Abstract
Various small molecule pharmacologic agents with different known functions produce similar outcomes in diverse Mendelian and complex disorders, suggesting that they may induce common cellular effects. These molecules include histone deacetylase inhibitors, 4-phenylbutyrate (4PBA) and trichostatin A, and two small molecules without direct histone deacetylase inhibitor activity, hydroxyurea (HU) and sulforaphane. In some cases, the therapeutic effects of histone deacetylase inhibitors have been attributed to an increase in expression of genes related to the disease-causing gene. However, here we show that the pharmacological induction of mitochondrial biogenesis was necessary for the potentially therapeutic effects of 4PBA or HU in two distinct disease models, X-linked adrenoleukodystrophy and sickle cell disease. We hypothesized that a common cellular response to these four molecules is induction of mitochondrial biogenesis and peroxisome proliferation and activation of the stress proteome, or adaptive cell survival response. Treatment of human fibroblasts with these four agents induced mitochondrial and peroxisomal biogenesis as monitored by flow cytometry, immunofluorescence and/or western analyses. In treated normal human fibroblasts, all four agents induced the adaptive cell survival response: heat shock, unfolded protein, autophagic and antioxidant responses and the c-jun N-terminal kinase pathway, at the transcriptional and translational levels. Thus, activation of the evolutionarily conserved stress proteome and mitochondrial biogenesis may be a common cellular response to such small molecule therapy and a common basis of therapeutic action in various diseases. Modulation of this novel therapeutic target could broaden the range of treatable diseases without directly targeting the causative genetic abnormalities.
INTRODUCTION
Many small molecules are under investigation as potential therapeutic agents for a spectrum of Mendelian and complex genetic disorders. Screens for small molecules typically target a specific cellular pathway related to, or implicated in, a particular disorder. However, various small molecules with different known mechanisms of action, including histone deacetylase inhibitors (HDACi) and those that do not inhibit histone deacetylases, produce similar favorable outcomes in a wide variety of heterogeneous diseases affecting different classes of proteins, different cell types and different molecular pathways (Table 1). This challenges our understanding of the role of the known mechanisms of action for this group of small molecules.
Table 1.
Similar therapeutic responses of diverse small molecules in various disease models
| Disease model studied | Small molecule | Therapeutic effect |
|---|---|---|
| Huntington's disease (2,62) | 4PBA, TSA | Reduce neurodegeneration; increase survival |
| Alzheimer's disease (63,64) | 4PBA, SFN | Reduce β-amyloid toxicity; prevent neuronal cell death |
| Diabetes (51,65,66) | 4PBA, TSA, SFN | Hyperglycemia normalization; improve glucose utilization |
| Sickle cell disease (57) | 4PBA, HU, TSA | Increase F-cell production |
| X-linked adrenoleukodystrophy(3) | 4PBA, TSA | Decrease very long-chain fatty acid levels |
| Spinal muscular atrophy (41) | 4PBA, HU, TSA | Increase percentage of full-length SMN2 transcript |
| Fragile X mental retardation (67,68) | 4PBA, HU, TSA | Increase transcription of FMR1 |
| Ischemia (69,70) | 4PBA, SFN | Protection via antioxidant pathway |
| Cystic fibrosis (71,72) | 4PBA, TSA | Increase proper protein trafficking |
4PBA, 4-phenylbutyrate; TSA, trichostatin A; SFN, sulforaphane; HU, hydroxyurea.
The class I and II HDACi, 4-phenylbutyrate (4PBA) and trichostatin A (TSA), have been extensively studied. In some cases, the beneficial effect of HDACi treatment was attributed to an increase in the expression of genes related to, or compensating for, the primary disease-causing gene (1,2).
Our studies of X-linked adrenoleukodystrophy, a neurological disorder, support the possibility that the observed therapeutic overlap of HDACi and other small molecules may be due to modulation of general cellular functions rather than direct targeting of compensatory targets. Both 4PBA and TSA treatment normalize the abnormally high levels of very long-chain fatty acids in human X-linked adrenoleukodystrophy fibroblasts and the X-linked adrenoleukodystrophy (Abcd1 -/Y) mouse model in vivo (3). 4PBA, but not TSA, increased the expression of the peroxisomal ATP-binding cassette transporter gene ABCD2, whose function overlaps that of the defective peroxisomal gene ABCD1 (4). Thus, ABCD2 induction cannot explain and is not necessary for the reduction in very long-chain fatty acid levels in all instances. However, 4PBA treatment of X-linked adrenoleukodystrophy fibroblasts also increased mitochondrial and peroxisomal biogenesis, organelles required for cellular detoxification and stress sensing (1,3). Furthermore, we have shown that peroxisome proliferation induced by 4PBA is dependent on peroxisome biogenesis factor 11 alpha (PEX11α) (5). Unlike PEX11β which is required for constitutive peroxisome abundance, PEX11α induces peroxisome proliferation in response to external stimuli or stress (6). Together, these data suggested the pharmacological involvement of cellular stress responses and prompted us to monitor the induction of the generalized cellular stress response, also known as the adaptive cell survival response, by small molecule treatment (7).
The cellular stress response protects against damage and promotes viability by adapting cells to their environment and has been conserved from archaea to eukaryotes (7). This response modulates the activity of molecular chaperones and proteins that affect reduction–oxidation regulation, sense and repair DNA damage, and are involved in protein degradation, and fatty acid, lipid and energy metabolism. Stimulation of these adaptive survival pathways readjusts the cell to various stressors and restores cellular homeostasis by inducing the heat shock response (HSR), the unfolded protein response (UPR), the autophagic response, the antioxidant response and mitochondrial and peroxisomal biogenesis. Activation of the UPR and autophagy, proteostatic components of the stress response and modulation of mitochondrial energetics can alleviate symptoms of neurodegenerative and aging disorders and extend the lifespan of model organisms (8–10). The work presented here and that done by others in assorted disease models expands upon this approach to therapy by suggesting that joint activation of the stress proteome and the subsequent reestablishment of homeostasis may be beneficial in a broad range of diseases and account for the observed therapeutic overlap of diverse small molecules.
We investigated the potential of four small molecules with overlapping therapeutic benefits (Table 1), but different known functions, to induce the adaptive cell survival response. The primary known modes of action and the clinical utility of these small molecules: 4PBA, TSA, hydroxyurea (HU) and sulforaphane (SFN) are listed in Table 2. Our studies show that at concentrations minimally affecting cellular proliferation, these four drugs increase mitochondrial biogenesis and peroxisome proliferation in normal, X-linked adrenoleukodystrophy, and spinal muscular atrophy human fibroblasts as well as human K562 erythroleukemic cells. We found that these four pharmacological agents induced primary pathways that constitute the cytoprotective stress proteome. SFN treatment of spinal muscular atrophy cells increased full-length expression of spinal motor neuron 2 (SMN2) mRNA and SMN protein levels. Inhibition of the c-jun N-terminal kinase (JNK) pathway, autophagy, mitochondrial biogenesis and sirtuin 1 (SIRT1) activity blocked the therapeutic induction of full-length SMN2 mRNA and SMN protein expression providing evidence that the therapeutic effects of these pharmacological agents result from stimulation of the adaptive cell survival response and the reestablishment of cellular homeostasis. Screening for agents that induce these common cellular responses could identify clinically efficacious molecules, expand the repertoire of currently treatable diseases to include those with unknown genetic etiology and shorten the time to treatment for some diseases.
Table 2.
Small molecules under investigation
| Small molecule | Known mode of action | Used clinically |
|---|---|---|
| 4-phenylbutyrate | Histone deacetylase inhibitor | Yes, orally |
| Trichostatin A | Histone deacetylase inhibitor | No |
| Hydroxyurea | Ribonucleotide reductase inhibitor | Yes, orally |
| Sulforaphane | Phase II detoxification enzyme inducer | Yes, clinical trials, orally |
RESULTS
HU and SFN do not inhibit class I and II HDACs
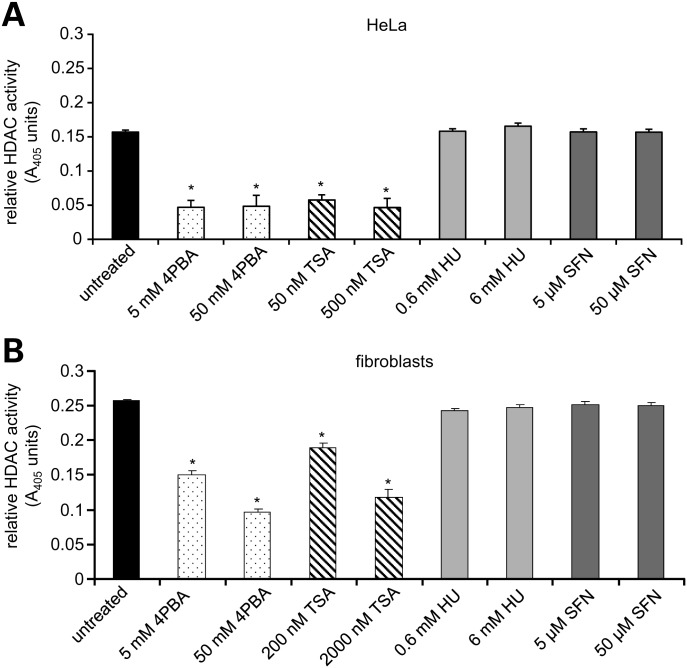
Since two of the small molecules under study, 4PBA and TSA, are known to directly inhibit histone deacetylase activity in vitro (11), we tested if the inhibition of class I and II HDAC activities is a shared biochemical function that could account for the common cellular effects of all four agents. Using lysates from HeLa cells and primary human fibroblasts, an in vitro colorimetric assay of HDAC activity was performed at two concentrations of each drug: that used to treat primary human fibroblasts in the experiments reported here, which minimally affects growth, and 10 times that concentration, which is lethal in cell culture. As expected, 4PBA and TSA significantly decreased HDAC activity compared with untreated HeLa and human fibroblast lysates at both concentrations (Fig. 1A and B). However, neither concentration of HU nor SFN reduced class I or II HDAC activity in lysates. Therefore, pharmacological inhibition of HDAC activity is not a common biochemical activity that accounts for the observed therapeutic overlap of these small molecules. This observation further supports the possibility that the overlapping therapeutic potential of these four small molecules is the induction of a common cellular response(s).
Figure 1.
HU and SFN do not inhibit class I and class II histone deacetylase (HDAC) activities. HDAC activity was measured by an in vitro colorimetric assay using whole cell extracts from HeLa cells or normal human fibroblasts. A decrease in A405 correlates with a reduction in HDAC activity. *P ≤ 0.00009; paired t-test. Bars = SEM for n ≥ 3 independent experiments performed in triplicate.
We note that Myzak et al. (12) demonstrated a physiological increase in histone acetylation after SFN treatment of whole cells or mice in vivo. However, this may reflect an indirect change in gene expression in response to drug treatment and not direct biochemical inhibition of HDAC enzymatic activity by SFN.
Small molecule induction of mitochondrial biogenesis and peroxisome proliferation
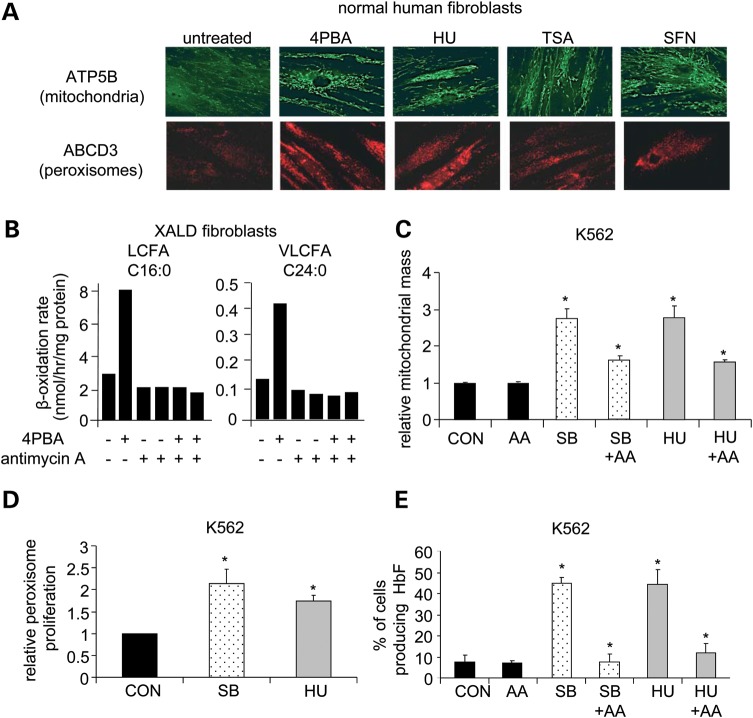
We previously showed that 4PBA treatment of fibroblasts from healthy individuals, X-linked adrenoleukodystrophy patients and patients with peroxisomal biogenesis disorders increased mitochondrial mass and peroxisome proliferation 2–3-fold (1,3,13). Since 4PBA, HU, TSA and SFN have overlapping therapeutic potential in a variety of disorders (Table 1), we broadened the examination of small molecule induction of mitochondrial biogenesis and peroxisome proliferation and treated normal human fibroblasts with 4PBA, HU, TSA or SFN for 5 days. Drug concentrations were titrated to allow 100% viability and minimally affect cellular proliferation. Both mitochondrial and peroxisomal biogenesis were monitored by immunofluorescence. Staining for a mitochondrial membrane protein, ATP synthase beta subunit (ATP5B) and for the 70 kDa peroxisomal membrane protein, ABCD3, revealed drug-induced increases in mitochondrial biogenesis and peroxisome proliferation by all four agents (Fig. 2A; see Fig. 3B for quantitative estimates of similar images). This identified mitochondrial biogenesis and peroxisome proliferation as a common cellular response to treatment with these small molecules. It is unclear how these cellular responses relate to direct HDAC inhibition or the known functions of HU and SFN.
Figure 2.
Pharmacological induction of mitochondrial function is necessary for beneficial responses in X-linked adrenoleukodystrophy and K562 cells. (A) Immunofluorescence staining for mitochondrial membrane protein ATP5B (top) and the peroxisomal membrane protein ABCD3 (bottom). Normal human fibroblasts were treated with each drug for 5 days and cells from the same treated cell population stained with either anti-ATP5B or anti-ABCD3. An increase in fluorescence indicates an increase in mitochondrial mass or peroxisome proliferation, respectively. ×400 magnification. In-cell western quantitation of induced mitochondrial biogenesis in normal human fibroblasts is shown in Figure 3B. (B) Very long-chain fatty acid analysis. X-linked adrenoleukodystrophy fibroblasts were treated with 4PBA in the presence or absence of antimycin A (AA), a cytochrome c reductase inhibitor, for 5 days and β-oxidation levels of long-chain fatty acid (LCFA; C16:0) and very long-chain fatty acid (VLCFA; C24:0) levels were measured in duplicate. (C–E) K562 cells were treated with SB, HU or PBS as a control (CON) in the presence or absence of antimycin A (AA). N ≥ 3 unless otherwise noted. (C) Flow cytometric analysis of mitochondrial mass. After 2–4 days of drug treatment, mitochondria were stained with Mitotracker, a mitochondrion-selective dye, and mitochondrial mass measured by flow cytometry. The fold change (drug treated/control) in mitochondrial mass is shown. (D) Flow cytometric analysis of peroxisome proliferation. After 8–10 days of drug treatment, peroxisome proliferation was measured by flow cytometry using an antibody against the peroxisomal membrane protein PEX14 and a FITC-labeled secondary antibody. The fold change (drug treated/control) in peroxisome staining is shown. (E) Analysis of HbF-producing cell production. After 4 days of drug treatment, cells were stained with 2,7-diaminofluorene (DAF) to measure HbF content. The percent of cells producing HbF (DAF-stained cells) is plotted. *A statistically significant increase in a measurement between drug-treated and control samples or a statistically significant decrease in a measurement between drug-treated and drug-treated samples in the presence of AA (P ≤ 0.05). Bars = SEM.
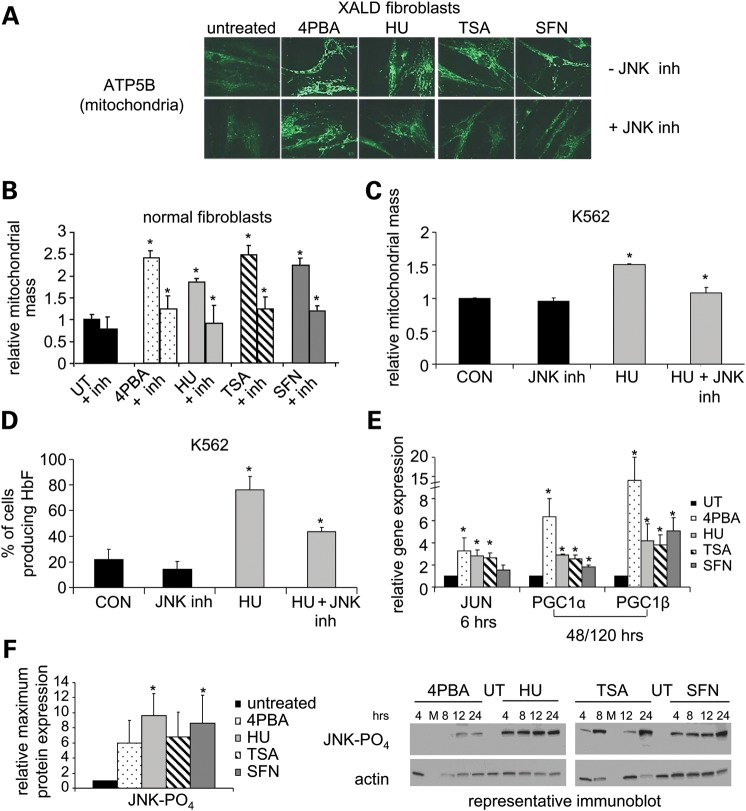
Figure 3.
Mitochondrial biogenesis induced by 4PBA, HU, TSA or SFN is JNK-dependent. (A–D) JNK inhibitor SP600125 (10 μm). (A) Immunofluorescence staining for mitochondrial membrane protein ATP5B. X-linked adrenoleukodystrophy (XALD) fibroblasts were treated with each drug for 4 days with (top row; -JNK inh) or without SP600125 (bottom row; +JNK inh). ×400 magnification. (B) Quantitation of pharmacological induction of mitochondrial biogenesis. Normal human fibroblasts were treated with each drug with (+ inh) or without SP600125 for 6 days. Mitochondrial mass was quantitated via in-cell western analyses using anti-ATP5B staining, a mitochondrial membrane marker. Untreated values were normalized to 1 and the relative mitochondrial mass plotted. Mitochondrial mass significantly increased 2.4-fold, 1.8-fold, 2.5-fold and 2.2-fold with 4PBA, HU, TSA or SFN treatment, respectively, compared with untreated cells. (C and D) JNK inhibition of mitochondrial biogenesis and HbF-containing cell production. K562 cells were treated for 4 days with HU in the presence (+ inh) or absence of SP600125 or PBS as a control (CON). (C) Mitochondrial mass (plotted as in Fig. 1C) and (D) the percent of cells producing HbF were determined by staining with Mitotracker and DAF, respectively (n = 2). (E) mRNA expression of JUN and mitochondrial transcription factors PGC1α and PGC1β by RT-PCR. The relative gene expression for each treatment compared with untreated normal human fibroblasts was calculated and the fold change (drug treated/untreated) is shown. Untreated values were normalized to 1. Measurements of PGC1α and PGC1β levels represent the average of two or more measurements after 5 days and one treatment after 48 h of treatment. Measurements of PGC1α and PGC1β after SFN treatment were performed twice in duplicate. (F) Immunoblot analysis of JNK phosphorylation (46 kDa). Treatment of normal human fibroblasts was initiated at the indicated times prior to cell collection. The mean of the maximum protein expression observed within a 24 h treatment interval for three or more independent experiments is shown as fold change (drug treated/untreated). Actin (43 kDa) was the loading control. M denotes the marker lane. 0.05 ≤ P ≤ 0.10 for 4PBA and TSA treated samples. *Statistically significant increase in a measurement between drug treated and untreated or control samples or a statistically significant decrease in a measurement between drug-treated and drug-treated samples in the presence of SP600125 (P ≤ 0.05). N ≥ 3 independent experiments unless otherwise noted. Bars = SEM. See also Supplementary Material, Table S1.
Beneficial effects of 4PBA in X-linked adrenoleukodystrophy cells require increased mitochondrial function
We previously found that 4PBA-induced peroxisomal very long-chain fatty acid (>C22:0) β-oxidation (degradation) is dependent on mitochondrial long-chain fatty acid (C16:0) β-oxidation (3). To determine whether 4PBA-induced peroxisomal very long-chain fatty acid β-oxidation is dependent on mitochondrial function, mitochondrial function was chemically inhibited with antimycin A, a cytochrome c reductase inhibitor that inhibits the mitochondrial electron transport chain, mitochondrial biogenesis and, thus, cellular respiration (14). The concentration of antimycin A was titrated to minimally affect basal levels of long-chain fatty acid and very long-chain fatty acid β-oxidation and did not exhibit any observable cellular toxicity or affect cellular proliferation. Treatment of X-linked adrenoleukodystrophy fibroblasts with 4PBA-induced long-chain fatty acid and very long-chain fatty acid β-oxidation (Fig. 2B) and increased carbon dioxide release (15). However, in the presence of antimycin A, the 4PBA induction of mitochondrial long-chain fatty acid β-oxidation was inhibited and this concomitantly blocked the induction of peroxisomal very long-chain fatty acid β-oxidation. Thus, pharmacologically induced reduction in peroxisomal very long-chain fatty acid levels by 4PBA treatment is not only dependent on mitochondrial long-chain fatty acid β-oxidation, but is more generally dependent on increased mitochondrial energy production. This evidence highlights the importance of small molecule-induced mitochondrial biogenesis for eliciting a potentially therapeutic disease outcome.
Beneficial effects of sodium butyrate and HU in a β-hemoglobinopathy model require induced mitochondrial biogenesis
The clinical severity of β-hemoglobinopathies, such as sickle cell disease and β-thalasemmia, is ameliorated by increasing the number of fetal hemoglobin (HbF)-containing cells (F-cells), thus elevating total HbF levels in peripheral blood (16). Sodium butyrate (SB, an analog of 4PBA) and HU have been shown to increase HbF levels in several contexts: in sickle cell disease and β-thalasemmia patients, in K562 cells, and in CD34+ derived hematopoietic stem cells (17). To further assess the pharmacological commonality of the induction of mitochondrial and peroxisomal biogenesis in a disease model unrelated to the neurological disease X-linked adrenoleukodystrophy, we studied these responses in K562 cells, an erythroleukemic cell line that produces HbF and is commonly used as a model of the induction of F-cell production.
K562 cells were treated with SB and HU for 4 to 10 days. Flow cytometric analyses demonstrated that both SB and HU significantly increased mitochondrial mass, peroxisomal proliferation and the number of HbF-producing cells (Fig. 2C–E). To determine whether the induction of mitochondrial biogenesis by SB or HU treatment is necessary for the induction of HbF-producing cells, K562 cells were treated with SB or HU in the presence or absence of antimycin A. The concentration of antimycin A was again titrated to minimally affect basal levels of HbF-producing cells, did not exhibit any observable cellular toxicity and did not affect cellular proliferation. The induction of mitochondrial biogenesis and the induction of HbF-producing cells by SB or HU treatment were significantly inhibited by antimycin A (Fig. 2C and E). This observation indicates dependence of the beneficial effect, elevated HbF levels, on induced mitochondrial biogenesis. Thus, pharmacological induction of mitochondrial biogenesis by SB or HU may also be central for the therapeutic effects observed in sickle cell disease patients.
Pharmacological induction of mitochondrial biogenesis is dependent on the JNK pathway
Since small molecule induction of mitochondrial biogenesis was necessary for the beneficial effects of 4PBA, SB or HU in models of X-linked adrenoleukodystrophy and sickle cell disease, we examined the known mitochondrial biogenesis pathways. Mitochondrial biogenesis can be stimulated by various signaling pathways that activate the transcription factors peroxisome proliferator-activated receptor gamma coactivator-1 alpha and beta (PGC1α and PGC1β, respectively) (18). To determine whether the induction of mitochondrial biogenesis by small molecules required the activation of a common kinase cascade or endothelial nitric oxide synthase, X-linked adrenoleukodystrophy fibroblasts were treated with at least one of the inducing drugs and inhibitors of various kinases or endothelial nitric oxide synthase (Supplementary Material, Table S1). Inhibition of pathways including p38 mitogen-activated kinase, extracellular-regulated kinase, mitogen-activated kinase kinases 1 and 2, adenosine monophosphate kinase, protein kinase C, phosphoinositide-3-kinase and endothelial nitric oxide synthase, had no effect on drug-induced mitochondrial biogenesis (data not shown). However, inhibition of the stress-activated protein kinase pathway, otherwise known as the JNK pathway, with SP600125 (19) reduced pharmacological induction of mitochondrial biogenesis by 4PBA, HU, TSA and SFN to levels not significantly different from untreated cells as demonstrated by immunofluorescence staining and quantitative in-cell western analyses in treated X-linked adrenoleukodystrophy or normal human fibroblasts, respectively (Fig. 3A and B). 4PBA, HU, TSA or SFN treatment alone significantly increased mitochondrial mass as indicated by an increase in ATP5B staining (Fig. 3B).
To determine whether activation of the JNK pathway was necessary for a therapeutic response, we assayed HbF production by HU in the presence and absence of the JNK inhibitor. In K562 cells, the JNK inhibitor SP600125 (19) also blocked HU-induced mitochondrial biogenesis and consequently HbF-producing cell production (Fig. 3C and D; SB not tested). In fully competent hematopoietic stem cells, HU-stimulated F-cell production was similarly dependent on the JNK pathway (data not shown). The JNK pathway is activated by external stressors and stimuli such as heat shock, osmotic shock and ultraviolet irradiation (20). Therefore, the induction of mitochondrial biogenesis is a common response to 4PBA, HU, TSA and SFN treatment and each induction is dependent on the same cytoprotective pathway, the JNK pathway.
Activation of the JNK pathway leads to activation of the transcription factor JUN via phosphorylation. JUN transcript levels were significantly increased after 6 h of treatment with 4PBA, HU or TSA (Fig. 3E). JUN transcript levels were not consistently changed after SFN treatment. However, JNK phosphorylation, a marker of JNK pathway activation, was increased within 24 h of treatment with each drug, including SFN (Fig. 3F). PGC1α and PGC1β maintain basal levels of mitochondria and their transcription was significantly increased after 48 h or longer of treatment with each of the four small molecules (Fig. 3E). Of note, PGC1α can induce mitochondrial biogenesis under physiological stresses such as during adaptive thermogenesis, endurance exercise or fasting (21). Thus, treatment with 4PBA, HU, TSA or SFN activated the stress-activated JNK pathway and increased transcription of the stress–responsive mitochondrial transcription factor PGC1α.
Small molecule activation of the adaptive cell survival response
In three different cell types (normal human fibroblasts, X-linked adrenoleukodystrophy fibroblasts, and K562 cells) inhibition of the stress-activated JNK pathway blocked the small molecule induction of mitochondrial biogenesis, a necessary response for the reduction of very long-chain fatty acid levels in X-linked adrenoleukodystrophy cells and for the increase in HbF levels in K562 cells. Mitochondria play a key role in cellular adaptation to stress, suggesting that a common cellular response to these pharmacological agents may be stimulation of the stress proteome or adaptive cell survival response. To test this hypothesis, expression of key components of the HSR, UPR, the autophagic response and the antioxidant response were monitored at the transcriptional, translational and/or post-translational levels. These responses are known to be transiently activated in response to various cellular stressors; and the degree of induction of each component can vary depending on the metabolic state of the cell (e.g. confluency, cell passage number) (7,22,23). Due to the transient nature of these responses, key components of each pathway were monitored at several time points within a 24 h treatment interval in at least two normal human fibroblast lines and in three or more independent experiments. We report the increase in mRNA expression after 2, 6 or 18 h of treatment and the average of the maximal increase in protein expression within a 24 h time frame after treatment compared with the untreated samples (n ≥ 3). Activation of each pathway was assessed by an increase in one or more components of each pathway.
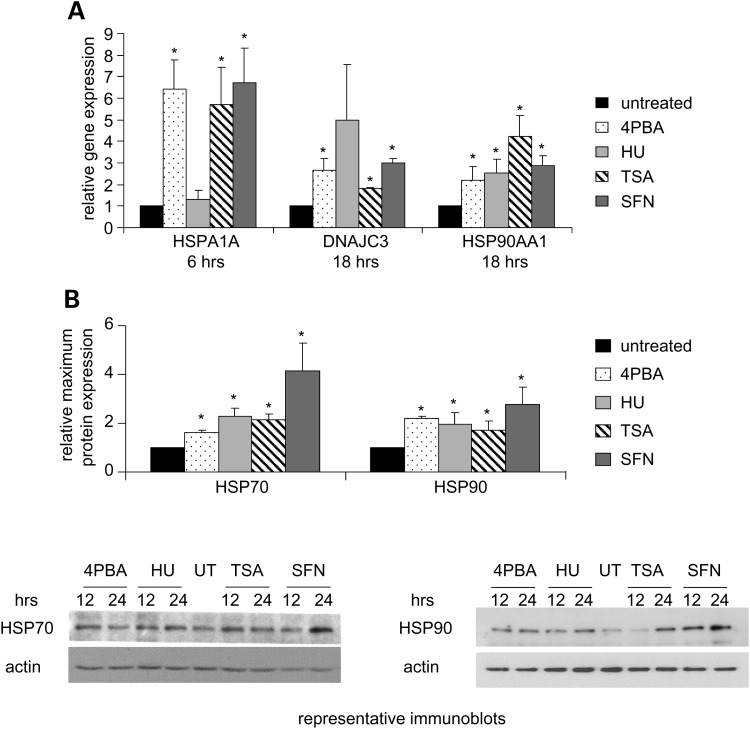
Activation of the HSR
Induction of the HSR is a hallmark of the adaptive cell survival response (24). To evaluate its activation, the expression of heat shock protein (HSP) 40, 70 and 90 family members was monitored. 4PBA, TSA or SFN increased transcription of HSP 70 kDa protein 1A (HSPA1A) and all four drugs increased transcription of DNAJ HSP40 homolog subfamily C member 3 (DNAJC3) and HSP 90 kDa class A member 1 (HSP90AA1;Fig. 4A). HSPA1A transcript expression was not stimulated by HU treatment at the time points examined. However, total HSP70 and HSP90 protein levels were significantly increased with all four agents (Fig. 4B). The pharmacological induction of HSR mRNA and protein expression was similar to that caused by mild heat shock (25). Thus, the HSR was activated by 4PBA, HU, TSA or SFN treatment.
Figure 4.
The HSR is induced by 4PBA, HU, TSA or SFN treatment. (A) RT-PCR analyses of mRNA expression of HSR genes. Normal human fibroblasts were treated and the mRNA expression of HSPA1A (HSP70), DNAJC3 (HSP40) and HSP90AA1 (HSP90) was measured and plotted as in Figure 2E. The P-value for the SFN DNAJC3 measurements is 0.10. (B) Immunoblot analyses of HSP expression. Normal human fibroblasts were treated at various time points and stained with antibodies that detect all HSP70 or all HSP90 family members. The mean of the maximum protein expression observed within a 24 h treatment interval for three or more independent experiments was calculated. The fold change (drug treated/untreated) is plotted for total HSP70 (70 kDa) and total HSP90 (90 kDa) protein levels. Actin (43 kDa) was used as a loading control. UT, untreated cells. *Statistical significance (P ≤ 0.05). Bars = SEM for n ≥ 3 independent experiments.
Activation of the UPR
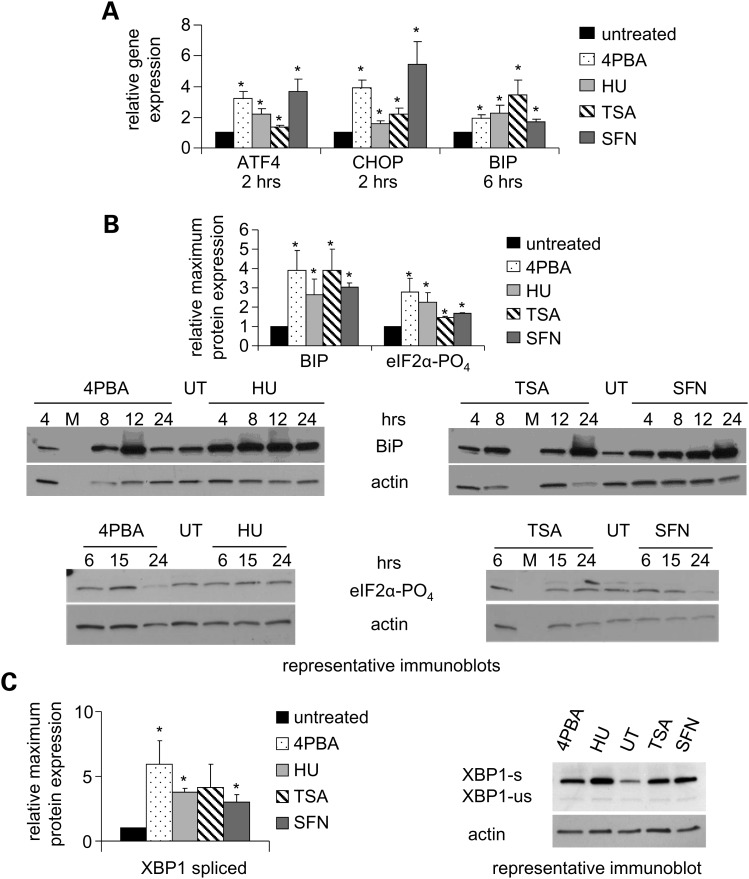
UPR markers evaluated included the central UPR regulator glucose regulated protein 78 (BIP) and key components that are stimulated after the activation of the three endoplasmic reticulum transmembrane receptors: PERK, inositol requiring 1 (IRE1) and activating transcription factor 6 (24). Elongation initiation factor 2 alpha (eIF2α) is phosphorylated by PERK, attenuates general translation and induces activating transcription factor 4 (ATF4). CHOP promotes reactivation of PERK. XBP1 mRNA is non-canonically spliced by the endonuclease activity of IRE1 upon UPR activation. BIP expression was significantly increased at the transcriptional and translational levels after treatment with 4PBA, HU, TSA or SFN (Fig. 5A and B). Treatment with these small molecules also increased ATF4 and CHOP mRNA expression (Fig. 5A), modestly increased eIF2α phosphorylation (Fig. 5B) and increased the total amount of XBP1 protein and the amount of activated XBP1 protein from the correctly spliced mRNA (Fig. 5C). Thus, the pro-survival capabilities of all three UPR branches were activated at the transcriptional, post-transcriptional, translational and/or post-translational levels by treatment with these agents.
Figure 5.
The UPR is activated by 4PBA, HU, TSA or SFN treatment. (A) RT-PCR analyses of mRNA expression of UPR genes. Normal human fibroblasts were treated with each drug as indicated. The expression of the UPR genes ATF4, CHOP and BIP was measured as described in Figure 2E. (B) Immunoblot analyses of UPR protein expression. Normal human fibroblasts were treated at various time points. The maximum protein expression observed within a 24 h interval was determined; and the mean of three or more independent experiments plotted as fold change (drug treated/untreated) for BIP (78 kDa) and phosphorylated eIF2α (38 kDa). Actin (43 kDa) was used as a loading control and is the same blot as Figure 2F. UT denotes untreated cells. M denotes marker lane. (C) XBP1 splicing. Normal human fibroblasts were treated. The expression of the unactivated/unspliced form (XBP1-us; 33 kDa) and the activated/spliced form of XBP1 (XBP1-s; 54 kDa) was analyzed by immunoblotting. XBP1-s is a larger protein than XBP1-us due to non-canonical mRNA splicing which results in a larger carboxy-terminal domain. The percentage of XBP1-s to XBP1-us increased with each treatment as shown (24 h time point). The mean of the maximum expression of XBP1-s within a 24 h treatment interval is plotted as in Figure 5B. Actin was used as a loading control. P = 0.08 for the TSA treated values. *Statistical significance (P ≤ 0.05). Bars = SEM for n ≥ 3 independent experiments.
Activation of autophagy
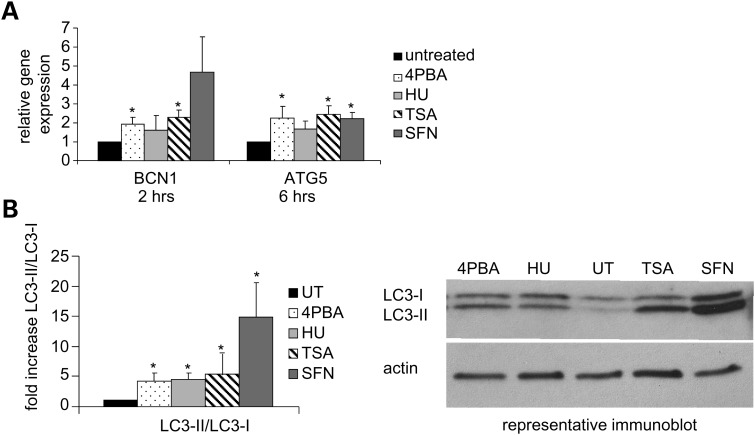
Adverse growth conditions increase the energetic demand on a cell stimulating the catabolic processes of autophagy to promote utilization of damaged and excess proteins and damaged organelles for cellular nutrients (26). To monitor activation of autophagy, we examined the expression of three classical autophagy markers, beclin-1 (BCN1), autophagy protein 5 (ATG5) and microtubule-associated protein 1 light chain 3 (LC3 or APG8). Autophagosome formation is signaled by the phosphorylation of BCN1 by JNK1 and is dependent on the conjugation of ATG5 and ATG12 and the cleavage (LC3-I) and phosphatidylethanolamine lipidation (LC3-II) of APG8. BCN1 and ATG5 mRNA levels increased with 4PBA, TSA or SFN treatment (Fig. 6A). After HU treatment, ATG5 mRNA levels also increased, but BCN1 mRNA levels were not reproducibly changed within 6h of treatment. Treatment with each of these small molecules significantly increased the proportion of LC3-II to LC3-I, a hallmark of autophagy activation (Fig. 6B). Therefore, the autophagy pathway is activated by treatment with each of the four agents.
Figure 6.
Autophagy is induced by 4PBA, HU, TSA or SFN treatment. (A) RT-PCR analyses of mRNA expression of autophagy genes. Normal human fibroblasts were treated as indicated. The expression of BCN1 and ATG5 was measured as described in Figure 2E. ATG5 mRNA levels after TSA treatment were measured in two independent experiments in duplicate. The P-value for the ATG5 HU-treated samples is 0.10. The P-values for the BCN1 HU and SFN samples are 0.23 and 0.06, respectively. (B) Immunoblot analyses of the cleavage and lipidation of autophagy protein APG8. Upon activation of autophagy, the APG8 protein (LC3-I; 17 kDa) is cleaved and lipidated (LC3-II; 13 kDa). Normal human fibroblasts were treated and the mean of the maximal increase in the proportion of LC-II to LC3-I within a 24 h treatment interval for three or more independent experiments is plotted as fold change (drug treated/untreated). A representative immunoblot is shown (24 h time point). Actin (43 kDa) was used as a loading control. UT denotes untreated cells. *Statistical significance (P ≤ 0.05). Bars = SEM for n ≥ 3 independent experiments unless otherwise noted.
Activation of the antioxidant response
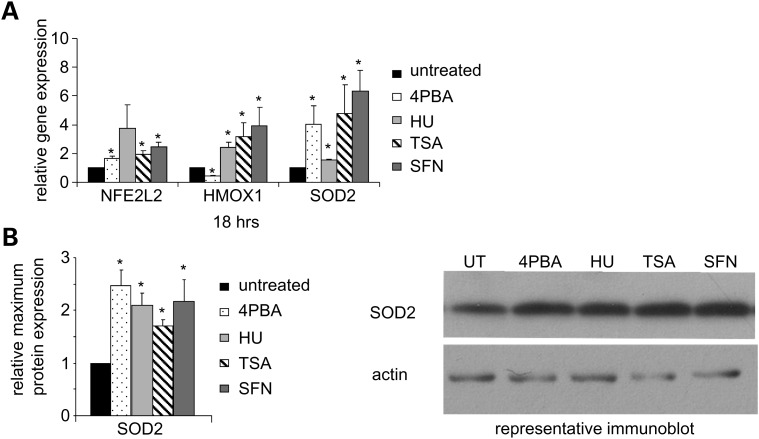
The antioxidant response detoxifies the cell and regulates reduction–oxidation (redox) homeostasis by neutralizing the effects of reactive oxygen and nitrogen species, second messengers of the adaptive cell survival response (24). We examined the expression of three key components, nuclear factor erythroid 2-like 2 (NFE2L2), heme oxygenase 1 (HMOX1) and superoxide dismutase 2 (SOD2). NFE2L2, a transcription factor that binds the antioxidant response element, is known to be involved in the chemoprotective response provided by SFN (12). After treatment with 4PBA, HU, TSA or SFN, NFE2L2 and SOD2 mRNA levels increased (Fig. 7A). HMOX1 mRNA levels significantly increased with HU, TSA or SFN treatment and significantly decreased with 4PBA treatment. SOD2 protein levels also significantly increased (Fig. 7B). Thus, cellular antioxidant defense mechanisms were induced at the transcriptional and translational levels by treatment with each of these small molecules.
Figure 7.
The antioxidant response is induced by 4PBA, HU, TSA or SFN treatment. (A) RT-PCR analyses of mRNA expression of antioxidant genes. Normal human fibroblasts were treated for 18 h. The expression of NFE2L2, HMOX1 and SOD2 was measured as described in Figure 2E. The P-value for the NFE2L2 HU measurements was 0.11. (B) Immunoblot analyses of SOD2 protein expression. Normal human fibroblasts were treated for 2 to 5 days. The mean of the relative expression of SOD2 (26 kDa) across three or more independent experiments is plotted for each treatment as fold change (drug treated/untreated). Actin (43 kDa) was used as a loading control. *Statistical significance (P ≤ 0.05). Bars = SEM for n ≥ 3 independent experiments.
Beneficial effects of SFN in spinal muscular atrophy cells are dependent on the JNK pathway, autophagy, mitochondrial biogenesis and SIRT1 activity
To determine which of the drug-induced stress pathways are necessary for the therapeutic effects of these small molecules, we studied their effects in spinal muscular atrophy fibroblasts. Ninety-five percent of spinal muscular atrophy patients have a homozygous deletion of the telomeric SMN1 (chr 5q13) gene or gene conversion at exon 7 or 8 (27). However, patients have one or more copies of a centromeric SMN1 pseudogene, SMN2. Compared with SMN1, SMN2 has a C to T transition within an exonic splice enhancer that results in the skipping of exon 7 (SMNΔ7) and an unstable protein that is degraded (28,29). Only 15–30% of SMN2 transcripts include exon 7 and are full length (FL-SMN). Since the clinical severity of spinal muscular atrophy patients inversely correlates with the levels of FL-SMN transcript and SMN protein, therapeutic strategies that increase FL-SMN and SMN protein production offer promise.
Others have previously shown that treatment of spinal muscular atrophy cell lines and/or mouse models with three of the four drugs studied here, 4PBA, HU or TSA, results in increased FL-SMN transcript expression and increased SMN protein expression (30–32). If these three small molecules and SFN share a common therapeutic mechanism, SFN treatment should also increase FL-SMN mRNA and SMN protein expression. To test the hypothesis that a common effect of treatment with these four small molecules is induction of the stress proteome, we examined the induction of mitochondrial biogenesis by these four small molecules in spinal muscular atrophy fibroblasts, the effect of SFN treatment on the expression of FL-SMN mRNA and SMN protein in spinal muscular atrophy and the ability of inhibitors of the various stress pathways to block induction of FL-SMN mRNA and SMN protein expression.
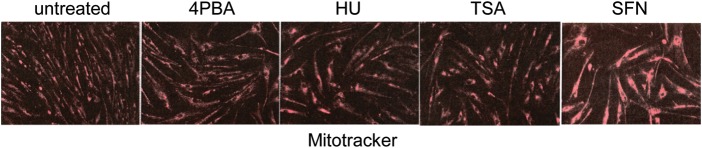
Similar to treatment of X-linked adrenoleukodystrophy fibroblasts, K562 cells and normal human fibroblasts, treatment of spinal muscular atrophy fibroblasts with each of the four small molecules increased mitochondrial biogenesis as monitored by Mitotracker staining, a cell-permeant mitochondrion-selective dye (Fig. 8).
Figure 8.
Mitochondrial biogenesis is induced in spinal muscular atrophy fibroblasts by 4PBA, HU, TSA or SFN treatment. Immunofluorescence staining for mitochondria using Mitotracker Red CMXROS. Spinal muscular atrophy fibroblasts were treated with 2.5 mm 4PBA, 300 μm HU, 100 nm TSA or 2.5 μm SFN for 5 days. ×80 magnification.
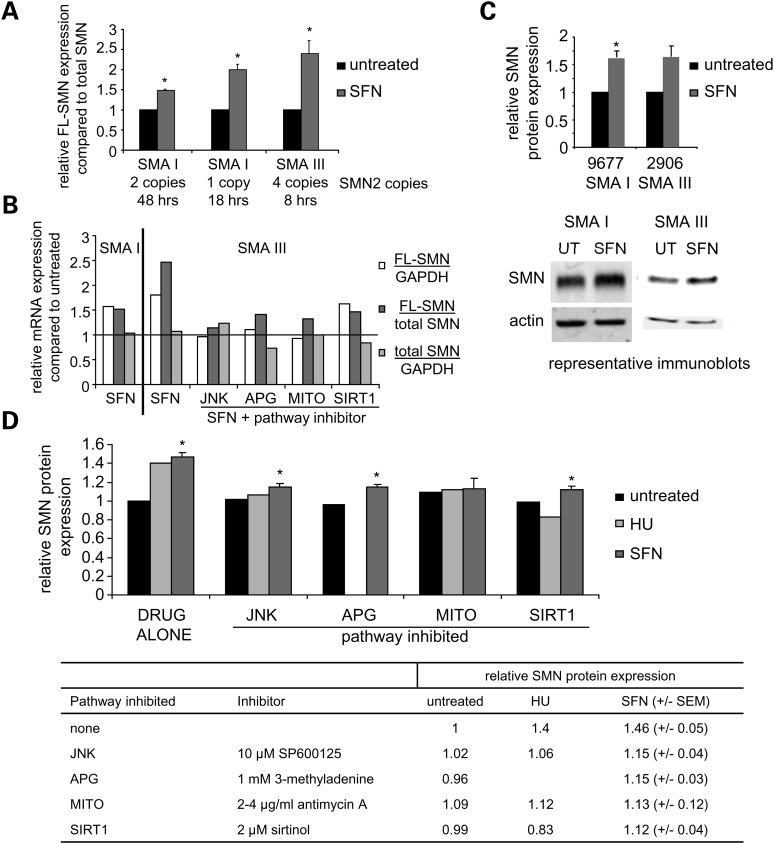
Similar to 4PBA, HU or TSA treatment (30–32), SFN treatment significantly increased FL-SMN mRNA expression compared with total SMN transcription (FL-SMN plus SMNΔ7) in two spinal muscular atrophy type I cell lines (GM09677 two copies SMN2 and GM00232 one copy SMN2) and one spinal muscular atrophy type III cell line (96–2906 four copies SMN2; Fig. 9A) (28). As demonstrated by others, cell lines with greater numbers of SMN2 gene copies respond better to treatment.
Figure 9.
The induction of FL-SMN mRNA expression and SMN protein expression by SFN is dependent on the JNK pathway, autophagy, mitochondrial biogenesis and SIRT1 activity. (A) RT-PCR analyses of FL-SMN mRNA expression compared with total SMN transcript expression. Type I and type III spinal muscular atrophy (SMA I or SMA III) fibroblasts were treated with SFN for the indicated times. The relative fold increase in the ratio of FL-SMN/total SMN transcripts compared with the untreated cell line (normalized to 1) is plotted. Cell lines from left to right are GM09677, GM00232 and 2906. N ≥ 2 independent experiments. (B) RT-PCR analyses of FL-SMN and total SMN mRNA expression. Spinal muscular atrophy fibroblasts were treated with 1.5 μm SFN alone for 51 h (SMA I, GM09677) or treated with 0.5 μm SFN for 8h (SMA III, 2906) with or without 12.5 µm SP600125 (JNK inhibitor), 2.5 mm 3-methyladenine (autophagy inhibitor, APG), 5 µg/ml antimycin A (mitochondrial inhibitor, MITO) or 3 µm sirtinol (SIRT1 inhibitor). The fold increases in FL-SMN expression compared with either GAPDH or total SMN expression and the fold increase in total SMN expression compared with GAPDH expression after SFN treatment is plotted. Untreated values were normalized to 1 and are indicated by the horizontal line. (n = 1 per cell line, performed in duplicate) (C) Immunoblot analyses of SMN protein expression. Spinal muscular atrophy type I (SMA I; GM09677, n = 7) or type III (SMA III; 2906, n= 2) fibroblasts were treated with either 1.5 µm SFN or 0.5–2 µm SFN, respectively, for 48–72 h. The average of the relative expression of SMN protein (39 kDa) is plotted. Actin (43 kDa) was used as a loading control. (D) Summary of the effects of stress pathway inhibitors on SMN protein expression. Spinal muscular atrophy type I (SMA I; GM09677) fibroblasts were treated with either 300 μm HU (n = 1) or 1.5 μm SFN (n ≥ 3) in the presence or absence of 10 μm SP600125 (JNK inhibitor), 1 mm 3-methyladenine (APG inhibitor), 2–4 μg/ml antimycin A (MITO inhibitor) or 2 μm sirtinol (SIRT1 inhibitor) for 48 h. SMN protein expression was measured by quantitative immunoblot analyses using actin as a loading control. Fold increases in SMN protein expression are plotted and represented numerically in the table below the graph. Untreated values are normalized to 1. *P ≤ 0.05; bars = SEM.
After SFN treatment, the relative expression of FL-SMN transcripts increased when normalized to either GAPDH transcript levels or to total SMN transcript levels (Fig. 9B). However, total SMN transcript levels remained unchanged when normalized to GAPDH transcript levels after treatment. Therefore, SFN treatment increases the quantity of FL-SMN transcripts by enhancing the inclusion of exon 7 rather than increasing overall transcriptional expression from the SMN2 gene.
We demonstrated that the reduction in very long-chain fatty acid levels in X-linked adrenoleukodystrophy fibroblasts by 4PBA was dependent on the induction of mitochondrial biogenesis and that the induction of HbF production in K562 cells by HU was dependent on the JNK pathway and mitochondrial function (Figs 2B, D and 3D). To determine which stress pathways are necessary for the increase in FL-SMN production, spinal muscular atrophy fibroblasts were co-treated with SFN and either SP600125, an inhibitor of the JNK pathway (19), 3-methyladenine, an inhibitor of the autophagy pathway (33), antimycin A, a mitochondrial inhibitor (14), or sirtinol, an inhibitor of SIRT1 activity (34). The concentration of each inhibitor did not affect cell growth or viability. SIRT1, an NAD+-dependent deacetylase, is known to regulate cellular stress responses, cellular metabolism and cellular survival (35). Specifically, SIRT1 inhibition reduces the induction of HSR genes (23); SIRT1 activity enhances SOD2 expression (36); SIRT1 negatively regulates the mammalian target of rapamycin triggering autophagy (37); and SIRT1 activates mitochondrial biogenesis via the stress–responsive transcription factor PGC1α (38). The yeast ortholog Sir2 is necessary for the induction of mitochondrial biogenesis by 4PBA or HU in Saccharomyces cerevisiae (39). There were no marked changes in SIRT1 protein expression after treatment of normal human fibroblasts with 4PBA, HU, TSA or SFN (data not shown). However, SIRT1 can be activated by various post-translational changes. Therefore, we examined the potential involvement of SIRT1 in the therapeutic effects of these drugs by inhibition studies. Biochemically blocking the JNK pathway, the autophagy pathway, induction of mitochondrial biogenesis and SIRT1 activity prevented the increase in FL-SMN transcript levels in SFN-treated spinal muscular atrophy fibroblasts indicating the necessity and potential concerted action of each of these pathways for the therapeutic response to SFN treatment (Fig. 9B).
To further demonstrate that induction of these various stress pathways is necessary for the therapeutic effects of the small molecules under study, we monitored the expression of total SMN protein expression after either SFN or HU treatment in the presence or absence of the stress pathway inhibitors. After 48–72 h of treatment, SFN increased SMN protein expression 1.57-fold in spinal muscular atrophy type I fibroblasts and 1.64-fold in spinal muscular atrophy type III fibroblasts (Fig. 9C). In separate experiments, HU or SFN treatment increased SMN protein expression 1.4- and 1.46-fold, respectively (Fig. 9D). The pharmacologically induced increase in SMN protein expression was significantly reduced by the various stress pathway inhibitors, indicating that the increase in SMN protein expression by both HU and SFN was dependent on activation of the JNK pathway, autophagy (not studied with HU), mitochondrial biogenesis and SIRT1 (Fig. 9D). At the concentrations used, the stress pathway inhibitors had no effect on basal levels of SMN protein expression. The magnitude of the increases in FL-SMN expression and SMN protein expression in SFN-treated spinal muscular atrophy cells compared with untreated cells is similar to that observed after 4PBA, HU or TSA treatment in SMA fibroblasts (30–32). Since SFN induction of FL-SMN transcript expression and SMN protein expression is dependent on the JNK pathway, autophagy pathway, mitochondrial biogenesis and SIRT1, the therapeutic potential of the activation of the stress proteome may require the concerted action of each of the individual stress pathways.
SIRT1 activation is not a common mechanism of action of 4PBA, HU, TSA and SFN
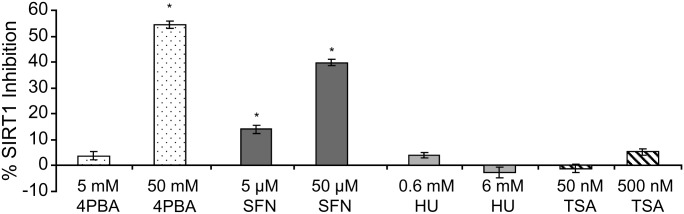
To determine whether SIRT1 activation was a common biochemical activity of the small molecules under study, a fluorescence-based in vitro SIRT1 activity assay was performed with two concentrations of each drug: that used to treat normal primary human fibroblasts in the experiments reported here (low), and 10 times that concentration, which is lethal in cell culture (high). HU and TSA had no effect on SIRT1 activity, whereas 4PBA (50 mm; high) and SFN (5 and 50 μm) significantly inhibited SIRT1 activity by 54, 14 and 40%, respectively (Fig. 10). This was a non-cell-based assay, and therefore, it is difficult to determine the effects of 4PBA and SFN on SIRT1 activity in vivo. Regardless, while SIRT1 activation may play a role in the induction of cellular stress responses (36), direct biochemical activation or inhibition of SIRT1 is not necessary for the common pharmacological induction of the adaptive cell survival response by all four small molecules.
Figure 10.
Biochemical SIRT1 activation is not necessary for the induction of the stress proteome by 4PBA, HU, TSA and SFN. SIRT1 activity assay. SIRT1 activity was measured by incubating an acetylated lysine substrate with human recombinant SIRT1, cosubstrate NAD+ and the indicated concentration of each small molecule. The percent inhibition of SIRT1 is plotted. Bars = SEM for triplicate measurements. *P ≤ 0.05; paired t-test.
DISCUSSION
Collectively, these findings demonstrate that pharmacological agents without known overlapping molecular functions, but with similar therapeutic potential in a variety of genetic disorders, induce mitochondrial and peroxisomal biogenesis and the transcription and translation of signature components of the adaptive cell survival response (HSR, UPR, autophagy and antioxidant response). Under adverse cellular conditions, mitochondria either induce the endogenous adaptive cell survival response reestablishing homeostasis (mild stress) or induce apoptosis (severe stress) (18). The drugs studied here exhibit a biphasic or hormetic dose response (40). At the low doses used they activate the stress proteome and produce beneficial effects; at higher doses they are cytotoxic. The pharmacological induction of mitochondrial biogenesis is consistent with the significant increase in energy expenditure required for metabolic adaptation to increased cellular stress (21,40).
Our results demonstrate the therapeutic importance of the pharmacological induction of mitochondrial biogenesis in X-linked adrenoleukodystrophy cells and K562 cells, a model of HbF induction. The reduction in very long-chain fatty acid levels in 4PBA-treated X-linked adrenoleukodystrophy fibroblasts and the increase in HbF-producing cell number in HU-treated K562 cells depends on the stress-activated JNK signaling cascade and the subsequent induction of mitochondrial biogenesis. Furthermore, the direct inhibition of other aspects of the cellular stress proteome, including the JNK pathway, autophagy pathway, mitochondrial biogenesis and SIRT1 activity, blocked the therapeutic increase in FL-SMN expression and SMN protein expression in SFN-treated spinal muscular atrophy fibroblasts. In these three disease models, we demonstrated that the therapeutic mechanism of these small molecules is induction of the cytoprotective adaptive cell survival response. Since inhibition of individual stress pathways blocked the therapeutic effects of SFN in spinal muscular atrophy cells, the collective induction of these stress pathways may be necessary for the therapeutic effects of these small molecules. The investigation of the therapeutic potential of these small molecules in additional genetic diseases may reveal if one stress pathway or the concerted induction of the entire stress proteome is necessary for the therapeutic effects in each responsive disease.
The therapeutic manipulation of the adaptive cell survival response or stress proteome explains the diverse number of agents that appear effective in studies of spinal muscular atrophy, including 4PBA, HU, TSA, resveratrol (a known activator of SIRT1) and, as shown here, SFN (41–43). The SMN protein facilitates the assembly of stress granules, vesicles which protect cellular mRNAs during cellular stress. Exon 7 of SMN1/2 is critical for stress granule formation (44). Changes in pH are able to elicit an increase in the proportion of FL-SMN transcripts compared with SMNΔ7 transcripts, demonstrating that the inclusion of exon 7 from SMN2 is a stress-dependent event (45). Therefore, small molecules that induce the adaptive cell survival response, as shown here with SFN, are expected to increase FL-SMN expression.
It is well documented that the induction of FL-SMN expression in spinal muscular atrophy cell lines by small molecules such as 4PBA, HU or TSA varies in magnitude, timing and within and between patients (32,46). The induction of the stress proteome as the therapeutic mechanism of 4PBA, TSA and HU may explain the variability in observed therapeutic effects in spinal muscular atrophy cell lines (30–32). Stress-induced splicing changes can be very sensitive to exogenously induced stress and occur quickly (47) which may account for the observed variability.
A therapeutic role for adaptive survival pathways has been observed in a range of diseases. Many of the disorders that have positively responded to induction of individual stress pathways have also responded favorably to treatment with one or more of the small molecules studied here. This suggests that activation of the stress proteome is the therapeutic mechanism of these small molecules. For example, overexpression or pharmacological induction of HSPs corrects the defect in Niemann-Pick patient cell lines, a lysosomal storage disorder, ameliorates the phenotype of spinal bulbar muscular atrophy mouse models and reduces protein aggregation in studies of Huntington's disease, a neurological disorder (48). Induction of the UPR proteins XBP1 and ATF6 protects against ischemia–reperfusion injury (49). Pharmacological induction of autophagy or overexpression of autophagy proteins protects against ischemia–reperfusion injury, improves mutant protein clearance and reduces protein toxicity in cell and animal models of Huntington's disease, Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, spinocerebellar ataxia and spinal bulbar muscular atrophy (26,50). Antioxidant and 4PBA treatment improve insulin sensitivity and glucose homeostasis in diabetic mouse models (51,52). 4PBA-induced mitochondrial biogenesis may also help normalize glucose levels because mitochondrial dysfunction is a key contributor to insulin resistance. 4PBA-induced expression of SOD2 increases neuroprotection in amyotrophic lateral sclerosis (53). Improved mitochondrial membrane potential and increased peroxisome proliferation induced by 4PBA treatment promotes neuronal integrity in Alzheimer's disease studies (54). These observations point to the significant therapeutic nature of the adaptive cellular responses which have traditionally been studied as a cell's response to acute stress.
Beneficial therapeutic effects that improve disease pathology and are independent of altering the specific genetic mutation have been well documented. Examples include TSA treatment of Duchenne's muscular dystrophy models and losartan treatment of Marfan's syndrome patients (41,55,56). Such examples provide an elegant illustration of corrective biological responses from indirect interventions.
The induction of the adaptive cell survival response and the subsequent reestablishment of cellular homeostasis may explain why these diverse pharmacological agents produce similar therapeutic effects in a myriad of disease models (Table 1). We note that the diseases responsive to these small molecules have mild cellular abnormalities, i.e. the cells are viable even though their suboptimal function may lead to severe clinical manifestations. We propose that pharmacological enhancement of the adaptive cell survival response and the subsequent reestablishment of cellular homeostasis beneficially alters disease-related metabolic stress, promotes cell viability and ameliorates some mild cellular genetic abnormalities without directly targeting the disease-causing gene. For example, enhancement of autophagy in the SOD1 transgenic amyotrophic lateral sclerosis mouse model clears protein aggregates and significantly increases lifespan (50). Also, TSA treatment of X-linked adrenoleukodystrophy (Abcd1 -/Y) mouse fibroblasts normalizes β-oxidation levels of very long-chain fatty acid levels independent of Abcd1 or Abcd2 expression (4). This indirect approach to therapy offers the advantage of pharmacologically modulating the adaptive capacity of the endogenous cellular machinery, specifically the stress proteome and mitochondrial function, to compensate for aberrant metabolic pathways and improve cellular function. This provides a rationale for the treatment of diseases whose specific genetic abnormalities are unknown, including many complex disorders, and may shorten the time to treatment for some currently untreatable diseases.
We have not tested if the known functions of each molecule, such as HDAC inhibition, are necessary for the activation of the stress proteome. These molecules may activate the stress proteome via their known biochemical activities or via uncharacterized molecular interactions within cells. For example, in K562 cells the production of reactive oxygen species, second messengers of the stress response, is necessary to increase HbF-containing cell production after SB and TSA treatment, but not after HU treatment (57). This result indicates that HbF-containing cell production can be stimulated by differing pathways. However, since nitric oxide is an intracellular metabolite of HU, reactive nitrogen species may act as second messengers to induce the stress response and increase HbF levels after HU treatment. In fact, nitric oxide release after HU treatment has been shown to enhance full-length SMN2 gene expression (58). Identification of the cellular responses (i.e. mitochondrial biogenesis, nitric oxide release, etc.) necessary for downstream therapeutic effects of interest may assist screening for small molecules with optimal clinical effects.
The agents investigated here are in clinical use with few if any side effects. They were initially identified by their effects on disease outcomes, not the underlying pathways responsible for their beneficial effects. Establishment of the therapeutic potential of the adaptive cell survival response by these small molecules provides targets for the identification of more efficacious small molecules with even lower toxicity. The exploitation of the innate cellular survival program may ameliorate disease symptoms in a spectrum of disorders with mild cellular phenotypes without targeting a specific molecule or signaling pathway for each individual disease. The amenable disorders not only include those with mitochondrial or peroxisomal defects, increased oxidative stress, or protein conformation or trafficking defects, but also include aging disorders and complex diseases with unknown genetic or environmental etiology and mild cellular phenotypes. Therapy that targets homeostatic regulation could have a profound effect on medicine.
MATERIALS AND METHODS
Cell culture
Primary human fibroblasts, HeLa cells (ATCC #CCL-2, a gift from Dr Hal Dietz, Johns Hopkins University, Baltimore, MD, USA), K562 cells (ATCC # CCL-243) and SMA type I fibroblasts GM09677 and GM00232 (Coriell Institute for Medical Research, Camden, NJ, USA) or SMA type III fibroblasts 96-2906 (Kennedy Krieger Institute, Baltimore, MD, USA) were grown in minimal Eagle medium (MEM; Mediatech, Manassas, VA, USA), RPMI medium (Mediatech) supplemented with 10 mm HEPES and 1 mm sodium pyruvate, RPMI or DMEM (Mediatech), respectively, and supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 U/ml). Fibroblasts (70–95% confluent) were treated with 5 mm 4PBA (1 mm for JNK inhibition studies), 600 µm HU, 200nm TSA or 5μm SFN unless otherwise specified. K562 cells were treated with 1.2 mm SB or 100 µm HU. All drugs were obtained from Sigma-Aldrich (St Louis, MO, USA). The concentration of each drug per cell type was titrated to allow 100% viability and to minimally affect growth rate. The duration of treatment varied by experiment as indicated in the figure legends.
For SFN treatment of SMA fibroblasts, the cells were treated 36 h after plating, the media changed 24 h before collection with media that were pre-warmed and equilibrated in a 37°C CO2 incubator overnight to minimize pH effects, and the cells collected on ice by scraping.
HDAC activity assay
A colorimetric HDAC activity assay was performed three times in triplicate using 100–200 μg of HeLa or fibroblast lysates following the manufacturer's protocol (K331; Biovision, Mountain View, CA, USA).
Indirect immunofluorescence
Performed as described in reference (59) using 4% formaldehyde, 1% Triton X-100, primary antibody anti-ATP5B (Millipore, Billerica, MA, USA) or anti-ABCD3 (Invitrogen, Carlsbad, CA, USA) and secondary antibody sheep anti-mouse IgG FITC-conjugated (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or goat anti-rabbit IgG rhodamine-conjugated (Jackson ImmunoResearch, West Grove, PA, USA). Staining for anti-ATP5B and anti-ABCD3 was carried out using the same cell populations, but on separate cover slips because of the imaging capabilities available at the time.
In-cell western analysis
The immunostaining of primary human fibroblasts with anti-ATP5B, DRAQ5 (Cell Signaling Technologies, Danvers, MA, USA; a cellular DNA stain to normalize cell number), and secondary antibody IRDye 800CW donkey anti-mouse IgG (Li-cor Biosciences, Lincoln, NE, USA) was identical to the indirect immunofluorescence procedure described above and performed in duplicate. Imaging and quantification were performed using the Odyssey Imager (Li-cor Biosciences).
Inhibition studies
Antimycin A (10 ng/ml), LY294002, PD98059, RO-31-8425, SB203580, SP600125 and L-NAME, 3-methyladenine were obtained from Sigma. Compound C was from EMD Biosciences (Darmstadt, Germany). Sirtinol was from Tocris Bioscience (Minneapolis, MN, USA). Human fibroblasts were treated with each inhibitor, 45–75 min later 4PBA, HU, TSA or SFN was added and cells were stained 4–6 days after treatment for mitochondrial studies and as indicated for SMN studies. As a control for inhibitors dissolved in DMSO, fibroblasts were treated with an equivalent amount of DMSO in the absence of inhibitor.
Real-time PCR analysis (RT-PCR)
Following the manufacturer's protocols, DNase I-treated cDNA was synthesized using Superscript III or Thermoscript reverse transcriptase (Invitrogen). PCR reactions were performed in duplicate on the Roche Lightcycler 3.5 (Basel, Switzerland) or the Bio-Rad iCycler (Hercules, CA, USA) using Quantitech SYBR green PCR mix (Qiagen, Valencia, CA, USA). Primer sequences are available upon request. GeNorm software calculated a normalization factor for each sample using the relative amounts of at least two of the following reference genes, β2-microglobulin, glyceraldehyde-6-phosphate, actin and/or eukaryotic elongation factor 1A (60). The following primer sequences were used to detect FL-SMN: forward primer 5′ GCGATGA TTCTGACATTTGG 3′ and reverse primer 5′ AAATGAAGCCACAGCTTTATCA 3′. Total SMN transcripts (SMN + exon 7 and SMNΔ7 transcripts) were detected using forward primer 5′ ATAATTCCCCCACCACCTC 3′ (61) and reverse primer 5′ CACCTTCCTTCTTTTTGATT TTGTC 3′ (30).
Western blot analysis
Cell lysates were collected in mammalian protein extraction reagent (m-PER; Thermo Scientific, Rockford, IL, USA) plus 1× protease inhibitor (Sigma-Aldrich) and 1× Halt phosphatase inhibitor (Thermo Scientific). Denatured SDS–PAGE and immunoblot analyses were performed using the following antibodies: phosphorylated JNK and β-actin from Santa Cruz Biotechnology, total HSP70, total HSP90, BIP, phosphorylated eIF2α and XBP1 from Cell Signaling, APG8 (Abgent, San Diego, CA, USA), SOD2 (Stressgen, Ann Arbor, MI, USA), β-actin (Sigma) and SMN (BD Transduction Laboratories, Franklin Lakes, NJ, USA). Quantitation for all proteins except SMN was performed using a Fuji Intelligent Dark box II, FUJI LAS-1000 Lite software and Image Gauge v4.22 software (Tokyo, Japan). SMN quantitation was performed using the Odyssey Imager (Li-Cor Biosciences).
Fatty acid β-oxidation
Fatty acid β-oxidation activity in XALD fibroblasts was determined by measuring their capacity to degrade 1-14C-labeled fatty acids to water-soluble products as described (4).
Flow cytometry
K562 cells were incubated with 50 nm Mitotracker Green FM, 300 nm Mitotracker Deep Red 633 (Invitrogen) or phosphate-buffered saline (PBS) as a negative control for 15 min at 37°C or stained with anti-Pex14 using Caltag Fix and Perm reagents (Invitrogen) following the manufacturer's instructions. Stained cells were suspended in 0.5 ml of 1% paraformaldehyde/PBS and subjected to flow cytometric analysis (FACScan machine, Becton Dickinson, Franklin Lakes, NJ, USA).
Measurement of hemoglobin
K562 cells were suspended in 1.1 ml media and 240 µl of DAF [working solution 50 μl 2,7 diaminofluorene (DAF) stock (50 µg DAF in 5 ml 90% glacial acetic acid), 50 µl 30% hydrogen peroxide, and 2.5 ml 200 mm Tris–HCl, pH 7.0]. DAF-stained hemoglobinized cells were scored using a hemocytometer.
Mitotracker staining
SMA type I (GM00232) fibroblasts were treated with 2.5 mm 4PBA, 300 μm HU, 100 nm TSA or 2.5 μm SFN for 5 days, live stained with 25 nm Mitotracker Red CMXROS (Invitrogen) in pre-warmed MEM media for 15 min at 37°C in a CO2 incubator. Stained cells were washed with pre-warmed MEM, washed with PBS and excess PBS was removed. The cells were examined under the microscope at ×80.
SIRT1 activity assay
A fluorescence-based SIRT1 activity assay was performed in triplicate following the manufacturer's protocol (10010401; Cayman Chemical Company, Ann Arbor, MI, USA). The percent developer interference was below 3% for each small molecule. The percent fluorophore interference was below 9% for each small molecule. The acceptable interference values were ≤10%.
Statistical analyses
Three or more independent experiments were performed for each technique unless otherwise noted and the standard error of the mean (SEM) calculated and graphed. A one-tailed Student's t-test was used to calculate P-values. P-values ≤0.05 were considered significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Johns Hopkins McKusick-Nathans Institute of Genetic Medicine and Public Health Service grants (GM077456, HD10981 and HD20961) from the National Institutes of Health.
Supplementary Material
Acknowledgements
We thank H. Dietz, G. Maegawa, A. McCallion, J. Mendell, S. Michaelis, B. Migeon, J. Pevsner and C. Thompson for reviewing the manuscript.
Conflict of Interest statement. None declared.
References
- 1.Kemp S., Wei H.M., Lu J.F., Braiterman L.T., McGuinness M.C., Moser A.B., Watkins P.A., Smith K.D. Gene redundancy and pharmacological gene therapy: implications for X-linked adrenoleukodystrophy. Nat. Med. 1998;4:1261–1268. doi: 10.1038/3242. [DOI] [PubMed] [Google Scholar]
- 2.Gardian G., Browne S.E., Choi D.K., Klivenyi P., Gregorio J., Kubilus J.K., Ryu H., Langley B., Ratan R.R., Ferrante R.J., et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J. Biol. Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 3.McGuinness M.C., Lu J.F., Zhang H.P., Dong G.X., Heinzer A.K., Watkins P.A., Powers J., Smith K.D. Role of ALDP (ABCD1) and mitochondria in X-linked adrenoleukodystrophy. Mol. Cell. Biol. 2003;23:744–753. doi: 10.1128/MCB.23.2.744-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuinness M.C., Zhang H.P., Smith K.D. Evaluation of pharmacological induction of fatty acid beta-oxidation in X-linked adrenoleukodystrophy. Mol. Genet. Metab. 2001;74:256–263. doi: 10.1006/mgme.2001.3239. [DOI] [PubMed] [Google Scholar]
- 5.Li X., Baumgart E., Dong G.X., Morrell J.C., Jimenez-Sanchez G., Valle D., Smith K.D., Gould S.J. PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 2002;22:8226–8240. doi: 10.1128/MCB.22.23.8226-8240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrader M., Reuber B.E., Morrell J.C., Jimenez-Sanchez G., Obie C., Stroh T.A., Valle D., Schroer T.A., Gould S.J. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- 7.Kultz D. Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J. Exp. Biol. 2003;206:3119–3124. doi: 10.1242/jeb.00549. [DOI] [PubMed] [Google Scholar]
- 8.Ong D.S., Kelly J.W. Chemical and/or biological therapeutic strategies to ameliorate protein misfolding diseases. Curr. Opin. Cell Biol. 2011;23:231–238. doi: 10.1016/j.ceb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 10.Durieux J., Wolff S., Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung M. Inhibitors of histone deacetylase as new anticancer agents. Curr. Med. Chem. 2001;8:1505–1511. doi: 10.2174/0929867013372058. [DOI] [PubMed] [Google Scholar]
- 12.Myzak M.C., Karplus P.A., Chung F.L., Dashwood R.H. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 13.Wei H., Kemp S., McGuinness M.C., Moser A.B., Smith K.D. Pharmacological induction of peroxisomes in peroxisome biogenesis disorders. Ann. Neurol. 2000;47:286–296. [PubMed] [Google Scholar]
- 14.Ranganathan S., Harmison G.G., Meyertholen K., Pennuto M., Burnett B.G., Fischbeck K.H. Mitochondrial abnormalities in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2009;18:27–42. doi: 10.1093/hmg/ddn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzer A.K., McGuinness M.C., Lu J.F., Stine O.C., Wei H., Van der Vlies M., Dong G.X., Powers J., Watkins P.A., Smith K.D. Mouse models and genetic modifiers in X-linked adrenoleukodystrophy. Adv. Exp. Med. Biol. 2003;544:75–93. doi: 10.1007/978-1-4419-9072-3_12. [DOI] [PubMed] [Google Scholar]
- 16.Perrine S.P. Fetal globin stimulant therapies in the beta-hemoglobinopathies: principles and current potential. Pediatr. Ann. 2008;37:339–346. doi: 10.3928/00904481-20080501-10. [DOI] [PubMed] [Google Scholar]
- 17.Keefer J.R., Schneidereith T.A., Mays A., Purvis S.H., Dover G.J., Smith K.D. Role of cyclic nucleotides in fetal hemoglobin induction in cultured CD34+ cells. Exp. Hematol. 2006;34:1151–1161. doi: 10.1016/j.exphem.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Lee H.C., Wei Y.H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int. J. Biochem. Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Bennett B.L., Sasaki D.T., Murray B.W., O'Leary E.C., Sakata S.T., Xu W., Leisten J.C., Motiwala A., Pierce S., Satoh Y., et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl Acad. Sci. USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J., Yu Y., Duerksen-Hughes P.J. Protein kinases and their involvement in the cellular responses to genotoxic stress. Mutat. Res. 2003;543:31–58. doi: 10.1016/s1383-5742(02)00069-8. [DOI] [PubMed] [Google Scholar]
- 21.Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- 22.Lallemand D., Ham J., Garbay S., Bakiri L., Traincard F., Jeannequin O., Pfarr C.M., Yaniv M. Stress-activated protein kinases are negatively regulated by cell density. EMBO J. 1998;17:5615–5626. doi: 10.1093/emboj/17.19.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerheide S.D., Anckar J., Stevens S.M., Jr, Sistonen L., Morimoto R.I. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedoroff N. Redox regulatory mechanisms in cellular stress responses. Ann. Bot. 2006;98:289–300. doi: 10.1093/aob/mcl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark M.S., Peck L.S. Triggers of the HSP70 stress response: environmental responses and laboratory manipulation in an Antarctic marine invertebrate (Nacella concinna) Cell Stress Chaperones. 2009;14:649–660. doi: 10.1007/s12192-009-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinet W., Agostinis P., Vanhoecke B., Dewaele M., De Meyer G.R. Autophagy in disease: a double-edged sword with therapeutic potential. Clin. Sci. (Lond.) 2009;116:697–712. doi: 10.1042/CS20080508. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 28.Sumner C.J., Huynh T.N., Markowitz J.A., Perhac J.S., Hill B., Coovert D.D., Schussler K., Chen X., Jarecki J., Burghes A.H., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 29.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 30.Andreassi C., Angelozzi C., Tiziano F.D., Vitali T., De Vincenzi E., Boninsegna A., Villanova M., Bertini E., Pini A., Neri G., et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 31.Grzeschik S.M., Ganta M., Prior T.W., Heavlin W.D., Wang C.H. Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann. Neurol. 2005;58:194–202. doi: 10.1002/ana.20548. [DOI] [PubMed] [Google Scholar]
- 32.Avila A.M., Burnett B.G., Taye A.A., Gabanella F., Knight M.A., Hartenstein P., Cizman Z., Di Prospero N.A., Pellizzoni L., Fischbeck K.H., et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seglen P.O., Gordon P.B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grozinger C.M., Chao E.D., Blackwell H.E., Moazed D., Schreiber S.L. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 35.Salminen A., Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell. Signal. 2009;21:1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Anastasiou D., Krek W. SIRT1: linking adaptive cellular responses to aging-associated changes in organismal physiology. Physiology (Bethesda) 2006;21:404–410. doi: 10.1152/physiol.00031.2006. [DOI] [PubMed] [Google Scholar]
- 37.Haigis M.C., Yankner B.A. The aging stress response. Mol. Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J., Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann. N. Y. Acad. Sci. 2009;1173((Suppl. 1):E10–E19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cha G. Baltimore, MD, USA: Johns Hopkins University; 2010. A conserved response to diverse small molecules: implications for therapy. Unpublished doctoral dissertation. [Google Scholar]
- 40.Calabrese V., Cornelius C., Dinkova-Kostova A.T., Calabrese E.J., Mattson M.P. Cellular stress responses, the hormesis paradigm, and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid. Redox Signal. 2010;13:1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lunn M.R., Wang C.H. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 42.Dayangac-Erden D., Bora G., Ayhan P., Kocaefe C., Dalkara S., Yelekci K., Demir A.S., Erdem-Yurter H. Histone deacetylase inhibition activity and molecular docking of (e)-resveratrol: its therapeutic potential in spinal muscular atrophy. Chem. Biol. Drug Des. 2009;73:355–364. doi: 10.1111/j.1747-0285.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 43.Sakla M.S., Lorson C.L. Induction of full-length survival motor neuron by polyphenol botanical compounds. Hum. Genet. 2008;122:635–643. doi: 10.1007/s00439-007-0441-0. [DOI] [PubMed] [Google Scholar]
- 44.Hua Y., Zhou J. Rpp20 interacts with SMN and is re-distributed into SMN granules in response to stress. Biochem. Biophys. Res. Commun. 2004;314:268–276. doi: 10.1016/j.bbrc.2003.12.084. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y.C., Yuo C.Y., Yang W.K., Jong Y.J., Lin H.H., Chang Y.S., Chang J.G. Extracellular pH change modulates the exon 7 splicing in SMN2 mRNA. Mol. Cell. Neurosci. 2008;39:268–272. doi: 10.1016/j.mcn.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Also-Rallo E., Alias L., Martinez-Hernandez R., Caselles L., Barcelo M.J., Baiget M., Bernal S., Tizzano E.F. Treatment of spinal muscular atrophy cells with drugs that upregulate SMN expression reveals inter- and intra-patient variability. Eur. J. Hum. Genet. 2011;19:1059–1065. doi: 10.1038/ejhg.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pleiss J.A., Whitworth G.B., Bergkessel M., Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans C.G., Chang L., Gestwicki J.E. Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toth A., Nickson P., Mandl A., Bannister M.L., Toth K., Erhardt P. Endoplasmic reticulum stress as a novel therapeutic target in heart diseases. Cardiovasc. Hematol. Disord. Drug Targets. 2007;7:205–218. doi: 10.2174/187152907781745260. [DOI] [PubMed] [Google Scholar]
- 50.Madeo F., Eisenberg T., Kroemer G. Autophagy for the avoidance of neurodegeneration. Genes Dev. 2009;23:2253–2259. doi: 10.1101/gad.1858009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R.O., Gorgun C.Z., Hotamisligil G.S. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Shen W., Zhao B., Wang Y., Wertz K., Weber P., Zhang P. Targeting mitochondrial biogenesis for preventing and treating insulin resistance in diabetes and obesity: hope from natural mitochondrial nutrients. Adv. Drug Deliv. Rev. 2009;61:1343–1352. doi: 10.1016/j.addr.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Petri S., Kiaei M., Kipiani K., Chen J., Calingasan N.Y., Crow J.P., Beal M.F. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2006;22:40–49. doi: 10.1016/j.nbd.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Santos M.J., Quintanilla R.A., Toro A., Grandy R., Dinamarca M.C., Godoy J.A., Inestrosa N.C. Peroxisomal proliferation protects from beta-amyloid neurodegeneration. J Biol. Chem. 2005;280:41057–41068. doi: 10.1074/jbc.M505160200. [DOI] [PubMed] [Google Scholar]
- 55.Minetti G.C., Colussi C., Adami R., Serra C., Mozzetta C., Parente V., Fortuni S., Straino S., Sampaolesi M., Di Padova M., et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat. Med. 2006;12:1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 56.Habashi J.P., Judge D.P., Holm T.M., Cohn R.D., Loeys B.L., Cooper T.K., Myers L., Klein E.C., Liu G., Calvi C., et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsiao C.H., Li W., Lou T.F., Baliga B.S., Pace B.S. Fetal hemoglobin induction by histone deacetylase inhibitors involves generation of reactive oxygen species. Exp. Hematol. 2006;34:264–273. doi: 10.1016/j.exphem.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 58.Xu C., Chen X., Grzeschik S.M., Ganta M., Wang C.H. Hydroxyurea enhances SMN2 gene expression through nitric oxide release. Neurogenetics. 2011;12:19–24. doi: 10.1007/s10048-010-0268-z. [DOI] [PubMed] [Google Scholar]
- 59.Watkins P.A., Gould S.J., Smith M.A., Braiterman L.T., Wei H.M., Kok F., Moser A.B., Moser H.W., Smith K.D. Altered expression of ALDP in X-linked adrenoleukodystrophy. Am. J. Hum. Genet. 1995;57:292–301. [PMC free article] [PubMed] [Google Scholar]
- 60.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034-34.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brichta L., Holker I., Haug K., Klockgether T., Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann. Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 62.Oliveira J.M., Chen S., Almeida S., Riley R., Goncalves J., Oliveira C.R., Hayden M.R., Nicholls D.G., Ellerby L.M., Rego A.C. Mitochondrial-dependent Ca2+ handling in Huntington's disease striatal cells: effect of histone deacetylase inhibitors. J. Neurosci. 2006;26:11174–11186. doi: 10.1523/JNEUROSCI.3004-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park H.M., Kim J.A., Kwak M.K. Protection against amyloid beta cytotoxicity by sulforaphane: role of the proteasome. Arch. Pharm. Res. 2009;32:109–115. doi: 10.1007/s12272-009-1124-2. [DOI] [PubMed] [Google Scholar]
- 64.Ricobaraza A., Cuadrado-Tejedor M., Perez-Mediavilla A., Frechilla D., Del Rio J., Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer's disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 65.Xue M., Qian Q., Adaikalakoteswari A., Rabbani N., Babaei-Jadidi R., Thornalley P.J. Activation of NF-E2-related factor-2 reverses biochemical dysfunction of endothelial cells induced by hyperglycemia linked to vascular disease. Diabetes. 2008;57:2809–2817. doi: 10.2337/db06-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun C., Zhou J. Trichostatin A improves insulin stimulated glucose utilization and insulin signaling transduction through the repression of HDAC2. Biochem. Pharmacol. 2008;76:120–127. doi: 10.1016/j.bcp.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Gorski G., Rozynkowa D. Increased expression of fragile site Xq by hydroxyurea. Preliminary communication. Acta Anthropogenet. 1985;9:71–76. [PubMed] [Google Scholar]
- 68.Chiurazzi P., Pomponi M.G., Pietrobono R., Bakker C.E., Neri G., Oostra B.A. Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum. Mol. Genet. 1999;8:2317–2323. doi: 10.1093/hmg/8.12.2317. [DOI] [PubMed] [Google Scholar]
- 69.Qi X., Hosoi T., Okuma Y., Kaneko M., Nomura Y. Sodium 4-phenylbutyrate protects against cerebral ischemic injury. Mol. Pharmacol. 2004;66:899–908. doi: 10.1124/mol.104.001339. [DOI] [PubMed] [Google Scholar]
- 70.Yoon H.Y., Kang N.I., Lee H.K., Jang K.Y., Park J.W., Park B.H. Sulforaphane protects kidneys against ischemia-reperfusion injury through induction of the Nrf2-dependent phase 2 enzyme. Biochem. Pharmacol. 2008;75:2214–2223. doi: 10.1016/j.bcp.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 71.Choo-Kang L.R., Zeitlin P.L. Induction of HSP70 promotes DeltaF508 CFTR trafficking. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2001;281:L58–L68. doi: 10.1152/ajplung.2001.281.1.L58. [DOI] [PubMed] [Google Scholar]
- 72.Hutt D.M., Herman D., Rodrigues A.P., Noel S., Pilewski J.M., Matteson J., Hoch B., Kellner W., Kelly J.W., Schmidt A., et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat. Chem. Biol. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.