Abstract
The G2019S leucine rich repeat kinase 2 (LRRK2) mutation is the most common genetic cause of Parkinson's disease (PD), clinically and pathologically indistinguishable from idiopathic PD. Mitochondrial abnormalities are a common feature in PD pathogenesis and we have investigated the impact of G2019S mutant LRRK2 expression on mitochondrial bioenergetics. LRRK2 protein expression was detected in fibroblasts and lymphoblasts at levels higher than those observed in the mouse brain. The presence of G2019S LRRK2 mutation did not influence LRRK2 expression in fibroblasts. However, the expression of the G2019S LRRK2 mutation in both fibroblast and neuroblastoma cells was associated with mitochondrial uncoupling. This was characterized by decreased mitochondrial membrane potential and increased oxygen utilization under basal and oligomycin-inhibited conditions. This resulted in a decrease in cellular ATP levels consistent with compromised cellular function. This uncoupling of mitochondrial oxidative phosphorylation was associated with a cell-specific increase in uncoupling protein (UCP) 2 and 4 expression. Restoration of mitochondrial membrane potential by the UCP inhibitor genipin confirmed the role of UCPs in this mechanism. The G2019S LRRK2-induced mitochondrial uncoupling and UCP4 mRNA up-regulation were LRRK2 kinase-dependent, whereas endogenous LRRK2 levels were required for constitutive UCP expression. We propose that normal mitochondrial function was deregulated by the expression of G2019S LRRK2 in a kinase-dependent mechanism that is a modification of the normal LRRK2 function, and this leads to the vulnerability of selected neuronal populations in PD.
INTRODUCTION
Parkinson's disease (PD) is the second most common neurodegenerative disease affecting 1.5% of the population over 65 years of age (1). Although the causes of PD have not been identified in the majority of cases, patients share the core clinical symptoms of bradykinesia, akinesia and rigidity, and the pathological features of dopaminergic neuronal loss and the presence of Lewy bodies. Various genetic mutations have been detected in PD, with the G2019S mutation of the leucine rich repeat kinase 2 (LRRK2) gene being the most common with a prevalence in PD patients of 1–2% in the UK and USA, rising to 10% in Ashkenazi Jews and 39% among the North African Berber population (2). These patients exhibit clinical symptoms and pathology typical of sporadic PD and therefore may share a similar mechanism of disease initiation and propagation. LRRK2 encodes a multi-domain protein belonging to the ROCO family characterized by an Ras of complex (ROC) GTPase domain, a C-terminus of ROC domain and a kinase domain (3–5). Although the G2019S mutation is in the kinase domain and leads to increased kinase activity (6) linked to its toxicity (7), mutations have also been described in other domains of the protein including the GTPase.
LRRK2 is widely expressed in the brain and peripheral tissues, with the highest mRNA abundance in kidneys, lungs and lymph nodes (8). High levels of LRRK2 protein have been detected in animal and human studies of these tissues in support of the RNA data (9–11). Although LRRK2 mRNA has been detected in dopaminergic areas of the brain (12), the corresponding analysis of LRRK2 protein is limited (13).
The function of LRRK2 is unknown but it has been suggested to play a role in a wide variety of cellular processes including vesicle endocytosis (14), neurite morphology (15), autophagy (16), Wnt signalling (17), microRNA (18) and transcription factor regulation (11,19). Several LRRK2 substrates and interacting proteins have been described, including ezrin, radixin, moesin proteins, mitogen-activated protein kinase, eukaryotic initiation factor 4E-binding protein, tubulin, futsch and 14-3-3 proteins. Some of these substrates link in with the proposed cellular functions of LRRK2 (20).
The majority of data suggest that LRRK2 is located predominantly in the cytosol, with a proportion associated with cellular membranes (21–23). LRRK2 has been suggested to exist as a dimer, which is more abundant when membrane-associated, where it has increased kinase activity (21). There is no suggestion that G2019S mutant (MT) LRRK2 has an altered subcellular location or segregation to membranes (23). However, there are contradictory data regarding with which subcellular membranes LRRK2 is associated, including synaptic vesicles (22,23), lysosomes (22), lipid rafts (23) and mitochondria (14,22,24).
The potential localization of a fraction of LRRK2 to mitochondria links this protein to a pathway suggested to play an important role in PD pathogenesis. In particular, decreased mitochondrial complex I function and increased levels of mitochondrial DNA deletions have been reported in PD substantia nigra (25). In addition, PD caused by autosomal recessive PINK1 and parkin mutations has been suggested to be caused by aberrant turnover of damaged mitochondria (26), further supporting the role of mitochondrial dysfunction in PD. The particular vulnerability of dopaminergic neurons to systemic exposure to the mitochondrial toxins MPTP and rotenone may explain their selective loss in the disease.
Fibroblasts from PD patients with the G2019S LRRK2 mutation showed evidence of mitochondrial dysfunction and increased mitochondrial elongation although a mechanism was not proposed (27). In contrast, although SHSY5Y cells expressing G2019S LRRK2 also showed decreased mitochondrial membrane potential, this was suggested to be associated with increased mitochondrial fragmentation linked with higher levels of DLP1 in the mitochondria (28).
We have used both fibroblasts from patients with the G2019S LRRK2 mutation and SHSY5Y cells with stable ectopic expression of wild-type (WT) and G2019S LRRK2 to examine the effect of LRRK2 upon mitochondrial function. We have demonstrated that both endogenous and ectopic G2019S LRRK2 caused increased mitochondrial proton leak caused by increased uncoupling protein (UCP) expression. This effect was LRRK2 kinase-dependent and may contribute to PD pathogenesis through compromised mitochondrial function.
RESULTS
LRRK2 expression levels
Western blot analysis of LRRK2 protein in a variety of samples was used to test four different anti-LRRK2 antibodies (NT2, 3514-1, 100–500 and C terminus). Although all the antibodies were able to detect over-expressed LRRK2 protein (280 kDa) in SHSY5Y cells, LRRK2 was not reliably detected in any other sample with the exception of control lymphoblasts by the NT2 antibody (Supplementary Material, Fig. S1A). LRRK2 was analysed using an immunoprecipitation (IP) protocol (29) followed by western blotting. The specificity of this approach was demonstrated using control and LRRK2 knockout mouse-brain samples (Supplementary Material, Fig. S1B), and this method could be used quantitatively as LRRK2 levels were proportional to the protein input (Supplementary Material, Fig. S1C). Although LRRK2 levels were highest in control human lymphoblasts followed by fibroblasts, and SHSY5Y cells (Fig. 1A), we were unable to detect it in post-mortem human brain samples (data not shown). In the mouse brain, LRRK2 levels were highest in the cerebellum, striatum and cortex (Supplementary Material, Fig. S1D), where they were comparable with the levels of endogenous protein in control SHSY5Y cells and 7-fold lower than those in fibroblasts (Supplementary Material, Fig. S1E).
Figure 1.
Analysis of LRRK2 protein expression. LRRK2 protein levels were analysed using western blot analysis (3514-1 antibody) of LRRK2 immunoprecipitates (IP, 100–500 antibody) from 1 mg of protein. (A) The relative LRRK2 levels were highest in control human lymphoblasts (l), which were greater than in fibroblasts (f), which were greater than in control SHSY5Y cells (control). However LRRK2 levels were much greater in SHSY5Y cells over-expressing WT LRRK2 (1:10 denotes loading of 10% of the IP sample). (B) LRRK2 expression levels were comparable in fibroblasts from control (c) and G2019S LRRK2 PD patients (p), with GAPDH demonstrating equivalent protein input.
Given that fibroblasts expressed LRRK2 protein to levels in excess of those detected in various mouse brain regions, it was concluded that fibroblasts were a useful system to evaluate how the expression of WT and G2019S LRRK2 influenced mitochondrial function. The G2019S LRRK2 mutation was confirmed in primary fibroblasts from eight PD patients by DNA sequencing, and LRRK2 protein levels were similar in the G2019S and age-matched control cell lines (Fig. 1B).
Mitochondrial function in fibroblasts
Mitochondrial respiratory chain function was assessed by determining the rates of oxygen utilization in the presence of glucose, using both phosphorescent and polarographic measurements. These methods identified significant increases of 128 and 46%, respectively, in oxygen utilization by the G2019S LRRK2 fibroblasts in comparison with control cells (Fig. 2A and B). These increases were not associated with an altered cellular mitochondrial content as assessed by western blot analysis of the levels of various mitochondrial proteins (Fig. 2C) or the percentage of the cell stained with tetramethylrhodamine methyl ester (TMRM) (Supplementary Material, Fig. S2A). However, mitochondrial membrane potential quantified using single-cell analysis of TMRM staining demonstrated a 20% decrease in intensity in the MT cells (Fig. 2D). There was no consistent alteration in mitochondrial morphology detectable in TMRM-stained cells (Supplementary Material, Fig. S2B).
Figure 2.
Mitochondrial function in G2019S LRRK2 patient fibroblasts. Mitochondrial function was assessed in fibroblast cell lines expressing MT G2019S LRRK2 (G2019S) and controls (control). Rates of cellular oxygen consumption in the presence of glucose were increased in the MT cells as determined by (A) an extracellular phosphorescent oxygen probe [relative fluorescence unit (RFU)/min/µg protein] and (B) a Clark-type oxygen electrode (nmole O/min/µg protein). (C) Mitochondrial content was comparable between control and MT fibroblasts as determined by western blot analysis, probing for mitochondrial markers (TFAM, mitochondrial transcription factor A; Core, complex III core subunit) relative to actin in fibroblasts from control and G2019S LRRK2 PD patients. (D) Mitochondrial membrane potential was reduced in the G2019S cells as assessed by live-cell imaging of TMRM fluorescence (RFU/cell, n = 1600 cells), with typical images of TMRM staining in control and G2019S fibroblasts showing the reduction in TMRM intensity. Cellular oxygen utilization measurements for control (open bars) and G2019S (closed bars) fibroblasts with a Clark-type oxygen electrode (nmole O/min/µg protein) in the absence or presence of: (E) oligomycin (2 µg/ml) showed an increased proton leak in G2019S LRRK2 fibroblasts; (F) FCCP (0.5 µm) demonstrated comparable maximal respiratory chain capacities between the two cohorts (n = 12,). (G) Cellular ATP content was decreased in the G2019S MT fibroblasts (RLU/µg protein) and (H) rates of cellular ROS generation were reduced in the MT cells as determined by evaluating the rate of DHE (100 nm) oxidation (RFU/min/cell, n = 300 cells). All values are expressed as mean ± SEM, using fibroblasts from eight patients with the G2019S LRRK2 mutation and seven controls, n = 3 per cell line unless stated otherwise. Statistical analyses were performed using unpaired Student's t-test; statistical significance: *P < 0.05, ***P < 0.0001 for G2019S versus control fibroblasts.
Maximal rates of mitochondrial respiratory chain function and proton leak were evaluated polarographically in control and G2019S LRRK2 MT fibroblasts following the addition of the mitochondrial uncoupler FCCP or oligomycin, respectively. Addition of oligomycin essentially inhibited respiration in control fibroblasts but a residual rate of oxygen utilization was observed in the MT cells (Fig. 2E). The maximal rates of oxygen utilization were similar for control and G2019S MT fibroblasts (Fig. 2F). This suggests that mitochondrial respiratory chain function was normal in the MT cells but there was a proton leak consistent with an increased uncoupling of the mitochondria in the G2019S LRRK2 fibroblasts.
Cellular ATP levels were significantly decreased in MT fibroblasts (12% decrease, P < 0.005, Fig. 2G) consistent with previous observations (27). The rate of DHE oxidation was significantly reduced in the MT LRRK2 fibroblasts compared with controls (46% decrease, P < 0.05, Fig. 2H), suggesting that free radical generation was decreased in the MT cells, a feature previously reported in uncoupled mitochondria (30).
SHSY5Y cells over-expressing LRRK2
To determine whether the effects observed in the patient-derived G2019S fibroblasts were extended to a neuronal model, we analysed mitochondrial function in SHSY5Y cells with stable over-expression of WT or G2019S LRRK2. The levels of G2019S LRRK2 were 50% of the levels of the WT LRRK2 protein (Fig. 3A). Comparison with other cell lines suggested the G2019S LRRK2 levels were ∼1.5-fold higher than those detected in control lymphoblasts and ∼25-fold higher than those in fibroblasts (Fig. 1A).
Figure 3.
Analysis of SHSY5Y cells over-expressing WT or G2019S LRRK2. (A) Western blot analysis of ectopic LRRK2 level (V5 antibody) in SHSY5Y cells with stable LRRK2 expression confirmed that the levels of WT LRRK2 was ∼2-fold greater than the MT G2019S (MT) LRRK2. (B) Rates of cellular oxygen utilization were increased in G2019S-over-expressing cells as revealed by oxygen consumption measurements, using a Clark-type oxygen electrode (nmole O/min/µg protein, n = 6). (C) Mitochondrial membrane potential was reduced in the MT cells as determined by live-cell imaging of TMRM fluorescence [relative fluorescence unit (RFU)/cell, n = 200 cells]. (D) Inhibition of LRRK2 kinase by IN-1 (1 µm, 90 min) restored oxygen consumption rates in the MT clones without affecting the rates in control or WT cells (nmole O/min/µg protein, n = 4); (E) LRRK2 kinase inhibition was confirmed at the end of the experiment in (D), as determined by the decrease in the phosphorylation of LRRK2 at serine 935 [IP followed by western blot analysis with P-Ser 935 relative to total LRRK2 (3514-1)]; (F) Mitochondrial membrane potential was restored in G2019S cells following kinase inhibition as determined by TMRM staining (RFU/cell, n = 200 cells). All values are expressed as mean ± SEM, unless stated otherwise. Statistical analyses were performed using ANOVA (the Bonferonni post hoc test); statistical significance: *P < 0.05, ***P < 0.0001 for G2019S versus control and WT over-expressing SHSY5Y cells or untreated versus LRRK2 IN1-treated cells.
Rates of oxygen utilization for WT LRRK2-over-expressing SHSY5Y cells were similar to untransfected cells; however, they were increased by 80% in cells expressing G2019S LRRK2 (Fig. 3B). TMRM analysis demonstrated that the mitochondrial membrane potential was unaffected in WT LRRK2-expressing cells but was significantly decreased (by 20%, P < 0.0001) in the cells expressing the G2019S LRRK2 mutation (Fig. 3C). There was no significant difference between the aconitase activities in the control, WT and G2019S LRRK2-expressing cells in agreement with the lack of free radical damage (Supplementary Material, Fig. S3).
The G2019S LRRK2 mutation has been linked to increased kinase activity (31). To determine whether the mitochondrial changes associated with MT LRRK2 were related to the altered kinase activity, we looked at the effects of the recently characterized LRRK2 kinase inhibitor, LRRK2-IN1 (32). In the presence of the kinase inhibitor (1 µm), oxygen utilization rates were unchanged in the untransfected and WT LRRK2-expressing cells; however, there was a significant decrease in the rate of oxygen utilization by the G2019S LRRK2-expressing cells resembling rates observed in control cells (Fig. 3D). To confirm inhibition of the LRRK2 kinase activity, the phosphorylation status of LRRK2 Serine 935 was evaluated at the end of the experiment. Phosphorylation of serine residue 935 (Ser 935) was markedly reduced for both the WT and G2019S LRRK2-expressing cells following incubation with LRRK2-IN1 (Fig. 3E), consistent with the inhibition of the LRRK2 kinase activity (32).
In response to kinase inhibition by IN-1 (LRRK2 kinase inhibitor IN-1), mitochondrial membrane potential increased in both WT and G2019S MT cells to 110% of control cells (Fig. 3F). This suggested that although LRRK2 kinase activity was not required for normal mitochondrial function, the mitochondrial uncoupling associated with G2019S LRRK2 was kinase-dependent. To confirm that the increased mitochondrial membrane potential was specifically related to the influence of IN-1 on LRRK2 kinase activity, a second LRRK2 kinase inhibitor, CZC25146, was tested. This inhibitor has been reported to be more potent and structurally unrelated to LRRK2 IN-1 with different off-target effects, in particular it does not inhibit ERK5, which has been reported to be inhibited by IN-1 (32). The minimal concentration of CZC25146 to maximally inhibit LRRK2 kinase was evaluated using SHSY5Y cells expressing WT LRRK2 (Supplementary Material, Fig. S4A). Treating cells with 0.1 µm CZC25146, a dose within the range used to rescue neurite defects in G2019S models (33), resulted in a significant increase in TMRM intensity in MT LRRK2-expressing SHSY5Y cells (Supplementary Material, Fig. S4B), confirming the improvement observed with IN-1. There was a mild decrease in TMRM staining in control and WT LRRK2-expressing cells following CZC25146 treatment (Supplementary Material, Fig. S4B).
Subcellular localization of LRRK2
The subcellular distribution of endogenous LRRK2 was analysed in control lymphoblasts and compared with the distribution in SHSY5Y cells over-expressing WT LRRK2, using differential centrifugation. In both cell types, the majority of the LRRK2 was located in the cytosolic and 250 k fractions with ∼40% of the total LRRK2 signal in each fraction (Fig. 4A and B). The remainder of LRRK2 was equally distributed in the other fractions which contained mitochondrial, lysosomal and ER markers. The G2019S MT LRRK2-expressing SHSY5Y cells showed a similar distribution pattern (Supplementary Material, Fig. S5A).
Figure 4.
LRRK2 intracellular and mitochondrial localization. The distribution of LRRK2 was analysed in the subcellular fractions isolated from SHSY5Y cells over-expressing WT LRRK2 (A) and control lymphoblasts (B). Disrupted cells were fractionated by differential centrifugation (1–250 000g) and 10% of each pellet and cytosol (cyt) fraction analysed by western blotting. LRRK2 distribution was determined by (A) V5 or (B) NT2 LRRK2 immunoreactivity. Subcellular distribution was characterized by probing blots with mitochondrial complex III core protein (Core), lysosomal (GCase), ER calreticulin (Cal) and vesicular synaptophysin (Synapto) markers. LRRK2 was most abundant in the cytosolic and vesicular fractions, with lower levels in the other subcellular compartments. Affinity-purified mitochondria (Mito) were isolated from the postnuclear supernatant (PNS) from WT and G2019S (MT) LRRK2-over-expressing SHSY5Y cells. These highly purified mitochondria were essentially free from other subcellular components, and similar levels of LRRK2 (V5 antibody) were detected in the mitochondria from WT (C) and MT (D) LRRK2-expressing cells. (E) LRRK2 was found to be associated with mitochondrial membranes as determined by mitochondrial fractionation experiments. Affinity-purified mitochondria from WT SHSY5Y cells were subjected to fractionation in hypotonic and sodium carbonate treatment (Na2CO3) with equivalent percentages of the soluble (s) and pellet (p) fractions analysed on western blots, using LRRK2 (V5) and SDHA as an inner-membrane and cytochrome c (cytc) as an inter-membrane space marker.
With our observations that G2019S LRRK2 expression was associated with altered mitochondrial respiration, we analysed whether this could be mediated by a direct interaction with the mitochondrion. The mitochondria-enriched fractions from the WT and G2019S LRRK2-expressing SHSY5Y cells and the control lymphoblasts clearly demonstrated the presence of LRRK2 protein (Fig. 4A and B, Supplementary Material, Fig. S5A). However, there was also evidence of both lysosomal and ER markers in these fractions. The generation of affinity-purified mitochondrial preparations from WT and G2019S LRRK2-over-expressing cells removed the ER and lysosomal contamination, whereas the LRRK2 cross-reactivity remained (Fig. 4C). This demonstrated that a small fraction of both WT and G2019S LRRK2 was present in highly purified mitochondrial preparations. The ratio to the mitochondrial complex III core protein was similar for both the WT and G2019S MT LRRK2 protein (Fig. 4D) and was not influenced by LRRK2-IN1 treatment (Supplementary Material, Fig. S5B). Endogenous LRRK2 protein was not readily detectable in highly purified lymphoblast mitochondrial preparations, using either western blot or IP followed by western blot (Supplementary Material, Fig. S5C).
The sub-mitochondrial distribution of LRRK2 was evaluated using the affinity-purified mitochondrial preparations from cells over-expressing WT LRRK2. Following hypotonic treatment, the LRRK2 signal remained in the pellet along with a marker of the mitochondrial inner membrane (SDHA, succinate dehydrogenase subunit A) while a mitochondrial soluble protein, cytochrome c, was released (Fig. 4E), suggesting that LRRK2 was associated with mitochondrial membranes. This was further supported by solubilization of non-integral membrane proteins with sodium carbonate, where cytochrome c was completely solubilized, although SDHA and LRRK2 were only partially solubilized (Fig 4E).
The levels of the pro-fission protein DLP1 were not significantly altered in G2019S fibroblasts (Supplementary Material, Fig. S6A). The SHSY5Y cells over-expressing WT or MT LRRK2 also showed similar levels of total (Supplementary Material, Fig. S6B) and mitochondrial (Supplementary Material, Fig. S6C) DLP1 protein, which was not influenced by IN-1 inhibition of the LRRK2 kinase (Supplementary Material, Fig. S6C). Furthermore, we were unable to detect DLP1 in either WT or G2019S LRRK2 immunoprecipitates (Supplementary Material, Fig. S6D). These data suggest that WT or G2019S LRRK2 did not associate with DLP1 or influence the levels of this mitochondrial fission protein consistent with the normal mitochondrial morphology.
Influence of LRRK2 on the expression of UCPs
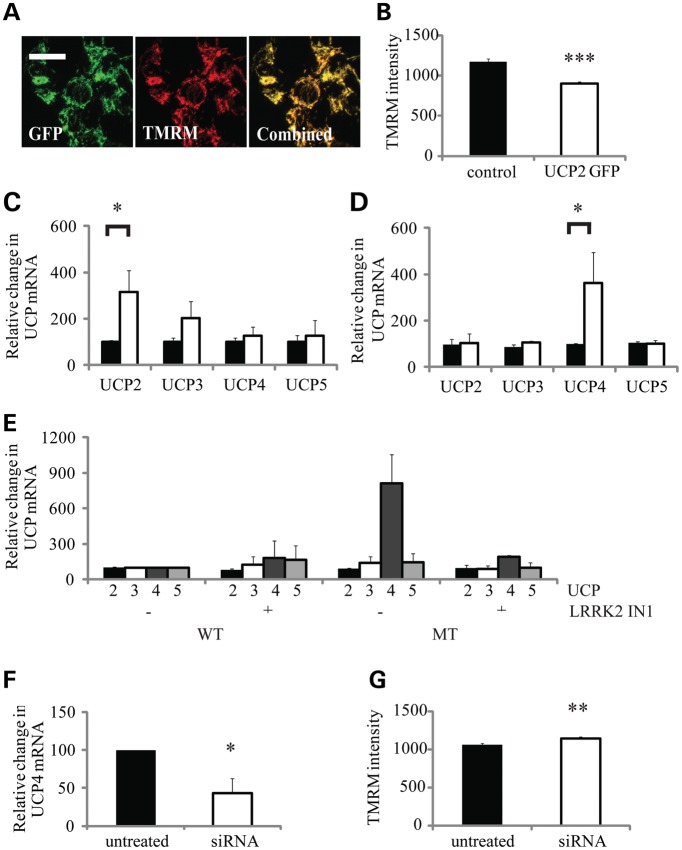
There are five proteins, known as UCPs, reported to be able to regulate mitochondrial permeability to protons (30). To compare the impact of UCP expression on mitochondrial membrane potential with that seen with G2019S LRRK2 expression, murine GFP-UCP2 cDNA was transiently transfected into SHSY5Y cells. These cells showed a high transfection efficiency and co-localization of the GFP signal with the mitochondrial TMRM signal (Fig. 5A). Analysis of the TMRM intensity demonstrated that increased UCP2 expression was associated with a 23% reduction in the mitochondrial membrane potential (Fig. 5B) comparable with that seen with G2019S expression in fibroblasts (Fig. 2D) and SHSY5Y cells (Fig. 3C).
Figure 5.
Analysis of the role of UCP expression in G2019S LRRK2-induced mitochondrial uncoupling. Confocal microscopy of SHSY5Y cells transiently transfected with murine GFP-UCP2 demonstrated: (A) co-localization of UCP2 with mitochondrial TMRM staining and (B) a decrease in mitochondrial membrane potential as quantified by TMRM fluorescence in GFP-positive cells [relative fluorescence unit (RFU)/cell, n = 100 cells]. G2019S LRRK2 expression was associated with increased UCP mRNA levels. mRNA for UCPs2-5 was analysed by RT-PCR in (C), control (black bar) and G2019S LRRK2 (open bar) fibroblasts and (D) SHSY5Y cells over-expressing WT LRRK2 (black bar) and G2019S LRRK2 (MT, open bar). Data are expressed relative to GAPDH mRNA levels and normalized to the control cells. (E) Inhibition of LRRK2 kinase by IN1 (1 µm, 90 min) decreased the significant increase in UCP4 mRNA levels (n = 4) observed in SHSY5Y cells expressing G2019S (MT) LRRK2. Data are expressed relative to GAPDH mRNA levels and normalized to the ratios detected in WT untreated cells. (F) Knockdown of LRRK2 in control SHSY5Y cells by siRNA decreased UCP4 mRNA levels as assessed by RT PCR (n = 3), which was (G) associated with an increased mitochondrial membrane potential as quantified by the TMRM signal intensity (RFU/cell, n = 200 cells). All values are expressed as mean ± SEM (n = 6), unless stated otherwise. Statistical analyses were performed using unpaired Student's t-test; statistical significance: *P < 0.05, **P < 0.005, ***P < 0.0005 for G2019S versus WT over-expressing SHSY5Y cells or control versus siRNA-transfected SHSY5Y cells.
To determine whether UCP expression levels were altered in the G2019S LRRK2 fibroblasts or LRRK2-over-expressing SHSY5Y cells, we quantified the mRNA levels of UCP 1, 2, 3, 4 and 5. Although UCP1 mRNA was not reliably detected in either of our cell models, UCP2–5 were detected in both fibroblasts and SHSY5Y cells, with mRNA for UCPs 4 and 5 being more abundant in fibroblasts and UCP 2 in SHSY5Y cells (Supplementary Material, Fig. S7A and B). In G2019S LRRK2 fibroblasts, there were increased mRNA levels for both UCP2 and 3 but these were only statistically significant for UCP2, which was increased by 215% of the levels in control fibroblasts (Fig. 5C). In SHSY5Y cells over-expressing G2019S LRRK2, the only UCP mRNA that was significantly increased was the neuronal UCP4 isoform, which increased by 265% of the levels in the WT LRRK2 cells (Fig. 5D). In G2019S SHSY5Y cells, the levels of MnSOD and catalase mRNAs were comparable with those of WT and untransfected SHSY5Y cells (Supplementary Material, Fig. S7C and D).
To investigate the possibility that LRRK2 regulated UCP activity via direct phosphorylation, we conducted an in vitro phosphorylation assay using G2019S LRRK2 and UCP2 protein. UCP2 was chosen as it was the most affected in G2019S LRRK2 fibroblasts and has the highest expression in SHSY5Y cells. We were unable to find any evidence of UCP2 phosphorylation by LRRK2 (Supplementary Material, Fig. S8).
Incubation with the LRRK2 kinase inhibitor IN-1 had no significant effect upon UCP mRNA levels in WT LRRK2 cells but restored UCP4 mRNA levels in G2019S LRRK2 cells to the levels observed in the WT LRRK2-expressing cells (Fig. 5E). Control SHSY5Y cells were treated with a mixture of four siRNAs (10 nm) and LRRK2 levels analysed after the 3- and 6-day siRNA protocols. LRRK2 protein levels were decreased by 65 and 85%, respectively (Supplementary Material, Fig. S9). The 6-day siRNA knockdown of endogenous LRRK2 in SHSY5Y cells significantly decreased UCP4 mRNA, suggesting that the normal expression of UCP4 required the expression of LRRK2 (Fig. 5F). Consistent with the decreased UCP4 mRNA levels, the decreased LRRK2 levels resulted in a significant increase in TMRM intensity (10% increase, Fig. 5G).
To determine whether increased UCP expression associated with G2019S LRRK2 mediated the mitochondrial uncoupling, we used genipin as an inhibitor of UCP activity (34). To confirm the inhibitory effect of genipin on the mitochondrial uncoupling activity of UCPs, and to determine its optimal concentration, it was titrated into SHSY5Y cells transiently transfected with UCP2 and the mitochondrial membrane potential analysed over time. At all three concentrations used, genipin increased the TMRM signal in UCP2-over-expressing cells (Fig. 6A), suggesting that it was inhibiting the mitochondrial uncoupling associated with increased UCP2. Using the lower genipin concentration (1.375 nm), we found that there was little influence upon the TMRM signal in control and WT LRRK2-expressing SHSY5Y cells. However, there was an increase in the TMRM signal in SHSY5Y cells over-expressing G2019S LRRK2 to values approaching those observed in control cells (Fig. 6B), consistent with the role of UCP up-regulation leading to the observed drop in mitochondrial membrane potential in MT cells. The addition of genipin (1.375 nm) to G2019S LRRK2 fibroblasts also caused a marked increase in TMRM intensity to levels approaching those observed for the control fibroblasts (Fig. 6C), confirming the role of UCP up-regulation in the uncoupling of these cells.
Figure 6.
Influence of genipin upon mitochondrial membrane potential. The influence of genipin upon the mitochondrial membrane potential as determined by live-cell imaging of TMRM fluorescence. (A) UCP2 transfected SHSY5Y cells were treated with increasing concentrations of genipin (solid line 1.375 nm, dashed line 2.5 nm, dotted line 5 nm).There was a time-dependent increase in mitochondrial membrane potential of cells. (B) Genipin treatment (1.375 nm) increased the mitochondrial membrane potential in the G2019S LRRK2 (solid line) expressing SHSY5Y cells, with only mild influence upon untransfected (dotted line) and WT LRRK2 (dashed line) over-expressing SHSY5Y cells. (C) Genipin treatment (1.375 nm) of control (C2, C4, solid lines) and G2019S LRRK2 fibroblasts (P1, P2, P7, dotted lines) caused an increase in mitochondrial membrane potential in all cell lines. This was more pronounced for the G2019S fibroblasts. The TMRM intensity data were expressed as the percentage of the untreated controls cells at t = 0. Data represent the mean values of four independent experiments focusing on a field containing approximately 30 cells.
DISCUSSION
LRRK2 has been associated with a variety of cellular functions, including synaptic vesicle endocytosis and transcription factor regulation (19,35). However, many of the proposed functions are not influenced by the most common PD-associated G2019S mutation, questioning their relevance to PD pathogenesis. In this study, we have not only confirmed that compromised mitochondrial function is associated with the G2019S mutation, but also demonstrated that this was caused by mitochondrial uncoupling mediated through the LRRK2 kinase-dependent up-regulation of UCP expression.
LRRK2 expression is widespread with mRNA levels found throughout the human brain and peripheral tissues (10–12). It is generally accepted that LRRK2 protein levels are highest in the kidney, lung, macrophages, B cells and monocytes (9–11). Using a combined IP and western blot approach, we confirmed that endogenous LRRK2 levels in control SHSY5Y cells were similar to those in the mouse cortex, but the levels in fibroblast and lymphoblast cultures were ∼7- and 100-fold higher than those seen in the brain. LRRK2 protein levels in our SHSY5Y cells over-expressing G2019S LRRK2 were 1.5-fold higher than control lymphoblasts. Consequently, although LRRK2 expression in the over-expressing cells was higher than that observed in normal human tissue, the expression in fibroblasts was between that seen for the brain and lymphoblasts, confirming their suitability for the analysis of the influence of G2019S MT LRRK2 upon generic cellular processes, including mitochondrial function.
Mitochondrial dysfunction is a recurrent theme in PD, suggesting that it may play an important role in pathogenesis (25). Our observation of decreased mitochondrial membrane potential in G2019S LRRK2 PD fibroblasts agrees with two previous studies (27,28) and is consistent with defective oxidative phosphorylation resulting in decreased cellular ATP levels (27). We have shown that the decreased cellular ATP levels were found in combination with increased basal oxygen utilization. The decrease in spare respiratory capacity and increased proton leak confirmed that the mitochondrial oxidative phosphorylation system was uncoupled from the respiratory chain activity. These observations were restricted to the fibroblasts and SHSY5Y cells over-expressing G2019S LRRK2. However, the normal maximal oxygen utilization capacity excluded a respiratory chain defect in contrast to the suggestion in previous reports (27,28).
The decrease in mitochondrial membrane potential combined with normal maximal oxygen utilization rates associated with G2019S expression implied an increased permeability of the mitochondrial inner membrane to protons. This may involve opening of the mitochondrial permeability pore, the presence of proteins forming a pore or the up-regulation/activation of UCPs. Opening of the mitochondrial permeability pore is associated with increased reactive oxygen species (ROS), increased mitochondrial calcium levels and apoptosis (36). There was no evidence of increased ROS or cell death in these cells, suggesting that mitochondrial permeability transition pore was not involved.
Regarding any direct interaction of LRRK2 with mitochondria leading to increased proton permeability of the inner membrane, we confirmed that the majority of LRRK2 protein was localized to the cytosol, with significant levels in the small vesicular compartment and the remainder associated with other membrane fractions (23). This distribution was similar for WT and G2019S LRKK2 and confirmed for endogenous WT protein in lymphoblasts. A small proportion of LRRK2 was present in highly purified mitochondria, and further mitochondrial sub-fractionation suggested LRRK2 associated with mitochondrial membranes; however, we were not able to restrict this to either the outer or inner membranes. Although these localization data were derived from over-expressing cells, it is consistent with previous reports suggesting that LRRK2 localizes to mitochondria (28) and specifically the outer membrane (22). Although our inability to detect endogenous LRRK2 in lymphoblasts in the purified mitochondrial fraction suggested that the mitochondrial localization could be a consequence of the over-expressing system, it could also reflect difficulties in detecting endogenous LRRK2. However, we are confident that the mitochondrial uncoupling associated with the G2019S LRRK2 expression was not merely a consequence of mitochondrial localization of the ectopic protein as uncoupling was apparent in G2019S fibroblasts with endogenous LRRK2 expression and absent in WT LRRK2-over-expressing SHSY5Y cells.
LRRK2 levels in the mitochondrial fraction were not influenced by the G2019S mutation or kinase inhibition by IN-1, suggesting that mitochondrial LRRK2 levels were not directly regulating mitochondrial uncoupling. LRRK2 is known to interact with 14-3-3, which is disrupted when LRRK2 kinase is inhibited (37). Interactions with 14-3-3 proteins are recognized mechanisms regulating the mitochondrial localization of certain proteins, including Bax (38). However, inhibition of LRRK2 kinase activity did not influence mitochondrial LRRK2 levels, suggesting that 14-3-3 binding was not regulating its mitochondrial localization. The inability of G2019S LRRK2 to phosphorylate UCP2 in vitro also supported the lack of any direct link between LRRK2 and the regulation of mitochondrial uncoupling by modification of UCP2 phosphorylation.
Previous studies have shown G2019S LRRK2 expression to influence mitochondrial morphology, although the data are contradictory, with both increased mitochondrial elongation (27) and fragmentation (28) reported. Mitochondrial fragmentation was linked to increased cellular and mitochondrial DLP1 levels in G2019S SHSY5Y cells, suggesting that LRRK2 plays a role in DLP1 mitochondrial recruitment (28). However, we were unable to replicate these findings in our G2019S fibroblast or SHSY5Y models and there were no detectable changes in mitochondrial morphology observed in either of these cell models.
Increased mitochondrial uncoupling has been associated with the presence of UCPs in the mitochondrial inner membrane. The best characterized is UCP1, which is predominantly found in brown fat (39); however, UCPs 2, 4 and 5 are expressed at relatively high levels in the brain. UCP levels can be regulated in the mitochondria by purine nucleotides, increased free radical generation and fatty acids (40), and degraded by the ubiquitin proteosome system with rapid turnover (41). Consequently, transcription and translation are likely to play an important role in UCP regulatory mechanisms.
The lack of suitable antibodies precluded the measurement of mitochondrial UCP protein levels, but increased UCP2 expression in SHSY5Y cells was sufficient to replicate the decreased mitochondrial membrane potential. Furthermore, genipin, which has been used to inhibit UCP protein activity (34), normalized the mitochondrial membrane potential in G2019S SHSY5Y cells and fibroblasts, confirming that UCP protein played an important role in the observed mitochondrial uncoupling. The G2019S LRRK2-dependent changes in UCP4 mRNA levels, oxygen utilization and mitochondrial membrane potential were all reversed at least in part by incubation with the LRRK2 kinase inhibitor IN-1, suggesting that the effects were LRRK2 kinase-dependent. However, although IN-1 is a relatively selective LRRK2 kinase inhibitor, it has also been reported to inhibit other kinases, notably ERK5 (32). Consequently, the demonstration that the alternative LRRK2 kinase inhibitor, CZC25146, which has a different inhibition profile (33), also significantly improved the G2019S-dependent decrease in mitochondrial membrane potential was further confirmation that these effects of G2019S LRRK2 were dependent upon its kinase activity. Our data suggest that the G2019S LRRK2-dependent increase in UCP mRNA levels is likely to lead to the other mitochondrial changes we have observed. Given that we also identified a reduced UCP transcript level with knockdown of endogenous LRRK2, it is possible that LRRK2 plays a role in the maintenance of normal UCP mRNA levels.
LRRK2 has been linked to the regulation of both NFAT and NFκβ transcription factors and therefore may play an important role in transcription more generally (11,19). In particular, G2019S LRRK2 expression activated NFκβ, resulting in increased transcription and knockdown of LRRK2-perturbed transcriptional activity (11) consistent with the pattern of UCP mRNA changes in our G2019S models. Several microarray analyses of cell lines and brain samples expressing G2019S have been published with evidence of both up-regulated and down-regulated mRNAs but UCP transcripts were not reported.
Increased expression of peroxisome proliferator-activated receptor gamma (PPARγ) co-activator 1 alpha (PGC1α) was sufficient to stimulate UCP expression and mitochondrial uncoupling (42). PGC1α, PPARγ and sterol-regulating-element-binding protein 1c have been shown to stimulate UCP mRNA expression in response to cellular bioenergetic requirements, stress and elevated ROS production as part of a global induction of transcriptional responses involved in adaptive thermogenesis and mitochondrial biogenesis (43,44). Although the expression of G2019S LRRK2 was associated with increased UCP expression, this was not in association with increased mitochondrial proteins or expression of the antioxidants MnSOD or catalase. This implies that the UCP changes were not in response to oxidative stress (45), nutrient deprivation (46) or mitochondrial biogenesis (47) and suggests a more specific influence of MT LRRK2.
We observed a differential expression of the four different UCPs in control fibroblasts and SHSY5Y cells which replicated the tissue-specific expression of these genes previously reported (48,49). Consequently, the variation of the influence of G2019S LRRK2 expression on UCP expression in the fibroblasts and SHSY5Y cells may reflect the impact upon a cell-specific regulatory mechanism. It is of interest that another PD-associated protein, DJ-1, has been reported to regulate UCP mRNA levels in a cell-specific manner. DJ1 knockout mice had normal levels of UCP2 mRNA, but reduced levels of UCP4 and 5 specifically in the substantia nigra pars compacta. These changes were linked to an increased oxidative stress response and deregulation of calcium-dependent pace-making activity (50).
Alternatively, LRRK2 has recently been shown to interact with argonaute, with G2019S LRRK2 leading to the repression of the miRNA down-regulation of protein expression, resulting in increased expression of selected genes (18). Consequently, the possible impact of G2019S LRRK2 upon miRNA function leading to increased UCP mRNA levels via repression of mRNA degradation warrants further analysis.
Increased UCP2 expression was neuroprotective in response to both MPTP toxicity (51) and excitotoxicity (52), suggesting that the increase in UCPs induced by G2019S LRRK2 cells may not be linked directly to PD pathogenesis. However, there is evidence that increased UCP2 expression leads to decreased cellular ATP and toxicity in pancreatic cells (53) and therefore the influence of UCPs upon cellular regulation may be more complex and cell-specific (54). UCP regulation plays an important role in regulating mitochondrial calcium uptake and release, which is particularly important in neuronal function and would be deregulated in G2019S cells. In this regard, the DJ1 regulation of mitochondrial uncoupling via UCPs plays an important role in pace-making in dopaminergic neurons influencing their vulnerability (50). If this pathway is deregulated in dopaminergic neurons in PD patients expressing G2019S LRRK2, this could account for the selective vulnerability of these neurons. A 2-fold increase in UCP2 transcript levels has been recently reported in the substantia nigra from patients with idiopathic PD (55), suggesting that it could play a broader role in the disease pathogenesis.
In conclusion, we have confirmed that the G2019S LRRK2 mutation is associated with mitochondrial uncoupling leading to a decreased mitochondrial membrane potential and decreased ADP phosphorylation. The mechanism is a kinase-dependent gain of function involving up-regulation of UCP expression. It is important to determine whether the mechanism involves the regulation of specific transcription factors and whether the deregulated mitochondrial uncoupling influences the function of selected neuronal populations affected in PD.
MATERIALS AND METHODS
pET-DEST51 plasmids containing the cDNA encoding full-length WT or G2019S LRRK2 with a C terminal V5 tag were kindly provided by Mark Cookson (NIH, Bethesda, MD, USA). pEGFPN1 plasmid with a GFP-tagged UCP2 insert was described previously (56). Details of the antibodies used are described in Supplementary Material, Table S1.
Brain samples
Brain stem, olfactory lobe, mid-brain, cerebellum, striatum and cortical regions were dissected from control adult mouse brains (C57BL/6 strain). Brain samples from adult LRRK2 knockout mice (57) were obtained from D. Alessi (Dundee).
Cell culture
Human skin fibroblasts established from skin biopsies taken from PD patients with G2019S LRRK2 mutation and controls (Supplementary Material, Table S2), with informed consent and ethical approval, were grown under standard conditions (58). Fibroblast analyses were carried out on passages 4–12. Peripheral blood mononuclear cells from control subjects were commercially transformed into lymphoblastoid cell lines, using Epstein–Barr virus transformation (ECACC, Salisbury, UK). Cells were grown in suspension in surface-ventilated 75 cm flasks (Nunc) in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin (50 U/ml), streptomycin (50 µg/ml) and 20 mm HEPES buffer. SHSY5Y cells were cultivated in DMEM/F12 (50/50) growth medium as previously described (59).
Transfections
SHSY5Y cells were transiently transfected with murine UCP2-GFP pEGFPN1 plasmid (1 µg) by electroporation (Neon, Invitrogen, 1000 V, 30 ms, two pulses), which typically generated >95% of cells expressing the ectopic protein. Analyses were carried out 3 days after transfection.
Transfection of SHSY5Y cells with WT or G2019S LRRK2 constructs linearized by digestion with the Fsp1 restriction enzyme was carried out as previously described (59), using Superfect reagents (Qiagen, Hilden, Germany). Colonies resistant to blasticisdin (400 µg/ml) were selected and characterized by immunocytochemistry, western blotting and DNA sequencing.
SHSY5Y cells (2.7 × 105) were transfected with HiPerfect transfection reagent (Qiagen), as described by the manufacturer and LRRK2 siRNA (10 nm final concentration, Dharmacon) containing a cocktail of the following siRNA sequences: GGAAGUUGCUGAUAGUAGA, GAGGACAGCUCUCAUUUCU, GCCCAGGUCUUUGACAUUU, AGACACUGCUC UCUAUAUU. For prolonged treatments, cells were re-transfected at day 3 and LRRK2 protein expression assessed by IP and western blotting.
Real-time PCR analysis
Total RNA was extracted from whole cells (RNeasy Mini Kit, Qiagen) according to the manufacturer's instructions and quantified using a nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). RNA (0.5 µg) was treated with deoxyribonuclease 1 and reverse-transcribed to produce cDNA (QuantiTech Reverse Transcriptase, Qiagen). For the quantification of gene expression, 1 µl of the first-strand cDNA sample was used as template for quantitative real-time PCR amplification with the TaqMan system or SybrGreen assay (Applied Biosystems, Paisley, UK), containing the appropriate primers (refer to Supplementary Material, Table S3). An initial polymerase activation step (95°C for 10 min) was followed by 40 cycles of standard PCR as described in the TaqMan (95°C for 15 s, 60°C for 1 min) or SybrGreen (95°C for 15 s, 56°C for 20 s, 72°C for 30 s) protocols. StepOne real-time PCR system (Applied Biosystems) was used for all analyses and the comparative cycle threshold (CT) method used to determine mRNA expression, correcting gene amplification to GAPDH CTs.
LRRK2 mutation screen
DNA amplification was carried out with Phusion Flash High Fidelity PCR mix (Fisher Scientific, Loughborough, UK) on 2 µl of cDNA in combination with 0.5 µm forward and reverse LRRK2 primers spanning G2019A/S on exon 41 (F5′-GGAATTTGAACAAGCTCCAGAG-3′, R5′-CTTTGTCCTGCTGAAGCAGG-3′), using the three-step protocol as suggested by the manufacturer (98°C for 10 min, 56°C for 20 s, 72°C for 30 s). Amplified PCR products were purified (QAIquick PCR Purification Kit, Qiagen) and sequenced commercially.
Protein extraction and IP
Whole-cell lysates were prepared for western blot analysis by solubilizing cells or tissues in 0.1% sodium dodecyl sulphate (SDS), 10 mm Tris–HCL-containing protease inhibitors (Thermo Scientific), followed by sonication (2 × 15 s pulses).
For IP procedures (29), cell pellets were disrupted in lysis buffer [LB: 50 mm Tris–HCl, pH 7.35, 0.27 m sucrose, 1 mm Na3VO4, 1 mm EDTA, 1 mm EGTA, 10 mm β-glycerophosphate, 5 mm sodium pyrophosphate, 50 mm NaF supplemented with 1% Triton X-100 (IP) or 1% NP-40 (for co-immunoprecipitation—co-IP), 1 mm benzamidine, 0.1 mm PMSF), retaining detergent-soluble fractions after centrifugation (4°C, 17 000g, 10 min). For brain IP, samples were homogenized in LB (2.5 mg tissue per 1 ml of buffer), using a Potter-type homogenizer, pulsed by sonication (15 s, two pulses) and cleared by centrifugation (4°C, 17 000g, 30 min).
IP required 1 mg of protein, incubated with 20 µl of protein G agarose beads (Invitrogen) pre-bound to antibody. Samples were rotated (4°C, 2 h), beads collected by centrifugation and non-specific interactions removed by washes in LB containing high salt (500 or 150 mm NaCl for IP or co-IP experiments, respectively), followed by two washes in LB buffer. Immunoprecipitated proteins were eluted by solubilization in 2× LDS sample buffer containing reducing agent (Invitrogen) for western blot analyses.
Western blot analysis
Samples were prepared for electrophoresis by addition of LDS sample buffer and reducing agent (Invitrogen), followed by heating (65°C, 5 min). Proteins were separated on precast Novex 12 or 4–12% polyacrylamide Bis–Tris gels, transferred to PVDF membranes, probed with various antibodies (Supplementary Material, Table S1) and processed by ECL detection as previously described (60).
Cellular fractionation
Cellular fractionation was carried out as previously described, with some modifications (61). Lymphoblasts or SHSY5Y cells over-expressing LRRK2 (20 × 106) were harvested and resuspended in homogenization buffer (HB: 320 mm sucrose, 10 mm Tris–HCl, 1 mm EDTA, pH 7) containing protease inhibitors (Thermo Scientific, Erembodegen, Belgium). Disruption of plasma membranes was achieved using nitrogen cavitation (Parr Instrument Company, Moline, IL, USA) at 1200 psi (4°C, 20 min). Subcellular fractions were obtained by collecting the pellets after sequential centrifugation at: 1000g for 10 min (P1); 12 000g for 10 min (P2); 27 000g for 10 min (P3); 47 000g for 15 min (P4) and 250 000g for 90 min (P5). The final supernatant (cytosolic fraction) was concentrated using concentrator columns (30 kDa cutoff, Millipore) to a final volume of 50 µl. Characterization of each fraction and the relative LRRK2 distribution was assessed by separation of 10% of each fraction by SDS–PAGE and western blotting.
Mitochondrial affinity purification and fractionation
Affinity-purified mitochondria were prepared from lymphoblasts or SHSY5Y cells (10 × 106) over-expressing WT LRRK2, using the mitochondrial isolation kit (Miltenyi, Surrey, UK). Cells were lysed by needle homogenization (27 gauge, 25 passes) in lysis buffer supplied with the kit. Mitochondria were isolated from the post-nuclear supernatant (1000g, 5 min) by affinity purification (4°C, 1 h) and centrifugation (14 000g, 2 min).
In mitochondrial fractionation experiments, the purified mitochondrial pellet was extracted in HB buffer, hypotonic buffer (10 mm Tris–HCl, 100 mm NaCl, pH 7.6) or 100 mm Na2CO3 (pH 11.5). Samples were separated into soluble and insoluble components by centrifugation (4°C, 17 000g, 10 min), pellets were washed in appropriate buffers and 10% of each fraction analysed by western blotting.
Oxygen utilization
For the phosphorescent analysis of oxygen utilization, fibroblasts were seeded on standard 96-well plates (Sarstedt) pre-coated with 0.01% collagen IV. Measurements were conducted in 100 µl of air-equilibrated DMEM supplemented with 1 mm pyruvate, 10 mm glucose, 20 mm HEPES (pH 7.4) and 100 nm of MitoXpressTM probe (Luxcel Biosciences) (62). Medium was overlaid with heavy mineral oil to prevent oxygen back diffusion. The plate was monitored at 37°C on a Victor 2 time-resolved fluorescence (TR-F) reader (PerkinElmer, Cambridge, UK) with a Samarium filter set (340 nm excitation, 642 nm emission). In each measurement, two intensity signals were recorded with delay times set at 30 and 70 µs and gate time 100 µs, at 2 min intervals over 90 min for each sample well and converted into phosphorescence lifetime values as follows: τ = (t1 − t2)/ln(F1/F2), where F1 and F2 are the TR-F intensity signals at delay times t1 and t2. Signal slope values were calculated as lifetime change/min reflecting oxygen consumption rates, with rates corrected for total protein content.
Cellular oxygen utilization rates were analysed polarographically using a micro-Clark-type oxygen electrode (YSI, Hampshire, UK) (63). The electrode was calibrated with air-saturated respiration buffer (HBSS: 156 mm NaCl, 3 mm KCl, 2 mm MgSO4, 1.25 mm KH2PO4, 2 mm CaCl2, 10 mm glucose, 10 mm HEPES, pH 7.35) thermostatically maintained at 37°C. Cells were resuspended in 250 µl of respiration medium with oxygen levels monitored over a period of 10 min and with the addition of oligomycin (2 µg/ml), FCCP (0.5 μm) or LRRK2 IN-1 inhibitor (1 µm, 90 min treatment prior to rate measurements), where stated. Rates were calculated using 406 nmol O per millilitre and corrected for protein content.
Single-cell analysis
Mitochondrial membrane potential was assessed as previously described (64) by staining cells with 25 nm TMRM (Molecular Probes) in HBSS buffer or phenol red free culture medium. Images were obtained using an inverted laser scanning confocal microscope (LSM510 Zeiss, Oberkochen, Germany) at a λexcitation of 543 nm and directing specimens with the HFT 485/540 short-pass filter. Emitted red fluorescence was captured with a 40×/1.2 oil objective, through a 560 nm long-pass filter. Images were projected through the 1.52 µm Z stacks for a maximum intensity value and quantified using the Metamorph software with a set threshold value, calculating the average signal intensity per cell.
Mitochondrial content was assessed by Image J. Live-cell TMRM images were collected before and after the addition of 1 µm FCCP, which released the TMRM dye into the cytosol to provide an image of the cell contour. Maximal intensity Z projections of the TMRM signal were binarized in Image J and the area of binarized signal quantified relative to the cell contour with data presented as the percentage of cellular area stained by TMRM.
Cellular ATP content
Fibroblasts were seeded at a density of 10 000 cells per well on 96-well plates (Nunc) and relative cellular ATP levels quantified using the CellTiter-Glo assay (Promega) according to manufacturer's instructions on a Victor 2 plate reader (PerkinElmer). Data were expressed as relative luminescence units (RLUs) corrected for protein content.
Array scanner
Fibroblasts were seeded on 96-well plates (Nunc) and stained with 1 µm 4′,6′-diamidino-2-phenylindole (DAPI) in culture medium (37°C, 20 min). Medium was then replaced with HBSS containing 100 nm dihydroethidium (DHE, Molecular Probes). Generation of ROS was monitored by exciting samples with LED light source [λexcitation of 543 nm (DHEox) and 361 nm (DAPI)] at 37°C, 5% CO2 (Array Scan VTI HCS Reader, Thermo Scientific, Pittsburg, PA, USA). Emitted Red fluorescence was captured within the nuclear contour by a 20× LD Plan-Neofluar objective through an automated filter wheel. Images were analysed with the Cellomics Target Activation Bio Application to assess the average DHE intensity per cell for each time point, allowing for rates of DHE oxidation to be quantified in real-time.
Aconitase activity
SHSY5Y cells (4 × 106) were harvested and immediately analysed spectrophotometrically for aconitase activity as previously described (58), correcting for protein content.
Protein quantification
Protein quantification was carried out using the bi-cinchoninic acid protocol (Pierce, Cramlington, UK) using bovine serum albumin as standard.
Statistical analysis
All values represent mean ± SEM. Statistical analyses were preformed using Student's t-test or ANOVA, followed by the Bonferronni post hoc test, using SPSS 16.0 (IBM, Hampshire, UK).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by an Eisai PhD studentship (T.P.), Parkinson's UK (G0910 to J.M.C.), Science Foundation Ireland (07/IN.1/BG1804), the Wellcome Trust/Medical Research Council Joint Call in Neurodegeneration (WT089698). Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the PD patients for the donation of their skin biopsies. We are grateful to Dr Dario Alessi (Dundee) for the LRRK2 antibodies, LRRK2 knockout mouse brains and LRRK2 IN1; Dr Malcolm Roberts (Eisai) for the NT2 antibody; Dr Mark Cookson (NIH) for supplying the LRRK2 cDNA plasmids; Dr Jan Willem Taanman for the generation of the fibroblast cultures; Alexander Zhdanov (Cork) for help with the phosphorescent oxygen utilization assays; Dr R Ramsden (Cellzone) for CZC25146; and Dr Gyorgy Szabadkai for help with the array scanner.
Conflict of Interest statement. J.H. is a paid consultant on Parkinson's disease genetics for Eisai, and J.S. is an employee of Eisai.
REFERENCES
- 1.Lees A.J., Hardy J., Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. doi:10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 2.Healy D.G., Wood N.W., Schapira A.H. Test for LRRK2 mutations in patients with Parkinson's disease. Pract. Neurol. 2008;8:381–385. doi: 10.1136/jnnp.2008.162420. doi:10.1136/jnnp.2008.162420. [DOI] [PubMed] [Google Scholar]
- 3.Marin I. The Parkinson disease gene LRRK2: evolutionary and structural insights. Mol. Biol. Evol. 2006;23:2423–2433. doi: 10.1093/molbev/msl114. doi:10.1093/molbev/msl114. [DOI] [PubMed] [Google Scholar]
- 4.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M.P., Beilina A., Blackinton J., Thomas K.J., et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. doi:10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Ito G., Okai T., Fujino G., Takeda K., Ichijo H., Katada T., Iwatsubo T. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson's disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. doi:10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 6.West A.B., Moore D.J., Choi C., Andrabi S.A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K.L., Dawson V.L., et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. doi:10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 7.Smith W.W., Pei Z., Jiang H., Dawson V.L., Dawson T.M., Ross C.A. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. doi:10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 8.Biskup S., Moore D.J., Rea A., Lorenz-Deperieux B., Coombes C.E., Dawson V.L., Dawson T.M., West A.B. Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci. 2007;8:102. doi: 10.1186/1471-2202-8-102. doi:10.1186/1471-2202-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herzig M.C., Kolly C., Persohn E., Theil D., Schweizer T., Hafner T., Stemmelen C., Troxler T.J., Schmid P., Danner S., et al. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum. Mol. Genet. 2011;20:4209–4223. doi: 10.1093/hmg/ddr348. doi:10.1093/hmg/ddr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakimi M., Selvanantham T., Swinton E., Padmore R.F., Tong Y., Kabbach G., Venderova K., Girardin S.E., Bulman D.E., Scherzer C.R., et al. Parkinson's disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J. Neural. Transm. 2011;118:795–808. doi: 10.1007/s00702-011-0653-2. doi:10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardet A., Benita Y., Li C., Sands B.E., Ballester I., Stevens C., Korzenik J.R., Rioux J.D., Daly M.J., Xavier R.J., et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J. Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. doi:10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galter D., Westerlund M., Carmine A., Lindqvist E., Sydow O., Olson L. LRRK2 expression linked to dopamine-innervated areas. Ann. Neurol. 2006;59:714–719. doi: 10.1002/ana.20808. doi:10.1002/ana.20808. [DOI] [PubMed] [Google Scholar]
- 13.Melrose H.L., Kent C.B., Taylor J.P., Dachsel J.C., Hinkle K.M., Lincoln S.J., Mok S.S., Culvenor J.G., Masters C.L., Tyndall G.M., et al. A comparative analysis of leucine-rich repeat kinase 2 (Lrrk2) expression in mouse brain and Lewy body disease. Neuroscience. 2007;147:1047–1058. doi: 10.1016/j.neuroscience.2007.05.027. doi:10.1016/j.neuroscience.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Shin N., Jeong H., Kwon J., Heo H.Y., Kwon J.J., Yun H.J., Kim C.H., Han B.S., Tong Y., Shen J., et al. LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. doi:10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 15.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. doi:10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Plowey E.D., Cherra S.J., III, Liu Y.J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. doi:10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sancho R.M., Law B.M., Harvey K. Mutations in the LRRK2 Roc-COR tandem domain link Parkinson's disease to Wnt signalling pathways. Hum. Mol. Genet. 2009;18:3955–3968. doi: 10.1093/hmg/ddp337. doi:10.1093/hmg/ddp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehrke S., Imai Y., Sokol N., Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. doi:10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Lee J., Krummey S., Lu W., Cai H., Lenardo M.J. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. doi:10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drolet R.E., Sanders J.M., Kern J.T. Leucine-rich repeat kinase 2 (LRRK2) cellular biology: a review of recent advances in identifying physiological substrates and cellular functions. J. Neurogenet. 2011;25:140–151. doi: 10.3109/01677063.2011.627072. doi:10.3109/01677063.2011.627072. [DOI] [PubMed] [Google Scholar]
- 21.Berger Z., Smith K.A., Lavoie M.J. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 2010;49:5511–5523. doi: 10.1021/bi100157u. doi:10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biskup S., Moore D.J., Celsi F., Higashi S., West A.B., Andrabi S.A., Kurkinen K., Yu S.W., Savitt J.M., Waldvogel H.J., et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. doi:10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 23.Hatano T., Kubo S., Imai S., Maeda M., Ishikawa K., Mizuno Y., Hattori N. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum. Mol. Genet. 2007;16:678–690. doi: 10.1093/hmg/ddm013. doi:10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- 24.West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A., Dawson V.L., Dawson T.M. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl Acad. Sci. USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. doi:10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schapira A.H. Mitochondrial dysfunction in Parkinson's disease. Cell Death Differ. 2007;14:1261–1266. doi: 10.1038/sj.cdd.4402160. doi:10.1038/sj.cdd.4402160. [DOI] [PubMed] [Google Scholar]
- 26.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortiboys H., Johansen K.K., Aasly J.O., Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75:2017–2020. doi: 10.1212/WNL.0b013e3181ff9685. doi:10.1212/WNL.0b013e3181ff9685. [DOI] [PubMed] [Google Scholar]
- 28.Wang X., Yan M.H., Fujioka H., Liu J., Wilson-Delfosse A., Chen S.G., Perry G., Casadesus G., Zhu X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. doi:10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols R.J., Dzamko N., Morrice N.A., Campbell D.G., Deak M., Ordureau A., Macartney T., Tong Y., Shen J., Prescott A.R., et al. 14–3–3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 2010;430:393–404. doi: 10.1042/BJ20100483. doi:10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim-Han J.S., Dugan L.L. Mitochondrial uncoupling proteins in the central nervous system. Antioxid. Redox Signal. 2005;7:1173–1181. doi: 10.1089/ars.2005.7.1173. doi:10.1089/ars.2005.7.1173. [DOI] [PubMed] [Google Scholar]
- 31.Greggio E., Cookson M.R. Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro. 2009;1:e00002. doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng X., Dzamko N., Prescott A., Davies P., Liu Q., Yang Q., Lee J.D., Patricelli M.P., Nomanbhoy T.K., Alessi D.R., et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat. Chem. Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. doi:10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsden N., Perrin J., Ren Z., Lee B.D., Zinn N., Dawson V.L., Tam D., Bova M., Lang M., Drewes G., et al. Chemoproteomics-based design of potent LRRK2-selective lead compounds that attenuate Parkinson's disease-related toxicity in human neurons. ACS Chem. Biol. 2011;6:1021–1028. doi: 10.1021/cb2002413. doi:10.1021/cb2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C.Y., Parton L.E., Ye C.P., Krauss S., Shen R., Lin C.T., Porco J.A., Jr, Lowell B.B. Genipin inhibits UCP2-mediated proton leak and acutely reverses obesity- and high glucose-induced beta cell dysfunction in isolated pancreatic islets. Cell Metab. 2006;3:417–427. doi: 10.1016/j.cmet.2006.04.010. doi:10.1016/j.cmet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Piccoli G., Condliffe S.B., Bauer M., Giesert F., Boldt K., De Astis S., Meixner A., Sarioglu H., Vogt-Weisenhorn D.M., Wurst W., et al. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J. Neurosci. 2011;31:2225–2237. doi: 10.1523/JNEUROSCI.3730-10.2011. doi:10.1523/JNEUROSCI.3730-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasola A., Bernardi P. The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis. 2007;12:815–833. doi: 10.1007/s10495-007-0723-y. doi:10.1007/s10495-007-0723-y. [DOI] [PubMed] [Google Scholar]
- 37.Dzamko N., Deak M., Hentati F., Reith A.D., Prescott A.R., Alessi D.R., Nichols R.J. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14–3–3 binding and altered cytoplasmic localization. Biochem. J. 2010;430:405–413. doi: 10.1042/BJ20100784. doi:10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuruta F., Sunayama J., Mori Y., Hattori S., Shimizu S., Tsujimoto Y., Yoshioka K., Masuyama N., Gotoh Y. JNK promotes Bax translocation to mitochondria through phosphorylation of 14–3–3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. doi:10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouillaud F., Raimbault S., Ricquier D. The gene for rat uncoupling protein: complete sequence, structure of primary transcript and evolutionary relationship between exons. Biochem. Biophys. Res. Commun. 1988;157:783–792. doi: 10.1016/s0006-291x(88)80318-8. doi:10.1016/S0006-291X(88)80318-8. [DOI] [PubMed] [Google Scholar]
- 40.Esteves T.C., Brand M.D. The reactions catalysed by the mitochondrial uncoupling proteins UCP2 and UCP3. Biochem. Biophys. Acta. 2005;1709:35–44. doi: 10.1016/j.bbabio.2005.06.002. doi:10.1016/j.bbabio.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Azzu V., Affourtit C., Breen E.P., Parker N., Brand M.D. Dynamic regulation of uncoupling protein 2 content in INS-1E insulinoma cells. Biochem. Biophys. Acta. 2008;1777:1378–1383. doi: 10.1016/j.bbabio.2008.07.001. doi:10.1016/j.bbabio.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St-Pierre J., Lin J., Krauss S., Tarr P.T., Yang R., Newgard C.B., Spiegelman B.M. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. doi:10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 43.Meirhaeghe A., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., et al. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valle A., Catala-Niell A., Colom B., Garcia-Palmer F.J., Oliver J., Roca P. Sex-related differences in energy balance in response to caloric restriction. Am. J. Physiol. Endocrinol. Metab. 2005;289:E15–E22. doi: 10.1152/ajpendo.00553.2004. doi:10.1152/ajpendo.00553.2004. [DOI] [PubMed] [Google Scholar]
- 45.Valle I., Alvarez-Barrientos A., Arza E., Lamas S., Monsalve M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc. Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. doi:10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 46.Olmos Y., Valle I., Borniquel S., Tierrez A., Soria E., Lamas S., Monsalve M. Mutual dependence of Foxo3a and PGC-1alpha in the induction of oxidative stress genes. J. Biol. Chem. 2009;284:14476–14484. doi: 10.1074/jbc.M807397200. doi:10.1074/jbc.M807397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. doi:10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 48.Mori S., Yoshizuka N., Takizawa M., Takema Y., Murase T., Tokimitsu I., Saito M. Expression of uncoupling proteins in human skin and skin-derived cells. J. Invest. Dermatol. 2008;128:1894–1900. doi: 10.1038/jid.2008.20. doi:10.1038/jid.2008.20. [DOI] [PubMed] [Google Scholar]
- 49.Smorodchenko A., Rupprecht A., Sarilova I., Ninnemann O., Brauer A.U., Franke K., Schumacher S., Techritz S., Nitsch R., Schuelke M., et al. Comparative analysis of uncoupling protein 4 distribution in various tissues under physiological conditions and during development. Biochem. Biophys. Acta. 2009;1788:2309–2319. doi: 10.1016/j.bbamem.2009.07.018. doi:10.1016/j.bbamem.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Guzman J.N., Sanchez-Padilla J., Wokosin D., Kondapalli J., Ilijic E., Schumacker P.T., Surmeier D.J. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. doi:10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews Z.B., Horvath B., Barnstable C.J., Elsworth J., Yang L., Beal M.F., Roth R.H., Matthews R.T., Horvath T.L. Uncoupling protein-2 is critical for nigral dopamine cell survival in a mouse model of Parkinson's disease. J. Neurosci. 2005;25:184–191. doi: 10.1523/JNEUROSCI.4269-04.2005. doi:10.1523/JNEUROSCI.4269-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan P.G., Dube C., Dorenbos K., Steward O., Baram T.Z. Mitochondrial uncoupling protein-2 protects the immature brain from excitotoxic neuronal death. Ann. Neurol. 2003;53:711–717. doi: 10.1002/ana.10543. doi:10.1002/ana.10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krauss S., Zhang C.Y., Scorrano L., Dalgaard L.T., St-Pierre J., Grey S.T., Lowell B.B. Superoxide-mediated activation of uncoupling protein 2 causes pancreatic beta cell dysfunction. J. Clin. Invest. 2003;112:1831–1842. doi: 10.1172/JCI19774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho P.W., Ho J.W., Liu H.F., So D.H., Tse H.M., Chan K.H., Ramsden D.B., Ho S.L. Mitochondrial neuronal uncoupling proteins: a target for potential disease-modification in Parkinson's disease. Transl. Neurodegen. 2012;1:1–9. doi: 10.1186/2047-9158-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin J.H., Ko H.S., Kang H., Lee Y., Lee Y.I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. doi:10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishio K., Qiao S., Yamashita H. Characterization of the differential expression of uncoupling protein 2 and ROS production in differentiated mouse macrophage-cells (Mm1) and the progenitor cells (M1) J. Mol. Histol. 2005;36:35–44. doi: 10.1007/s10735-004-2915-x. doi:10.1007/s10735-004-2915-x. [DOI] [PubMed] [Google Scholar]
- 57.Andres-Mateos E., Mejias R., Sasaki M., Li X., Lin B.M., Biskup S., Zhang L., Banerjee R., Thomas B., Yang L., et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J. Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. doi:10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradley J.L., Homayoun S., Hart P.E., Schapira A.H., Cooper J.M. Role of oxidative damage in Friedreich's ataxia. Neurochem. Res. 2004;29:561–567. doi: 10.1023/b:nere.0000014826.00881.c3. doi:10.1023/B:NERE.0000014826.00881.c3. [DOI] [PubMed] [Google Scholar]
- 59.Chau K.Y., Cooper J.M., Schapira A.H. Rasagiline protects against alpha-synuclein induced sensitivity to oxidative stress in dopaminergic cells. Neurochem. Int. 2010;57:525–529. doi: 10.1016/j.neuint.2010.06.017. doi:10.1016/j.neuint.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chau K.Y., Ching H.L., Schapira A.H., Cooper J.M. Relationship between alpha synuclein phosphorylation, proteasomal inhibition and cell death: relevance to Parkinson's disease pathogenesis. J. Neurochem. 2009;110:1005–1013. doi: 10.1111/j.1471-4159.2009.06191.x. doi:10.1111/j.1471-4159.2009.06191.x. [DOI] [PubMed] [Google Scholar]
- 61.Graham J.M. Isolation of mitochondria from tissues and cells by differential centrifugation. Curr. Protoc. Cell Biol. 2001;3 doi: 10.1002/0471143030.cb0303s04. 3.3.1–3.3.15. [DOI] [PubMed] [Google Scholar]
- 62.Zhdanov A.V., Dmitriev R.I., Papkovsky D.B. Bafilomycin A1 activates respiration of neuronal cells via uncoupling associated with flickering depolarization of mitochondria. Cell Mol. Life Sci. 2011;68:903–917. doi: 10.1007/s00018-010-0502-8. doi:10.1007/s00018-010-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. doi:10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kouri K., Duchen M.R., Lemmens-Gruber R. Effects of beauvericin on the metabolic state and ionic homeostasis of ventricular myocytes of the guinea pig. Chem. Res. Toxicol. 2005;18:1661–1668. doi: 10.1021/tx050096g. doi:10.1021/tx050096g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.