Abstract
Inbreeding depression (i.e. negative fitness effects of inbreeding) is central in evolutionary biology, affecting numerous aspects of population dynamics and demography, such as the evolution of mating systems, dispersal behaviour and the genetics of quantitative traits. Inbreeding depression is commonly observed in animals and plants. Here, we demonstrate that, in addition to genetic processes, epigenetic processes may play an important role in causing inbreeding effects. We compared epigenetic markers of outbred and inbred offspring of the perennial plant Scabiosa columbaria and found that inbreeding increases DNA methylation. Moreover, we found that inbreeding depression disappears when epigenetic variation is modified by treatment with a demethylation agent, linking inbreeding depression firmly to epigenetic variation. Our results suggest an as yet unknown mechanism for inbreeding effects and demonstrate the importance of evaluating the role of epigenetic processes in inbreeding depression.
Keywords: DNA methylation, epigenetics, inbreeding depression, Scabiosa columbaria, 5-azacytidine
1. Introduction
Negative effects of inbreeding have until now been exclusively explained by classic genetic theories: the partial dominance hypothesis, i.e. the expression of deleterious recessive alleles owing to increased homozygosity in inbred individuals [1]; and the overdominance hypothesis, i.e. the reduced frequency of superior heterozygote genotypes [1]. In addition, interacting effects between alleles at different loci (i.e. epistasis) have been suggested to contribute to inbreeding depression [2], as is interpreted from nonlinear relationships between inbreeding level and fitness [3]. There is consensus that partial dominance is the most likely explanation in most cases.
Inbreeding effects are often found to be environmentally dependent [4–8]. Environment-dependent inbreeding depression can be explained by different mechanisms [5,7], including the conditional expression of deleterious alleles, and the different levels of phenotypic plasticity expressed by outbred and inbred individuals [9]. We know that epigenetic modifications, chemical modifications to the DNA or histones that alter or regulate gene activity, are affected by environmental conditions and may modulate plasticity [10]. Some of these environmentally induced modifications are heritable over multiple generations [11] and variation in these modifications among individuals and populations [12,13] has been related to differences in phenotype [14], development and even mortality [15]. It has therefore been suggested that epigenetic processes are involved in the regulation of inbreeding effects [16,17]. However, empirical proof for this is, as yet, not available.
To investigate the link between inbreeding, epigenetic processes and inbreeding depression, we studied the perennial plant Scabiosa columbaria, a species known to suffer severely from inbreeding depression [18]. We tested whether inbred and outbred plants differ in levels of DNA methylation and whether modifying DNA methylation of inbred and outbred plants affects the phenotypic differences observed between inbred and outbred individuals.
2. Methods
Scabiosa columbaria is a self-compatible but predominantly outcrossing species with outcrossing rates close to 1 in natural populations [19]. In 2009, seeds were collected from a large French population (more than 100 000 individuals). These seeds were germinated and grown until flowering. The individual plants were both selfed and outcrossed by pollinating at least four flower heads per plant to create inbred and outbred siblings. From the F1 generation, 75 plants (38 outbred and 37 inbred siblings from six families, ca six to eight replicates per family with inbreeding coefficients of 0 and 0.5, respectively) were grown until flowering on a 1:3 compost:sand mixture, day/night temperatures of 24°C/18°C, 16 h photoperiod and 60 per cent relative humidity. To investigate the effect of inbreeding on DNA methylation, a key epigenetic mechanism [20], we experimentally demethylated half of our plants (ca three to four replicates per family) by germinating seeds on filter paper saturated with a daily-refreshed 50 µM 5-azacytidine [21] solution, applied for 9 days. The demethylating effect of 5-azacytidine has been demonstrated by others [21–23].
After three months, plant biomass was estimated non-destructively as the product of leaf number and the length and width of the largest leaf, which was highly correlated with actual biomass in a separate set of plants (Pearson correlation, R2 = 0.79, n = 96, p < 0.001). Photosynthesis light responses from 1500 to 0 photosynthetically active radiation (PAR) were measured between 10.00 and 15.00 on two and a half month-old plants using a LiCor LI-6400 (Lincoln, NB, USA), at 20°C, 60 per cent relative humidity, and 400 µmol mol−1 CO2 as reference. Photosynthetic efficiency was determined by fitting the slope between 0 and 60 PAR. Inbreeding depression coefficients were calculated as δ = (WO − WS)/WO, with WO and WS the mean values of a particular trait of outcrossed and selfed offspring, respectively. Inbreeding depression coefficients for bolting time were multiplied by −1 because high trait values indicate poor performance.
Mixed effect models (R-2.14.1; lme-package) with family included as random effect were used to test for effects of crossing type, demethylation and their interaction on the response variables. Model validation gave no indication of nonlinearity. Effects on bolting time were analysed using a Poisson distribution (R-2.14.1; lmer-package). Significances of explanatory factors were assessed by comparing the minimum adequate model with a reduced model using the likelihood ratio test.
DNA methylation of cytosine was analysed in leaf samples, as a measure of epigenetic variation, using a methylation-sensitive amplification polymorphism (MSAP) technique modified after Keyte et al. [12]. MSAP analyses were performed on 200 ng gDNA on a subset of 24 plants (in duplo) using six selective primer combinations (EcoR1-AAC/ACA+MspI/HpaII-TAC/TCA, EcoR1-ACA+MspI/HpaII-TAG, EcoR1-AAC+MspI/HpaII-TGC) on a Beckman CEQ8800 sequencer (Beckman Coulter). Fragments were scored as ‘non-methylated’ (fragment in EcoR1-HpaII and EcoR1-MspI) or ‘methylated’ (fragment in EcoR1-HpaII or EcoR1-MspI) using GeneMarker (SoftGenetics) (see electronic supplementary materials).
3. Results
As expected [24], F1 inbred plants showed a reduced performance. Inbreeding depression was observed for leaf number, biomass and photosynthetic efficiency, but not for bolting time (table 1). In outbred plants, a mean methylation percentage of 42 per cent (range 25–60%) was found, similar to the percentages observed in other plant species [12,25,26]. Methylation levels of inbred plants, however, were higher by almost 10 per cent compared with outbred plants (F1,19 = 6.36, p = 0.021; figure 1). 5-Azacytidine reduced DNA methylation in S. columbaria by 11 per cent, measured on a separate group of similar-aged plants and grown under similar conditions to the plants in our experiment (F1,22 = 8.11, p = 0.009), comparable with the reduction observed in other species [22,23].
Table 1.
Inbreeding depression in Scabiosa columbaria. Inbreeding depression coefficients in different plant traits. Positive numbers indicate inbreeding depression.
| inbreeding depression coefficient | statistics |
|||
|---|---|---|---|---|
| mean ± s.e.m. | ttwo-tailed | nfamilies | p-value | |
| leaf number | 0.23 ± 0.04 | 5.11 | 6 | 0.004 |
| biomass | 0.46 ± 0.05 | 9.40 | 6 | <0.001 |
| photosynthetic efficiency | 0.25 ± 0.04 | 6.15 | 6 | 0.002 |
| bolting time | 0.11 ± 0.27 | 0.81 | 4 | 0.476 |
Figure 1.
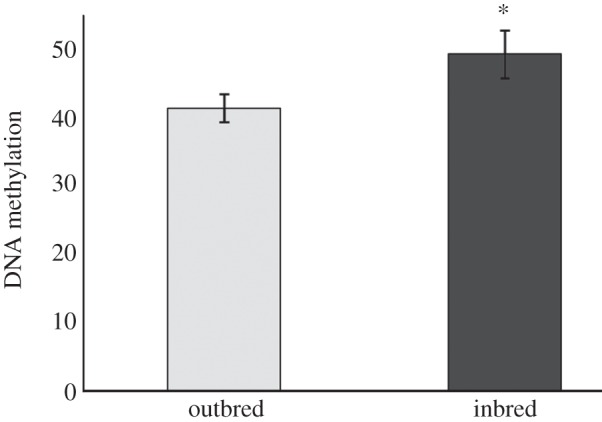
Epigenetic variation in outbred and inbred plants. Percentage of DNA methylation (mean ± s.e.m.), estimated by the percentage of methylated fragments in 198 MSAP-fragments.
Treatment with 5-azacytidine reduced the mean values of some, but not all, traits in outbred plants. Demethylated outbred plants were on average 29 per cent smaller and bolted 10 days later when compared with control outbred plants (figure 2). Intriguingly, while 5-azacytidine decreased biomass in outbred plants, it increased the average biomass of inbred plants (table 2). In addition, partial demethylation did not affect photosynthetic efficiency and leaf number of outbred plants, but it restored the inbred trait values to the level of the control outbred plants (figure 2). Although biomass in demethylated inbred plants was not completely restored to the level of control outbred plants (figure 2), partial demethylation did significantly increase biomass of inbred plants compared with control inbred plants (F1,28 = 6.65, p = 0.016).
Figure 2.

Effects of demethylation on different plant traits. Photosynthetic efficiency (solid lines), biomass (dashed lines), leaf number (square-dotted lines), and bolting time (long-dashed-dotted lines) of outbred (circles with grey lines) and inbred (triangles with black lines) plants treated with and without 5-azacytidine (mean ± s.e.m.). For statistical results, see table 2.
Table 2.
Summary of mixed effect models. Effects of crossing type, demethylation and their interaction on different plant traits. d.f.1,2: degrees of freedom of explanatory factor and error term, respectively. Significant values (α = 0.05) are indicated in bold (see electronic supplementary materials).
| d.f.1,2 | crossing |
demethylation |
crossing × demethylation |
||||
|---|---|---|---|---|---|---|---|
| F | p-value | F | p-value | F | p-value | ||
| leaf number | 1,64 | 2.307 | 0.134 | 7.162 | 0.009 | 3.993 | 0.050 |
| biomass | 1,64 | 11.982 | 0.001 | 0.102 | 0.751 | 10.229 | 0.002 |
| photosynthetic efficiency | 1,64 | 6.534 | 0.013 | 11.199 | 0.001 | 8.672 | 0.005 |
| bolting timea | 1.247 | 0.264 | 29.504 | <0.001 | 2.474 | 0.116 | |
aFor bolting time, χ2-values were shown (see §2).
4. Discussion
Strong phenotypic effects were observed in response to experimental demethylation, consistent with observed effects of demethylation in other studies [22,27]. Intriguingly, our results show a direct effect of inbreeding on epigenetic markers: Inbreeding resulted not only in inbreeding depression for fitness-related traits, but also increased DNA methylation. Most interestingly, when the increased methylation level in inbreds was restored to the outbred level, inbreeding depression was completely (photosynthetic efficiency, leaf number) or almost completely (biomass) eliminated. This strongly suggests that in these cases DNA methylation mediates the negative effects of inbreeding. This suggestion is further enhanced by the observation that, in a trait without inbreeding depression (bolting time), demethylation did not change relative performance. These results underline the potentially important role of DNA methylation in determining the level of inbreeding depression. At the same time, the incomplete restoration of biomass demonstrates that other genetic and/or epigenetic factors contribute to inbreeding effects.
In addition, the observed link between epigenetic variation and inbreeding depression suggests that environment-dependent inbreeding depression may at least partly be explained via epigenetic processes, providing a new explanation for the interaction between environment and inbreeding depression. Since this experiment was conducted in a single environment, we are not able to distinguish plasticity effects from conditionally expressed alleles and fitness [6]. Our results inspire further research on this.
To our knowledge, there is no clear-cut mechanistic explanation for the observed interplay between epigenetic variation, inbreeding and inbreeding depression. It has been suggested that the increased homozygosity that results from inbreeding may disrupt epigenetic crosstalk between alleles at the same locus [16,17]. This could cause partial inappropriate silencing of genes and consequently result in inbreeding depression. In mammals, for example, one of the two copies of the X-chromosome in females is silenced. This inactivation is regulated epigenetically [28]. Another potential explanation may be that inbreeding disrupts the enzymatic machinery involved in the induction and maintenance of cytosine methylation. This machinery builds on complex genomic and transcriptomic interactions in which methylation maintenance enzymes such as MET1 and CMT3, enzymes for de novo methylation of cytosines such as domains-rearranged methyltransferase (DRM), and regulation via small RNAs and DICER-like enzymes are involved [29]. In addition, interactions with histone modifications are thought to play a role [29]. If deleterious recessive alleles are present at loci that code for enzymes involved in this machinery, inbreeding would result in non-adaptive methylation patterns. However, at present it remains unknown whether a possible disruption of the enzymatic epigenetic machinery can lead to increased methylation as a consequence of inbreeding, and whether this is compatible with the observation that different traits respond differently to inbreeding.
Our results provide new insights into the mechanisms of inbreeding effects. The relative importance of epigenetic compared with genetic effects, however, needs to be tested in future studies. Other studies have shown that Mendelian factors such as recessive deleterious alleles contribute to inbreeding depression (see references in [16]). In addition, other species, such as Drosophila, show high levels of inbreeding depression without significant methylation of their DNA ([30], but see [31]). In these species, genetic or other epigenetic processes [17] may contribute more to inbreeding effects.
To conclude, in addition to genetic effects that are known to contribute strongly to inbreeding depression, our results provide strong evidence that epigenetic processes may play an important role.
Acknowledgments
We thank C. Biémont, R. K. Butlin, O. Bossdorf, J. Eygensteyn, H. de Kroon, E. S. Pierson, V. Latzel, K. J. F. Verhoeven and E. J. W. Visser for help. This work, as part of the ESF EUROCORES Programme EuroEEFG, was supported by the Dutch National Science Foundation (Veni-NWO-863.08.029).
References
- 1.Charlesworth B., Charlesworth D. 1999. The genetic basis of inbreeding depression. Genet. Res. 74, 329–340 10.1017/s0016672399004152 (doi:10.1017/s0016672399004152) [DOI] [PubMed] [Google Scholar]
- 2.Carr D. E., Dudash M. R. 2003. Recent approaches into the genetic basis of inbreeding depression in plants. Phil. Trans. R. Soc. Lond. B 358, 1071–1084 10.1098/rstb.2003.1295 (doi:10.1098/rstb.2003.1295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pray L. A., Goodnight C. J. 1995. Genetic variation in inbreeding depression in the red flour beetle Tribolium castaneum. Evolution 49, 176–188 10.2307/2410303 (doi:10.2307/2410303) [DOI] [PubMed] [Google Scholar]
- 4.Kristensen T. N., Pedersen K. S., Vermeulen C. J., Loeschcke V. 2010. Research on inbreeding in the ‘omic’ era. Trends Ecol. Evol. 25, 44–52 10.1016/j.tree.2009.06.014 (doi:10.1016/j.tree.2009.06.014) [DOI] [PubMed] [Google Scholar]
- 5.Armbruster P., Reed D. H. 2005. Inbreeding depression in benign and stressful environments. Heredity 95, 235–242 10.1038/sj.hdy.6800721 (doi:10.1038/sj.hdy.6800721) [DOI] [PubMed] [Google Scholar]
- 6.Cheptou P. O., Donohue K. 2011. Environment-dependent inbreeding depression: its ecological and evolutionary significance. New Phytol. 189, 395–407 10.1111/j.1469-8137.2010.03541.x (doi:10.1111/j.1469-8137.2010.03541.x) [DOI] [PubMed] [Google Scholar]
- 7.Fox C. W., Reed D. H. 2011. Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution 65, 246–258 10.1111/j.1558-5646.2010.01108.x (doi:10.1111/j.1558-5646.2010.01108.x) [DOI] [PubMed] [Google Scholar]
- 8.Reed D. H., Fox C. W., Enders L. S., Kristensen T. N. 2012. Inbreeding–stress interactions: evolutionary and conservation consequences. Ann. N. Y. Acad. Sci. 1256, 33–48 10.1111/j.1749-6632.2012.06548.x (doi:10.1111/j.1749-6632.2012.06548.x) [DOI] [PubMed] [Google Scholar]
- 9.Auld J. R., Relyea R. A. 2010. Inbreeding depression in adaptive plasticity under predation risk in a freshwater snail. Biol. Lett. 6, 222–224 10.1098/rsbl.2009.0726 (doi:10.1098/rsbl.2009.0726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossdorf O., Richards C. L., Pigliucci M. 2008. Epigenetics for ecologists. Ecol. Lett. 11, 106–115 10.1111/j.1461-0248.2007.01130.x (doi:10.1111/j.1461-0248.2007.01130.x) [DOI] [PubMed] [Google Scholar]
- 11.Johannes F., et al. 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5, e1000530. 10.1371/journal.pgen.1000530 (doi:10.1371/journal.pgen.1000530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keyte A. L., Percifield R., Liu B., Wendel J. F. 2006. Intraspecific DNA methylation polymorphism in cotton (Gossypium hirsutum L.). J. Hered. 97, 444–450 10.1093/jhered/es1023 (doi:10.1093/jhered/es1023) [DOI] [PubMed] [Google Scholar]
- 13.Vaughn M. W., et al. 2007. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5, 1617–1629 10.1371/journal.pbio.0050174 (doi:10.1371/journal.pbio.0050174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jablonka E., Lamb M. J. 2005. Evolution in four dimensions: genetic, epigenetic, behavioral, and symbolic variation in the history of life. Cambridge, MA: MIT Press [Google Scholar]
- 15.Li E., Bestor T. H., Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 10.1016/0092-8674(92)90611-f (doi:10.1016/0092-8674(92)90611-f) [DOI] [PubMed] [Google Scholar]
- 16.Charlesworth D., Willis J. H. 2009. Fundamental concepts in genetics. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 10.1038/nrg2664 (doi:10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 17.Biemont C. 2010. Inbreeding effects in the epigenetic era. Nat. Rev. Genet. 11, 234. 10.1038/nrg2664-c1 (doi:10.1038/nrg2664-c1) [DOI] [PubMed] [Google Scholar]
- 18.Vantreuren R., Bijlsma R., Ouborg N. J., Vandelden W. 1993. The significance of genetic erosion in the process of extinction. IV. Inbreeding depression and heterosis effects caused by selfing and outcrossing in Scabiosa columbaria. Evolution 47, 1669–1680 10.2307/2410211 (doi:10.2307/2410211) [DOI] [PubMed] [Google Scholar]
- 19.Vantreuren R., Bijlsma R., Ouborg N. J., Kwak M. M. 1994. Relationships between plant-density, outcrossing rates and seed set in natural and experimental populations of Scabiosa columbaria. J. Evol. Biol. 7, 287–302 10.1046/j.1420-9101.1994.7030287.x (doi:10.1046/j.1420-9101.1994.7030287.x) [DOI] [Google Scholar]
- 20.Jaenisch R., Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 10.1038/ng1089 (doi:10.1038/ng1089) [DOI] [PubMed] [Google Scholar]
- 21.Jones P. A. 1985. Altering gene-expression with 5-azacytidine. Cell 40, 485–486 10.1016/0092-8674(85)90192-8 (doi:10.1016/0092-8674(85)90192-8) [DOI] [PubMed] [Google Scholar]
- 22.Fieldes M. A., Schaeffer S. M., Krech M. J., Brown J. C. L. 2005. DNA hypomethylation in 5-azacytidine-induced early-flowering lines of flax. Theor. Appl. Genet. 111, 136–149 10.1007/s00122-005-2005-9 (doi:10.1007/s00122-005-2005-9) [DOI] [PubMed] [Google Scholar]
- 23.Tatra G. S., Miranda J., Chinnappa C. C., Reid D. M. 2000. Effect of light quality and 5-azacytidine on genomic methylation and stem elongation in two ecotypes of Stellaria longipes. Physiol. Plantarum 109, 313–321 10.1034/j.1399-3054.2000.100313.x (doi:10.1034/j.1399-3054.2000.100313.x) [DOI] [Google Scholar]
- 24.Husband B. C., Schemske D. W. 1996. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution 50, 54–70 10.2307/2410780 (doi:10.2307/2410780) [DOI] [PubMed] [Google Scholar]
- 25.Cervera M. T., Ruiz-Garcia L., Martinez-Zapater J. M. 2002. Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol. Genet. Genomics 268, 543–552 10.1007/s00438-002-0772-4 (doi:10.1007/s00438-002-0772-4) [DOI] [PubMed] [Google Scholar]
- 26.Salmon A., Clotault J., Jenczewski E., Chable V., Manzanares-Dauleux M. J. 2008. Brassica oleracea displays a high level of DNA methylation polymorphism. Plant Sci. 174, 61–70 10.1016/j.plantsci.2007.09.012 (doi:10.1016/j.plantsci.2007.09.012) [DOI] [Google Scholar]
- 27.Bossdorf O., Arcuri D., Richards C. L., Pigliucci M. 2010. Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evol. Ecol. 24, 541–553 10.1007/s10682-010-9372-7 (doi:10.1007/s10682-010-9372-7) [DOI] [Google Scholar]
- 28.Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 10.1038/nrg887 (doi:10.1038/nrg887) [DOI] [PubMed] [Google Scholar]
- 29.Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 10.1101/gad.947102 (doi:10.1101/gad.947102) [DOI] [PubMed] [Google Scholar]
- 30.Lyko F., Ramsahoye B. H., Jaenisch R. 2000. Development: DNA methylation in Drosophila melanogaster. Nature 408, 538–540 10.1038/35046205 (doi:10.1038/35046205) [DOI] [PubMed] [Google Scholar]
- 31.Redchuk T. A., Rozhok A. I., Zhuk O. W., Kozeretska I. A., Mousseau T. A. 2012. DNA methylation in Drosophila melanogaster may depend on lineage heterogeneity. Cytol. Genet. 46, 58–61 10.3103/s0095452712010094 (doi:10.3103/s0095452712010094) [DOI] [PubMed] [Google Scholar]


