Abstract
Monitor lizards are emblematic reptiles that are widely distributed in the Old World. Although relatively well studied in vertebrate research, their biogeographic history is still controversial. We constructed a molecular dataset for 54 anguimorph species, including representatives of all families with detailed sampling of the Varanidae (38 species). Our results are consistent with an Asian origin of the Varanidae followed by a dispersal to Africa 41 (49–33) Ma, possibly via an Iranian route. Another major event was the dispersal of monitors to Australia in the Late Eocene–Oligocene 32 (39–26) Ma. This divergence estimate adds to the suggestion that Australia was colonized by several squamate lineages prior to the collision of the Australian plate with the Asian plate starting 25 Ma.
Keywords: biogeography, squamates, Varanus, Cenozoic
1. Introduction
Anguimorph lizards (203 species) belong to the Toxicofera clade of squamate reptiles together with iguanians and snakes [1]. They comprise two major lineages with different geographical distributions: the Old World Paleoanguimorpha, and the primarily New World Neoanguimorpha [2]. The Neoanguimorpha includes Anguidae (67 sp.), Anniellidae (two sp.), Diploglossidae (51 sp.), Helodermatidae (two sp.) and Xenosauridae (six sp.); while Paleoanguimorpha includes Shinisauridae (one sp.), Lanthanotidae (one sp.) and Varanidae (73 sp.) [2,3]. Anguimorphs have a Laurasian origin [2], but the biogeographic history of the largest family, the Varanidae, is controversial.
Extant Varanus are distributed in Africa, the Arabian Peninsula and central Asia (six ‘African’ species); southern mainland Asia and Malaysian and Indonesian islands (40 ‘Indo-Asian’ species); and New Guinea, Solomon Islands and Australia (27 ‘Indo-Australian’ species). The oldest known Varanus is from the Late Eocene–Early Oligocene of Africa ([4,5], J. C. Rage 2012, personal communication). Three different biogeographic scenarios have been proposed for Varanus: (i) an Asian origin followed by dispersals to Africa and Australasia in the Cenozoic [6–8], (ii) an African origin followed by a dispersal to Asia and Australia in the Cenozoic [5], and (iii) a Gondwanan origin followed by vicariant events tied to Jurassic and Early Cretaceous plate movements [9]. To address these questions, we infer a timetree of anguimorphs from a molecular dataset including nuclear and mitochondrial genes to overcome the inherent limitation of mitochondrial chronograms for inferring evolutionary histories.
2. Material and methods
We constructed a molecular genetic dataset for 54 anguimorph species including representatives of all families, with detailed sampling of the Varanidae (38 species). The dataset comprised three nuclear protein-coding genes (brain-derived neurotrophic factor: (BDNF), bone morphogenetic protein 2: (BMP2) and neurotrophin 3: (NT3); 1914 nuclear gene sites) and two mitochondrial protein-coding genes (ND1 and ND2; 1995 mtDNA sites) for 55 taxa (the 140 nuclear sequences that were newly determined have been deposited in GenBank under accession numbers JQ844905–JQ845044). Phylogenies were built using probabilistic approaches (maximum-likelihood (ML) and Bayesian inferences). Dating analyses were performed according to the Bayesian relaxed molecular clock approach. We used BEAST v. 1.7 [10] with uncorrelated lognormal rate model and Yule speciation prior. A series of uniform and exponential dating calibration priors were used. The dates presented in the paper are mean and 95% confidence interval from the ‘inclusive’ calibration analysis (see electronic supplementary material for details).
3. Results and discussion
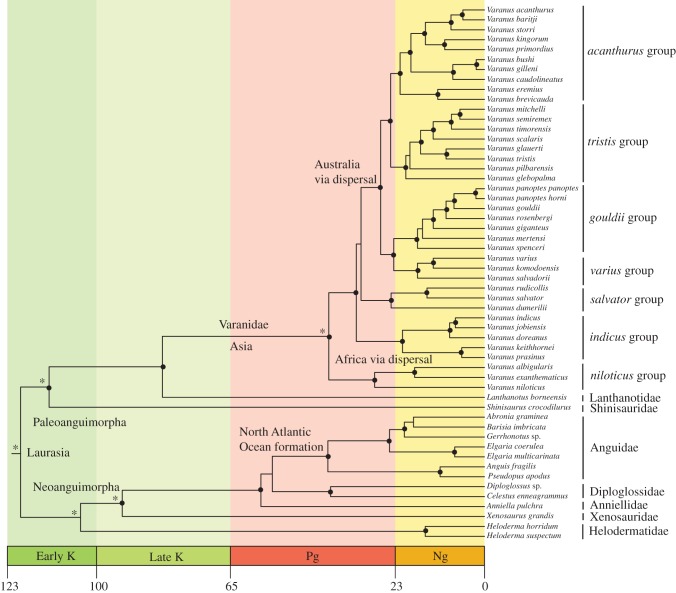
The ML and Bayesian trees are identical (figure 1). The major split between Paleoanguimorpha and Neoanguimorpha is retrieved here. Within Neoanguimorpha, the following successive branching order is supported: Helodermatidae, Xenosauridae, Anniellidae, Diploglossidae and Anguidae, a result in accordance with Wiens et al. [11]. The two Anguidae subfamilies, Anguinae and Gerrhonotinae, are each found to be monophyletic as in Macey et al. [12]. Within Paleoanguimorpha, Shinisauridae and Lanthanotidae are successive sister lineages to the Varanidae.
Figure 1.
A BEAST timetree of anguimorph lizards based on analysis of DNA sequences from three nuclear protein-coding genes and two mitochondrial protein-coding genes. Inferred biogeographic events are indicated at nodes on the timetree. Nodes with black circles are supported by PP >95% and ML BP >70%. Nodes with asterisks are calibrating nodes. Labels on timescale are J, Jurassic; K, Cretaceous; Pg, Paleogene; Ng, Neogene.
Within Varanidae, the traditionally recognized groups are recovered: an African ‘niloticus group’ that is sister to the Indo-Asian and the Indo-Australian groups. Their relationships are strongly supported with one exception: the clustering of the ‘salvator group’ with the Indo-Australian group (ML bootstrap proportion (BP): 58%, Bayesian posterior probability (PP): 66%). Within the Indo-Australian group, four major groups are found: the ‘varius group’ which clusters with the ‘gouldii group’ and the ‘acanthurus group’ which clusters with the ‘tristis group’. These relationships within Varanus are congruent with studies based on mitochondrial data only [6,13–16]. It is interesting to note that independent evidence derived from genital morphological characters supports several molecular nodes both at the interfamilial (Shinisauridae/Xenosauridae) and intrageneric (Varanus) level [17].
The timetree shows that the divergence between the Paleoanguimorpha and the Neoanguimorpha happened in the Early Cretaceous, 121 (138–107) Ma, possibly reflecting a Laurasian vicariant event. A striking result is the presence of four relict anguimorph lineages dating back to the Cretaceous between 133 and 69 Myr ago: the Shinisauridae and the Lanthanotidae in the Old World and the Helodermatidae and the Xenosauridae in the New World. The remaining anguimorph divergences have happened much later, in the Cenozoic. Among Anguidae, the split between the Anguinae that originated in the Old World and the American Gerrhonotinae is dated at 41 (51–32) Ma, which corresponds to the formation of the North Atlantic Ocean, a hypothesis already proposed by Macey et al. [12].
As their two successive outgroups, the Lanthanotidae and the Shinisauridae, are Asian, we infer an Asian origin of the Varanidae (and Varanus) followed by a dispersal to Africa 41 (49–33) Ma, probably via an Iranian route that was the filtering precursor of the definitive connection between Eurasia and Africa [18]. The lack of monitor lizards in Madagascar adds biogeographical support to this relatively recent immigration to Africa. Moreover, this time estimate is very similar to the one obtained by Portik & Papenfuss [19], who used ND2 and RAG1, and dated the split between African and Asian Varanus at 40 Ma. We therefore refute the Gondwanan vicariant hypothesis proposed by Schulte et al. [9], and agree with the Cenozoic dispersal hypothesis of Hugall & Lee [20], Sweet & Pianka [7] and Amer & Kumazawa [8]. Our time estimates are younger than those obtained in the latter study based on mitochondrial genes only, which proposed dispersal to Africa between 60 and 47 Myr ago. In any case, an Asian origin for varanid monitor lizards implies that all of the lineages older than the extant dispersal to Africa must have gone extinct.
Another major event is the dispersal of monitors to Australia in the Late Eocene–Oligocene 32 (39–26) Ma. These divergence estimates are similar to those obtained for blindsnakes, pythons, agamid lizards and the gekkonid lizard Gehyra [20–23], and suggest that Australia was colonized by these groups prior to the collision of the Australian plate with the Asian plate which started 25 Ma [24].
Acknowledgments
This work was funded by grants from the Service de Systématique Moléculaire du Muséum National d'Histoire Naturelle to N.V., J.M. and J.S., and by the Consortium National de Recherche en Génomique, Genoscope. We thank the president of the Muséum National d'Histoire Naturelle for support to J.S. We thank those persons and institutions who contributed some of the tissue and DNA: K. Daoues, R. Macey and T. Papenfuss. We thank J.-C. Rage for help with palaeontological issues.
References
- 1.Vidal N., Hedges S. B. 2005. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C. R. Biologies 328, 1000–1008 10.1016/j.crvi.2005.10.001 (doi:10.1016/j.crvi.2005.10.001) [DOI] [PubMed] [Google Scholar]
- 2.Vidal N., Hedges S. B. 2009. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C. R. Biologies 332, 129–139 10.1016/j.crvi.2008.07.010 (doi:10.1016/j.crvi.2008.07.010) [DOI] [PubMed] [Google Scholar]
- 3.Uetz P., Hosek J., Hallerman J. 2011. The reptile database. See http://www.reptile-database.org (accessed 30 December 2011) [Google Scholar]
- 4.Smith K. T., Bhullar B.-A. S., Holroyd P. A. 2008. Earliest African record of the Varanus stem-clade (Squamata: Varanidae) from the early Oligocene of Egypt . J. Vert. Pal. 28, 909–913 10.1671/0272-4634(2008)28[909:EAROTV]2.0.CO;2 (doi:10.1671/0272-4634(2008)28[909:EAROTV]2.0.CO;2) [DOI] [Google Scholar]
- 5.Holmes R. B., Murray A. M., Attia Y. S., Simons E. L., Chatrath P. 2010. Oldest known Varanus (Squamata: Varanidae) from the Upper Eocene and Lower Oligocene of Egypt: support for an African origin of the genus. Palaeontology 53, 1099–1110 10.1111/j.1475-4983.2010.00994.x (doi:10.1111/j.1475-4983.2010.00994.x) [DOI] [Google Scholar]
- 6.Fuller S., Baverstock P., King D. 1998. Biogeographic origins of goannas (Varanidae): a molecular perspective. Mol. Phylogenet. Evol. 9, 294–307 10.1006/mpev.1997.0476 (doi:10.1006/mpev.1997.0476) [DOI] [PubMed] [Google Scholar]
- 7.Sweet S. S., Pianka E. R. 2007. Monitors, mammals, and Wallace's line. Mertensiella 16, 79–99 [Google Scholar]
- 8.Amer S. A. M., Kumazawa Y. 2008. Timing of a mtDNA gene rearrangement and intercontinental dispersal of varanid lizards. Genes Genet. Syst. 83, 275–280 10.1266/ggs.83.275 (doi:10.1266/ggs.83.275) [DOI] [PubMed] [Google Scholar]
- 9.Schulte J. A., II, Melville J., Larson A. 2003. Molecular phylogenetic evidence for ancient divergence of lizard taxa on either side of Wallace's line. Proc. R. Soc. Lond. B 270, 597–603 10.1098/rspb.2002.2272 (doi:10.1098/rspb.2002.2272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond A. J., Suchard M. A., Xie D., Rambaut A. In press. Bayesian phylogenetics with BEAUTi and the BEAST v. 1.7. Mol. Biol. Evol. 10.1093/molbev/mss075 (doi:10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiens J. J., Kuczynski C. A., Townsend T., Reeder T. W., Mulcahy D. G., Sites J. W., Jr 2010. Combining phylogenomics and fossils in higher-level squamate reptile phylogeny: molecular data change the placement of fossil taxa. Syst. Biol. 59, 674–688 10.1093/sysbio/syq048 (doi:10.1093/sysbio/syq048) [DOI] [PubMed] [Google Scholar]
- 12.Macey J. R., Schulte J. A., II, Larson A., Tuniyev B. S., Orlov N., Papenfuss T. J. 1999. Molecular phylogenetics, tRNA evolution, and historical biogeography in anguid lizards and related taxonomic families. Mol. Phylogenet. Evol. 12, 250–272 10.1006/mpev.1999.0615 (doi:10.1006/mpev.1999.0615) [DOI] [PubMed] [Google Scholar]
- 13.Ast J. C. 2001. Mitochondrial DNA evidence and evolution in Varanoidea (Squamata). Cladistics 17, 211–226 [DOI] [PubMed] [Google Scholar]
- 14.Fitch A. J., Goodman A. E., Donnellan S. C. 2006. A molecular phylogeny of the Australian monitor lizards (Squamata: Varanidae) inferred from mitochondrial DNA sequences. Aust. J. Zool. 54, 253–269 10.1071/ZO05038 (doi:10.1071/ZO05038) [DOI] [Google Scholar]
- 15.Thompson G. G., Clemente C. J., Withers P. C., Fry B. G., Norman J. A. 2008. Is body shape of varanid lizards linked with retreat choice? Aust. J. Zool. 56, 351–362 10.1071/ZO08030 (doi:10.1071/ZO08030) [DOI] [Google Scholar]
- 16.Collar D. C., Schulte J. A., Losos J. B. 2011. Evolution of extreme body size disparity in monitor lizards (Varanus). Evolution 65, 2664–2680 10.1111/j.1558-5646.2011.01335.x (doi:10.1111/j.1558-5646.2011.01335.x) [DOI] [PubMed] [Google Scholar]
- 17.Böhme W., Ziegler T. 2009. A review of iguanian and anguimorph lizard genitalia (Squamata: Chamaeleonidae; Varanoidea, Shinisauridae, Xenosauridae, Anguidae) and their phylogenetic significance: comparisons with molecular data sets. J. Zool. Syst. Evol. Res. 47, 189–202 10.1111/j.1439-0469.2008.00495.x (doi:10.1111/j.1439-0469.2008.00495.x) [DOI] [Google Scholar]
- 18.Gheerbrant E., Rage J. C. 2006. Paleobiogeography of Africa: how distinct from Gondwana and Laurasia? Palaeogeogr. Palaeoclimatol. Palaeoecol. 241, 224–246 10.1016/j.palaeo.2006.03.016 (doi:10.1016/j.palaeo.2006.03.016) [DOI] [Google Scholar]
- 19.Portik D. M., Papenfuss T. J. 2012. Monitors cross the Red Sea: the biogeographic history of Varanus yemenensis. Mol. Phylogenet. Evol. 62, 561–565 10.1016/j.ympev.2011.09.024 (doi:10.1016/j.ympev.2011.09.024) [DOI] [PubMed] [Google Scholar]
- 20.Hugall A. F., Lee M. S. Y. 2004. Molecular claims of Gondwanan age for Australian agamid lizards are untenable. Mol. Biol. Evol. 21, 2102–2110 10.1093/molbev/msh219 (doi:10.1093/molbev/msh219) [DOI] [PubMed] [Google Scholar]
- 21.Rawlings L. H., Rabosky D. L., Donnellan S. C., Hutchinson M. N. 2008. Python phylogenetics: inference from morphology and mitochondrial DNA. Biol. J. Linn. Soc. 93, 603–619 10.1111/j.1095-8312.2007.00904.x (doi:10.1111/j.1095-8312.2007.00904.x) [DOI] [Google Scholar]
- 22.Vidal N., et al. 2010. Blindsnake evolutionary tree reveals long history on Gondwana. Biol. Lett. 6, 558–561 10.1098/rsbl.2010.0220 (doi:10.1098/rsbl.2010.0220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinicke M. P., Greenbaum E., Jackman T. R., Bauer A. M. 2011. Phylogeny of a trans–Wallacean radiation (Squamata, Gekkonidae, Gehyra) supports a single early colonization of Australia. Zool. Scripta 40, 584–602 10.1111/j.1463-6409.2011.00495.x (doi:10.1111/j.1463-6409.2011.00495.x) [DOI] [Google Scholar]
- 24.Hall R. 2002. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer–based reconstructions, model and animations. J. Asian Earth Sci. 20, 353–431 10.1016/S1367-9120(01)00069-4 (doi:10.1016/S1367-9120(01)00069-4) [DOI] [Google Scholar]