Abstract
In multicellular organisms, developmental changes to replication timing occur in 400–800 kb domains across half the genome. While examples of epigenetic control of replication timing have been described, a role for DNA sequence in mammalian replication-timing regulation has not been substantiated. To assess the role of DNA sequences in directing developmental changes to replication timing, we profiled replication timing in mice carrying a genetically rearranged Human Chromosome 21 (Hsa21). In two distinct mouse cell types, Hsa21 sequences maintained human-specific replication timing, except at points of Hsa21 rearrangement. Changes in replication timing at rearrangements extended up to 900 kb and consistently reconciled with the wild-type replication pattern at developmental boundaries of replication-timing domains. Our results are consistent with DNA sequence-driven regulation of Hsa21 replication timing during development and provide evidence that mammalian chromosomes consist of multiple independent units of replication-timing regulation.
INTRODUCTION
Eukaryotic DNA replication proceeds in a defined temporal order known as the replication-timing program. Though its significance is unknown, replication timing is evolutionarily conserved and is closely associated with the organization and function of chromatin (1,2). Accordingly, replication timing in multicellular organisms is cell-type specific and changes coordinately with gene expression during development (1–10). Programmed developmental changes in replication timing involve over half the genome and occur consistently in 400–800 kb increments (1,8). The discrete size of developmental regulatory units suggests that chromosomes consist of multiple domains that are independently regulated. An independent regulatory unit would be predicted to maintain its regulation in an ectopic genomic context. However, previous work in mammals has demonstrated that small DNA fragments containing single genes or replication origins do not generally transfer cis regulatory information to ectopic loci (11,12). Some combinations of strong transcriptional regulatory elements can affect replication timing in ectopic locations (13–15), but the importance of these elements to replication in their native context is unclear (16,17). Thus, the existence of cis elements that define domain-sized units of timing regulation remains unsubstantiated.
One possible explanation for the inability to identify cis elements of replication-timing control is that regulation requires DNA sequence information integrated across large segments of chromosomes. Alternatively, replication timing might be regulated entirely by epigenetic features that can only be transmitted in a chromatin context, as is suggested by the asynchronous replication of homologous loci during X inactivation (18,19) and imprinting (20), as well as the consequences of chromosome translocations (2,21–26). To assess the role of DNA sequences in the regulation of replication timing, we profiled replication timing in the Tc1 mouse model of Down syndrome, which had previously been used to evaluate the role of DNA sequence in directing species-specific transcription (27). The Tc1 strain was created by introducing Human Chromosome 21 (Hsa21) into mouse embryonic stem cells using irradiation microcell-mediated chromosome transfer, followed by chimeric mouse generation and germline transmission (28). Although the resulting Tc1 mice contain a single, freely segregating Hsa21, sequencing revealed the irradiated trans-chromosome underwent extensive genetic rearrangement during the establishment of the Tc1 mouse strain (S. Gribble, F. Wiseman and N. Carter, manuscript available upon request, ENA database Study Accession number: ERP000439). Thus, in addition to allowing single-copy analysis and direct comparison between syntenic mouse and human sequences in the same nucleus (27), Tc1 mice provided the opportunity to observe cis effects of ectopic genomic contexts on the replication timing of Hsa21 fragments in different mouse tissues.
RESULTS
Tc1 mouse replication-timing program is unperturbed by the presence of Hsa21
To verify normal replication-timing control in Tc1 mice, we generated genome-wide replication-timing profiles in Tc1 mouse fibroblasts and compared them with unrelated control mouse fibroblasts. Nascent DNA in asynchronously growing fibroblasts was first labeled with 5-bromo-2-deoxyuridine (BrdU), after which labeled fibroblasts were sorted into early and late S-phase fractions by flow cytometry, and BrdU-labeled DNA was purified by immunoprecipitation (Supplementary Material, Fig. S1). Purified DNA fractions were differentially labeled and co-hybridized to a comparative genomic hybridization (CGH) microarray with 3.5 kb median probe spacing across the entire mouse genome. Hybridization data were then Loess normalized and smoothed to provide a genome-wide profile with relative replication-timing values for each probe position (29). We found a strong, genome-wide correlation between Tc1 and control mouse fibroblast samples (r= 0.92, also see Supplementary Material, Fig. S2), demonstrating that the overall replication-timing program in Tc1 mice was not disrupted by the presence of a foreign chromosome.
DNA sequence variation accounts for divergent replication timing across regions of conserved Hsa21 synteny
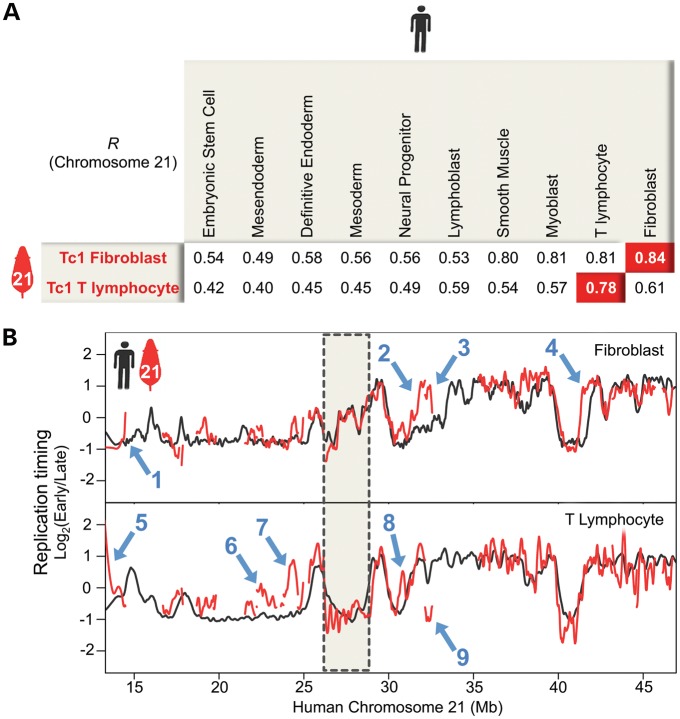
To determine the extent to which the Tc1 trans-chromosome (Tc1-21) maintains its human replication-timing regulation, we generated chromosome-wide profiles in two Tc1 mouse cell types (fibroblasts and T lymphocytes) and corresponding human controls (fibroblasts and T lymphocytes) using a Hsa21 tiling array with 70 bp median probe spacing across the chromosome. Since duplicated segments could replicate at different times but could not be distinguished by DNA sequence, array probes for Tc1-21 regions with copy number variation were omitted from our analysis (Supplementary Material, Fig. S3). Timing profiles of the remaining 25 Mb exhibit general maintenance of human replication-timing regulation in Tc1 mouse fibroblasts and T lymphocytes (Fig. 1). Tc1-21 replication timing in each mouse cell type is most highly correlated with Hsa21 replication timing in the matching human cell type (Fig. 1A). High correlation between Tc1-21 and Hsa21 in non-matching cell types, such as between fibroblast and myoblast, is also consistent with previous studies (8,30). Cell-type-specific regulation is clearly evident between Hsa21 coordinates 27 and 28 Mb, which contain a late-replicating domain that shifts to early replication specifically in T lymphocytes (Fig. 1B).
Figure 1.
Developmental regulation of Hsa21 replication timing is maintained in Tc1 mice. (A) Pearson's correlations are shown between Tc1-21 and Hsa21 in various cell types. (B) Tc1-21 (red) and control Hsa21 (black) replication-timing profiles are shown in fibroblasts and T-lymphocytes. Regions of Tc1-21 copy number variation were omitted from analysis and correspond to gaps in Tc1-21 profiles. The gray box highlights a developmentally regulated replication domain between Hsa21 coordinates 27 and 28 Mb, which shifts to early replication specifically in fibroblasts. Blue arrows 1–9 indicate the regions in which Tc1-21 replication timing deviates from Hsa21 controls.
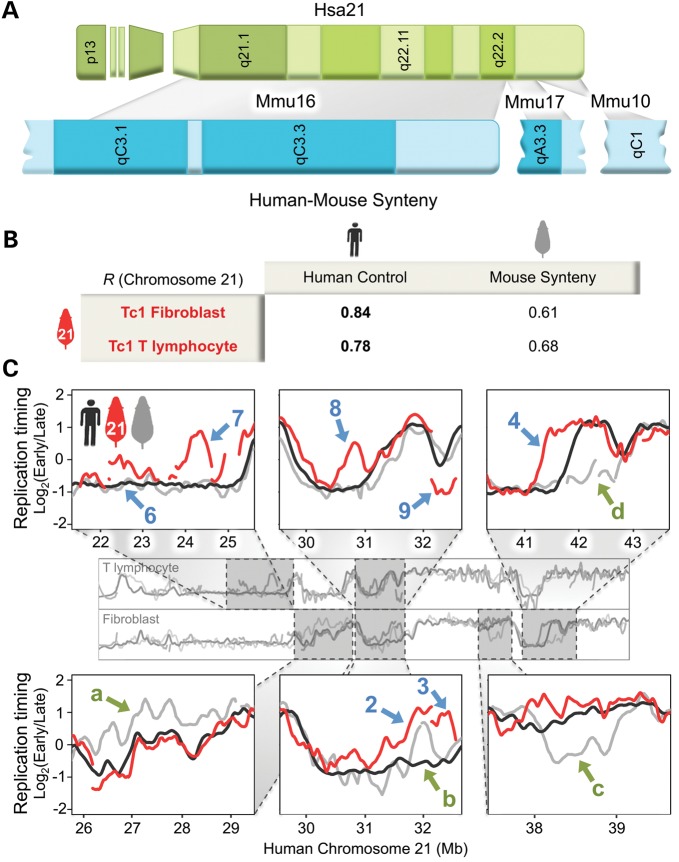
Despite the general maintenance of human replication timing, nine Tc1-21 regions deviated considerably from Hsa21 controls (indicated by blue arrows 1–9 in Fig. 1B). To determine whether these replication-timing differences were due to the divergence of mouse and human trans-acting factors, we examined the conservation of replication-timing regulation across the regions of conserved Hsa21 synteny in mouse (Fig. 2A). Syntenic regions of mouse and human replication-timing profiles were compared by converting genomic coordinates of each mouse probe to human coordinates using the UCSC genome lift annotation tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver, last accessed date on 2 July 2012, see Materials and methods). Using this method, we obtained correlations between Hsa21 and syntenic mouse replication-timing profiles (r= 0.87 for T lymphocytes and 0.77 for fibroblasts) consistent with previous studies (1,2), which demonstrate a high degree of conservation of the replication-timing program between similar human and mouse cell types. In addition, we identified four regions (indicated by green arrows a–d in Fig. 2C) at which profiles for Hsa21 (black curve in Fig. 2C) and mouse synteny (gray curve in Fig. 2C) diverge. However, of the nine replication-timing differences between Tc1-21 and Hsa21, only one (blue arrow 2 in Fig. 2C) fell within a region of evolutionarily divergent mouse replication timing (green arrow b in Fig. 2C), indicating that differences between mouse and human trans-acting factors were not likely responsible for the replication-timing differences between Tc1-21 and Hsa21. Consistent with this conclusion, Tc1-21 exhibited human-specific replication timing at all other regions of divergent mouse replication timing in both fibroblasts and T lymphocytes (green arrows a, c and d in Fig. 2C) and overall aligned more closely with Hsa21 than syntenic mouse sequences throughout the trans-chromosome (Fig. 2B), implicating DNA sequences in the regulation of replication timing at these loci.
Figure 2.
Tc1-21 exhibits human-specific replication timing across syntenic regions in multiple mouse cell types. (A) Major regions of conserved Hsa21 (green) synteny are distributed across mouse chromosomes 10, 16 and 17 (blue). Chromosome bands are represented by different color shades. Gray boxes delineate syntenic regions. (B) Pearson's correlations of replication-timing profiles are shown between Tc1-21, Hsa21 and the corresponding mouse syntenic regions in fibroblasts and T lymphocytes. (C) Zoomed-in profiles of Tc1-21 (red), control Hsa21 (black) and the corresponding mouse synteny (gray) replication timing are shown in fibroblasts and T lymphocytes. The same numbered blue arrows in Figure 1B are shown to indicate regions in which Tc1-21 replication timing deviates from Hsa21 controls. Green arrows a–d indicate syntenic regions in which human and mouse replication timing diverge. Only one region in which Tc1-21 replication timing deviates from Hsa21 controls (blue arrow 2) coincides with a region in which human and mouse replication timing diverge (green arrow b).
Intact domains maintain replication timing independent of genomic context while fragmented domains are replicated with flanking sequences
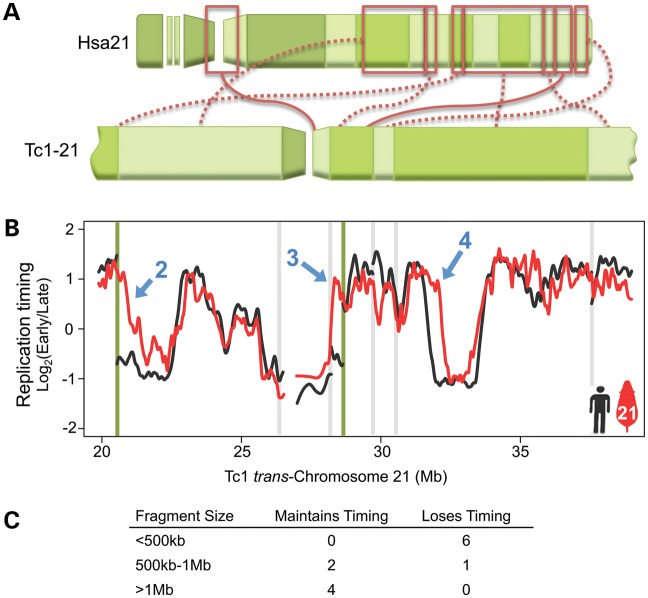
Because chromosome rearrangement is often associated with disrupted replication timing (2,21–26), we reconstructed Tc1-21 replication-timing profiles to reflect the trans-chromosome's actual sequence (S. Gribble, F. Wiseman and N. Carter, manuscript available upon request, ENA database Study Accession number: ERP000439; Fig. 3A). The rearranged nature of Tc1-21 provided numerous opportunities to evaluate the consequences of chromosome rearrangement to replication-timing regulation. Of the nine replication-timing differences observed between Tc1-21 and control Hsa21, seven were immediately proximal to rearrangement points (including the one indicated by blue arrow 2 in Fig. 2C) while the remaining two (blue arrows 4 and 8 in Fig. 2C) were within 2 Mb of rearrangement points. A majority (five) of Tc1-21 changes occurred at rearrangements that fused early- and late-replicating regions, consistent with studies of evolutionary chromosome rearrangement (2). The remaining four changes occurred at fusions of regions with similar replication timing (three) or at a region fused to repetitive sequences excluded from our microarray. Figure 3B shows fibroblast replication-timing profiles across two early–late fusions (indicated by green lines). In each of these cases, the early side remained unchanged while the late side exhibited advanced replication timing (green lines in Fig. 3B). A third early–late fusion resulted in delayed replication timing of the early side (Supplementary Material, Fig. S4B). This raises the intriguing question of what molecular mechanisms determine whether early or late replication dominates at fusion points. A comparison of GC content and repetitive DNA sequences surrounding each of the five early–late fusions did not reveal any clear relationship between sequence composition and dominance of replication timing (Supplementary Material, Fig. S4E), however, additional examples are required for a conclusive analysis. Taken together, the coincidence of Tc1-21 rearrangement points and shifts in replication timing strongly suggests that the cis regulation of replication timing in these regions was disrupted by genetic rearrangement.
Figure 3.
Large Tc1-21 fragments maintain replication timing independent of genomic context, while small fragments are replicated with flanking sequences. (A) Diagram of rearranged Tc1-21 segments plotted in (B) and the corresponding Hsa21 segments (boxed in red) are displayed. Solid red lines connect regions in Hsa21 orientation, while dotted lines indicate inversions. (B) Tc1-21 (red) and control Hsa21 (black) fibroblast replication-timing profiles are plotted across a 20-Mb region of rearranged Tc1-21. Vertical green lines mark fusion points of early- and late-replicating control regions, while vertical gray lines mark fusions of regions with similar replication timing. The same numbered blue arrows in Figure 1B are shown to indicate regions in which Tc1-21 replication timing deviates from Hsa21 controls. Differences between Hsa21 and Tc1-21 profiles are seen at both early–late fusion points (blue arrows 2 and 3) and between Tc1-21 coordinates 32 and 32.5 Mb (blue arrow 4), which is 2 Mb from an early–early fusion point. (C) Comparison of Tc1-21 fragment size versus maintenance of replication timing in foreign timing context relative to Hsa21 controls.
Because small, integrated DNA constructs generally replicate at times consistent with their surrounding genomic context (11,12), we next looked at whether the size of rearranged fragments on Tc1-21 correlated with preservation of replication-timing control at early–late fusions. Table 1 lists all fragments of the trans-chromosome by size and indicates whether native replication timing was maintained for fragments involved in early–late fusion events. All fragments smaller than 500 kb lost timing control, while all fragments larger than 1 Mb maintained timing control independent of chromosomal context (Fig. 3C). Of the three fragments between 500 kb and 1 Mb, two (650 and 960 kb) maintained timing while one (850 kb) did not, suggesting that regulation may not be determined by size alone but rather that larger size corresponds with greater likelihood that sufficient elements are contained within a given fragment.
Table 1.
Developmental timing features of Tc1-21 fragments
| Hsa21 coordinates (hg18) | Size (kb) | Timing maintained | Developmental timing features | Name in Figure 4 and Supplementary Material, Figure S4 |
|---|---|---|---|---|
| 22215973–22228788 | 13 | — | — | — |
| 9730110–9761585 | 31 | No | Interior piece of constitutive late domain | — |
| 17795451–17864961 | 70 | No | Interior piece of developmentally regulated late domain | L7 |
| 19678800–19815865 | 137 | No | Interior piece of constitutive late domain | — |
| 23647747–23802829 | 155 | — | — | L8 |
| 19435895–19678793 | 243 | — | — | — |
| 9761586–10041451 | 280 | No | Piece of developmentally regulated domain boundary | L6 |
| 24649893–24960295 | 310 | — | — | — |
| 22228789–22570341 | 342 | No | Interior piece of constitutive late domain | — |
| 32145444–32562221 | 417 | No | Developmentally regulated domain boundary | L3 |
| 21534106–22120244 | 586 | — | — | — |
| 46146529–46741200 | 595 | — | — | — |
| 35292160–35939108 | 647 | Yes | Developmentally regulated early domain | E1 |
| 18683561–19435894 | 752 | — | — | — |
| 22570342–23382690 | 812 | — | — | — |
| 23802830–24649883 | 847 | No | Constitutive late domain | L5 |
| 25298129–26185187 | 887 | — | — | — |
| 16693182–17656030 | 963 | Yes | Developmentally regulated late domain with boundaries | L4 |
| 44620047–45692741 | 1073 | Yes | Constitutive early domain with upstream boundary | E2 |
| 13260011–14483921 | 1224 | Yes | Constitutive late domain with boundaries | L2 |
| 43187508–44620046 | 1433 | Yes | Constitutive early domain downstream boundary | — |
| 26185188–32145307 | 5960 | Yes | Eight domains and upstream boundary | L1 |
| 35939109–43187507 | 7248 | — | — | — |
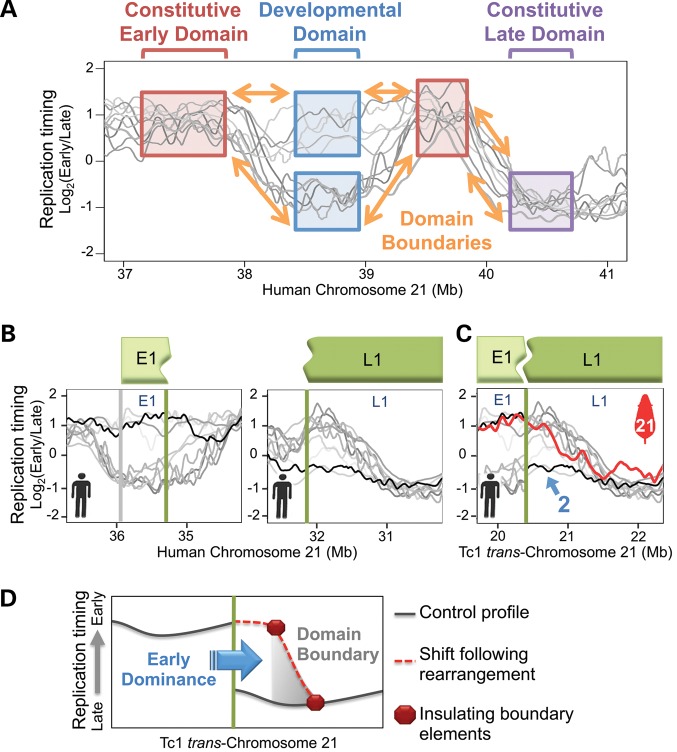
Given that the size range of the minimal Tc1-21 fragments that maintained timing regulation (500 kb–1 Mb) closely matches the sizes of developmentally regulated replication domains (400–800 kb), we examined whether particular developmental timing features could be found within the Tc1-21 fragments with preserved timing. Using previously published replication-timing profiles from a variety of human cell types (1,10: http://www.replicationdomain.org, last accessed date on 2 July 2012), we classified each Tc1-21 fragment based on its developmental regulation (Fig. 4A, also see Materials and methods). With one exception, all fragments that maintained replication timing at early–late fusions contained at least one intact developmental domain and some part of the boundaries between domains with differential timing (Table 1). The exception, an intact domain without any of the surrounding boundaries (647 kb fragment in Table 1), maintained its early replication time, suggesting that boundaries may only play a role in establishing late replication. Indeed, the largest fragment that lost timing regulation (847 kb fragment in Table 1) was a constitutive late domain without any of the surrounding boundaries. Further, fragments containing isolated boundaries (417 and 280 kb fragments in Table 1) or partial late domains (342, 137, 70 and 32 kb fragments in Table 1) were insufficient to maintain replication timing when repositioned. The sufficiency of partial early domains to direct replication timing could not be addressed because none were isolated among the analyzed Tc1-21 fragments. Together, these results suggest developmentally regulated replication domains retaining at least part of their surrounding boundaries represent sufficient cis regulatory units of replication timing.
Figure 4.
Replication boundaries insulate domains from position effects. (A) Replication-timing profiles from various human cell types (shades of gray, see Materials and methods) are shown across 4 Mb of Hsa21 to exemplify the method employed to classify Tc1-21 fragments in Table 1. Examples of constitutive and developmentally regulated replication-timing domains are boxed and labeled with brackets. Boundaries connecting adjacent domains are indicated with orange arrows. (B) Control Hsa21 replication-timing profiles from fibroblasts (black) and other various human cell types (shades of gray, see Materials and methods) are plotted across both regions involved in an early–late fusion in Figure 3A. Fragments E1 and L1 correspond, respectively, to 647 and 5960 kb fragments from Table 1. Vertical green lines mark the point of early–late fusion at which two developmental domains are adjoined without a domain boundary in between. The vertical gray line marks the opposite end of fragment E1. The opposite end of fragment L1 is not shown due to its distance from the early–late fusion point. (C) Fragments E1 and L1 from (B) are plotted in Tc1-21 arrangement. Tc1-21 fibroblast (red) replication-timing profile is overlaid on profiles from control human fibroblasts (black) and other human cell types (shades of gray, see Materials and methods). The same numbered blue arrow in Figure 1B is shown to indicate a region in which Tc1-21 replication timing deviates from Hsa21 controls. (D) Illustrated model depicts a shift to early replication on the late side of an early–late fusion (vertical green line). The dominant position effect (early replication) spreads up to the nearest developmental domain boundary (gray triangle), which contains insulating elements (red octagons) that prevent further spreading.
Replication boundaries insulate domains from position effects
The previous data also suggest that replication boundaries allow independent regulation of adjacent domains by insulating against position effects. This hypothesis would predict that, in cases where early and late replication domains become juxtaposed, position effects on replication timing imposed by the dominant domain should spread to the next developmental boundary. Indeed, a comparison of early–late fusions revealed that replication-timing shifts in all five cases did not spread beyond the closest detected developmental boundary. One example is shown in Figure 4B and C. In this example, replication of fragment L1, which occurs in late S in its normal Hsa21 context, shifted to the earlier replication time of its fusion partner, fragment E1. This shift extended into fragment L1 up to a developmental boundary that can be detected in other cell types. The remaining four examples were more complex, each involving multiple smaller translocated fragments. Still in each case, timing shifts extended through regions without detected developmental boundaries and ended near boundaries found in other cell types (Supplementary Material, Fig. S4A–D). These results indicate that developmental boundaries could retain their ability to function as boundaries in cis, independent of cell type or species background.
DISCUSSION
Our analysis of Tc1-21 suggests that the divergence of human and mouse replication timing across Hsa21 regions of conserved synteny has been primarily driven by differences in the DNA sequence. After several generations of meiotic transmission and epigenetic reprogramming in mice (31–33), syntenic Tc1-21 sequences continued to replicate in human-specific patterns in two distinct mouse tissues. The possibility that human-specific epigenetic marks determine the replication timing of these regions independent of the underlying DNA sequence would require such marks to be consistently preserved on Tc1-21 by the mouse epigenetic machinery. In addition, analysis of rearranged Tc1-21 fragments suggests that sufficient replication-timing regulatory information is contained within chromosome fragments 500 kb–1 Mb in length.
Our results also provide evidence that boundaries of developmentally regulated replication-timing domains contain insulating cis regulatory elements (Fig. 4D). Rearranged Tc1-21 fragments that contained developmental domains and their boundaries maintained replication-timing control, while fragments lacking boundaries experienced shifts in replication timing that extended 500–900 kb, up to the nearest detected replication boundary. This result is consistent with previous observations that deletions of a well-defined replication-timing boundary at the mouse immunoglobulin heavy-chain (Igh) locus create a new late domain boundary 500 kb downstream of the deletion (34). In addition, replication-timing aberrations in acute lymphoblastic leukemia patients are also bounded by normal replication boundaries found in different cell types (35).
Altogether, this study gives unprecedented insight into the organization and regulation of mammalian chromosome domains, supporting the replication-domain model of replication-timing regulation. It also raises a number of intriguing questions. For example, given the striking similarity of genome-wide replication-timing profiles and chromatin interaction maps (1,36), what cis elements might coordinately regulate replication timing and chromatin interactions? Conserved non-coding sequences have been shown to interact with each other and play important regulatory roles in a variety of processes including enhancement of gene expression (37). Could similar elements cluster to form domains for coordinate replication? Interestingly, Forkhead transcription factors in budding yeast bind to replication origins, organize these origins into spatial clusters and are required for their early replication timing (38). In addition, recent genome-wide analysis in human and mouse has identified conserved topologically associating domains (TADs) flanked by boundaries enriched in insulating elements (36). Although TADs were reported to be independent from replication domains (36), the relatively abrupt transitions at TAD boundaries could mark insulating elements within the more gradual transitions at replication-domain boundaries. These results implicate cis elements that dictate chromatin interactions as candidate elements for directing replication-timing regulation during mammalian development.
MATERIALS AND METHODS
Cell culture
Tc1 mice with Hsa21 and wild-type littermates used in this study were bred by crossing female Tc1 mice to a male (129S8 × C57BL/6J)F1 mouse. Tc1 mouse fibroblasts were isolated using a modified protocol (39) from E13.5 post coitum embryos in Leibovitz's L-15 GlutaMAX I medium (Gibco, UK, Invitrogen). The head and visceral organs were removed and remaining tissues were minced and suspended in 1.5 ml of 0.05% Trypsin (Gibco, Invitrogen) in Dulbecco's phosphate buffered saline (with Ca2+/Mg2+) supplemented with DNase I (200 U, Invitrogen). Tissues were incubated at 37°C for 20 min, transferred into warm culturing medium [D-MEM GlutaMAX I supplemented with 10% fetal bovine serum (FBS) and 1/100 (v/v) penicillin/streptomycin]. After settling, the cell suspension was cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% FBS and maintained at ambient oxygen. Experiments were performed between population doubling (PD) 2 and PD 3. Tc1 mouse T lymphocytes (CD4+ splenocytes) were obtained by the mechanical disaggregation of whole spleens in RPMI-1640 medium (Invitrogen) supplemented with 2 mm l-glutamine, 10% fetal calf serum (Gibco), 100 IU/ml penicillin and 0.1 mg/ml streptomycin (Invitrogen). After passing through a 100-μm filter, CD4+ splenocytes were obtained by centrifugation over Lymphoprep and then negative selection using the murine CD4+ isolation kit II (Miltenyi Biotec) according to the manufacturers' instructions. Mice were maintained in compliance with UK Home Office regulations.
Human control fibroblasts (fetal lung, IMR90, ATCC CCL-186) were cultured in DMEM supplemented with 10% FBS and were maintained in physiological 5% oxygen. Human control T lymphocytes were obtained from healthy volunteers after written informed consent and with the approval of the Cambridge (UK) Research Ethics Committee. None gave a history of chronic illness or intravenous drug usage. CD4+ peripheral blood mononuclear cells (PBMCs) were obtained by the centrifugation of citrated blood over Lymphoprep (Nycomed, Roskilde, Denmark) before negative selection using the human CD4+ isolation kit II (Miltenyi Biotec) according to the manufacturers' instructions. CD4+ purity routinely exceeded 90% in both human and murine preparations.
CD4+ PBMCs or splenocytes were cultured in RPMI-1640 medium (Invitrogen ) supplemented with 2 mm l-glutamine, 10% fetal calf serum (Gibco), 100 IU/ml penicillin and 0.1 mg/ml streptomycin (Invitrogen). Human and murine CD4+ T-cells were stimulated with 5 μg/ml of phytohaemagglutinin in a humidified 5% CO2 atmosphere for 3 days prior to harvesting.
Genome-wide replication-timing profile generation
Replication-timing profiles were generated as described (29) using a whole-genome microarray with 3.5 kb median probe spacing (Roche NimbleGen Inc., 100718_MM9_WG_CGH; 713,358 oligonucleotide probes) for mouse whole-genome data or an Hsa21-specific tiling array with 70 bp median probe spacing (Roche NimbleGen Inc., 2006-09-22_HG18_CHR21_FT; 385,183 oligonucleotide probes) for Tc1-21 and Hsa21 data. Sample labeling, microarray hybridization and data extraction were performed according to the standard procedures recommended by NimbleGen. A complete replication-timing data set for all probes is downloadable and graphically displayed at http://www.replicationdomain.org, last accessed date on 2 July 2012 (40).
Cross-hybridization of mouse sequences to Hsa21 array probes was addressed by calculating the similarity of each Hsa21 array probe (50–75 bp) to sequences in the mouse genome. Less than 3% of all probes had greater than 80% homology to mouse sequences making significant interference from cross-hybridization unlikely (41). In addition, no significant signal was detected on Hsa21 arrays hybridized with samples from Tc1 littermates lacking Hsa21.
Replication-timing profile analysis
Replication-timing data sets were normalized, averaged from at least two biological replicates and scaled together using the limma package in R as described previously (5,29). Loess smoothing was applied across a span of 300 kb to normalized replication-timing ratios (log2early/late) at each probe to generate a genome-wide profile. The Tc1-21 replication-timing profile displayed in all figures was obtained by smoothing Hsa21 array data ordered according to the Tc1-21 sequence (S. Gribble, F. Wiseman and N. Carter, manuscript available upon request, ENA database Study Accession number: ERP000439). Syntenic regions of mouse and human timing profiles were compared by converting genomic coordinates of each mouse probe to human coordinates using the UCSC genome lift annotation tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver, last accessed date on 2 July 2012) with accepted default parameters. Previously published human replication-timing data sets used for developmental analysis in Figures 1 and 4 and Table 1 included lymphoblast [male lymphoblastoid cell line with normal (46, XY) karyotype, CO202 ECCAC no. 94060845], ESC (BG01, BG02, H7, H9), BG01-derived NPC (1), ESC-derived mesendoderm, mesoderm, definitive endoderm and smooth muscle (10) and primary myoblast profiles (30). Hsa21 and Tc1-21 timing profiles ordered with respect to the Tc1-21 sequence (S. Gribble, F. Wiseman and N. Carter, manuscript available upon request, ENA database Study Accession number: ERP000439), and syntenic mouse replication-timing profiles converted to human coordinates have been deposited in the GEO database (Study Accession number: GSE38472).
DNA sequence composition analysis
DNA sequence composition characteristics listed in Supplementary Material, Figure S4E, were calculated using the UCSC Table Browser tool (http://genome.ucsc.edu/cgi-bin/hgTables, last accessed date on 2 July 2012) for the 500 kb on either side of each fusion point or for the entire fused fragment if smaller than 500 kb. GC content was calculated by averaging 5 base window GC Percent (table: gc5Base) values across each region, and repeat class densities were calculated by summing the length of each repeat identified by RepeatMasker (table: rmsk), dividing by the total length of each region and multiplying by 100.
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by the National Institutes of Health (GM083337 to D.M.G.), European Research Council (D.T.O.), the EMBO Young Investigator Programme (D.T.O.), Hutchinson Whampoa (D.T.O.), Cancer Research, UK (T.C., M.D.W., M.H.) and the University of Cambridge (T.C., M.D.W.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elizabeth Fisher and Victor Tybulewicz for providing Tc1 mouse resources, Ruth Didier for assistance with flow cytometry, Susan Gribble and Nigel Carter for sharing Tc1 trans-chromosome sequencing data prior to publication and Masashi Narita, Ichiro Hiratani, Matthias Merkenschlager and Shin-ichiro Takebayashi for helpful discussions.
Conflict of Interest statement. None declared.
References
- 1.Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., Schulz T.C., Robins A.J., Dalton S., Gilbert D.M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yaffe E., Farkash-Amar S., Polten A., Yakhini Z., Tanay A., Simon I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiratani I., Leskovar A., Gilbert D.M. Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. Proc. Natl Acad. Sci. USA. 2004;101:16861–16866. doi: 10.1073/pnas.0406687101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry P., Sauer S., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle. 2004;3:1645–1650. [PubMed] [Google Scholar]
- 5.Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C.-W., Lyou Y., Townes T.M., Schübeler D., Gilbert D.M. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwaiger M., Stadler M.B., Bell O., Kohler H., Oakeley E.J., Schübeler D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desprat R., Thierry-Mieg D., Lailler N., Lajugie J., Schildkraut C., Thierry-Mieg J., Bouhassira E.E. Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 2009;19:2288–2299. doi: 10.1101/gr.094060.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D.P., Plath K., Dalton S., et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen R.S., Thomas S., Sandstrom R., Canfield T.K., Thurman R.E., Weaver M., Dorschner M.O., Gartler S.M., Stamatoyannopoulos J.A. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryba T., Hiratani I., Sasaki T., Battaglia D., Kulik M., Zhang J., Dalton S., Gilbert D.M. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput. Biol. 2011;7:e1002225. doi: 10.1371/journal.pcbi.1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert D.M., Cohen S.N. Position effects on the timing of replication of chromosomally integrated simian virus 40 molecules in Chinese hamster cells. Mol. Cell. Biol. 1990;10:4345–4355. doi: 10.1128/mcb.10.8.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Z., Hughes C.M., Kosiyatrakul S., Norio P., Sen R., Fiering S., Allis C.D., Bouhassira E.E., Schildkraut C.L. Decreased replication origin activity in temporal transition regions. J. Cell Biol. 2009;187:623–635. doi: 10.1083/jcb.200905144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon I., Tenzen T., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H. Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C.M., Fu H., Martinovsky M., Bouhassira E., Aladjem M.I. Dynamic alterations of replication timing in mammalian cells. Curr. Biol. 2003;13:1019–1028. doi: 10.1016/s0960-9822(03)00382-8. [DOI] [PubMed] [Google Scholar]
- 15.Goren A., Tabib A., Hecht M., Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimbora D.M., Schübeler D., Reik A., Hamilton J., Francastel C., Epner E.M., Groudine M. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol. Cell. Biol. 2000;20:5581–5591. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bender M.A., Byron R., Ragoczy T., Telling A., Bulger M., Groudine M. Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for beta-globin transcription. Blood. 2006;108:1395–1401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagi N., Sugawara O., Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- 19.Hansen R.S., Canfield T.K., Fjeld A.D., Gartler S.M. Role of late replication timing in the silencing of X-linked genes. Hum. Mol. Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- 20.Gribnau J., Hochedlinger K., Hata K., Li E., Jaenisch R. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 2003;17:759–773. doi: 10.1101/gad.1059603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Disteche C.M., Eicher E.M., Latt S.A. Late replication in an X-autosome translocation in the mouse: correlation with genetic inactivation and evidence for selective effects during embryogenesis. Proc. Natl Acad. Sci. USA. 1979;76:5234–5238. doi: 10.1073/pnas.76.10.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guinta D.R., Tso J.Y., Narayanswami S., Hamkalo B.A., Korn L.J. Early replication and expression of oocyte-type 5S RNA genes in a Xenopus somatic cell line carrying a translocation. Proc. Natl Acad. Sci. USA. 1986;83:5150–5154. doi: 10.1073/pnas.83.14.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karube T., Watanabe S. Analysis of the chromosomal DNA replication pattern using the bromodeoxyuridine labeling method. Cancer Res. 1988;48:219–222. [PubMed] [Google Scholar]
- 24.State M.W., Greally J.M., Cuker A., Bowers P.N., Henegariu O., Morgan T.M., Gunel M., DiLuna M., King R.A., Nelson C., et al. Epigenetic abnormalities associated with a chromosome 18(q21-q22) inversion and a Gilles de la Tourette syndrome phenotype. Proc. Natl Acad. Sci. USA. 2003;100:4684–4689. doi: 10.1073/pnas.0730775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breger K.S., Smith L., Thayer M.J. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum. Mol. Genet. 2005;14:2813–2827. doi: 10.1093/hmg/ddi314. [DOI] [PubMed] [Google Scholar]
- 26.Chang B.H., Smith L., Huang J., Thayer M. Chromosomes with delayed replication timing lead to checkpoint activation, delayed recruitment of Aurora B and chromosome instability. Oncogene. 2007;26:1852–1861. doi: 10.1038/sj.onc.1209995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson M.D., Barbosa-Morais N.L., Schmidt D., Conboy C.M., Vanes L., Tybulewicz V.L.J., Fisher E.M.C., Tavaré S., Odom D.T. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Doherty A., Ruf S., Mulligan C., Hildreth V., Errington M.L., Cooke S., Sesay A., Modino S., Vanes L., Hernandez D., et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryba T., Battaglia D., Pope B.D., Hiratani I., Gilbert D.M. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat. Protoc. 2011;6:870–895. doi: 10.1038/nprot.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pope B.D., Tsumagari K., Battaglia D., Ryba T., Hiratani I., Ehrlich M., Gilbert D.M. DNA replication timing is maintained genome-wide in primary human myoblasts independent of D4Z4 contraction in FSH muscular dystrophy. PLoS One. 2011;6:e27413. doi: 10.1371/journal.pone.0027413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 32.Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 33.Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., Ermakova O.V., Riblet R., Birshtein B.K., Schildkraut C.L. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryba T., Battaglia D., Chang B.H., Shirley J.W., Buckley Q., Pope B.D., Devidas M., Druker B.J., Gilbert D.M. Abnormal developmental control of replication timing domains in pediatric acute lymphoblastic leukemia. Genome Res. 2012 doi: 10.1101/gr.138511.112. doi:10.1101/gr.138511.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012 doi: 10.1038/nature11082. doi:10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robyr D., Friedli M., Gehrig C., Arcangeli M., Marin M., Guipponi M., Farinelli L., Barde I., Verp S., Trono D., et al. Chromosome conformation capture uncovers potential genome-wide interactions between human conserved non-coding sequences. PLoS One. 2011;6:e17634. doi: 10.1371/journal.pone.0017634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knott S.R.V., Peace J.M., Ostrow A.Z., Gan Y., Rex A.E., Viggiani C.J., Tavaré S., Aparicio O.M. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reznikoff C.A., Brankow D.W., Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973;33:3231–3238. [PubMed] [Google Scholar]
- 40.Weddington N., Stuy A., Hiratani I., Ryba T., Yokochi T., Gilbert D.M. ReplicationDomain: a visualization tool and comparative database for genome-wide replication timing data. BMC Bioinformatics. 2008;9:530. doi: 10.1186/1471-2105-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renn S.C.P., Machado H.E., Jones A., Soneji K., Kulathinal R.J., Hofmann H.A. Using comparative genomic hybridization to survey genomic sequence divergence across species: a proof-of-concept from Drosophila. BMC Genomics. 2010;11:271. doi: 10.1186/1471-2164-11-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.