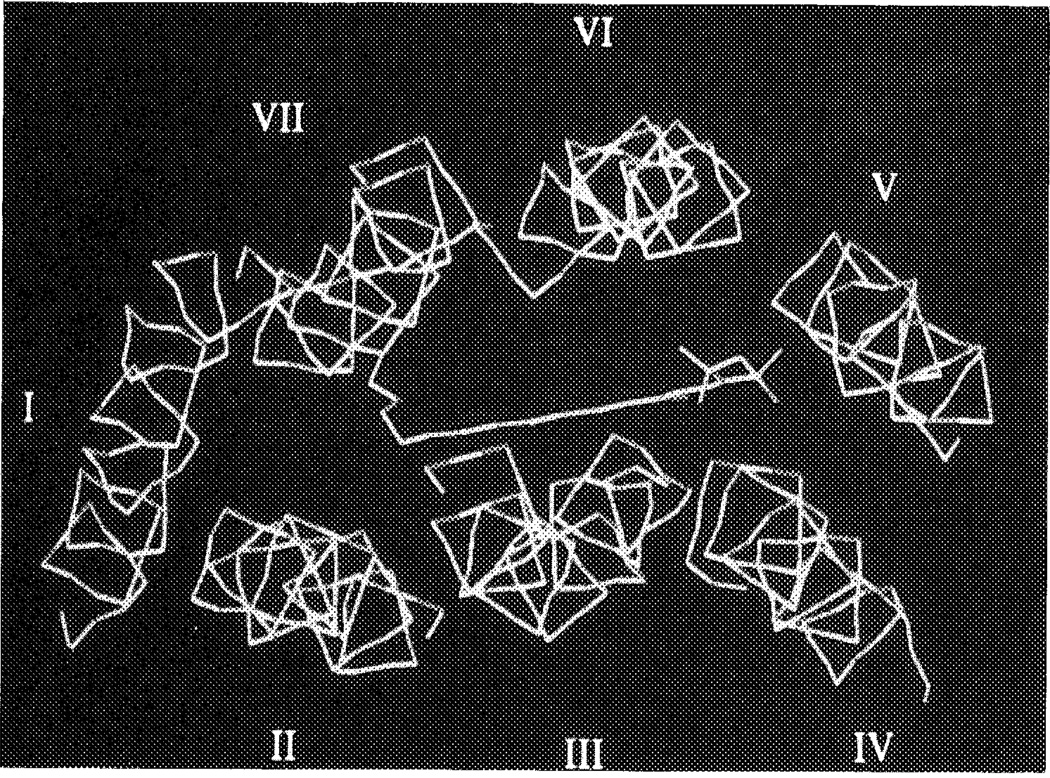
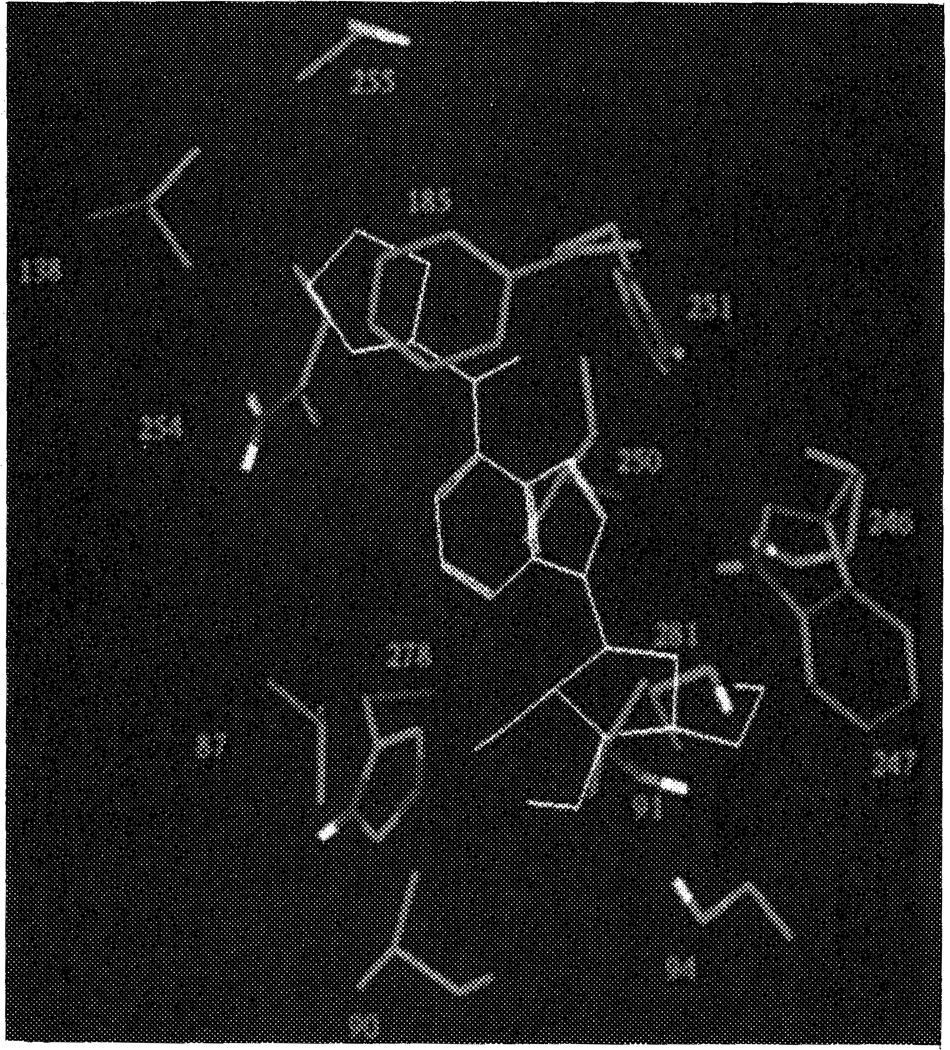
CPA was docked in several orientations into the A1 receptor pore, as described in Methods. The two orientations of CPA with the cyclopentyl substituents between the helices led to an overall distortion of receptor architecture, and were not considered further. Of the other three possibilities, with CPA entirely within the pore, it appeared that their gain in interaction energy between ligand and receptor was not identical. The two less favorable ‘complexes’ differed by 3 (gain in energy 75 kcal/mol) and 8 kcal/mol (gain 70 kcal/mol), respectively, from the best (gain 78 kcal/mol, see Computational Methods), according to the Dreiding force field. The most favorable orientation was with the cyclopentyl substituent pointing to the extracellular side of the protein, close to helices IV, V and VI. Figure 5 is a representation of CPA surrounded by the seven alpha-helices (main chains shown only). For comparison the analogous topology of bacteriorhodopsin with its endogenous ligand, retinal, binding to it, as retrieved from the Bookhaven Protein Data Bank, is shown in Figure 6. The ligand binding site on the A1 receptor (arbitrarily defined as the amino acid residues within 4.5 Å from N6-cyclopentyladenosine) is represented as a stereo drawing in Figure 7, showing more hydrophilic residues in close proximity to the hvdrophilic purine and ribose moieties of CPA. The two histidine residues (251 and 278) may form hydrogen bonds with CPA, via N6–H, and 2′- and 3′-OH, respectively, as does Ser281 with 5′-OH. It should be stressed that a slightly different orientation of CPA probably would lead to other hydrogen bonds formed, e.g. with the other alcoholic amino acid residues (Thr91, Ser94 and Ser246). Nevertheless, all hydrophilic amino acid residues in Figure 7 remain an integral part of the ligand binding site. The presence of Asn254 at the interface between the purine ring and the cyclopentyl substituent could also be important for interaction (see also below). The hydrophobic cyclopentyl substituent, in contrast, is surrounded by two hydrophobic amino acid side chains, viz. Val138 and Phe185, together with Cys255. Upon binding, both the receptor and CPA change their conformations, in order to maximize the interaction strength. Since we started the docking procedure with the receptor and the ligand in their energetically optimized conformations, these changes led to increases in intramolecular energy content, namely 2 kcal/mol for the receptor and 7 kcal/mol for CPA. The interaction energy of the complex is −78 kcal/mol, resulting in a decrease in total van der Waals repulsion energy of 69 kcal/mol.
FIGURE 5.
The binding of N6-cyclopentyladenosine (CPA) to the canine adenosine A1 receptor (direction of view from outside to inside). CPA is in yellow. The receptor alpha-helical structure is represented by main chains (green) only, thus yielding the peptide helical backbone (I–VII) without side chains. See color plate at back of issue.
FIGURE 6.
The binding of retinal (yellow) to bacteriorhodopsin (direction of view from outside to inside). The alpha-helical structure is represented by main chains (green) only, thus yielding the peptide helical backbone (I–VII) without side chains, except for Lys216 (also in yellow) to which retinal is covalently bound. Atomic coordinates were retrieved from the Brookhaven Protein Data Bank.20 See color plate at back of issue.
FIGURE 7.
Stereo representation of the proposed N6-cyclopentyladenosine (CPA) binding site on the canine A1 receptor. CPA is represented in yellow. The amino acids, of which the side chains are shown only, have been numbered according to their appearance in the canine A1 receptor sequence. Colors represent carbon (grey), nitrogen (blue), oxygen (red), and hydrogen atoms (white, bound to heteroatoms only). Residues involved are: valine (87, 138), leucine (90, 250), threonine (91), serine (94, 246, 281), phenylalanine (185), tryptophan (247), histidine (251, 278), asparagine (254), and cysteine (255). All amino acids are within 4.5 Å from CPA. Ser246 and Leu 250 are located between helices VI and VII, and are thus not highlighted in Figure 2. See color plate at back of issue.
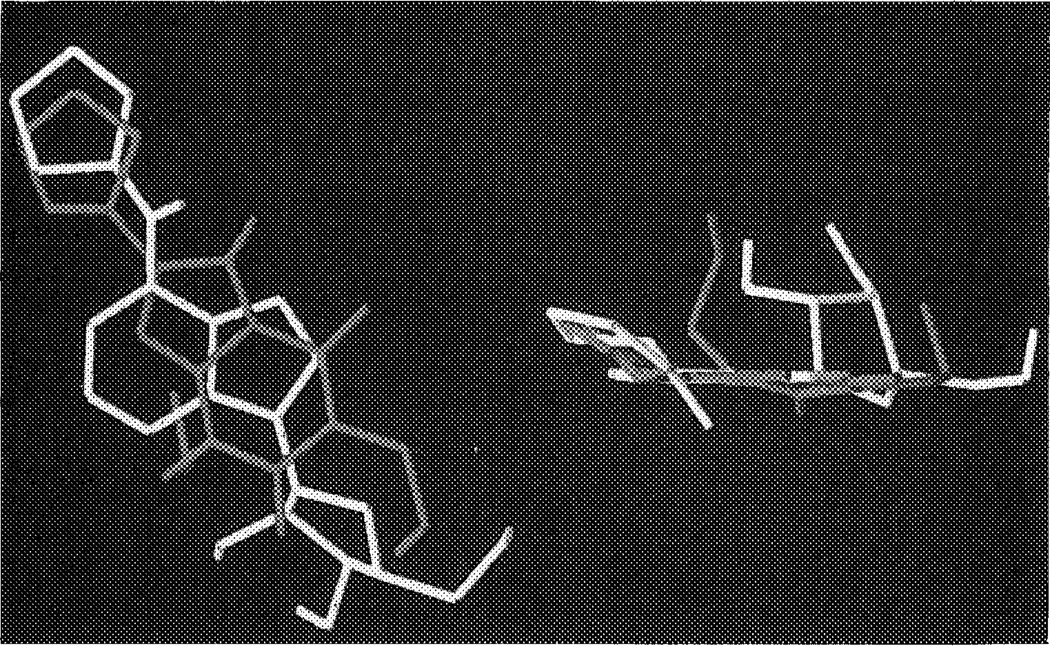
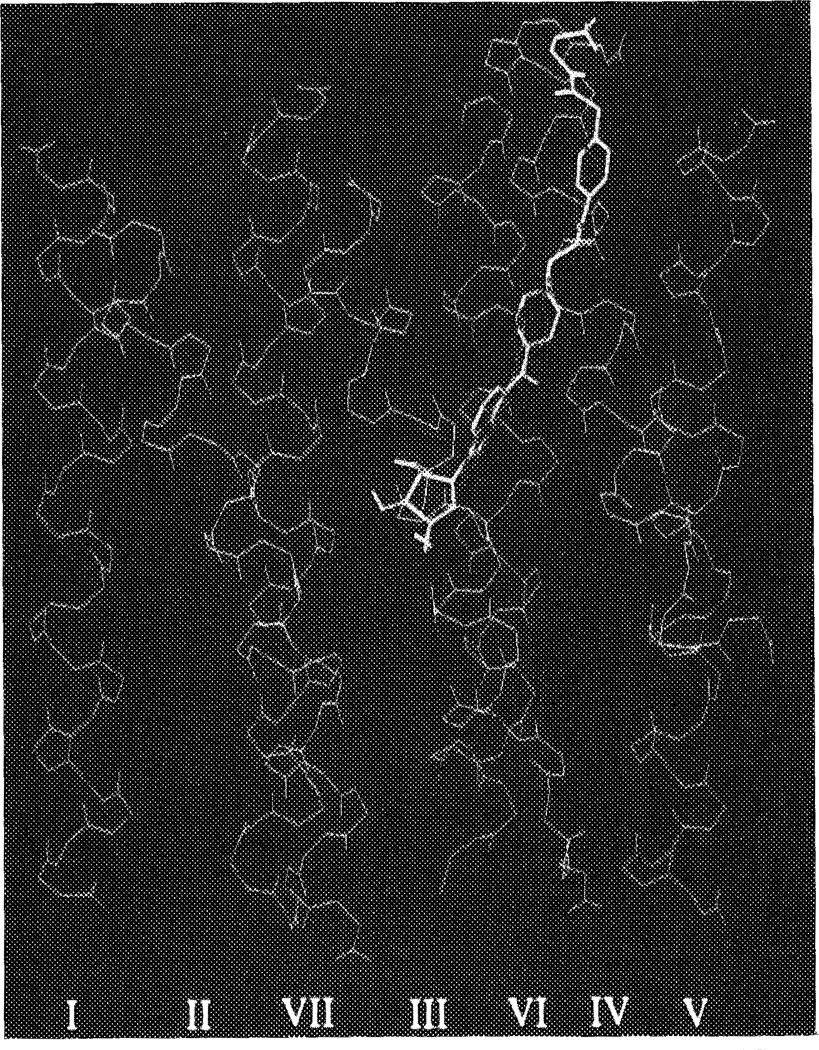
How might the potent and A1 selective antagonist DPCPX (l,3-dipropyl-8-cyclopentylxanthine) fit in this binding pocket? As discussed in the Introduction there are in principle three ways of superimposing agonists and antagonists. From our present modeling studies it appeared that the mode in which both cyclopentyl substituents of DPCPX and CPA are more or less overlapping (the N6/C8 model) is the most obvious possibility (see also Figure 8); this particular orientation appears to have more general validity, since longer C8 substituents, like in XAC (xanthine amine congener), can also be accommodated. Indeed, the extended substituents in the ‘functionalized congeners’30 ADAC (agonist) and XAC (antagonist) easily ‘climb up’ the cavity to reach the extracellular space (Figure 9). The two n-propyl substituents on the xanthine ring system in both DPCPX and XAC can be accommodated by the A1 receptor upon docking. The propyl group on N3 has significant van der Waals interactions with Val87 and Leu90, whereas the N1-substituent is close to Trp247, Leu250 and Ser281, a somewhat less hydrophobic environment.
FIGURE 8.
Receptor bound conformations of N6-cyclopentyladenosine (CPA) and l,3-dipropyl-8-cyclopentylxanthine (DPCPX). CPA is in yellow, DPCPX in red. The representation on the left is in the XY-plane, whereas the one on the right is rotated 90°, to show the coplanarity of both ligands. See color plate at back of issue.
FIGURE 9.
The binding of ADAC and XAC to the canine A1 receptor (overview). ADAC is in yellow, XAC in red. The receptor alpha-helical structure is represented by main chains (green) only, thus yielding the peptide helical backbone (I–VII) without side chains. Top of figure represents the extracellular side. See color plate at back of issue.
Finally, the topic of stereoselectivity should be addressed. If the ligand binding site as defined for CPA is used as a template to dock R- and S-N6-phenylisopropyladenosine (PIA), it appears that the methyl group in S-PIA is piercing into a ‘forbidden’ area. This region is occupied by amino acid residue Asn254, which is very close to the purine heterocycle and the cyclopentyl group. The same methyl group in R-PIA, however, overlaps with (part of) the cyclopentyl substituent in CPA, and does not interfere unfavorably with the asparagine residue.
DISCUSSION
A1 Receptor Topology
The alignment of the adenosine and other receptors as represented in Figure 1 slightly differs from the one presented by Libert et al.,3 due to reasons explained in the Methods section. These differences occur in helices I, IV, V, and VII. It should be noted that the Kyte–Doolittle algorithm used can at best yield an approximation of the transrnembrane domains, so future adjustments may be warranted.31 This uncertainty is also reflected in the different alignments (for the other receptors) by Hibert and coworkers in their comprehensive study on the modeling of G protein-coupled receptors.21 Since it is not likely that solid (X-ray crystallographic or NMR) structural data on G protein-coupled receptors will become available soon, indirect evidence, e.g. from proteolysis studies with or without crosslinkers, or experiments with antibodies directed against putative extracellular domains will be needed to assess the correctness of these alignments.
With respect to the orientation of the seven helices, it has been noted in many other receptors that the transmembrane domains are often amphipathic in nature, i.e. hydrophobic and hydrophilic amino acids line up on opposite parts of the helix. It is assumed that the hydrophobic phase is directed toward the lipophilic cell membrane, whereas the more hydrophilic parts face each other, forming a relatively hydrophilic center.
What is the relative orientation of the seven helices? Again, three-dimensional atomic coordinates are not known for any of the G protein-coupled receptors. Henderson and coworkers have been able to generate a model for bacteriorhodopsin by electron cryo-microscopy experiments.20 This protein serves as a proton pump in the bacterial cell wall of Halobacterium halobium, and is not linked to a G protein. However the seven helical structure of the protein, together with the fact that retinal, its endogenous ligand, is also found in the G protein-coupled mammalian rhodopsin, render the protein a suitable starting point for attempts to model the G protein-coupled receptors.21 Caution should be stressed here, since overall amino acid homology between bacteriorhodopsin and the receptors is very low, and no alignment could be obtained. Moreover, the structural map of bacteriorhodopsin has a resolution of 3.5 Å at the best.
Bearing these caveats in mind, we used the relative orientation of the seven helices in bacteriorhodopsin only as a framework for the positioning of the seven helices in the canine adenosine A1 receptor. We used the canine sequence for our modeling studies, since at the time of the experiments the rat A1 receptor sequence was not known yet. In the transmembrane regions of A1 receptor of both species only three residues are different, all not forming part of the central cavity.5 Further energy minimizations and molecular dynamics runs revealed that a bacteriorhodopsin-like structure is very well compatible with all the different amino acids that are present in the A1 receptor. Hibert et al.21 discussed the possible relevance of the several (conserved) receptor proline residues, and argued that these residues as such lead to a minor distortion only of the alpha-helical backbone. We agree on this topic, and would like to add that in bacteriorhodopsin the proline in helix II does not lead to changes in the helical structure, whereas the two in helix III appear to do so, although other factors seem to play a role as well. Moreover, only the proline in helix II forms part of the pore, whereas the other four seem to be distributed elsewhere. Thus, in our opinion, the role of prolines (conserved or not) in defining receptor architecture is not very clear, and does not fit their putative characterization as alpha-helix breakers. Also in molecular dynamics calculations, the overall structure of the seven helical configuration remains fairly unaffected, whereas the amino acid side chains are relatively flexible (and may span considerable distances within the protein) over the observed time periods. As is evident from Figure 1 (and discussed in a sequence analysis of adenosine receptors)10 the largest homology between the respective receptor sequences (indicated by the shaded areas) occurs in the lower half of the transmembrane domains (i.e. near the cytoplasmic side) and in those parts of the cytoplasmic domains that are close to the membrane. These areas are generally thought to be important for interaction with G proteins, the feature that all these receptors have in common. On the other hand, the respective ligands for the various receptors differ greatly, and much less homology occurs in the upper half of the helices and in the extracellular domains. Thus, these may well be the areas where ligand binding occurs.
The Ligand Binding Site
The binding site for the highly hydrophobic retinal in bacteriorhodopsin has been shown to be buried in the membrane, just above the center of the cavity formed by the seven transrnembrane domains.20 Each of the helices, by virtue of at least one amino acid residue, appears to be involved in the binding of retinal. (Positively) charged, much more hydrophilic endogenous ligands such as (nor) epinephrine and acetylcholine interact with a (negatively) charged aspartate residue on helix III, present in all receptors for biogenic amines, again suggesting that the binding site for these ligands is also somewhere within the cavity.32,33
How do adenosine and derivatives (Figure 3) bind to the receptor? Unlike the biogenic amines, adenosine is uncharged at physiologic pH, and the aspartate residue on helix III is substituted by a valine in both adenosine receptor subtypes. The affinity-profile of a homologous series of N6-alkyl, -alkylamine and -alkyladenosine substituted adenosines suggests that the adenosine binding site may be located away from the outer membrane surface.34 Klotz et al.22 and Garritsen et al.23 have shown for the adenosine A1 receptor that at least one and possibly two histidine residues in the binding pocket may be involved in interactions with adenosine receptor agonists and antagonists. Similar observations have been made for A2 receptors.24 Within the transmembrane domains there are only two conserved histidine residues, viz. in helix VI and helix VII (His251 and His278 in the A1 receptor). Hence, these two residues are probably near or within the ligand binding site. Interestingly, His278 is homologous to Lys216 in bacteriorhodopsin, the residue that is thought to form a Schiff base with its ligand retinal,20 whereas His251 (in helix VI) appears to be conserved in some peptide receptors, notably those for endothelin and the tachykinins substance P (see Figure 1), substance K and neuromedin K (alignments not shown). This residue might serve a similar function as the aspartic acid (helix III) in the biogenic amine receptors, i.e. the coordination of ligands. From Figure 4 it is apparent that His278 on helix VII is present in a largely hydrophilic environment, and as such this region may play a role in ion movements as a possible trigger for receptor activation.35 The other histidine residue, His251 on helix VI, is at the interface between hydrophobicity and -philicity. Could it therefore be that His278 is particularly involved in the coordination of the ribose moiety in agonists that appears to be essential for receptor activation,10 and that His251 is in close proximity to the N6-region of agonists, which is hydrophobic in nature and critical for enhanced receptor affinity ?10
For our docking purposes we reasoned that any model for the ligand binding site should involve at least one of the histidine residues. With respect to the ligand we decided not to use the endogenous ligand adenosine itself, but a synthetic derivative, N6-cyclopentyladenosine (CPA). CPA is a reference ligand for adenosine A1 receptors for which it has high affinity and selectivity.10 The receptor bound orientation of the N6-substituent in CPA was previously determined in a rather detailed way via the ‘active analog approach’.11,12 Of particular relevance was the relative orientation of the cyclopentyl substituent toward the purine base moiety, as specified by the torsion angle N1-C6-N6-C (cyclopentyl) of approximately −75°. Other studies addressed the relative orientation of the ribose group.18,19 All available evidence suggests that an anti conformation is essential for receptor binding, although more precise data is lacking.
With these prerequisites in mind we docked CPA in the A1 receptor model in several ways. Herbette et al36 have suggested that ligands could approach their receptors via the lipid bilayer, i.e. from the side. Thus, we initially explored whether conformations in which the hydrophobic cyclopentyl substituent was oriented between two helices (helix I/helix VII, and helix V/helix VI) with the rest of the molecule within the pore were feasible. The rationale here was that more extended N6-substituents could then interact with the phospholipids of the membrane.37 Although such orientations are energetically possible after full relaxation of the receptor molecule, the overall helical structure becomes drastically distorted, since the helices involved have to bend away from each other. Considering the bacteriorhodopsin template we argued that these options probably do not represent the adenosine binding site under equilibrium conditions, although they cannot be ruled out as transient states in the binding sequence. The other possibilities with CPA entirely within the pore favored the one with the cyclopentyl substituent directed to the extracellular side close to helices IV, V and VI, although the energy differences between these latter complexes are relatively small (< 10 kcal/mol) compared to the gain in interaction energies that are obtained with each of the complexes (approximately 70–80 kcal/mol). The best orientation of CPA was used once more, but now with the A1 receptor backbone kept fixed to maintain the bacteriorhodopsin-like configuration of all seven helices as much as possible (except for the differing prolines). CPA was allowed to relax now, but its geometry did not change greatly. The purine–cyclopentyl torsion angle was −60°, whereas the ribose moiety adopted a definite anti conformation, fully in line with the earlier findings described above. The hydrogen bond formed between His251 and N6-H might well explain the fact that N6-disubstitution of adenosine derivatives is detrimental for affinity,8 imparting the disruption of this energetically favorable bond with additional steric hindrance. Semi-empirical calculations using the MOPAC AM1 Hamiltonian led to a similar hydrogen bond, with possibly an additional (but weaker) hydrogen bond between N7 in adenosine and C2 of the imidazole ring of the histidine residue, which is somewhat negatively charged.38
Similarly, the hydrogen bonds formed by His278 and the 2′- and 3′-OH groups of the sugar in CPA may well explain the lower affinities of 3′-deoxy-R-PIA and, in particular, 2′-deoxy-R-PIA.39 Removal of both hydroxyl groups as in 2′,3′-dideoxy-N6-cyclohexyladenosine, yields an antagonist with only moderate affinity.40 Furthermore, the inactivity of 2′-F,2′-deoxyadenosine,19 in which the fluoro substituent may act as a hydrogen bond acceptor, suggests that the 2′-hydroxy group acts as a hydrogen bond donor to initiate the agonist response. It is appropriate to mention here that water molecules could serve as a ‘glue’ between the hydrophilic groups of the receptor and the ligand. This has recently been shown the case in the crystal structure of adenosine deaminase interacting with an adenosine-like transition-state analog.41 According to the Gibbs free energy function (ΔG° = RT ln Ki), the binding of CPA to the (dog) A1 receptor (Ki = 3.5 nM)4 involves a total energy decrease of approximately 12 kcal/mol. This value greatly deviates from our calculated value of the van der Waals repulsion energy (69 kcal/mol). Once more, it should be stressed that the calculated energy values in BIOGRAF do not represent absolute values, and, moreover, that ΔG° values comprise both changes in enthalpy and entropy (ΔG° = ΔH° − T ΔS°); unfortunately, the latter term cannot be included in the energy calculations. Furthermore, solvation energies are also neglected in these calculations.
A comparison between the CPA binding site of the A1 receptor (Figure 5) and the retinal binding site of bacteriorhodopsin (Figure 6) reveals that both ligands in their bound conformations occupy approximately the same space in the pore. Thus, the lack of overall homology between the two proteins and the entirely different chemical characteristics of both proteins and ligands appear to somehow compensate for each other, allowing a similar location of the binding site.
From the modeling efforts with the agonist ADAC (Figure 9) it appeared that the relative freedom of substitution on N6 is easily explained by the fact that extended N6-substituents can be aligned along the pore wall to eventually leave the receptor cavity, also enabling further interactions. As an example, it might be hypothesized that the positively charged terminal amino group in ADAC interacts with the almost extracellular glutamic acid residue in helix V (Glu178) of the A1 receptor. A distal electrostatic interaction between this amino group and a membrane component has been proposed to account for enhanced affinity.42 Similarly, the affinities of adenosine derivatives with N6-alkyl (hydrophobic), and N6-alkylamine as well as N6-alkyladenosine (hydrophilic) substituents are in excellent agreement with this proposal.34 Up to alkyl chain lengths of 8 or 9 carbon atoms (the length necessary to reach the extracellular boundaries of the receptor helices) affinities increase, whereas larger chains lead to less active compounds. In particular the hydrophobic alkyl chains (now in an aqueous environment) are not well tolerated. As an example, N6-dodecyladenosine is almost 100-fold less active than the corresponding N6-12-aminoalkyladenosine. Thus, the general concept of ‘functionalized congeners’ appears to have its biochemical correlate here.
If this is also true for the xanthine receptor antagonists with long C8-substituents like in XAC, this imposes the xanthine core structure in the ‘N6/C8’ orientation, as represented in Figure 8 for DPCPX and in Figure 9 for XAC. This orientation would also explain the similar increases in affinity for very bulky substituents, such as adamantyl, in N6-substituted agonists and C8-substituted antagonists.10 Further-more, it was shown by Barrington et al.43 that agonist and antagonist photoaffinity labels (in which an azido group was placed on similarly extended N6- or C8-substituents) interacted with the same region of the receptor. It remains, however, possible that smaller antagonist structures adopt other orientations such as in the ‘flipped’ model, thereby still impeding the binding of the endogenous agonist adenosine, and thus acting as competitive antagonists.
A striking feature of the N6-region is its stereoselectivity, at least for A1 receptors. Thus, R-PIA is some 50-fold more potent than its stereoisomer S-PIA.10 Similar values are found for other pairs of stereoisomers. In mechanistic terms this could mean that stereoselectivity is induced by one common factor, e.g. an identical steric hindrance. In our earlier studies on the mapping of the N6-region it became apparent that all S-stereoisomers (with lower affinity) were piercing in a ‘forbidden’ area to some extent.11 From the present work this forbidden area could well be explained by the presence of an asparagine residue (Asn254) here. Site-directed mutagenesis studies in which this residue would be exchanged for a smaller one could provide a more definite proof.
A final issue is the A1 receptor selectivity of CPA. Indeed, the cyclopentyl substituent induces this preference for the A1 receptor, as do other (bi) cycloalkyl substituents.10 However, other hydrophobic substituents of similar and larger size are equally well accommodated by the A2 receptor, e.g. N6-fluorenyladenosine.44 Moreover, further exploration of the N6-region even led to the design of N6-substituted agonists with significant A2 receptor selectivity.45 Clearly, rather subtle differences between A1 and A2 receptors govern this selectivity, and it is questionable whether molecular modeling studies without the precise knowledge of receptor subtype architecture will provide definite clues in this respect. However, the amino acids that are in the vicinity of the cyclopentyl substituent (Figure 7) are also found in the A2 receptor in corresponding positions, except for Val138. This hydrophobic, aliphatic amino acid residue is replaced by a larger isoleucine in the A2 receptor, which could indeed disrupt the rather favorable hydrophobic interaction between carbon/hydrogen atoms in both the non-flat cyclopentyl substituent and the amino acid in position 138.
In conclusion, in this report we describe a model for the ligand binding site on the adenosine A1 receptor. Two histidine and several serine and threonine residues appear to play a role in the coordination of the hydrophilic purine and ribose moieties in the agonist CPA. The N6-region is much more hydrophobic in nature, allowing van der Waals interactions with the cyclopentyl substituent. The model could serve as a starting point for site-directed mutagenesis studies. Combined efforts in computational chemistry and molecular biology may lead to validation and optimization of the proposed model, eventually enabling the rational design of new chemical entities.
References
- 1.Van Calker D, Müller M, Hamprecht B. Adenosine regulates via two different types of receptors, the accumulation of cyclic AMP in cultured brain cells. J. Neurochem. 1979;33:999–1005. doi: 10.1111/j.1471-4159.1979.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 2.Londos C, Cooper DMF, Wolff J. Subclasses of external adenosine receptors. Proc. Natl. Acad. Sci. USA. 1980;77:2551–2554. doi: 10.1073/pnas.77.5.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Libert F, Parmentier M, Lefort A, Dinsart C, Van Sande J, Maenhaut C, Simons MJ, Dumont JE, Vassart G. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science. 1989;244:569–572. doi: 10.1126/science.2541503. [DOI] [PubMed] [Google Scholar]
- 4.Libert F, Schiffmann SN, Lefort A, Parmentier M, Gerard C, Dumont JE, Vanderhaeghen JJ, Vassart G. The orphan receptor cDNA RDC7 encodes an A1 adenosine receptor. EMBO J. 1991;10:1677–1682. doi: 10.1002/j.1460-2075.1991.tb07691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahan LC, McVittie LD, Smyk-Randall EM, Nakata H, Monsma FJ, Gerfen CR, Sibley DR. Cloning and expression of an A1 adenosine receptor from rat brain. Mol. Pharmacol. 1991;40:1–7. [PubMed] [Google Scholar]
- 6.Maenhaut C, Van Sande J, Libert F, Abramowicz M, Parmentier M, Vanderhaeghen JJ, Dumont JE, Vassart G, Schiffmann SN. RDC8 codes for an adenosine A2 receptor with physiological constitutive activity. Biochem. Biophys. Res. Commun. 1990;173:1169–1178. doi: 10.1016/s0006-291x(05)80909-x. [DOI] [PubMed] [Google Scholar]
- 7.Williams M. Purine receptors in mammalian tissues: Pharmacology and functional significance. Ann. Rev. Pharmacol. Toxicol. 1987;27:315–345. doi: 10.1146/annurev.pa.27.040187.001531. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson KA. Adenosine (P1) and ATP (P2) receptors. In: Hansch C, et al., editors. Comprehensive Medicinal Chemistry. Vol. 3. Oxford: Pergamon Press; 1990. pp. 601–642. [Google Scholar]
- 9.Sattin A, Rall TW. The effect of adenosine and adenine nucleotides on the cyclic adenosine 3′,5′-phosphate content of guinea pig cerebral cortex slices. Mol. Pharmacol. 1970;6:13–23. [PubMed] [Google Scholar]
- 10.Van Galen PJM, Stiles GL, Michaels G, Jacobson KA. Adenosine A1 and A2 receptors: Structure-function relationships. Med. Res. Rev. 1992 doi: 10.1002/med.2610120502. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Galen PJM, Leusen FJJ, IJzerman AP, Soudijn W. Mapping the N6-region of the adenosine A1 receptor with computer graphics. Eur. J. Pharmacol. – Mol. Pharmacol. Sect. 1989;172:19–27. doi: 10.1016/0922-4106(89)90041-2. [DOI] [PubMed] [Google Scholar]
- 12.Marshall GR, Motoc I. Approaches to the conformation of the drug bound to the receptor. In: Burgen ASV, et al., editors. Topics in Molecular Pharmacology. Vol. 3. Amsterdam: Elsevier; 1986. pp. 115–156. [Google Scholar]
- 13.Van der Wenden EM, Van Galen PJM, IJzerman AP, Soudijn W. Mapping the xanthine C8-region of the adenosine A1 receptor with computer graphics. Eur. J. Pharmacol. – Mol. Pharmacol. Sect. 1991;206:315–323. doi: 10.1016/0922-4106(91)90116-y. [DOI] [PubMed] [Google Scholar]
- 14.Van der Wenden EM, IJzerman AP, Soudijn W. A steric and electrostatic comparison of three models for the agonist/antagonist binding site on the adenosine A1receptor. J. Med. Chem. 1992;35:629–635. doi: 10.1021/jm00082a003. [DOI] [PubMed] [Google Scholar]
- 15.Van Galen PJM, Van Vlijmen HWT, IJzerman AP, Soudijn W. A model for the antagonist binding site on the adenosine A1 receptor, based on steric, electrostatic, and hydrophobic properties. J. Med. Chem. 1990;33:1708–1713. doi: 10.1021/jm00168a027. [DOI] [PubMed] [Google Scholar]
- 16.Peet NP, Lentz NL, Meng EC, Dudley MW, Ogden AML, Demeter DA, Weintraub HJR, Bey P. A novel synthesis of xanthines: Support for a new binding mode for xanthines with respect to adenosine at adenosine receptors. J. Med. Chem. 1990;33:3127–3130. doi: 10.1021/jm00174a004. [DOI] [PubMed] [Google Scholar]
- 17.Van Galen PJM, Nissen P, Van Wijngaarden I, IJzerman AP, Soudijn W. lH-Imidazo[4.5-c]quinolin-4-amines: Novel non-xanthine adenosine antagonists. Med. Chem. 1991;34:1202–1206. doi: 10.1021/jm00107a046. [DOI] [PubMed] [Google Scholar]
- 18.Van Galen PJM, IJzerman AP, Soudijn W. Xanthine-7-ribosides as adenosine A1 receptor antagonists: Further evidence for adenosine’s anti mode of binding. Nucleosides & Nucleotides. 1990;9:275–291. [Google Scholar]
- 19.Bruns RF. Adenosine receptor activation in human fibroblasts: Nucleoside agonists and antagonists. Can. J. Physiol. Pharmacol. 1980;58:673–691. doi: 10.1139/y80-110. [DOI] [PubMed] [Google Scholar]
- 20.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 21.Hibert MF, Trumpp-Kallmeyer S, Bruinvels A, Hoflack J. Three-dimensional models of neurotransmitter G-binding protein-coupled receptors. Mol. Pharmacol. 1991;40:8–15. [PubMed] [Google Scholar]
- 22.Klotz K-N, Lohse MJ, Schwabe U. Chemical modification of A1 adenosine receptors in rat brain membranes. J. Biol. Chem. 1988;263:17522–17526. [PubMed] [Google Scholar]
- 23.Garritsen A, IJzerman AP, Beukers MW, Soudijn W. Chemical modification of adenosine A1 receptors. Implications for the interaction with R-PIA, DPCPX and amiloride. Biochem. Pharmacol. 1990;40:835–842. doi: 10.1016/0006-2952(90)90324-e. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson KA, Stiles GL, Ji X-D. Chemical modification and irreversible inhibition of A2 adenosine receptors. Mol. Pharmacol. 1992 in press. [PMC free article] [PubMed] [Google Scholar]
- 25.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Schuler GD, Altschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 28.Mayo SL, Olafson BD, Goddard WA., III DREIDING: A generic force field for molecular simulations. J. Phys. Chem. 1990;94:8897–8909. [Google Scholar]
- 29.Allen FH, Bellard S, Brice MD, Cartwright BA, Doubleday A, Higgs H, Hummelink T, Hummelink-Peters BG, Kennard O, Motherwell WDS, Rodgers JR, Watson DG. The Cambridge Crystallographic Data Base. Acta Crystallogr. 1979;B35:2331–2339. [Google Scholar]
- 30.Jacobson KA, Daly JW. Purine functionalized congeners as molecular probes for adenosine receptors. Nucleosides & Nucleotides. 1991;10:1029–1038. [Google Scholar]
- 31.Fasman GD, Gilbert WA. The prediction of transmembrane protein sequences and their conformation: An evaluation. Trends Biochem. Sci. 1990;15:89–92. doi: 10.1016/0968-0004(90)90187-g. [DOI] [PubMed] [Google Scholar]
- 32.IJzerman AP, Van Vlijmen HWT. A molecular graphics study exploring a putative ligand binding site of the (β-adrenoceptor. J. Comp.-aided Mol. Design. 1988;2:43–53. doi: 10.1007/BF01532052. [DOI] [PubMed] [Google Scholar]
- 33.Van Vlijmen HWT, IJzerman AP. Molecular modeling of a putative antagonist binding site on helix III of the β-adrenoceptor. J. Comp.-aided Mol. Design. 1989;3:165–174. doi: 10.1007/BF01557726. [DOI] [PubMed] [Google Scholar]
- 34.Van Galen PJM, IJzerman AP, Soudijn W. Adenosine derivatives with N6-alkyl, -alkylamine or -alkyladenosine substituents as probes for the A1 receptor. FEBS Lett. 1987;223:197–201. doi: 10.1016/0014-5793(87)80535-5. [DOI] [PubMed] [Google Scholar]
- 35.Venter JC, Fraser CM, Kerlavage AR, Buck MA. Molecular biology of adrenergic and muscarinic cholinergic receptors. Biochem. Pharmacol. 1989;38:1197–1208. doi: 10.1016/0006-2952(89)90325-0. [DOI] [PubMed] [Google Scholar]
- 36.Mason RP, Rhodes DG, Herbette LG. Reevaluating equilibrium and kinetic binding parameters for lipophilic drugs based on a structural model for drug interaction with biological membranes. J. Med. Chem. 1991;34:869–877. doi: 10.1021/jm00107a001. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson KA, Zimmet J, Schulick R, Barone S, Daly JW, Kirk KL. Adenosine analogs with covalently attached lipids have enhanced potency at A1-adenosine receptors. FEBS Lett. 1987;225:97–102. doi: 10.1016/0014-5793(87)81138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Wenden EM, Van Galen PJM, IJzerman AP, Soudijn W. A model for the hydrogen-bonding interactions between adenosine receptor ligands and histidyl residues in the adenosine A1 receptor binding site, based on AM1 calculations. J. Molec. Struct. (Theochem) 1991;23:175–184. [Google Scholar]
- 39.Taylor MD, Moos WH, Hamilton HW, Szotek DS, Patt WC, Badger EW, Bristol JA, Bruns RF, Heffner TG, Mertz TE. Ribose-modified adenosine analogues as adenosine receptor agonists. J. Med. Chem. 1986;29:346–353. doi: 10.1021/jm00153a008. [DOI] [PubMed] [Google Scholar]
- 40.Lohse NJ, Klotz K-N, Diekmann E, Friedrich K, Schwabe U. 2′,3′-dideoxy-N6-cyclohexyladenosine: An adenosine derivative with antagonist properties at adenosine receptors. Eur. J. Pharmacol. 1988;156:157–160. doi: 10.1016/0014-2999(88)90158-6. [DOI] [PubMed] [Google Scholar]
- 41.Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: Understanding catalysis and immunodeficiency mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 42.Jacobson KA, Ukena D, Padgett W, Kirk KL, Daly JW. Molecular probes for extracellular adenosine receptors. Biochem. Pharmacol. 1987;36:1697–1707. doi: 10.1016/0006-2952(87)90056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrington WW, Jacobson KA, Stiles GL. Demonstration of distinct agonist and antagonist conformations of the A1 adenosine receptor. J. Biol. Chem. 1989;264:13157–13164. [PMC free article] [PubMed] [Google Scholar]
- 44.Trivedi BK, Bristol JA, Bruns RF, Haleen SJ, Steffen RP. N6-(Arylalkyl)adenosines. Identification of N6-(9-fluorenylmethyl)adenosine as a highly potent agonist for the adenosine A2 receptor. J. Med. Chem. 1988;31:271–273. doi: 10.1021/jm00396a044. [DOI] [PubMed] [Google Scholar]
- 45.Trivedi BK. Structure activity relationships for adenosine agonists. In: Jacobson KA, et al., editors. Purities in Cellular Signaling. Targets for New Drugs. New York: Springer-Verlag; 1990. pp. 136–145. [Google Scholar]