Abstract
Although Stat1 is essential for cells to respond fully to IFN-γ, there is substantial evidence that, in the absence of Stat1, IFN-γ can still regulate the expression of some genes, induce an antiviral state and affect cell growth. We have now identified many genes that are regulated by IFN-γ in serum-starved Stat1-null mouse fibroblasts. The proteins induced by IFN-γ in Stat1-null cells can account for the substantial biological responses that remain. Some genes are induced in both wild-type and Stat1-null cells and thus are truly Stat1-independent. Others are subject to more complex regulation in response to IFN-γ, repressed by Stat1 in wild-type cells and activated in Stat1-null cells. Many genes induced by IFN-γ in Stat1-null fibroblasts also are induced by platelet-derived growth factor in wild-type cells and thus are likely to be involved in cell proliferation. In mouse cells expressing the docking site mutant Y440F of human IFN-γ receptor subunit 1, the mouse Stat1 is not phosphorylated in response to human IFN-γ, but c-myc and c-jun are still induced, showing that the Stat1 docking site is not required for Stat1-independent signaling.
Interferon γ, a pluripotent cytokine produced by activated T lymphocytes and natural killer cells, helps to regulate many biological functions, including antiviral responses, cell proliferation, immune surveillance, and tumor suppression. IFN-γ plays an important role in immune responses involved in host defenses against infectious agents and tumors by up-regulating MHC class I and MHC class II, effecting IgG heavy chain switching, and stimulating the production of immunomodulatory cytokines such as IL-12 and tumor necrosis factor and of antiviral proteins such as 2–5A synthetase and RNase L (1, 2). Genetic and biochemical analyses have revealed the importance of the protein tyrosine kinases Jak1 and Jak2 and of the transcription factor Stat1 in IFN-γ-dependent signaling (3, 4). Upon ligand binding, the receptor oligomerizes and Jak1 and Jak2 are activated, leading to the phosphorylation of tyrosine 440 of the IFN-γ receptor subunit 1 (IFNGR1) of the receptor, which provides a docking site for Stat1 (5). Stat1 then is phosphorylated on tyrosine 701, leading to its dimerization and translocation to the nucleus, where it binds to the gamma-activated sequence (GAS) elements of promoters to regulate expression of the downstream genes (6). Recently, another DNA sequence, the gamma-activated transcription element (GATE), and its cognate transcription factor (C/EBP-β) were shown to be regulated by IFN-γ (7).
Many IFN-γ-regulated genes have been identified in different cell types (8, 9). The essential role of Stat1 in IFN-γ-dependent signaling has been demonstrated in cells that do not express this factor (10–12). However, there is also evidence for Stat1-independent responses to IFN-γ. Expression of kinase-negative Jak1 mutants in wild-type cells inhibits the development of the antiviral response without affecting the activation of Stat1 or Stat1-dependent gene expression in response to IFN-γ (13). Although Stat1 is important for the antiviral response, Stat1-null mice are 100 times more resistant to murine cytomegalovirus and Sindbis virus than are mice lacking expression of both IFN-γ and the IFN-α/β receptor (14). Furthermore, IFN-γ inhibits the proliferation of wild-type but not Stat1-null fibroblasts (15), and lymphocytes or fibroblasts from Stat1-null mice showed enhanced proliferation and reduced apoptosis in comparison with cells from wild-type mice (16). We have shown that the immediate-early genes c-myc and c-jun are induced transiently and rapidly in response to IFN-γ in serum-starved Stat1-null fibroblasts, but not in wild-type cells (17). These studies indicate that Stat1-independent regulation of gene expression has a role in the full range of biological responses to IFN-γ.
Materials and Methods
Reagents and Cell Culture.
Recombinant human and mouse IFN-γ were from Roche Molecular Biochemicals, and platelet-derived growth factor (PDGF)-BB was from GIBCO/BRL. Wild-type and Stat1-null mouse embryo fibroblasts (MEFs) and mouse cells expressing wild-type human IFNGR1 or the Y440F Stat1 docking site mutant of IFNGR1 were described by Meraz et al. (11) and Farrar et al. (18). All cells were grown in DMEM, supplemented with 5% FBS. Subconfluent cells were serum-starved for 48 h in DMEM containing 0.1% serum before treatment with IFN-γ or PDGF.
Oligonucleotide Microarrays.
Total RNA was prepared from fibroblasts treated with IFN-γ (1,000 units/ml) for 0, 0.5, 1, 3, or 6 h by using the TRIzol method (GIBCO/BRL). Poly(A) RNA was prepared from control and IFN-γ-treated Stat1-null fibroblasts by using the Oligotex method (Qiagen, Chatsworth, CA). cRNA was prepared and the murine genome array (MG-U74A) was hybridized as directed by Affymetrix, San Jose, CA. The washed arrays were stained with phycoerythrin-streptavidin (Molecular Probes) and read by using an Affymetrix GeneChip scanner and gene expression software.
RNA and Protein Analyses.
Northern transfers were analyzed with random-primed cDNA probes for c-myc, c-jun, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (17). RNase protection analyses were performed by using a kit (Ambion). The fos-jun templates were from PharMingen, and the chemokine templates were described by Lane et al. (19). Expression levels were quantified by using a PhosphorImager (Molecular Dynamics). Western analyses were performed as described by Chernov et al. (20). Antibodies against Tyr-701-phosphorylated Stat1 (Cell Signaling Technology, Beverly, MA), C/EBP-β, actin, and early growth response 1 (EGR-1) (Santa Cruz Biotechnology) were used.
Cell Cycle and Gel-Shift Analyses.
Cell cycle analyses were performed as described by Bromberg et al. (15), and the data were analyzed by using the lysis ii program. Electrophoretic mobility-shift assays were performed as described by Ramana et al. (17). Wild-type and mutant double-stranded oligonucleotides corresponding to the binding sites for EGR and NF-Y (core binding factor) were from Santa Cruz Biotechnology.
Results
Identification of Genes Regulated by IFN-γ in Serum-Starved Stat1-Null Fibroblasts by Oligonucleotide Array Analysis.
Poly(A) RNAs were isolated from Stat1-null MEFs treated with IFN-γ for 0, 0.5, 1, 3, or 6 h. Labeled cRNA probes were hybridized to murine genome U74A arrays (Affymetrix), which contain sequences corresponding to ≈6,000 genes and ≈6,000 expressed sequence tags (ESTs). The fold induction or repression of each gene was the ratio of intensities in the IFN-γ-treated and control samples. Only those genes induced or repressed by more than 3.5-fold were tabulated. We do not present data for ESTs where functional information is not available. The temporal profiles of gene expression were quite diverse, with the peaks of induction from 0.5 to 6 h (Table 1). Eighteen genes were induced maximally by IFN-γ in Stat1-null fibroblasts in 0.5–1 h. An additional 10 genes were induced maximally in 3–6 h, suggesting that their induction may be due to a secondary rather than primary effect of IFN-γ. The induced genes can be classified broadly into those encoding immediate-early proteins, transcription factors, cytoplasmic regulatory proteins, and secreted proteins. Among the immediate-early proteins and transcription factors are the zinc-finger factors EGR-1, EGR-2, and TIS11 and the leucine zipper and heterodimerization factors c-jun, LRG-21, and helix–loop–helix. It is interesting to note that many of the immediate-early genes (i.e., pip92, Gly96, and EGR-1) originally were cloned in screens for growth-regulated genes (21–23). Among the induced cytoplasmic regulatory proteins are SOCS-2 and SOCS-3, which are involved in the negative regulation of IFN-dependent signal transduction, K-ras and RhoB, involved in growth factor-dependent signaling, and metallothionein and PGHS-2, involved in stress responses. Secreted proteins induced in Stat1-null fibroblasts include both growth factors and cytokines (HB-EGF, PDGF-α, IL-11, IFN-β, and GDF-15), which play important roles in altering the microenvironment of adjacent tissues, in angiogenesis and in altering the metastatic potential of tumor cells. Approximately half of the genes induced by IFN-γ in serum-starved Stat1-null fibroblasts (Table 1) were originally identified as PDGF-responsive genes (24), suggesting that the stimulation of cell growth in Stat1-null fibroblasts in response to IFN-γ is likely to involve components that also participate in PDGF-dependent signaling. About a third of the genes induced by IFN-γ in Stat1-null fibroblasts also are induced by IFN-γ in wild-type cells. These include the transcription factors EGR-1, C/EBP-β, and LRG-21 (7, 25, 26), the cytoplasmic regulatory proteins SOCS-2 and SOCS-3 (27), the stress-response proteins metallothionein and PGHS-2 (28, 29), osteopontin (30), and the secreted autocrine factor HB-EGF (29). Several genes are suppressed by IFN-γ in Stat1-null fibroblasts (see Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). Many of the encoded proteins are involved in the negative regulation of cell growth (Mad-4, Gas1, and transforming growth factor β) or apoptosis [phospholipid scramblase, which is induced by IFN-γ in wild-type cells (8)].
Table 1.
Genes induced by IFN-γ in serum-starved Stat1-null MEFs
| Gene | GenBank number | Function | Fold induction | Peak of induction, h | Induced by PDGF in wild-type cells | Induced by IFN-γ in wild-type cells |
|---|---|---|---|---|---|---|
| c-Jun | J04115 | Transcription | 6.6 | 0.5 | ||
| EGR1 | NM007913 | Transcription | 7.4 | 0.5 | + | + |
| EGR2 | X06746 | Transcription | 8.7 | 0.5 | + | |
| TIS11 (ERF1) | X14678 | Transcription | 12.6 | 0.5 | ||
| C/EBP-β | M61007 | Transcription | 14.2 | 3 | + | |
| HLH (eip1) | Y07836 | Transcription | 20.5 | 1 | ||
| GIF | AF064088 | Transcription | 11.9 | 0.5 | ||
| LRG-21 | U19118 | Transcription | 9.4 | 0.5 | + | + |
| CBF-1α (PEBP2α1) | D14636 | Transcription | 8.0 | 3 | ||
| Fra-2 | X83971 | Transcription | 5.3 | 3 | + | |
| GADD45 | U00937 | Transcription | 3.8 | 0.5 | + | |
| ID1 | M31885 | Transcription | 17.8 | 1 | + | |
| 3CH134 | X61940 | Cytoplasmic, regulation | 6.5 | 0.5 | + | |
| PIP92 | M59821 | Cytoplasmic, regulation | 11.0 | 0.5 | + | |
| Metallothionein | V00835 | Cytoplasmic, regulation | 10.0 | 0.5 | + | |
| PGHS | M88242 | Cytoplasmic, regulation | 13.3 | 3 | + | |
| k-Ras | X02452 | Cytoplasmic, regulation | 5.7 | |||
| RHO-B | X99963 | Cytoplasmic, regulation | 3.5 | 0.5 | + | |
| SOCS-2 | U88327 | Cytoplasmic, regulation | 6.9 | 1 | + | |
| SOCS-3 | AF117732 | Cytoplasmic, regulation | 11.4 | 1 | + | |
| Pyruvate dehydrogenase-like protein | AJ001418 | Cytoplasmic, metabolism | 5.1 | 3 | ||
| Hexokinase II | Y11666 | Cytoplasmic, metabolism | 11.0 | |||
| GLY96 | X67644 | Transmembrane | 4.5 | 1 | + | |
| Osteopontin | X13986 | Cell matrix | 27.5 | 6 | + | |
| PAI-1 | M33960 | Secreted | 35.1 | 3 | + | |
| IFN-β | NM010510 | Secreted | 4.6 | 3 | + | |
| IL-11 | U03421 | Secreted | 23.1 | 3 | + | |
| HB-EGF | U39192 | Secreted | 3.6 | 0.5 | + | + |
| PDGF-α | M29464 | Secreted | 4.7 | 3 | ||
| GDF15 (MIC1) | AJ011967 | Secreted | 7.5 | 1 |
Regulation of Immediate-Early and Chemokine Gene Expression by IFN-γ in Stat1-Null Cells.
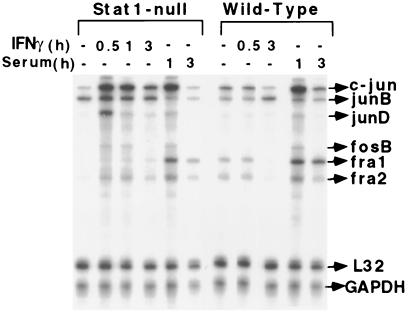
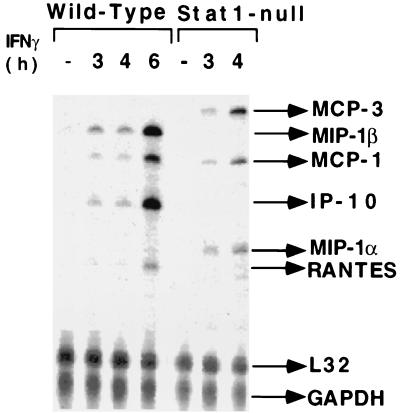
An RNase protection assay, using the fos-jun template set, revealed that both c-jun and junD were induced transiently and rapidly (within 30 min) in Stat1-null fibroblasts but not in wild-type cells. In addition, fosB and fra2 were induced in Stat1-null fibroblasts, but to a lesser extent. However, junB and c-fos were not induced significantly by IFN-γ in either cell type (Fig. 1). In contrast, adding serum to serum-starved cells induced c-jun, junD, fra1, and fra2 in both cell types. Northern analysis revealed that, although IFN-γ and IFN-β both induced c-jun expression in Stat1-null fibroblasts, only IFN-γ repressed c-jun expression (by 2.5-fold) in wild-type cells (data not shown). IFN-γ-induced Stat1 dimers may be required for this repression. RNase protection analyses (Fig. 2) indicated that the IFN-γ-dependent induction of MIP-1β, IP-10, and RANTES occurs only in wild-type cells. In contrast, MCP-3 and MIP-1α are induced only in Stat1-null fibroblasts, and MCP-1(JE) is induced by IFN-γ in both cell types. These three patterns of expression (induced only in wild-type cells, induced only in Stat1-null cells, induced in both) are seen for many other genes, and a mechanistic explanation for each pattern is presented in Discussion.
Figure 1.
Regulation of immediate-early gene expression in response to IFN-γ in Stat1-null and wild-type fibroblasts. Total RNA, prepared from serum-starved cells, untreated or treated with mouse IFN-γ (1,000 units/ml) or 10% serum for the times indicated, was subjected to RNase protection analysis.
Figure 2.
IFN-γ-dependent regulation of chemokine gene expression in serum-starved Stat1-null and wild-type fibroblasts. Total RNA was prepared from MEFs, untreated or treated with mouse IFN-γ (1,000 units/ml). RNase protection analysis was used.
The Transcription Factors EGR and C/EBP-β Are Induced by IFN-γ Independently of Stat1.
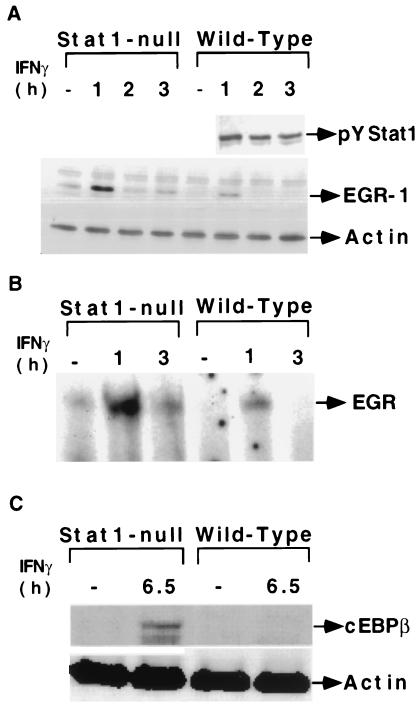
EGR-1 and EGR-2 are induced by IFN-γ in Stat1-null fibroblasts (Table 1). Members of the EGR family of transcription factors contain highly homologous zinc-finger DNA binding domains and regulate transcription through a common DNA response element (25). Western analyses and electrophoretic mobility-shift assays confirmed that both EGR protein and DNA binding activity were induced in both cell types, although the induction was higher in Stat1-null fibroblasts (Fig. 3 A and B). The specificity of EGR-1 DNA binding activity was demonstrated by competition with wild-type but not mutant unlabeled probes (data not shown). Recent studies have revealed that C/EBP-β, a member of the CCAAT family of transcription factors, is induced by IFN-γ in different tissues (7). Western analysis revealed that C/EBP-β is induced by IFN-γ in Stat1-null fibroblasts. Another CCAAT transcription factor [core binding factor or NF-Y (31)] was not induced (data not shown). The induction of C/EBP-β was late (6.5 h), indicating that a delayed transcriptional response probably is involved (Fig. 3C). There may be as many as three waves of transcriptional activation in response to IFN-γ in Stat1-null cells. The primary response leads to the rapid induction of transcription factors (i.e., EGR-1 and 2, see Table 1) that drive the secondary response. Additional transcription factors are induced in these secondary responses (i.e., C/EBP-β), and these may in turn drive a tertiary response, manifest at times later than those examined in the present study.
Figure 3.
The transcription factors EGR and C/EBP-β are induced by IFN-γ independently of Stat1. (A) Western analysis of tyrosine-phosphorylated Stat1 and EGR-1. (B) Electrophoretic mobility-shift assays with an EGR probe. Whole-cell extracts were prepared from serum-starved Stat1-null or wild-type fibroblasts treated with IFN-γ. (C) Western analysis of C/EBP-β in Stat1-null and wild-type fibroblasts.
PDGF Induces c-jun in Stat1-Null MEFs but Not in Wild-Type Cells.
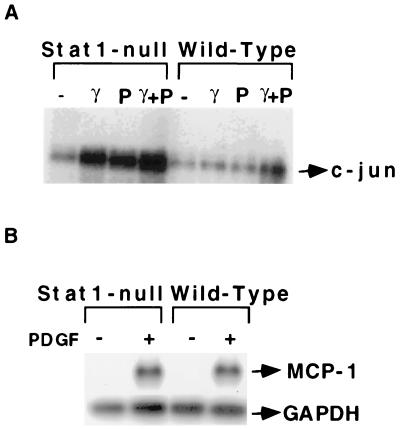
Many genes found here to be induced by IFN-γ in serum-starved Stat1-null fibroblasts originally were identified as PDGF-responsive genes. IFN-γ potentiates PDGF-mediated mitogenesis under pathological conditions, i.e., in arteriosclerosis (32). Furthermore, a switch from PDGF-dependent signaling to IFN-γ-dependent signaling was observed in 3T3 cells expressing the chimeric PDGF receptor Chi R (Y771), which retains only a single tyrosine residue, at the RAS-GAP site, in its cytoplasmic domain (24). Fambrough et al. (24) proposed that the activation of Stat1 by ChiR (Y771) may be responsible for the induction of IFN-γ-responsive genes by PDGF. Northern analysis revealed that c-jun is induced by PDGF in Stat1-null cells but not in wild-type cells (Fig. 4A). The induction is maximal at 30 min and begins to decline by 90 min (data not shown). This effect is similar to the induction of c-jun by IFN-γ in Stat1-null but not in wild-type cells and may be due similarly to the activation by PDGF of a repressor involving Stat1. Stimulation with serum of serum-starved cells induced c-jun expression in both cell types (Fig. 1), indicating that growth factors other than PDGF have different effects. In our previous study (17), PDGF induced the expression of the immediate-early gene c-myc in both Stat1-null and wild-type MEFs, although the induction was higher in Stat1-null cells. Although PDGF or IFN-γ alone each induced c-jun expression in Stat1-null cells, combined treatment was not additive or synergistic (Fig. 4A). In contrast to c-jun, MCP-1(JE) is induced by PDGF in both cell types (Fig. 4B).
Figure 4.
PDGF-dependent induction of gene expression in Stat1-null and wild-type fibroblasts. (A) Stat1-null or wild-type MEFs were untreated or treated for 30 min with IFN-γ or PDGF alone, or with PDGF plus IFN-γ. c-Jun mRNA levels were determined by Northern analysis. (B) Stat1-null or wild-type MEFs were untreated or treated with PDGF for 2 h. MCP-1 mRNA levels were determined by Northern analysis.
Mutation of the Stat1 Docking Site of IFNGR1 Permits IFN-γ to Induce the Expression of c-myc and c-jun.
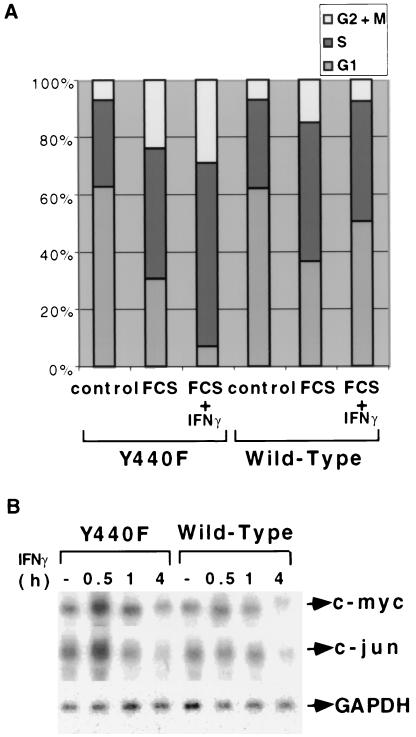
Both c-myc and c-jun are induced rapidly and transiently by IFN-γ in Stat1-null fibroblasts (17). The phosphorylation of Y440 of IFNGR1 is required for Stat1 to be recruited to the receptor and activated (5). To test the effect of mutating this Stat1 docking site, we used mouse cell lines expressing either wild-type human IFNGR1 (hGR) or the docking site mutant Y440F (18). Treatment with human IFN-γ induced Stat1-dependent luciferase activity in hGR but not in Y440F cells, whereas in the control experiment, mouse IFN-γ activated Stat1-dependent reporter activity in both (data not shown). Human IFN-γ suppressed the serum-induced proliferation of hGR cells but not of Y440F cells, where it potentiated the serum response (Fig. 5A). Northern analysis revealed that the expression of c-myc and c-jun was suppressed by human IFN-γ in hGR cells and that, in contrast, c-myc and c-jun were induced transiently and rapidly by human IFN-γ in Y440F cells (Fig. 5B). Thus, when Stat1 is present but cannot be activated by the receptor, IFN-γ is still capable of signaling to c-myc and c-jun.
Figure 5.
Mutation of the Stat1 docking site of IFNGR1 allows IFN-γ to induce the expression of c-myc and c-jun. (A) Cell cycle analysis. Cells grown to 20% confluence in 5% serum were serum-starved for 48 h and then not treated (control) or treated with 10% serum, alone or in combination with human IFN-γ. The cells were stained with propidium iodide 24 h later, and the DNA content was analyzed by flow cytometry. The percentages of cells in the G1, S, and G2 phases of the cell cycle are indicated. (B) Induction of immediate-early genes. Subconfluent, serum-starved hGR or Y440F cells were untreated or treated with 1,000 units/ml of human IFN-γ for 30 min, 1 h, or 4 h. c-Myc, c-jun, and GAPDH mRNA levels were determined by Northern analysis.
Discussion
Stat1 plays an essential and major role in the responses to the IFNs (11, 12), but there is now compelling evidence for an important Stat1-independent pathway in IFN-dependent signaling. Studies with kinase-negative mutants of Jak1 reveal a Jak1-dependent signal, which, in addition to Stat1 activation, is required for a full antiviral response to IFN-γ (13). Furthermore, Stat1-null mice are significantly more resistant to viral infection than are mice lacking both the IFN-γ and IFN-α/β receptors (14). IFN-γ potentiates the proliferation of fibroblasts or bone marrow cells derived from Stat1-null mice (14, 17). Thus, we can distinguish Stat1-independent responses to IFN-γ that mediate antiviral activities from those that stimulate cell proliferation. Immediate-early genes, induced by IFN-γ in Stat1-null fibroblasts, play important roles in the G1-S transition and cell proliferation (33–35). The chemokines induced by IFN-γ are involved in antiviral responses, stimulating the directional migration of leukocytes and the localization, rapid amplification, and coordination of the cytokine-to-chemokine-to-cytokine cascade (36). The immediate-early genes c-myc, c-jun, and junD, the transcription factor gene C/EBP-β, and the chemokine genes MIP-1α and MCP-3 are induced in response to IFN-γ in Stat1-null fibroblasts, but not in wild-type cells. In contrast, the EGR-1 and MCP-1(JE) genes are induced by IFN-γ in both wild-type and Stat1-null MEFs. The versatile MCP-1 promoter is induced in most examples of trauma, infection, or inflammation (37). Gil et al. (14) also have noted complex patterns of gene expression in response to IFN-γ. The IL-1β and arginase genes were induced in Stat1-null but not wild-type macrophages, the CXCR4 gene was suppressed by IFN-γ in both, and the MCP-1 and MIP-1α genes were induced in both types of macrophages (14). An IFN-stimulated response element is essential for the transcription of chemokines that are not induced by IFN-γ in Stat1-null cells (38, 39). The three patterns of gene expression are interpreted as follows: (i) induced in wild-type and not Stat1-null cells; a Stat1-dependent (gamma-activated sequence, GAS) element drives the gene; (ii) induced in both wild-type and in Stat1-null cells, a Stat1-independent element drives the gene, and (iii) induced in Stat1-null and not in wild-type cells, a Stat1-independent element drives the gene and a Stat1-dependent (GAS) element represses the gene. Interestingly, quite a few genes behave similarly to c-myc, responding both to Stat1-independent activation and Stat1-dependent repression. The biological relevance of simultaneous positive and negative signals emanating from the same receptor is as yet unclear.
Most genes are induced rapidly (0.5–1 h) by IFN-γ in Stat1-null cells. It is likely that transcription factors encoded by genes induced early are responsible for the later induction of other genes. EGR-1 is involved in the transcription of the PDGF-α gene (40). IFN-γ-dependent induction of PGHS-2 involves an autocrine loop mediated by the synthesis of HB-EGF (29) and core binding factor α is involved in transactivating the osteopontin gene (41). The genes encoding pip92, gly96, SOCS-3, C/EBP-β, MCP-1, and MIP-1α are induced by IFN-γ in both Stat1-null fibroblasts (this paper) and macrophages (14). We have used serum starvation to discern expression profiles in the absence of mitogenic stimuli, to detect genes involved in cell growth, the expression of which would otherwise be masked by the presence of serum. Note that our experiments differ from those of Gil et al. (14) in which the responses of macrophages to IFN-γ were evaluated in the presence of serum. Many genes induced by IFN-γ in Stat1-null fibroblasts also are induced by PDGF, a potent mitogen that drives cell proliferation. Because deregulated expression of PDGF is involved in tumor development (42–44), the stimulation of PDGF-dependent signaling by IFN-γ may contribute to this process in Stat1-null mice. Interestingly, chemokines induced by IFN-γ only through Stat1 (i.e., IP-10) have been associated with direct or indirect antitumor effects (45). In contrast, MCP-1 is often produced by neoplastic cells, and its expression has been associated with tumor growth (46). Although either IFN-γ or PDGF induced c-jun in Stat1-null fibroblasts, the combination was not additive, consistent with the idea that the growth stimulation of Stat1-null fibroblasts in response to IFN-γ may involve signaling components that also are activated in response to PDGF.
Jak1 is required for the induction of Stat1-independent gene expression by IFN-γ (14). Jak2 also may be important, because it is required for Stat1-dependent signal transduction in response to IFN-γ (13). These two kinases are involved in many growth factor- and cytokine-dependent signaling pathways, and their essential roles have been demonstrated by gene targeting studies (47, 48). Recent intensive analyses of protein expression patterns have revealed that many tyrosine-phosphorylated substrates are involved in growth factor-dependent signaling, and some of these may require Jaks for their phosphorylation (49, 50). Therefore, we speculate that both Jak1 and Jak2 are involved in Stat1-independent pathways. IFN-γ activates Stat3 in wild-type and Stat1-null MEFs (12). We have observed (data not shown) that this activation of Stat3 is transient, peaking 15 min after treatment, whereas the stronger activation of Stat1 is maintained for much longer. PDGF activates Stats 3, 5, and 6, which have been implicated in stimulating cell proliferation in a variety of signal transduction pathways (51). Whether the deregulation of cell growth in Stat1-null cells in response to IFN-γ involves the activation of other Stat family members remains under investigation. An important general goal is to identify the molecules and mechanisms responsible for Stat1-independent signaling.
Supplementary Material
Acknowledgments
We thank Drs. Lesleyann Hawthorne for the microarray analyses and Mikhail Chernov for help in formatting data. This work was supported by Grants P01 CA 62220 from the National Institutes of Health (to G.R.S.) and CA 43059 (to R.D.S.).
Abbreviations
- PDGF
platelet-derived growth factor
- MEF
mouse embryo fibroblast
- IFNGR1
IFN-γ receptor subunit 1
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- EGR
early growth response
References
- 1.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan D H, Shankaran V, Dighe A S, Stockert E, Aguet M, Old L J, Schreiber R D. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 4.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.Greenlund A C, Morales M O, Viviano B L, Yan H, Krolewski J, Schreiber R D. Immunity. 1995;2:677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 6.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 7.Roy S K, Wachira S J, Weihua X, Hu J, Kalvakolanu D V. J Biol Chem. 2000;275:12626–12632. doi: 10.1074/jbc.275.17.12626. [DOI] [PubMed] [Google Scholar]
- 8.Der S D, Zhou A, Williams B R, Silverman R H. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm U, Klamp T, Groot M, Howard J C. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 10.Müller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meraz M A, White J M, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 12.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 13.Briscoe J, Rogers N C, Witthuhn B A, Watling D, Harpur A G, Wilks A F, Stark G R, Ihle J N, Kerr I M. EMBO J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 14.Gil M P, Bohn E, O'Guin A K, Ramana C V, Levine B, Stark G R, Virgin H W, Schreiber R D. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C K, Smith E, Gimeno R, Gertner R, Levy D E. J Immunol. 2000;164:1286–1292. doi: 10.4049/jimmunol.164.3.1286. [DOI] [PubMed] [Google Scholar]
- 17.Ramana C V, Grammatikakis N, Chernov M, Nguyen H, Goh K C, Williams B R G, Stark G R. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrar M A, Campbell J D, Schreiber R D. Proc Natl Acad Sci USA. 1992;89:11706–11710. doi: 10.1073/pnas.89.24.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane T E, Asensio V C, Yu N, Paoletti A D, Campbell I L, Buchmeier M J. J Immunol. 1998;160:970–978. [PubMed] [Google Scholar]
- 20.Chernov M V, Ramana C V, Adler V V, Stark G R. Proc Natl Acad Sci USA. 1998;95:2284–2289. doi: 10.1073/pnas.95.5.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charles C H, Simske J S, O'Brien T P, Lau L F. Mol Cell Biol. 1990;10:6769–6774. doi: 10.1128/mcb.10.12.6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charles C H, Yoon J K, Simske J S, Lau L F. Oncogene. 1993;8:797–801. [PubMed] [Google Scholar]
- 23.Christy B A, Lau L F, Nathans D. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fambrough D, McClure K, Kazlauskas A, Lander E S. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 25.Cao X M, Guy G R, Sukhatme V P, Tan Y H. J Biol Chem. 1992;267:1345–1349. [PubMed] [Google Scholar]
- 26.Drysdale B E, Howard D L, Johnson R J. Mol Immunol. 1996;33:989–998. doi: 10.1016/s0161-5890(96)00043-0. [DOI] [PubMed] [Google Scholar]
- 27.Dogusan Z, Hooghe-Peters E L, Berus D, Velkeniers B, Hooghe R. J Neuroimmunol. 2000;109:34–39. doi: 10.1016/s0165-5728(00)00300-3. [DOI] [PubMed] [Google Scholar]
- 28.De S K, McMaster M T, Andrews G K. J Biol Chem. 1990;265:15267–15274. [PubMed] [Google Scholar]
- 29.Asano K, Nakamura H, Lilly C M, Klagsbrun M, Drazen J M. J Clin Invest. 1997;99:1057–1063. doi: 10.1172/JCI119233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Cai C Q, Schroeder R A, Kuo P C. J Immunol. 2001;166:1079–1086. doi: 10.4049/jimmunol.166.2.1079. [DOI] [PubMed] [Google Scholar]
- 31.Bi W, Wu L, Coustry F, de Crombrugghe B, Maity S N. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- 32.Tellides G, Tereb D A, Kirkiles-Smith N C, Kim R W, Wilson J H, Schechner J S, Lorber M I, Pober J S. Nature (London) 2000;403:207–211. doi: 10.1038/35003221. [DOI] [PubMed] [Google Scholar]
- 33.Kovary K, Bravo R. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wisdom R, Johnson R S, Moore C. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obaya A J, Mateyak M K, Sedivy J M. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 36.Cook D N, Beck M A, Coffman T M, Kirby S L, Sheridan J F, Pragnell I B, Smithies O. Science. 1995;269:1583–1585. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 37.Ransohoff R M, Tani M. Trends Neurosci. 1998;21:154–159. doi: 10.1016/s0166-2236(97)01198-3. [DOI] [PubMed] [Google Scholar]
- 38.Ohmori Y, Hamilton T A. J Immunol. 1995;154:5235–5244. [PubMed] [Google Scholar]
- 39.Lin R, Heylbroeck C, Genin P, Pitha P M, Hiscott J. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svaren J, Ehrig T, Abdulkadir S A, Ehrengruber M U, Watson M A, Milbrandt J. J Biol Chem. 2000;275:38524–38531. doi: 10.1074/jbc.M005220200. [DOI] [PubMed] [Google Scholar]
- 41.Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, Shimizu H, Yasui T, Ogihara H, Yasui N, et al. Oncogene. 1998;17:1517–1525. doi: 10.1038/sj.onc.1202064. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu A, O'Brien K P, Sjoblom T, Pietras K, Buchdunger E, Collins V P, Heldin C H, Dumanski J P, Ostman A. Cancer Res. 1999;59:3719–3723. [PubMed] [Google Scholar]
- 43.Heldin C H, Ostman A, Ronnstrand L. Biochim Biophys Acta. 1998;1378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 44.Heldin C H, Westermark B. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 45.Tannenbaum C S, Tubbs R, Armstrong D, Finke J H, Bukowski R M, Hamilton T A. J Immunol. 1998;161:927–932. [PubMed] [Google Scholar]
- 46.Balkwill F, Mantovani A. Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 47.Rodig S J, Meraz M A, White J M, Lampe P A, Riley J K, Arthur C D, King K L, Sheehan K C, Yin L, Pennica D, et al. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 48.Parganas E, Wang D, Stravopodis D, Topham D J, Marine J C, Teglund S, Vanin E F, Bodner S, Colamonici O R, van Deursen J M, et al. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 49.Pandey A, Mann M. Nature (London) 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 50.Pandey A, Fernandez M M, Steen H, Blagoev B, Nielsen M M, Roche S, Mann M, Lodish H F. J Biol Chem. 2000;275:38633–38639. doi: 10.1074/jbc.M007849200. [DOI] [PubMed] [Google Scholar]
- 51.Kriebel P, Patel B K, Nelson S A, Grusby M J, LaRochelle W J. Oncogene. 1999;18:7294–7302. doi: 10.1038/sj.onc.1203148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.