Abstract
Characteristic hydrogen bonding (H-bonding) patterns define secondary structure in proteins and nucleic acids. We show that similar patterns apply for α2-8 linked SiA (sialic acid) in H2O; and that H-bonds define its structure. A 15N,13C α2-8 SiA tetramer was used as a model system for the polymer. At 263K, we detected intra-residue through-bond J couplings between 15N and C8 for residues I, II and III of the tetramer, which indicate H-bonds between the 15Ns and the O8s of these residues. Additional J couplings between the 15Ns and C2s of the adjacent residues confirm the putative H-bonds. The NH groups showing this long-range correlation also experience slower 1H/2H exchange rate. Additionally, detection of couplings between H7 and C2 for residues II and III imply that the conformations of the linkers between these residues are different than the monomer's. These structural elements are consistent with two left handed helical models: 1) two residues per turn (24 helix) and 2) four residues per turn (14 helix). To discriminate between models, we resorted to 1H, 1H NOEs. The 24 helical model is in better agreement with the experimental data, for which all of the observed NOEs are predicted. On the other hand, for the 14 helix 87% of the observed intra-residue NOEs are predicted. In this report we provide direct evidence of H-bonding for a tetramer of α2-8 linked SiA and show how H-bonds can be a determining factor for shaping the carbohydrate three-dimensional structure.
The capsular PSs (polysaccharides) of Neisseria meningitidis and Escherichia coli K1 are crucial virulence factors that enable the organisms to survive in human serum1 and to colonize the meninges,2,3 resulting in the development of meningococcal disease. Among N. meningitidis serogroups (A, B, C, W-135, X and Y) the capsular PS of serogroup B, composed of α2-8 linked SiA (sialic acid), shows unique immunogenic properties. The N. meningitidis serogroup B PS elicits a weak immune response compared to the A, C, W-135 and Y PS’s.4–6 Binding to its cognate antibody requires 10 or more SiA residues6 as opposed to the usual 6–7,7 therefore α2-8 PSA (polysialic acid) is thought to adopt a helical conformation in solution, yielding a structural epitope.7 There is widespread agreement that α2-8 PSA forms a helix in solution, however there is no consensus on the type of helix.8–12 Proposed helical models are based on complementarity of the PS with its cognate antibody,11 sparse NMR data8,9 and/or simulations.12 However, to-date, there is no direct evidence for a helical structure. Herein, we report direct observation of H-bonds (hydrogen bonds), which alone supports a helix in a tetramer of α2-8 sialic acid, (SiA)4, at 263K. Experimental hetero-nuclear J couplings and inter-residue NOEs reinforce this structure.
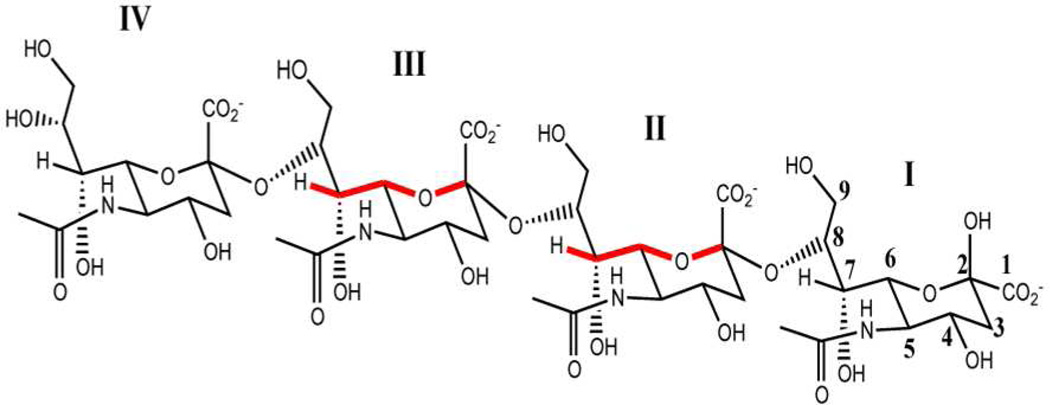
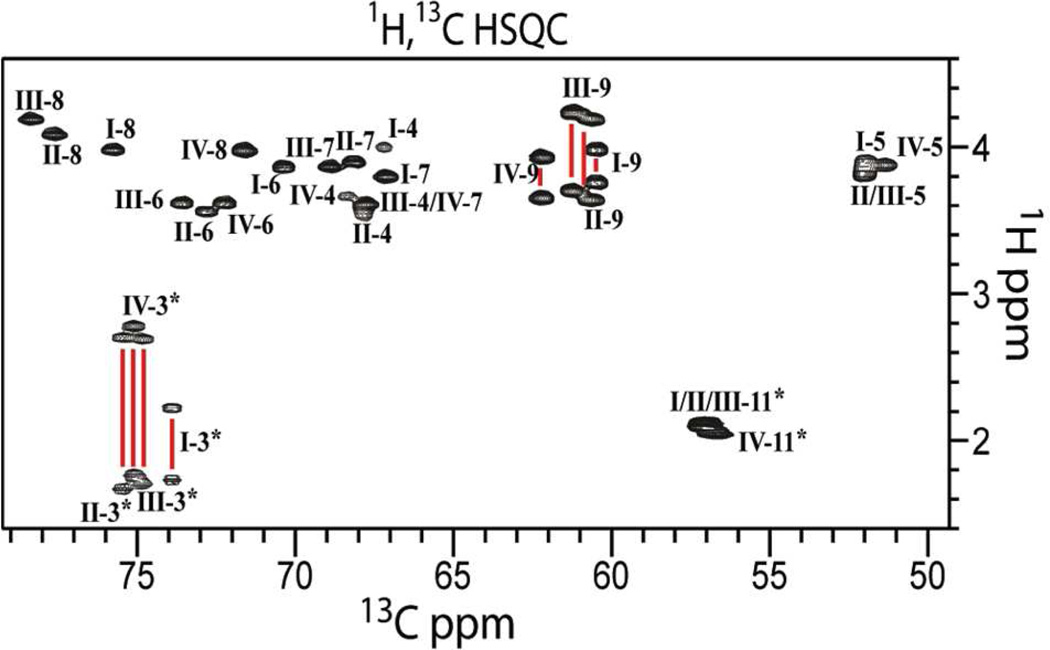
Solution NMR structure determination of α2-8 PSA, is hampered by signal degeneracy13 and the general lack of suitable tools for carbohydrate structural characterization. To overcome these difficulties, and to increase the number of NMR experiments that can be used in structure determination, we enriched the sample in 15N and 13C. We chose 15N, 13C labeled (SiA)4 (Chart 1) in our structural studies because nearly all its signals are well resolved, enabling unambiguous resonance assignment. The 1H,13C-HSQC spectrum of (SiA)4 (Figure 1), provides a set of well resolved cross-peaks, especially those corresponding to atoms at positions 3, 6, 8 and 9. Thus, NMR experiments on (SiA)4 enable the detection and unambiguous assignment of through-bond and through-space correlations. We further increased the number of NMR experiments available to us by collecting NMR data of (SiA)4 in 1H2O (as opposed to 2H2O) to utilize the amide HN in SiA as a probe for structural information.
Chart 1.
α2-8 Sialic acid tetramer (SiA)4 chemical structure. W-coupling is highlighted in red for residues for which it was detected.
Figure 1.
1H,13C-HSQC spectrum of (SiA)4. The spectrum, collected at 263K and pH 6.5 (20 mM Na2HPO4), displays excellent dispersion and signal-to-noise. Assignments are labeled with Roman numerals, to indicate residue position, followed by Arabic numerals to indicate atom position.
Secondary structure in proteins and nucleic acids is stabilized via characteristic H-bond patterns. It is plausible that H-bonding shapes carbohydrate structures as well. We hypothesized that by searching for H-bonds, we would identify important structural elements for carbohydrate structure determination. Secondary structure can be inferred from NMR based on direct evidence, via the detection of a long-range scalar coupling between the H-bonded nuclei,14,15 or indirect evidence, such as NOEs16 and slow 1H/2H exchange rates.17 Direct detection of H-bonds was reported for proteins and nucleic acids.14,15,18,19 Indirect evidence for H-bonding in carbohydrates has been presented,20–23 but to our knowledge no through-H-bond correlation was directly detected by NMR, for carbohydrates in H2O.
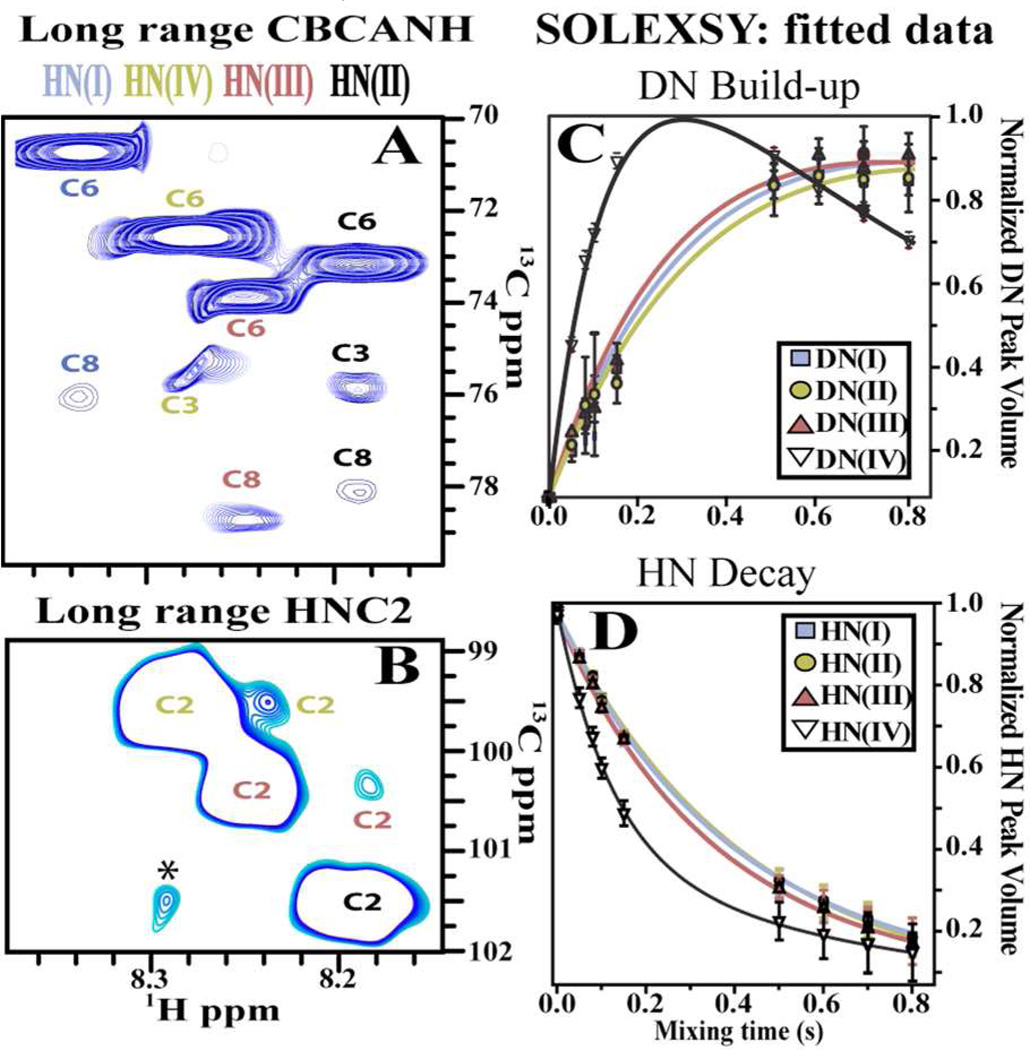
SiA residues have several functional groups for H-bonding: one N-acetyl group, in which the HN group can be a donor, and two carbonyl groups that can be acceptors: one at C1 and other at C10. Internal SiA residues in (SiA)4 have five oxygen atoms that can participate in H-bonds: 3 hydroxyl groups, at C4, C7 and C9, one forming the glycosidic linkage at C8 and one part of the pyranose ring at C6. All these carbons also bear hydrogen atoms (Chart 1). We therefore used a CBCANH experiment,24 modified to detect small J couplings to probe the tetramer for long-range a 13C-15N J couplings (Figure 2A).
Figure 2.
NMR experiments for hydrogen bond detection (263K pH* 6.5). A) Long range CBCANH: Correlations to different (SiA)4 residues are color coded based on residue position. B) Long range HNC2 on (SiA)4. Asterisk denotes an impurity. SOLEXSY DN build-up (C) and HN decay (D) traces obtained for each (SiA)4 residue. The data were fitted to extract exchange rate constants presented on Table 1.
The 15N’s of residues I, II and III show an unexpected intra-residue correlation to C8. A similar long-range correlation is absent for residue IV (Figure 2A) and for the monomer (Figure S1), suggesting that the proposed H-bond is a structural feature intrinsic to the tetramer. Thus, we hypothesized that the 15N to C8 coupling implies the presence of a H-bond.
If such an intra residue H-bond is present, there should also exist an inter-residue correlation between the HN and the C2 of a contiguous residue. We were able to detect such correlations for residues II and III (Figure 2B) via a long-range HNC2 experiment adapted from an HNCO experiment by exciting C2 resonances rather than CO.14 Thus, the existence of a H-bond between the HN and O8 is reinforced.
H-bonds in the HN groups of residues I–III should reduce the 1H/2H exchange rates for these HN's. Indeed, we observe decreased 1H/2H exchange rates in (SiA)4, via SOLEXSY experiments,25,26 ideal for (SiA)4 because it targets 1H/2H exchange rates in the 0.5–20 s−1 range. The measurement of 1H/2H exchange rates helps identify nascent helices in disordered proteins,26 where secondary structure elements are transient. It should also help in carbohydrates, which are thought to have partly disordered structures. Residues involved in secondary structure typically yield HN PFs (protection factors)27 between 2 and 5.26 We monitored peak intensities as a function of mixing time and fitted them to obtain exchange rate constants25 (Figures 2C, 2D S1, and Table 1). HN’s of residues I–III exchange ca. 3 fold slower than residue IV, as expected for H-bonded HN’s. We used SiA monomer (β configuration) 1H/2H exchange rate, as an external control, to obtain a PF for each SiA residue (Table 1). Residue IV in the tetramer displays a PF close to unity, and therefore behaves as a free SiA monomer, whose HN, is less protected from exchange. Conversely, HN’s for internal residues in (SiA)4 are more protected from exchange. Thus, we use residue IV’s HN as an internal reference for the PFs in the remaining HN’s in different sections of the (SiA)4 chain, as it is in the α configuration and is therefore more representative of similar residues in the oligomer. Together, the results from CBCANH and SOLEXSY experiments indicate that the structure of (SiA)4 stems from an intra-residue H-bonding pattern that repeats for each of the first three residues of the molecule (Figure S1).
Table 1.
Exchange rate, protection factors and "W" coupling constants measured at 263K
| kHD (Hz) | β PF* | α PF** | 4JH7-C2 (Hz) | |
|---|---|---|---|---|
| Residue I | 0.90 ± 0.12 | 3.33 | 3.79 | NO |
| Residue II | 1.28 ±0.33 | 2.34 | 2.66 | 0.8 ± 0.5 |
| Residue III | 0.77 ± 0.28 | 3.90 | 4.43 | 1.5 ± 0.5 |
| Residue IV | 3.41 ± 0.06 | 0.88 | 1.00 | NO |
Protection factor with respect to SiA monomer;
Protection factor with respect to Residue IV; NO means that no correlation was observed
An H-bond between a HN and O8 implies that conformations favoring the proximity between the lone pair of electrons of O8 and HN are preferred. The fulfillment of this requirement should translate into a restricted flexibility of the glycerol chain. Rigidification of the chain is further supported by heteronuclear coupling constants between the 1H at position 7 (H7) and the 13C at position 2 (4JH7-C2, Table 1) analogous to 1H−1H W coupling.28 W-couplings are present in rigid systems, where the arrangement of the bonds separating the pair of coupled nuclei is coplanar and resembles a "W" (Chart 1). W couplings are common in pyranose rings where the chair conformation enables the "W" arrangement to be adopted.29 However, conformations that break this coplanar arrangement can decrease or obliterate this long-range coupling.29
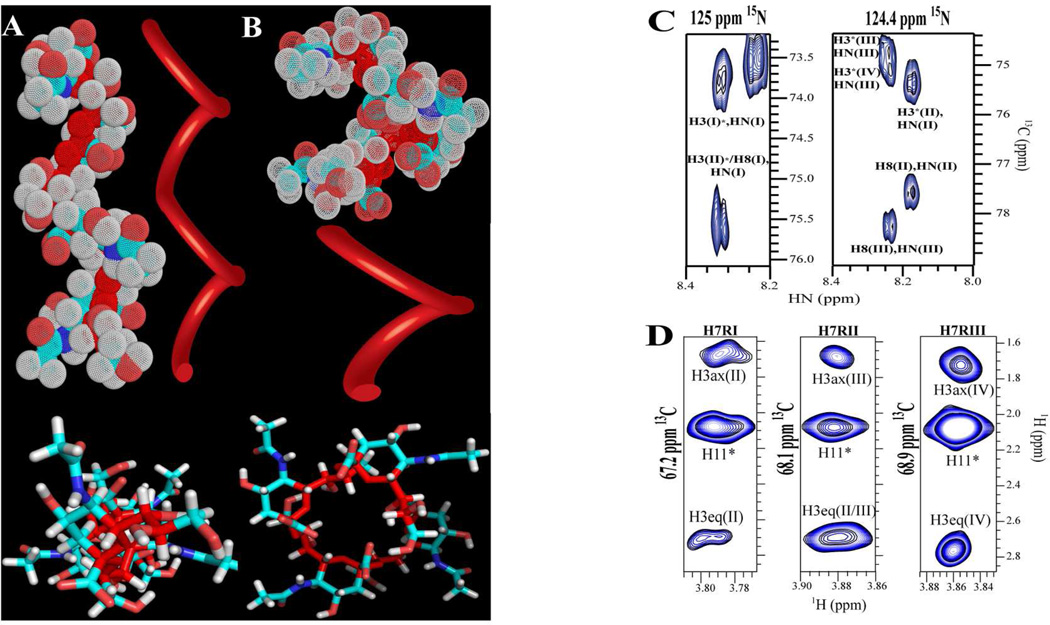
Such 4JH7-C2 couplings, have been proposed from DFT calculations,30 but have not been measured experimentally in αSiA. In contrast, we observed W-couplings only for residues II and III via HSQMBC31 in unlabeled (SiA)4. This suggests that the monomer’s glycerol chain (atoms 7, 8 and 9) adopts a different conformation than residue I in (SiA)4, and that it may be modulated by anomeric configuration. This coupling indicates that the H7-C7-C6-H6 torsion is ca. 90 degrees, as H7 must adopt a coplanar or quasi-coplanar W conformation with C2 to be measurable.30 Based on the above results, we generated, non-energy-minimized, static models for (SiA)4 (Figure 3), in which O8 of residues I, II and III, is H-bonded intra-residually to the HN, and H7s are W coupled to their own C2 in a quasi-coplanar arrangement. Interestingly, forcing the linker between residues, C6(i)-C7(i)-C8(i)-O8(i)-C2(i+1), into a conformation required to both, form a W arrangement between H7-C2, and a H-bond (O8-HN distance <3Å apart), rendered two out of three of the torsional angles of the linker chain fixed. Rotations about the C2(i+1)-O8(i) bond led to conformations that resulted in atom clashes. These restraints also imply two possible structural models for (SiA)4 with no clashes: a left-handed helix with two (helix 24) (Figure 3A) or four residues (helix 14) per turn (Figure 3B). It is also noteworthy that glycosidic torsions in the 14 model are consistent NO with the exo anomeric effect, while those in the 24 helix are not. Nonetheless, this finding indicates that the tetramer forms a helix.
Figure 3.
Panels A and B show two different static models for (SiA)4 consistent with the experimental data. Top figures depict side views of helical models with two (Panel A) and 4 residues per turn (Panel B). Bottom figures show a top view of the models in stick representation. Oligosaccharide backbone is depicted in dark red. A ribbon was placed proximal to each model to depict the trace followed by the oligosaccharide backbone. C) CNH-NOESY spectrum: though-space intra and inter-residue correlations observed between amide hydrogen and ring hydrogen atoms. D) 1H,13C-HSQC-NOESY: NOE correlations between H7 of each of the first three residues to H3s of a contiguous residue.
We used NOEs to probe for helical structure, and distinguish between the two helical models. The CNH-NOE32 and 1H,13C HSQC-NOESY experiments (Figure 3) yielded intra- and inter-residue correlations. The results of these experiments agree with (SiA)4 adopting a helical conformation, and support the 24 more strongly than the 14 helix. We observed NOEs only for residues I, II and III between H7, H8 and H9s and both H3s of the following residue. We also observed cross-peaks between HN and the intra-residue H8 and the inter-residue H3s (Table S1). These NOEs indicate that the oligomer is not in a fully extended conformation, but in a more compact one, where two contiguous residues twist, bringing H3 and H7, H8 and H9 less than 5Å apart. Moreover, the H7s of residues I, II and III show a strong intra-residue correlation to the H11s. This correlation is consistent with a conformation required to observe a W coupling between H7-C2 and the H3 to H8 inter-residue NOEs are consistent with H-bonding between O8 and the HN. The fact that 29 inter-residue NOEs (~7 for each residue) were observed is very unusual for carbohydrates and provides strong evidence for structure. Additionally, these sequential NOEs recur in residues I–III, and therefore, reinforce the notion of a restricted glycosidic linkage of a defined and periodic structure. Comparison of experimental and predicted NOEs yield: for the 24 helix, 71% of the predicted inter-residues NOEs were observed experimentally, but all of the experimental NOEs agree with the model. On the other hand, for the 14 helix, only 53% of the predicted intra-residue NOEs were observed and as many as four out of the 29 NOEs disagree with this model (Tables S1 and S2). The agreement between the NOE data and the 24 helix is striking.
Equally striking is the small number of experimental constraints necessary to generate this helical model: merely three H-bonds, and two W couplings. These atomic constraints induce a left-handed helix in solution. This conclusion is supported by previously unreported 1H, 1H NOEs and a slower 1H/2H exchange rate for the three first residues in the tetramer.
The presented results for (SiA)4 at 263K have several important implications: First, even short oligomers, such as the one used in these experiments, can adopt a helical structure in solution. Second, H-bonding in α2-8 PSA may be of great importance in defining the overall polysaccharide’s structure by stabilizing the glycosidic conformation and consequently defining the observed helix. By limiting the degrees of freedom of the glycosidic linkage, H-bonding appears to be a determining factor for shaping the α2-8 (SiA)4 molecular topology. Third, the 24 left-handed helix contains two residues per turn. While the left-handedness of this helix is consistent with predictions, the pitch is not.9–12 Only one study identifies a 2.2 residue per turn helix as a subpopulation of other conformations in a α2-8 (SiA)3 (trimer). Woods and co-workers12 predicted a model using NMR data (NOEs and J couplings) collected from a trimer, and selected a cluster of conformations from MD simulations that agreed with their experimental measurements. A helix was obtained when propagating the trimer structure, in good agreement with the 24 (SiA)4 model. This agreement highlights the potential for NMR-MD symbiosis for structure determination in carbohydrates. However, as previously observed, ten residues are required to span the antibody combining site, which would require longer stretches of oligomers to adopt a similar conformation as the polymer.13 Thus, it is possible that the helix observed in the tetramer may differ from that of larger oligomers. Nonetheless, we propagated both helices to a decamer and docked them with mAb73511 (Figure S2). Preliminary data suggests that the 24 model fits better in the binding site because it establishes more contacts than the 14 model. Affinity of IgMNOV7 for poly(A) and PSA is not explained with the 24 helix proposed herein because it disagrees with the poly(A) model. This may be due to structural reorganization on binding, which is suggested by experiments that show a higher entropy cost for mAb735 binding to longer oligomers. Therefore, models derived in the absence of mAb735 may differ from those of the free form. More research will verify if the (SiA)4 conformation resembles the decamer’s.
The original work on α2-8 linked SiAs9,11 set the stage for the present structural characterization of α2-8 linked SiAs, however, the results presented here disagree with the previously proposed models. The models derived from prior work were based on sparse NMR data. One model was built from torsional angles extracted from NMR (3JHH couplings) and using the mAb735 binding site as a target where the helical model contained 6 residues per turn.11 Another helical model9 based mainly on coupling constants and NOE data from the polymer suggests 3–4 residues per turn. These types of studies are complicated because no restraints can be extracted for individual residues as a result of spectral overlap. NOEs can result from intra-residue correlations, from a residue that is contiguous or from a sequentially remote residue, but it is difficult to distinguish intra- from inter-residue NOEs due to signal degeneracy. Moreover, angles extracted from coupling constants are not only a result of conformational averaging of a single residue, but may result from conformational averages of different residues. One major difference between the present and previous studies is the availability of fully 15N, 13C enriched SiA oligomers. These enabled us to utilize protein-type NMR methods and optimize them for use in determining carbohydrate structure. These methods could not have been used in previous studies since many were not developed at the time. In addition, isotopic labeling increased the sensitivity and resolution of the present NMR experiments and is currently catalyzing the development of carbohydrate-tailored NMR experiments.33
In summary, we present direct evidence of H-bonding, via through H-bond 3JN-C correlations, in an oligosaccharide chain in solution. These results agree with (SiA)4 adopting a left-handed helix in solution. H-bonding and W coupling data are consistent with two helical models obtained by varying the O6(i)-C8(i-1) angle: 1) A left-handed helix with 2 residues per turn (24) and 2) same helix chirality but with 4 residues per turn (14). NOE’s obtained from 1H,13C HSQC-NOESY yield distances that are in agreement with a 24 helix and rule out the 14 helix under the current experimental conditions. This conclusion is supported by previously unreported 1H, 1H NOEs and a slower 1H/2H exchange rate for the three first residues in the tetramer. We hope that this discovery will facilitate the determination of carbohydrate structures in solution via the detection of H-bonding. Our results could also provide information that was previously unavailable, should aid in the refinement of carbohydrate force fields, and provide clues regarding secondary structure in carbohydrates.
Secondary structure in carbohydrates may facilitate protein recognition by optimizing carbohydrate-protein contacts, and favor binding by pre-organizing the bound conformation. If the H-bond pattern presented in this report can be extrapolated to larger oligomers, H-bonds can have a cooperative effect in stabilizing the structure. The evidence presented for (SiA)4 and preliminary temperature dependent studies (unpublished results) suggest that the H-bonds reported herein are transient and may induce dynamic formation of helices. This behavior, on one hand, will ensure a structural epitope formed along the chain at a given time, and on the other, may increase the avidity of the ligand by having multiple propagating helical stretches.
The data presented herein demonstrate that helical (SiA)4 is a low energy conformation at 263K; at higher temperatures, this helix is likely in equilibrium with other conformations and will be present in solution even if not directly observed. If it resembles the structural epitope, the antibody will selectively bind the helix.
H-bond detection is of extreme importance in a structural world that appears to be dominated by averages. It may lead to unveiling structural patterns in glycans, to discovering secondary structure and to structure-function studies that could catalyze the deciphering of the “sugar code”.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Dr. D. A. Torchia and Dr. W. F. Vann for helpful discussions and V. Chevelkov help with SOLEXSY.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental procedures, SOLEXSY spectra, SiA CBCANH spectra, inter- and intra-residue NOE correlations and (SiA)4 structure with H-bonding pattern. This material is available free of charge at http://pubs.acs.org.
REFERENCES
- 1.Hill DJ, Griffiths NJ, Borodina E, Virji M. Clin. Sci. 2010;118:547. doi: 10.1042/CS20090513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Echenique-Rivera H, Muzzi A, Tordello ED, Seib KL, Francois P, Rappuoli R, Pizza M, Serruto D. PLOS Pathogens. 2011;7 doi: 10.1371/journal.ppat.1002027. e10020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KJ, Elliott SJ, Cello FD, Stins MF, Kim KS. Cell Microbiol. 2003;5:245. doi: 10.1046/j.1462-5822.2003.t01-1-00271.x. [DOI] [PubMed] [Google Scholar]
- 4.Wyle FA, Tramont EC, Lowentha Jp, Berman SL, Brandt BL, Altieri PL, Artenste Ms, Kasper DL. J Infect Dis. 1972;126:514. doi: 10.1093/infdis/126.5.514. [DOI] [PubMed] [Google Scholar]
- 5.Finne J, Leinonen M, Makela PH. Lancet. 1983;2:355. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 6.Jennings HJ, Roy R, Michon F. J Immunol. 1985;134:2651. [PubMed] [Google Scholar]
- 7.Kabat EA, Liao J, Osserman EF, Gamian A, Michon F, Jennings HJ. J Exp Med. 1988;168:699. doi: 10.1084/jem.168.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson TJ, Venable RM, Egan W. J Am Chem Soc. 2003;125:2930. doi: 10.1021/ja0210087. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki R, Bacon B. Biochemistry-US. 1991;30:851. doi: 10.1021/bi00217a039. [DOI] [PubMed] [Google Scholar]
- 10.Brisson JR, Baumann H, Imberty A, Perez S, Jennings HJ. Biochemistry-US. 1992;31:4996. doi: 10.1021/bi00136a012. [DOI] [PubMed] [Google Scholar]
- 11.Evans SV, Sigurskjold BW, Jennings HJ, Brisson JR, To R, Tse WC, Altman E, Frosch M, Weisgerber C, Kratzin HD, Klebert S, Vaesen M, Bittersuermann D, Rose DR, Young NM, Bundle DR. Biochemistry-US. 1995;34:6737. doi: 10.1021/bi00020a019. [DOI] [PubMed] [Google Scholar]
- 12.Yongye AB, Gonzalez-Outeirino J, Glushka J, Schultheis V, Woods RJ. Biochemistry-US. 2008;47:12493. doi: 10.1021/bi800431c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michon F, Katzenellenbogen E, Kasper DL, Jennings HJ. Biochemistry-US. 1987;26:476. doi: 10.1021/bi00376a020. [DOI] [PubMed] [Google Scholar]
- 14.Cordier F, Grzesiek S. J Am Cheml Soc. 1999;121:1601. [Google Scholar]
- 15.Dingley AJ, Grzesiek S. J Am Cheml Soc. 1998;120:8293. [Google Scholar]
- 16.Wagner G, Kumar A, Wuthrich K. Eur. J. Biochem. 1981;114:375. doi: 10.1111/j.1432-1033.1981.tb05157.x. [DOI] [PubMed] [Google Scholar]
- 17.Wagner G, Wuthrich K. J Mol Biol. 1979;134:75. doi: 10.1016/0022-2836(79)90414-5. [DOI] [PubMed] [Google Scholar]
- 18.Dingley AJ, Masse JE, Peterson RD, Barfield M, Feigon J, Grzesiek S. J Am Cheml Soc. 1999;121:6019. [Google Scholar]
- 19.Wang YX, Jacob J, Cordier F, Wingfield P, Stahl SJ, Lee-Huang S, Torchia D, Grzesiek S, Bax A. J Biomol NMR. 1999;14:181. doi: 10.1023/a:1008346517302. [DOI] [PubMed] [Google Scholar]
- 20.Sheng SQ, Vanhalbeek H. Biochem Bioph Res Co. 1995;215:504. doi: 10.1006/bbrc.1995.2493. [DOI] [PubMed] [Google Scholar]
- 21.Norris SE, Landström J, Bull AWTE, Widmalm G, Freedberg DI. Bioplolymers. 2011;97:145. doi: 10.1002/bip.21710. [DOI] [PubMed] [Google Scholar]
- 22.Sandström C, Baumann H, Kenne L. J Chem Soc Perk T 2. 1998:2385. [Google Scholar]
- 23.Langeslay DJ, Young RP, Beni S, Beecher CN, Mueller LJ, Larive CK. Glycobiology. 2012 doi: 10.1093/glycob/cws085. [DOI] [PubMed] [Google Scholar]
- 24.Grzesiek S, Bax A. J Magn Reson. 1992;91:201. [Google Scholar]
- 25.Chevelkov V, Xue Y, Rao DK, Forman-Kay JD, Skrynnikov NR. J Biomol NMR. 2010;46:227. doi: 10.1007/s10858-010-9398-8. [DOI] [PubMed] [Google Scholar]
- 26.Yao J, Chung J, Eliezer D, Wright PE, Dyson HJ. Biochemistry-US. 2001;40:3561. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 27.Molday RS, Englander SW, Kallen RG. Biochemistry-US. 1972;11:150. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 28.Sternhell S. Reviews of Pure and Applied Chemistry. 1964;14:15. [Google Scholar]
- 29.Pan QF, Klepach T, Carmichael I, Reed M, Serianni AS. J Org Chem. 2005;70:7542. doi: 10.1021/jo050615k. [DOI] [PubMed] [Google Scholar]
- 30.Klepach T, Zhang WH, Carmichael I, Serianni AS. J Org Chem. 2008;73:4376. doi: 10.1021/jo702204x. [DOI] [PubMed] [Google Scholar]
- 31.Williamson RT, Marquez BL, Gerwick WH, Kover KE. Magn Reson Chem. 2000;38:265. [Google Scholar]
- 32.Diercks T, Coles M, Kessler H. J Biomol NMR. 1999;15:177. doi: 10.1023/A:1008367912535. [DOI] [PubMed] [Google Scholar]
- 33.Barb A, Freedberg D, Battistel M, Prestegard J. J Biomol NMR. 2011;51:163. doi: 10.1007/s10858-011-9550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.