Abstract
The fitness landscape of a locus, the array of fitnesses conferred by its alleles, can be affected by allele replacements at other loci, in the presence of epistatic interactions between loci. In a pair of diverging homologous proteins, the initially high probability that an amino acid replacement in one of them will make it more similar to the other declines with time, implying that the fitness landscapes of homologous sites diverge. Here, we use data on within-population non-synonymous polymorphisms and on amino acid replacements between species to study the dynamics, after an amino acid replacement, of the fitness of the ancestral amino acid, and show that selection against its restoration increases with time. This effect can be owing to increase of fitness conferred by the new amino acid occupying the site, and/or to decline of fitness conferred by the replaced amino acid. We show that the fitness conferred by the replaced amino acid rapidly declines, reaching a new lower steady-state level after approximately 20 per cent of amino acids in the protein get replaced. Therefore, amino acid replacements in evolving proteins are routinely involved in negative epistatic interactions with currently absent amino acids, and chisel off the unused parts of the fitness landscape.
Keywords: evolution, fitness landscape dynamics, absent allele, reversing replacement, epistatic interactions
1. Introduction
The local fitness landscape, i.e. the function that relates the sequence of a particular portion of the genome to organismal fitness, can change as a result of evolution of the rest of the genome [1,2]. Consider an amino acid site (locus) x that has recently experienced a replacement of an amino acid (allele) A with an amino acid B. What will be the subsequent dynamics of the fitnesses conferred by different alleles at this site? Subsequent evolution at other sites of the same protein and, perhaps, of other proteins will affect these fitnesses in the following two ways (see the electronic supplementary material, figure S1). First, the fitness conferred by the new amino acid B may increase, because a fraction of these subsequent allele replacements may be adaptive, and any adaptive allele replacement increases the fitnesses of all currently present alleles throughout the genome. Second, the fitness conferred by the old amino acid A may decline, because evolution outside x now proceeds regardless of any possible side effects on the fitness conferred by this replaced amino acid A, and is thus likely to reduce its initially high fitness. It has been observed that the initially high [3–6] rate of convergent evolution, i.e. of amino acid replacements such that the new amino acid coincides with the one occupying the homologous site in a homologous protein, declines as the evolutionary distance between the two proteins increases [6, fig. 5]. This effect can be owing both to the increase of fitness of the new amino acids, or to the decline of fitness of the replaced amino acids, but no attempt has been made to estimate the relative contributions of these two effects.
The fitness conferred by an allele at the locus is the average fitness of all genotypes, within the population, which carry this allele. Therefore, the fitness landscape at a locus can evolve as a result of allele replacements at other loci, as long as there are epistatic interactions between loci [7–10].
2. Material and methods
We used genome-size multiple alignments of genomes of vertebrates and insects from the University of California, Santa Cruz (UCSC) genome database [11] (figure 1). Using the canonical splicing variants of 21 018 UCSC hg19 KnownGenes [12] for vertebrates, or of 13 300 FlyBase genes (BDGP release 5) [13] for insects, we extracted the alignment slices of protein coding regions for the orthologous genes. Single-nucleotide polymorphism data were obtained from dbSNP (release 134) (http://www.ncbi.nlm.nih.gov/projects/SNP/docs/build134.txt) for human, and from the Drosophila Genetic Reference Panel website (http://www.hgsc.bcm.tmc.edu/projects/dgrp/freeze1_July_2010/sequences/) for Drosophila melanogaster. Codon sites with gaps or missing data in any of the species were excluded from the analysis. The total numbers of genes and codons analysed are given in table 1. Lengths of segments of phylogenetic trees were taken from the UCSC Genome Bioinformatics site. All lengths are measured in the genome-average numbers of nucleotide replacements per site. The nucleotides in the internal nodes of the phylogenies were reconstructed using maximum likelihood as implemented in the CodeML program of the PAML package [14]. We mapped the nucleotide replacements to the phylogenetic trees as follows: whenever the nucleotides ascribed to the neighbouring nodes differed, a nucleotide replacement was inferred to have occurred at the edge that connected these two nodes. Throughout this paper, A and B refer to the two amino acids separated by a single-nucleotide substitution in the second position of a codon, and C is either of the one or two remaining amino acids different from both A and B and separated from them by a single-nucleotide substitution in the second position of the codon. For the analysis of polymorphisms, we counted the numbers of second-position nucleotides that experienced a replacement in one of the internal segments of the Homo sapiens (D. melanogaster) lineage, and are currently polymorphic in H. sapiens (D. melanogaster). The replacements were analysed similarly, except instead of the polymorphism in H. sapiens (D. melanogaster), we used the number of replacements in the H. sapiens (D. melanogaster) lineage after its divergence from Mus musculus (Drosophila sechellia), as inferred by CodeML.
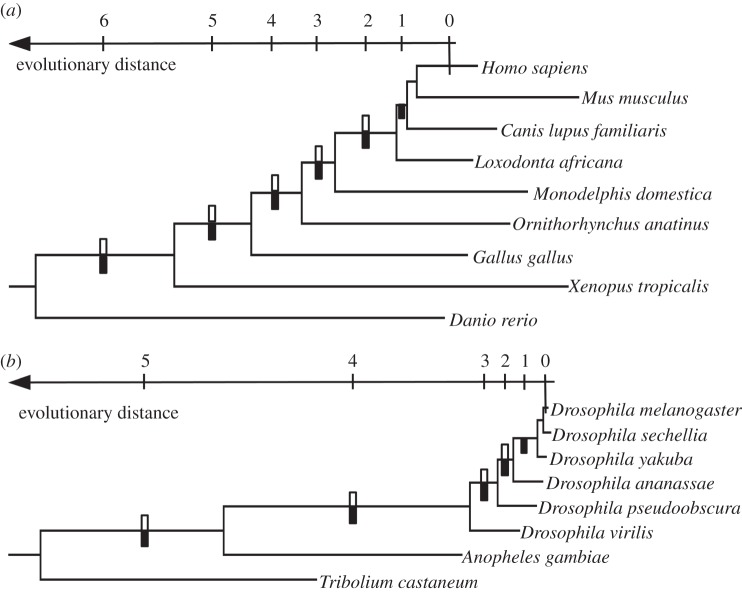
Figure 1.
Phylogenetic trees of species of (a) vertebrates and (b) insects used in the analysis. The horizontal axis shows the evolutionary age of the allele B that arose as a result of an A → B amino acid replacement, assuming that the replacement occurred at the middle of the corresponding phylogenetic branch. Bars denote the branches at which the A → B replacement occurred, for the analysis of polymorphism (black, below the corresponding branches) or divergence (white, above the branches). Six (5) evolutionary distances for the analyses of polymorphism, and 5 (4) evolutionary distances for analyses of reversals, were considered for vertebrates (insects). At sites of A → B replacements, polymorphism in Homo sapiens (Drosophila melanogaster), and the rate of replacements in the H. sapiens lineage after its divergence from Mus musculus (in the D. melanogaster lineage after its divergence from Drosophila sechellia), were measured.
Table 1.
Numbers of analysed genes and codons.
| vertebrates | insects | |
|---|---|---|
| species | 9 | 8 |
| genes | 7967 | 8477 |
| codons | 10 441 107 | 8 838 651 |
The extended methods are presented in the electronic supplementary material.
3. Results
We studied, in vertebrates and in insects, the dependence of the level of polymorphisms and of the rate of replacements on the evolutionary age of the currently present amino acid (figure 1). For a site where a ‘direct’ amino acid replacement A → B occurred at some moment in the past, we compared the frequencies of the ‘reversing’ B → A and ‘orthogonal’ B → C polymorphisms (or replacements), where C is one of the ‘side’ amino acids, i.e. is different from both A and B. The rate of the reversing B → A polymorphisms or replacements can change owing to changes in fitness of either A or B. By contrast, side amino acids C were absent at the site throughout its traceable evolutionary history, and there is no reason to expect the A → B replacement and subsequent evolution to systematically change their fitnesses in either direction; therefore, the rate of orthogonal B → C replacements should change owing to changes only in the fitness of B (see the electronic supplementary material, figure S1).
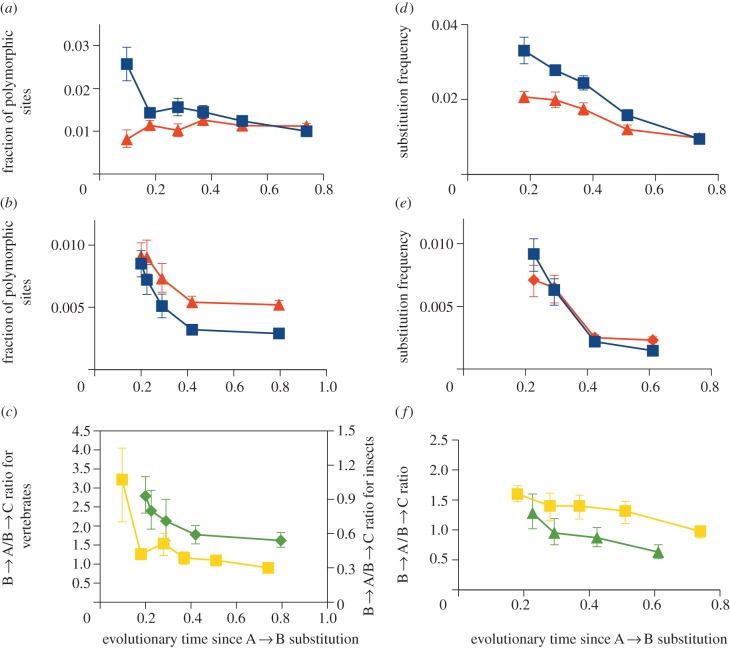
Data on polymorphisms (figure 2a–c) show that the within-population prevalence of the derived ‘old’ allele A, which is recreated by recurrent recent mutations, declines with the age of the currently (nearly) fixed allele B. Within-population prevalence of the derived ‘side’ alleles C is either independent of the age of B (in H. sapiens; figure 2a), or declines with this age (in Drosophila; figure 2b). However, the prevalence of the ancestral allele A declines more steeply than that of the side alleles C. As a result, the ratio of the B → A to B → C polymorphisms drops by a factor of approximately three when the age of B increases from a very young one to such that approximately one nucleotide substitution occurred per synonymous site since the moment of the A → B replacement (figure 2c). Data on amino acid replacements (figure 2d–f) show essentially the same patterns as the polymorphism data. The observed patterns were qualitatively similar when the nucleotide substitution corresponding to the A → B amino acid replacement was a replacement of a ‘weak’ base pair (AT) with a ‘strong’ one (GC), or vice versa (see the electronic supplementary material, figures S2 and S3), arguing against any involvement of biased gene conversion.
Figure 2.
Dependence of the levels of polymorphism (a–c) and of the rates of replacements (d–f) on the age of the currently prevalent allele B, which has become fixed as a result of an A → B amino acid replacement. The horizontal axis corresponds to the time since the A → B replacement, measured in the numbers of nucleotide replacements per fourfold synonymous site. (a,b,d,e) The fraction of the sites, among the sites with an A → B replacement, that carry the A allele (blue) or one of the two C alleles (red) (a,b), or experienced a B → A (blue) or B → C (red) replacement in the terminal lineage (d,e). (c,f) The ratio of the B → A and B → C polymorphisms (c) and replacements (f): yellow, vertebrates; green, insects. (a,d) vertebrates; (b,e) insects. Error bars are 95% CI estimated by bootstrapping.
The patterns were also qualitatively similar when A → B was a biochemically conservative versus radical amino acid replacement, judging by Miyata's distance [15] (see the electronic supplementary material, figures S4 and S5). As expected, when the A → B replacement was conservative, the reversals were more frequent, and the orthogonal polymorphisms or replacements were less frequent, compared with the radical A → B replacements; nevertheless, in all cases, the B → A/B → C ratio declined with time both for polymorphism and for divergence.
4. Discussion
By itself, the fact that the rate of polymorphisms and replacements of B is the highest in the sites in which B has fixed only recently does not necessarily imply any changes in the fitness landscape. Indeed, assume that different sites have different but invariant local fitness landscapes. Then, the set of sites in which an A → B replacement has been observed will be biased towards rapidly evolving sites, and therefore will also have an excess of replacements of B. However, the declining B → A/B → C ratio (figure 2) would be consistent only with different but invariant local fitness landscapes of individual sites if those sites that evolve rapidly allow only a small repertoire of amino acids (which include A and B), and slowly evolving sites can accept a wide variety of amino acids. In fact, the opposite is true [16], because rapidly evolving sites are mostly under relaxed selection [17] and can accept a wider variety of amino acids.
Therefore, the patterns observed in figure 2 do suggest that the relative fitness of different alleles at a site changes with time since an allele replacement. Decline in the rate of the B → C polymorphisms and replacements with the evolutionary time since fixation of the allele B implies that the fitness of the current allele B increases with its age. However, if this increase were the only change in the fitness landscape after an A → B replacement, and the fitnesses of both A and C remained invariant, then the B → A/B → C ratio for replacements and polymorphisms would increase, and not decline, with time, under the reasonable assumption that at the moment of its replacement, the incumbent amino acid A conferred a higher fitness than a never-fixed side amino acid C (see the electronic supplementary material and figure S6). Conversely, if the fitness conferred by the replaced allele A declines with the time since its replacement, then the B → A/B → C ratio is also expected to decline (see the electronic supplementary material and figure S7). Therefore, the observed decline of the B → A/B → C ratio suggests that, while both effects, i.e. the increase of the fitness of B and the decline of the fitness of A, do occur after an A → B replacement, the second effect is stronger, and must be responsible for the bulk of the decline of the rate of convergent evolution with evolutionary distance [6, fig. 5], and of the level of reversing polymorphism and replacements with the age of a direct replacement (figure 2). After time corresponding to approximately one nucleotide substitution per synonymous site lapses since the moment of the A → B replacement, the fitness of the replaced amino acid A reaches a new, lower steady-state value.
The change of the local fitness landscape is a slow process, and probably occurs owing to changes at other sites of the protein, or of other proteins. Because, on average, the ratio of the numbers of non-synonymous replacements and replacements at non-functional genomic sites is approximately 0.1, the patterns in figure 2 imply that a protein ‘forgets’ about a replaced amino acid after roughly 20 per cent of its amino acids get replaced. Of course, these ‘second-site’ replacements do not directly target the fitness of the replaced amino acid A. Instead, their negative impact on the fitness of A must stem from the preponderance of negative epistasis: a replacement is more likely to reduce, than to increase, the fitness of a random high-fitness genotype. This is not surprising: most genotypes have low (zero) fitnesses, and a random perturbation of a local fitness landscape is expected to reduce the fitness conferred by alleles when these alleles are no longer present. Further analysis will be needed to find out whether second-site replacements that reduce the fitnesses of the replaced amino acids are mostly adaptive or nearly neutral.
Acknowledgements
This work was supported by the Russian Ministry of Science and Education (grant no. 11.G34.31.0008 and contract no. P916) and the ‘Molecular and Cellular Biology’ Programme of the Russian Academy of Sciences.
References
- 1.Mustonen V., Lassig M. 2007. Adaptations to fluctuating selection in Drosophila. Proc. Natl Acad. Sci. USA 104, 2277–2282 10.1073/pnas.0607105104 (doi:10.1073/pnas.0607105104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carniero M., Hartl D. L. 2010. Adaptive landscapes and protein evolution. Proc. Natl Acad. Sci. USA 107, 1747–1751 10.1073/pnas.0906192106 (doi:10.1073/pnas.0906192106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazykin G., Kondrashov F., Brudno M., Poliakov A., Dubchak I., Kondrashov A. 2007. Extensive parallelism in protein evolution. Biol. Direct 2, 20. 10.1186/1745-6150-2-20 (doi:10.1186/1745-6150-2-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogozin I., Thomson K., Csürös M., Carmel L., Koonin E. 2008. Homoplasy in genome-wide analysis of rare amino acid replacements: the molecular-evolutionary basis for Vavilov's law of homologous series. Biol. Direct 3, 7. 10.1186/1745-6150-3-7 (doi:10.1186/1745-6150-3-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rokas A., Carroll S. B. 2008. Frequent and widespread parallel evolution of protein sequences. Mol. Biol. Evol. 25, 1943–1953 10.1093/molbev/msn143 (doi:10.1093/molbev/msn143) [DOI] [PubMed] [Google Scholar]
- 6.Povolotskaya I. S., Kondrashov F. A. 2010. Sequence space and the ongoing expansion of the protein universe. Nature 465, 922–926 10.1038/nature09105 (doi:10.1038/nature09105) [DOI] [PubMed] [Google Scholar]
- 7.Poelwijk F. J., Kivet D. J., Weinreich D. M., Tans S. J. 2007. Empirical fitness landscapes reveal accessible paths. Nature 445, 383–386 10.1038/nature05451 (doi:10.1038/nature05451) [DOI] [PubMed] [Google Scholar]
- 8.Khan A. I., Dinh D. M., Schneider D., Lenski R. E., Cooper T. F. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332, 1193–1196 10.1126/science.1203801 (doi:10.1126/science.1203801) [DOI] [PubMed] [Google Scholar]
- 9.Chou H. H., Chiu H. C., Delaney N. F., Segrè D., Marx C. J. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192 10.1126/science.1203799 (doi:10.1126/science.1203799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salverda M. L. M., Dellus E., Gorter F. A., Debets A. J. M., van der Oost J., Hoekstra R. F., Tawfik D. S., de Visser J. A. G. M. 2011. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 7, e1001321. 10.1371/journal.pgen.1001321 (doi:10.1371/journal.pgen.1001321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita P. A., et al. 2010. The UCSC genome browser database: update 2011. Nucleic Acids Res. 39, D876–D882 10.1093/nar/gkq963 (doi:10.1093/nar/gkq963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu F., Kent W. J., Clawson H., Kuhn R. M., Diekhans M., Haussler D. 2006. The UCSC known genes. Bioinformatics 22, 1036–1046 10.1093/bioinformatics/btl048 (doi:10.1093/bioinformatics/btl048) [DOI] [PubMed] [Google Scholar]
- 13.Tweedie S., et al. 2009. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 37, D555–D559 10.1093/nar/gkn788 (doi:10.1093/nar/gkn788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 10.1093/molbev/msm088 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 15.Miyata T., Miyazawa S., Yasunaga T. 1979. Two types of amino acid substitutions in protein evolution. J. Mol. Evol. 12, 219–236 10.1007/bf01732340 (doi:10.1007/bf01732340) [DOI] [PubMed] [Google Scholar]
- 16.Naumenko S., Kondrashov A. 2012. Rate and breadth of protein evolution are only weakly correlated. Biol. Direct 7, 8. 10.1186/1745-6150-7-8 (doi:10.1186/1745-6150-7-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura M. 1983. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press [Google Scholar]