Abstract
Variation in gene expression has been found to be important in disease susceptibility and pharmacogenomics. Local and distant expression quantitative trait loci (eQTLs) have been identified via genome-wide association study (GWAS); yet the functional analysis of these variants has been challenging. The aim of this study was to unravel the functional consequence of a gene with a local SNP with evidence for local and distant regulatory roles in cellular sensitivity to cisplatin, one of the most widely used chemotherapeutic drugs. To this end, we measured cellular susceptibility to cisplatin in 176 HapMap lymphoblastoid cell lines derived from Yoruba individuals from Ibadan, Nigeria. The 276 cytotoxicity-associated SNPs at the suggestive threshold of P ≤ 0.0001 were significantly enriched for eQTLs. Of these SNPs, we found one intronic SNP, rs17115814, that had a significant relationship with the expression level of its host gene, PRPF39 (P= 0.0007), and a significant correlation with the expression of over 100 distant transcripts (P ≤ 0.0001). Successful knockdown of PRPF39 expression using siRNA resulted in a significant increase in cisplatin resistance. We then measured the expression of 61 downstream targets after PRPF39 knockdown and found 53 gene targets had significant (P ≤ 0.05) expression changes. Included in the list of genes that significantly changed after PRPF39 knockdown were MAP3K4 and TFPD2, two important signaling genes previously shown to be relevant in cisplatin response. Thus, modulation of a local target gene identified through a GWAS was followed by a downstream cascade of gene expression changes resulting in greater resistance to cisplatin.
INTRODUCTION
Platinum compounds comprise one of the most widely used group of chemotherapeutics and are used in the treatment of head and neck, testicular, ovarian, cervical, lung, endometrial and colorectal cancer (1,2). These agents interact with DNA to form both inter- and intra- strand crosslinks (1). Although the mechanism of action is similar for cisplatin and carboplatin, cisplatin's toxicity profile is distinct from that of carboplatin. For example, ototoxicity, resulting in permanent sensorineural hearing loss and/or tinnitus, is one of the most common side effects of cisplatin chemotherapy, affecting more than half of patients (3). Sensory neuropathies, which also emerge during treatment, can be debilitating and permanent, affecting 30–40% of patients (4). There is no treatment for these debilitating toxicities, nor are preventive measures available. Because the toxicities associated with cisplatin are quite serious, an understanding of the genetic variation that contributes to their manifestation would significantly benefit patients.
To date, our understanding of genetic variation contributing to cisplatin-induced toxicities is incomplete. Using candidate gene approaches, glutathione S-transferase (GST) genotypes were identified as risk factors for ototoxicity in one survey of testicular cancer survivors (5). Additional candidate studies found genes such as ERCC2 (6) and ATP7A/B (7) to be important in cisplatin response. Later, in a study of 224 candidate genes among 54 children treated with cisplatin, Ross et al. (3) found 8- to 16-fold increased risk of ototoxicity for genetic variants within COMT and TPMT, respectively. However, despite these successes, the ability to predict cisplatin response and toxicity remains extremely limited.
Genetic variants can impact pharmacologic phenotypes by various means, including through their effect on protein structure or on the level of gene expression. We have previously used a whole-genome, cell-based approach to identify SNPs associated with cytotoxicity as a result of their association with baseline levels of gene expression (8–10). Significantly associated cisplatin SNPs and genes were identified in the International HapMap Caucasian (CEU) and Yoruba (YRI) populations (9,10) or identified within Asian (ASN) International HapMap populations and then validated in other populations (11). Interestingly, we found disproportionately more expression quantitative trait loci (eQTLs) in the SNPs associated with cisplatin-induced cytotoxicity in the CEU population than expected by chance matched on minor allele frequency (12).
Genome-wide discovery strategies have resulted in the identification of both local and distant eQTLs (8,13–17). Although some studies have begun to evaluate the functional effects of distant eQTLs (17), the majority of functional follow-up remains focused on local acting eQTLs because methods are readily available to measure local effects (for example, reporter gene assays and allelic imbalance assays). The evaluation of distant effects remains challenging.
The present study extends our understanding of the role of regulation of distant target genes, using a pharmacological phenotype. We identified local and distant eQTLs associated with cisplatin-induced cytotoxicity in 176 lymphoblastoid cell lines (LCLs) derived from YRI individuals residing in Ibadan, Nigeria. An intronic SNP of PRPF39 that was a local eQTL for its host gene as well as a distant eQTL to over 100 target genes was chosen for further study. We functionally evaluated the host gene, PRPF39, for its role in cisplatin-induced cytotoxicity as well as its effect on 61 distant target genes.
RESULTS
Genome-wide association study of cisplatin-induced cytotoxicity
Genome-wide association study (GWAS) using 2.5 million SNPs with cisplatin IC50 in 176 YRI LCLs revealed 296 SNPs that were associated at P ≤ 0.0001, a suggestive significance threshold. No SNPs reached traditional genome-wide significance. Supplementary Material, Figure S1 contains a Manhattan plot of the results.
Each of the SNPs associated with cytotoxicity at P ≤0.0001 was evaluated as either local or distant eQTLs. Local was defined as an SNP within a gene or within 2 kb of the 5′ end of the gene or within 500 bp of the 3′ end of the gene with a P-value <0.01 and distant was all other locations with P ≤0.0001. We observed a significant enrichment of all eQTLs with a Z-score of 3.34 for cisplatin-associated SNPs at P ≤0.0001 (Supplementary Material, Fig. S2). We prioritized SNPs with both local and distant target genes for two reasons. Local relationships are more straightforward for functional follow-up, whereas distant relationships suggest a larger role in the regulatory architecture.
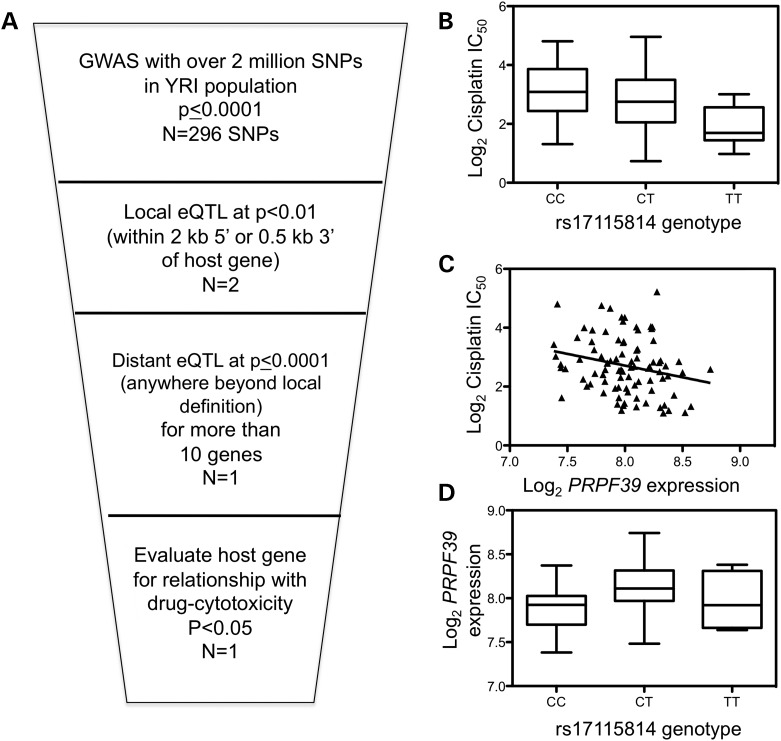
Using the fact that cisplatin IC50 GWAS results were enriched for eQTLs, we identified local and distant eQTLs in the GWAS results. Figure 1A illustrates the schema that identifies local eQTLs that also had distant regulatory relationships. Figure 1B displays one intronic SNP, rs17115814, associated with cisplatin IC50 that was a local eQTL for its host gene PRPF39 and the SNP was also associated with over 100 distant target genes at (P ≤ 0.0001). Baseline PRPF39 expression based on previously published exon array expression data on the first YRI panel (8) was correlated with cisplatin IC50 with a P-value of 0.01 and a q-value of 0.15 (Fig. 1C). Figure 1D illustrates the SNP association with host gene expression (P= 0.0007). This local eQTL was the strongest eQTL (within 10kb) for PRPF39 and each copy of the minor allele decreases the cisplatin IC50, on average, by 55% (P= 0.018). Furthermore, modeling the relationship of PRPF39 with cisplatin IC50 with rs17115814 genotype considered reduces somewhat the significance of the PRPF39 relationship (P= 0.059) but does not account for it entirely.
Figure 1.
Cisplatin IC50 significantly correlates with the local eQTL rs17115814 and PRPF39 expression. (A) Genome-wide association results of 176 YRI LCLs were analyzed to identify local eQTLs at P< 0.01, defined as being within the gene, 2 kb of the 5′ end of the gene, or 0.5 kb of the 3′ end of the gene. Local eQTLs were then analyzed as to whether they were distant eQTLs for more than 10 target genes at P< 0.0001. Finally, the host gene's relationship was evaluated for correlation with cisplatin cytotoxicity at P< 0.05. Cisplatin IC50 is significantly correlated with rs17115814 with an empirical P= 9.99 × 10−5 (B) as well as with PRPF39 expression at P= 0.01 (C). The intronic SNP is associated with PRPF39 expression at P= 0.0007 (D).
Effect of knockdown of PRPF39 on cisplatin cytotoxicity
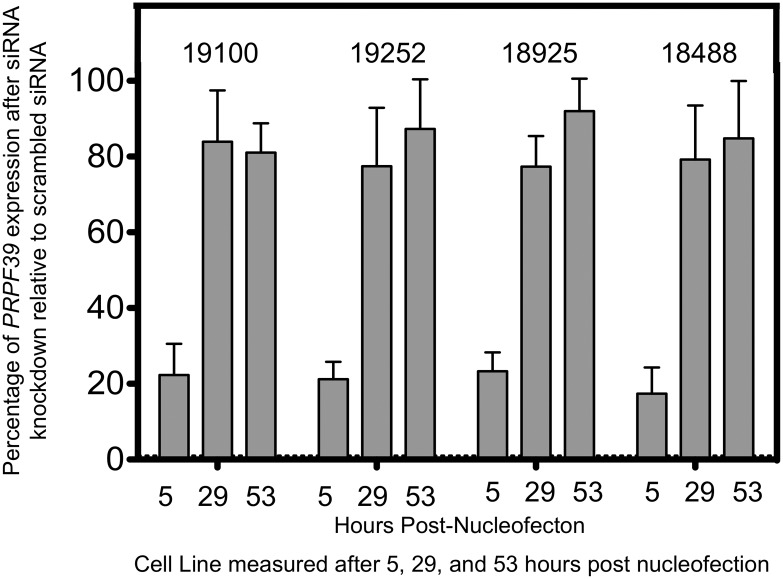
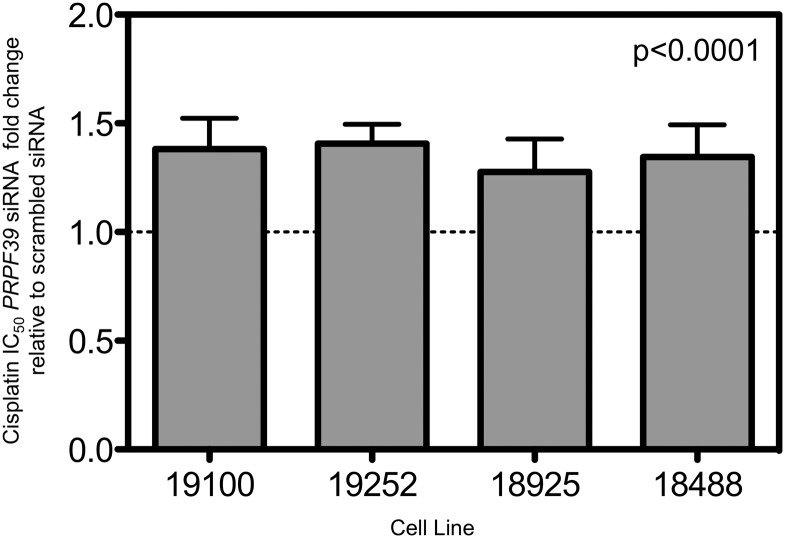
In order to validate the relationship of PRPF39 and cisplatin, knockdown of PRPF39 was assessed in four different YRI LCLs. Knockdown was assessed at 5 (time of drug addition), 29 and 53 h (corresponding to the measurement of cytotoxicity) after nucleofection. As seen in Figure 2, the knockdown of PRPF39 expression was 20% relative to control at 5 h and by 29 and 53 h was back to over 80% relative to control levels. Despite this short-term knockdown, the cytotoxic effect of cisplatin was significantly modulated as illustrated in Figure 3. The LCLs were significantly more resistant to cisplatin following PRPF39 knockdown. Using a combined mixed effects model, the P-value is significant (P< 0.0001) with an average IC50 increase of 35% relative to control. We also assessed the effect of the knockdown on carboplatin sensitivity; however, no significant change in IC50 using both a 48 and 72 h assay was observed.
Figure 2.
PRPF39 expression knockdown after siRNA nucleofection. Four different LCLs were evaluated for PRPF39 knockdown after siRNA treatment. Approximately 20% of PRPF39 expression remained 5 h after siRNA was introduced through nucleofection relative to a non-targeting scrambled control siRNA. Expression of PRPF39 rebounded after 29 and 53 h to >75% relative to a non-target control. All values are based on a minimum of three different nucleofections.
Figure 3.
PRPF39 knockdown significantly increases the resistance of four cell lines to cisplatin. After successful PRPF39 knockdown using siRNA, cisplatin cytotoxicity was assessed using the alamarBlue growth inhibition assay. For four different LCLs, IC50 values after successful knockdown of PRPF39 are significantly higher relative to a non-targeting control (19100 P= 0.025; 19252 P= 0.017; 18925 P= 0.069; 18488 P= 0.067), demonstrating that after knockdown cell lines are more resistant to cisplatin. P-values are from a Student's one-tailed t-test based on a minimum of three separate knockdown experiments. When incorporating the four different cell lines into a single mixed effects model, the P-value drops to 0.0001, with the IC50 of the knockdown being ∼34% above the non-targeted control.
Distant genes' association with rs17115814
After confirming the role of PRPF39 in modulating cisplatin response, we assessed expression changes of 61 distant target genes of rs17115814 in three YRI LCLs. Since the SNP was identified through association with cisplatin-induced cytotoxicity, we selected target genes that either were significantly (q-value < 0.16) correlated with cisplatin cytotoxicity or had the strongest eQTL relationship with rs17115814 (P ≤0.00002) regardless of cisplatin correlation. Gene expression of these target genes (Supplementary Material, Table S1 and Fig. S3) was measured at 5, 12, 18 and 24 h after PRPF39 knockdown by siRNA.
The downstream targets were evaluated within each cell line individually as well as in a mixed effects model combining all three cell lines at each time point (referred to as combined cell line model). PRPF39 showed the most significant knockdown at 5 h in preliminary experiments (Fig. 2) and this was confirmed with the TaqMan low-density array (TLDA) cards (data not shown). Of the 59 targets successfully assayed, 53 (90%) showed nominal significant (P< 0.05) change in expression after PRPF39 knockdown compared with control for at least one time point for either a single cell line or the combined cell line model (Supplementary Material, Fig. S3). No gene changed expression significantly (P< 0.05) in an identical pattern across all three cell lines, although there were genes where the direction of fold change in expression was consistent, despite lack of significance for some lines.
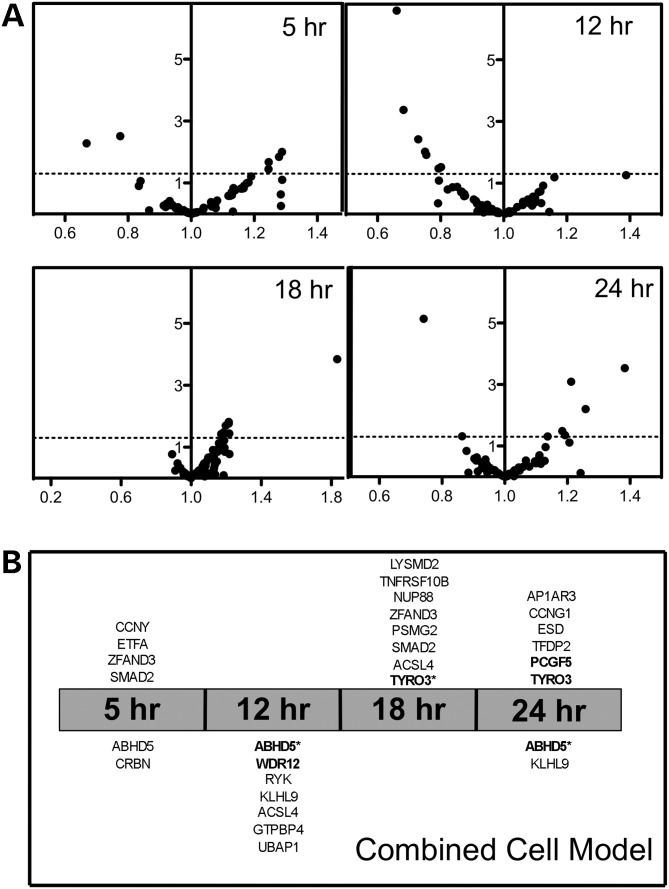
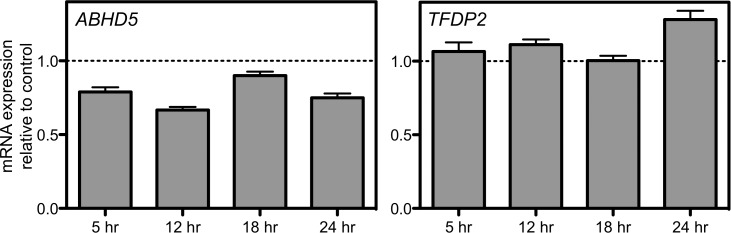
Figure 4A illustrates volcano plots of the combined cell line model evaluating the three cell lines. Using a P-value of 0.05 (uncorrected), we found that six, seven, eight and eight expression changes are significant at the 5, 12, 18 and 24 h time point, respectively. The genes and the direction of change are displayed in Figure 4B. However, when correcting for the number of genes tested at each time point, in the combined cell line model, the expression of six target genes (four unique genes) significantly changed at 12, 18 and/or 24 h (Fig. 4B). An example is ABHD5, a gene that was significantly lower compared with control at multiple time points (Fig. 5). More commonly, significant changes were seen for only one time point such as increased expression of TFDP2 at 24 h after transfection across all three cell lines (Fig. 5). Finally, using a strict Bonferroni-corrected P< 0.05 for the number of genes and time point evaluated, half of these relationships remain, representing two different genes at multiple time points (Fig. 4B).
Figure 4.
Gene expression changes combined across three cell lines. (A) Change in gene expression 5, 12, 18 and 24 h after PRPF39 knockdown was assessed for each of the 61 target genes, using a mixed effects model that combines all expression data from three cell lines. The X-axis shows fold change relative to a scrambled control, with identical expression having a value of 1. The Y-axis shows the negative log10 of the P-value from the combined model, with P= 0.05 shown with a dashed line. Identification of each gene and the direction of effect are found in Supplementary Material, Table S2. (B) The panel shows that above the time point, the genes significantly (P< 0.05) increased in expression, with the gene closest to the center being the most significant change. Below each time point are the genes significantly (P< 0.05) decreased in expression with the gene, with the most significant decrease being the closest to the center. Genes in bold achieve a P-value <0.05 corrected for the number of genes tested at each time point and those with an asterisk remain significant when corrected for number of genes and time points.
Figure 5.
Combined cell-line model reveals different expression change patterns. After PRPF39 knockdown, ABHD5 expression is also significantly (P< 0.004) knocked down at three time points (5, 12 and 24 h after PRPF39 knockdown). In contrast, TFDP2 shows significant (P= 0.006) increase of expression only at 24 h after PRPF39 knockdown.
In addition to an evaluation of combining the results of the three cell lines, we also evaluated each cell line independently. For cell lines GM19252 and GM19100, 59 genes were evaluated, and GM18488 had 29 genes evaluated. Correcting for the number of genes tested, five, two and eight genes significantly (P< 0.05) changed expression in cell lines 19252, 19100 and 18488, respectively, at one or more time points (Supplementary Material, Table S2). Using a Bonferroni correction for the number of genes and time points evaluated, one, none and four genes significantly (P< 0.05) changed gene expression levels in 19252, 19100 and 18488, respectively.
Different patterns emerged for significant changes, with some genes showing a similar trend in expression change across all time points for individual cell lines. When combining the multiple cell lines, differences in the time course for individual cell lines prevent the overall model from showing significance. For example, INPP5A shows increased expression relative to control across all time points in GM19100 and a similar trend of increase in expression in the other two cell lines, just missing significance in the combined cell line model. MAP3K4 and SLC38A2 showed a sharp knockdown at 5 h (P ≤0.0001) relative to control in 18488 with a trend of decreased expression at the earlier time point in the other two cell lines (Supplementary Material, Fig. S4).
DISCUSSION
In this study, we identified rs17115814, an intronic SNP of PRPF39, through a genome-wide approach evaluating over 2.5 million SNPs. We observed a significant enrichment of eQTLs among cisplatin-induced cytotoxicity SNPs at P ≤0.0001. Of the 296 SNPs associated with cisplatin-induced cytotoxicity at P ≤0.0001, rs17115814 was the only SNP that was both a local eQTL and a distant eQTL to over 100 genes. To begin to elucidate the function of rs17115814, we studied PRPF39, the host gene with a significant mRNA association with the SNP. We evaluated the effect of the knockdown of PRPF39 on: (i) the expression of PRPF39 over time; (ii) cellular sensitivity to cisplatin; (iii) the expression of 61 distant target genes that were significantly associated with rs17115814. Knockdown of the host gene resulted in a significant increase in cellular resistance to cisplatin, as predicted by the initial discovery. In one or more cell lines, 90% of the 61 distant target genes showed changes in gene expression at 5, 12, 18 or 24 h. Thus, the association of rs17115814 with resistance to cisplatin is likely due to its effect on PRPF39 gene expression and subsequent effects on multiple downstream targets.
Because of the challenges in evaluating distant eQTL relationships, few have functionally assayed distant eQTLs and their target genes. However, Cheung et al. (17) studied expression changes of 25 distant target genes 24 h after the knockdown of a host gene. They selected distant eQTLs located within host genes that spanned a range of significance and confirmed expression changes of the distant genes for 72% of the successfully knocked down host genes. Smirnov et al. (16) evaluated distant target genes after radiation treatment. In contrast, our study evaluates a single eQTL that targets a local and multiple distant target genes. Unlike previous studies, we simultaneously evaluate multiple distant target genes for changes in expression as a result of modulation of a host gene. This represents an important step forward in the annotation and evaluation of SNPs by recognizing the contribution of a polymorphism that is not located within the coding region of a gene. Our study is also novel in evaluating changes over time of multiple potential targets following the modulation of the host gene. The varied time points allowed us to identify different target genes that may have been missed in evaluating a single time point and provided a more thorough investigation of the sequential cascade of downstream changes.
The PRP family of proteins is believed to interact with the spliceosome and play a role in pre-RNA processing (18). Many of the genes in this family have not been studied in great detail. Three members, PRPF3, PRPF31 and PRPF8, have consistently been associated with retina diseases (19). Although the genes are expressed throughout many different tissues, mutations in PRPF3 are associated with autosomal retinitis pigmentosa (20) and have also been implicated in large pedigrees (21). Studies in mice and zebrafish have ruled out haploinsufficiency caused by mutation in PRPF3 as a mechanism causing retinitis pigmentosa (22). Studies have since then focused on looking for downstream targets that could be influenced by variants in these PRP genes. Mordes et al. (23) have identified a subset of photoreceptor-specific genes and found mutations in PRPF31 lead to defective pre-mRNA processing and splicing. Further work by Song et al. (24) has identified specific spliceosome components that interact with PRPF3. Beyond retinitis pigmentosa, studies in mice and human heptocellular carcinomas have identified PRPF3 as a disease gene whose expression is regulated by HNF4alpha (25). These studies suggest that PRPF39 could have impact on a number of different downstream targets, consistent with the results presented here.
Among the target genes that have shown significant gene expression changes, there are some interesting candidates. TFDP2, transcription factor Dp-2 (E2F dimerization partner 2), is from the family of transcription factors that is important in cell-cycle regulation (26). Multiple isoforms of this gene have also been described, which could suggest an important role for pre-mRNA processing. TFDP2 has been identified in thyroid carcinoma cell lines as a gene whose expression is significantly reduced after cisplatin treatment (27), further suggesting that expression patterns of TFDP2 are important for cisplatin response. In our study, TFDP2 significantly increased in expression 24 h after PRPF39 knockdown across three cell lines. The role of PRPF39 as a potential regulator of TFDP2 represents an interesting area of further study regarding the role of TFDP2 and cisplatin response.
Additionally, the significantly lower expression of MAP3K4, mitogen-activated protein kinase kinase kinase 4, as a result of PRPF39 knockdown is an interesting candidate for a role in modulating cisplatin sensitivity. MAP3K4 is part of the activation sequence for p38 and JNK activity, pathways that can lead to apoptosis (28). Because cisplatin is known to induce apoptosis (10,29), lowered expression of MAP3K4 could alter the activation sequence, which could explain the observed increased resistance to cisplatin. This study identified a potential upstream activator of the JNK and p38 MAPK signaling pathway through the target genes of a distant-eQTL SNP. Functional effect was observed through the knockdown of PRPF39, a novel candidate that has no known link to the MAPK signaling pathway. This relationship could further be explored in future studies.
In summary, this study used a GWAS to identify a SNP, rs17115814, associated with cisplatin sensitivity in the YRI population that was both a local and distant eQTL. No other associated SNP was both a local and multiple distant eQTL. The expression of host gene PRPF39 was also correlated with cisplatin sensitivity, and siRNA knockdown of PRPF39 resulted in a significant increase in resistance to cisplatin, functionally confirming its role in cisplatin sensitivity. We successfully evaluated 59 potential target genes for expression changes after PRPF39 knockdown, and 90% of the target genes showed significant (P ≤0.05) changes. Some of these target genes, including MAP3K4 and TFDP2, have a prior relationship with cisplatin sensitivity. Therefore, our novel approach can be applied to better understand trans-regulation of genes relevant in disease, response to drug treatment or other phenotypes.
MATERIALS AND METHODS
Cell lines
All LCLs were cultured in RPMI 1640 media containing 15% fetal bovine serum (Hyclone, Logan, UT, USA) and 3.7 mm l-glutamine. Cell lines were diluted three times per week at a concentration of 300 000–350 000 cells/ml and were maintained in a 37°C, 5% CO2 humidified incubator. Medium and components were purchased from Cellgro (Herndon, VA, USA). All cell lines (HAPMAPT03 and HAPMAPT04) were purchased from Coriell Institute for Medical Research (www.coriell.org). All genotyped cell lines were used from both panels with the exception of GM18871, which did not sustain viability over 85%.
Cytotoxicity phenotype measurement
Cisplatin was prepared as described previously (9,10). The cytotoxic effect of cisplatin was determined using a short-term cellular growth inhibition assay. LCLs in exponential growth and of >85% viability, as determined using the Vi-Cell XR viability analyzer (Beckman Coulter, Fullerton, CA, USA), were plated in triplicate at a density of 1 × 105 cells per milliliter in 96-well round-bottom plates (Corning, Inc., Corning, NY, USA) 24 h prior to drug treatment. The drug was immediately added to cells after preparation of stock for a final concentration of 1, 2.5, 5, 10, 20 µm cisplatin. AlamarBlue was added 24 h before absorbance reading at wavelengths of 570 and 600 nm using the Synergy-HT multi-detection plate reader (BioTek, Winooski, VT, USA) 48 h after the drug was added. Percent survival was quantified relative to a control well without drug addition. Final percent survival was ascertained by averaging at least six replicates from two independent experiments. We then determined an IC50 for each cell line, the concentration of drug at which 50% cellular growth inhibition occurred.
Drug treatment after nucleofection
Drug treatment was identical as described above, with the exception of the timing of the plating of the cells. Cells were plated in triplicate at a density of 1 × 105 cells per milliliter in 96-well round-bottom plates (Corning, Inc.) after a 15 min resting period following nucleofection. Five hours after plating, cisplatin was added to the cells as described for cytotoxicity experiments.
Genome-wide association
We used the total association model of the quantitative trait disequilibrium test (30) for 176 YRI cell lines. Because a different number of SNPs have been genotyped in the two panels, we used imputation to increase the number of SNPs in the second YRI panel. Non-genotyped markers were imputed using the BEAGLE software (31). The first YRI panel (HapMap r22) was used as the reference population. Beagle imputes non-genotyped markers for unrelateds, parent-offspring pairs and parent-offspring trios by modeling the family structure in the analysis. To measure the accuracy of the imputation at each SNP locus, r2 was calculated as described following 100 imputations of the data. Imputed SNP genotypes with r2 > 0.8, minor allele frequency >0.05, no Mendelian errors and in Hardy–Weinberg equilibrium (P> 0.001) were carried through the rest of the analysis. Owing to previously described growth rate differences (32), a covariate for HapMap panel was used for each LCL to reflect whether it was included in the first or second YRI panel. As was observed previously (33), gender was a significant predictor of IC50 and was also used as a covariate. To attain normality in the phenotype data, each cell's IC50 values were log2-transformed before data analysis.
eQTL enrichment analysis
Using eQTLs previously described in the YRI population (34), we identified the number of eQTLs associated with cisplatin IC50. We then calculated the minor allele frequency for each of the eQTLs and generated 1000 randomized SNP sets, matching minor allele frequency to the original eQTL list and determined the number of eQTLs in each random list as described previously to calculate an empirical P-value (12).
siRNA nucleofection
LCLs were diluted to 500 000 cells/ml 1 day prior to nucleofection. The cells were nucleofected using the SF Cell Line 96-well Nucleofector Kit (Lonza, Inc., Basel, Switzerland). Cells were centrifuged at 90g for 10 min at room temperature and resuspended at a concentration of 1 000 000 cells/20 µl in SF/supplement solution (included in SF Kit Lonza Catalog V4SC2096) and 2000 nm final concentration of AllStars Negative Control siRNA labeled with AlexaFluor488 (Qiagen, Inc., Valencia, CA, USA) or a pool of Hs_PRPF39 FlexiTube siRNA (SI04209695, SI04224262, SI04235126, SI04293919) (Qiagen). The cells were nucleofected using the DN-100 program on Lonza's Nucleofector 96-well shuttle system. Cells were allowed to rest for 10 min prior to the addition of pre-warmed (37°C water bath) RPMI media and then for another 5 min in the warm RPMI media. Cells were then plated for mRNA harvest or drug treatment. Cells were harvested at 5, 29 and 53 h post-nucleofection for PRPF39 expression measurement.
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to measure the level of expression of PRPF39. A total of 3 million cells were pelleted at 5 (corresponds to T = 0 h for cisplatin cytotoxicity assay), 29 and 53 h after nucleofection, washed in ice-cold PBS and centrifuged to remove PBS. All pellets were flash-frozen and stored at −80°C until RNA isolation. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) following the manufacturer's protocol. RNA quality assessment and quantification were conducted using the optical spectrometry 260/280 nm ratio. Subsequently, mRNA was reverse-transcribed to cDNA using Applied Biosystems High Capacity Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The final concentration of cDNA was 25 ng/µl. qRT-PCR was performed for PRPF39 and huB2M (beta-2-microglobulin; NM_004048.2) as an endogenous control using TaqMan Gene Expression Assays (Applied Biosystems) on the Applied Biosystems Viia7 RT-PCR system. Total reaction was carried out in 12 µl volume, which consisted of 6 µl of TaqMan Gene Expression PCR Master Mix or TaqMan Fast Advanced Master Mix, 0.6 µl of primers and probe mix (final of 900 nm forward and reverse primers and 250 nm of probe), and 0.4 µl of water, along with 5 µl of 1.25 ng/µl cDNA. The PRPF39 (Hs01547148_m1or Hs00215626_m1) TaqMan primer and probe mixtures were labeled with the FAM reporter dye and the MGB quencher dye. Linear regression was performed between the PRPF39/huB2M quantities obtained from each primer, with an r2 value of 0.99 (P< 0.0001). The huB2M primer/probe mixture was labeled either with the FAM or VIC reporter dye and the MGB quencher dye. The standard thermocycler parameters were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s then 60°C for 1 min, all with ramping speeds of 6°C/s. The fast thermocycler parameters were 95°C for 20 s, 40 cycles of 95°C for 1 s and then 60°C for 20 s, with ramping speeds of 1.6– 1.9°C/s. Viia7 automatically set the CT threshold and baseline for each experiment. A relative standard curve method was used to obtain the relative PRPF39 expression for samples treated with PRPF39 or scrambled siRNA. Each knockdown experiment was conducted three separate times, with the RT-PCR run in duplicate and individual samples run in triplicate on the RT-PCR plate.
TaqMan low-density arrays
Custom TaqMan Low Density Array Cards were created in either a 32- or 64-gene format by Applied Biosystems. Each card contained huB2M as the control. The genes present on the 64-gene format card are listed in Supplementary Material, Table S1. The genes present on the 32-gene format card are listed in Supplementary Material, Table S1, with an asterisk next to the name. Three YRI LCLs were evaluated: GM19100, GM19252 and GM18488. These cell lines were selected based on the most significant increase in resistance to cisplatin observed (Fig. 3). Two of the cell lines, GM19100 and GM19252, were heterozygous for rs17115814, and GM18488 was homozygous for the minor allele, TT. GM19100 and GM19252 were evaluated once on the 64 card and once on the 32 card, with the exception of GM19252 5 h time point, which was evaluated twice on the 64 card as well as on the 32 card. GM18488 was evaluated three times on the 32 card only. The nucleofection, RNA isolation and reverse transcription were conducted as listed previously. Time points analyzed by the TLDA cards were 5, 12, 18 and 24 h after nucleofection. The reaction consisted of Applied Biosystems's Gene Expression Master Mix and 500 ng cDNA. The thermocycle parameters were 95°C for 20 s, 40 cycles of 95°C for 1 s and then 60°C for 20 s, with ramping speeds of 1.0°C/s. For each card, Viia7 automatically set the CT threshold and baseline. The gene of interest expression was determined using the comparative delta-delta CT method, using the scrambled siRNA treated LCL as the reference sample and huB2M as the endogenous control. PRPF39 was included as a control on each card to ensure knockdown was occurring. Two genes, IDI2 and FGF13, did not have mRNA levels high enough to be detected and were dropped from the analysis.
Statistical analysis of the effect of PRPF39 knockdown
Within each cell line, statistical significance was assessed based on a Student's t-test. Single-tailed t-test was used to assess the significance of the difference in cisplatin response between PRPF39 knockdown and scrambled control since an increase of IC50 was expected. For the effect of PRPF39 knockdown on target genes, a two-tailed t-test was used.
Significance across multiple cell lines for the effect of PRPF39 knockdown on cisplatin resistance as well as on the target genes' expression was evaluated using mixed effects models. For cisplatin resistance, logarithm of IC50 was used as response variable and knockdown status (0 if control and 1 if knockdown) as covariate. Cell line and experiment (each of the paired experiments, knockdown and scrambled, was labeled differently) were included in the model as random effects. Log of the IC50 was used because it improved the fit substantially with respect to the untransformed response.
For target genes' expressions, each gene and time points were modeled separately. CT from the real-time PCR was used as response, and knockdown status, gene status (1 if the gene of interest and 0 if a housekeeping gene) and the interaction between gene and knockdown status were used as fixed effects. Cell lines and cards were included as random effects to take into account the clustering induced by them. The goodness of fit of the model was assessed inspecting the residuals. The R statistical software and the nlme package were used to fit the mixed effects model (35,36).
SUPPLEMENTARY MATERIAL
FUNDING
This Pharmacogenetics of Anticancer Agents Research (PAAR) Group (http://pharmacogenetics.org) study was supported by NIH/NIGMS grant UO1GM61393. In addition, A.L.S. was supported by TL1 RR25001. H.K.I. was partially funded by UC Cancer Center Support Grant P30 CA014599-36.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the PAAR Cell Line Core and Genotyping Core for their assistance on this work. We are grateful to Drs Nancy Cox and Stephanie Huang for helpful discussions and to Mr Steven Stark for his technical assistance in data analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rabik C.A., Dolan M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travis L.B., Beard C., Allan J.M., Dahl A.A., Feldman D.R., Oldenburg J., Daugaard G., Kelly J.L., Dolan M.E., Hannigan R., et al. Testicular cancer survivorship: research strategies and recommendations. J. Natl Cancer Inst. 2010;102:1114–1130. doi: 10.1093/jnci/djq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross C.J., Katzov-Eckert H., Dube M.P., Brooks B., Rassekh S.R., Barhdadi A., Feroz-Zada Y., Visscher H., Brown A.M., Rieder M.J., et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat. Genet. 2009;41:1345–1349. doi: 10.1038/ng.478. [DOI] [PubMed] [Google Scholar]
- 4.Brydoy M., Oldenburg J., Klepp O., Bremnes R.M., Wist E.A., Wentzel-Larsen T., Hauge E.R., Dahl O., Fossa S.D. Observational study of prevalence of long-term Raynaud-like phenomena and neurological side effects in testicular cancer survivors. J. Natl Cancer Inst. 2009;101:1682–1695. doi: 10.1093/jnci/djp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oldenburg J., Kraggerud S.M., Cvancarova M., Lothe R.A., Fossa S.D. Cisplatin-induced long-term hearing impairment is associated with specific glutathione S-transferase genotypes in testicular cancer survivors. J. Clin. Oncol. 2007;25:708–714. doi: 10.1200/JCO.2006.08.9599. [DOI] [PubMed] [Google Scholar]
- 6.Caronia D., Patino-Garcia A., Milne R.L., Zalacain-Diez M., Pita G., Alonso M.R., Moreno L.T., Sierrasesumaga-Ariznabarreta L., Benitez J., Gonzalez-Neira A. Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J. 2009;9:347–353. doi: 10.1038/tpj.2009.19. [DOI] [PubMed] [Google Scholar]
- 7.Fukushima-Uesaka H., Saito Y., Maekawa K., Kurose K., Sugiyama E., Katori N., Kaniwa N., Hasegawa R., Hamaguchi T., Eguchi-Nakajima T., et al. Genetic polymorphisms of copper- and platinum drug-efflux transporters ATP7A and ATP7B in Japanese cancer patients. Drug Metab. Pharmacokinet. 2009;24:565–574. doi: 10.2133/dmpk.24.565. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Duan S., Kistner E.O., Bleibel W.K., Huang R.S., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang R.S., Duan S., Shukla S.J., Kistner E.O., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Dolan M.E. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla S.J., Duan S., Badner J.A., Wu X., Dolan M.E. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet. Genomics. 2008;18:253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donnell P.H., Gamazon E., Zhang W., Stark A.L., Kistner-Griffin E.O., Stephanie Huang R., Eileen Dolan M. Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants. Pharmacogenet. Genomics. 2010;20:327–337. doi: 10.1097/FPC.0b013e3283396c4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamazon E.R., Huang R.S., Cox N.J., Dolan M.E. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl Acad. Sci. USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 14.Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smirnov D.A., Morley M., Shin E., Spielman R.S., Cheung V.G. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung V.G., Nayak R.R., Wang I.X., Elwyn S., Cousins S.M., Morley M., Spielman R.S. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 2010;8:e1000480. doi: 10.1371/journal.pbio.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang A., Forman-Kay J., Luo Y., Luo M., Chow Y.H., Plumb J., Friesen J.D., Tsui L.C., Heng H.H., Woolford J.L., Jr., et al. Identification and characterization of human genes encoding Hprp3p and Hprp4p, interacting components of the spliceosome. Hum. Mol. Genet. 1997;6:2117–2126. doi: 10.1093/hmg/6.12.2117. [DOI] [PubMed] [Google Scholar]
- 19.Cao H., Wu J., Lam S., Duan R., Newnham C., Molday R.S., Graziotto J.J., Pierce E.A., Hu J. Temporal and tissue specific regulation of RP-associated splicing factor genes PRPF3, PRPF31 and PRPC8—implications in the pathogenesis of RP. PLoS One. 2011;6:e15860. doi: 10.1371/journal.pone.0015860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comitato A., Spampanato C., Chakarova C., Sanges D., Bhattacharya S.S., Marigo V. Mutations in splicing factor PRPF3, causing retinal degeneration, form detrimental aggregates in photoreceptor cells. Hum. Mol. Genet. 2007;16:1699–1707. doi: 10.1093/hmg/ddm118. [DOI] [PubMed] [Google Scholar]
- 21.Chakarova C.F., Hims M.M., Bolz H., Abu-Safieh L., Patel R.J., Papaioannou M.G., Inglehearn C.F., Keen T.J., Willis C., Moore A.T., et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 22.Graziotto J.J., Inglehearn C.F., Pack M.A., Pierce E.A. Decreased levels of the RNA splicing factor Prpf3 in mice and zebrafish do not cause photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 2008;49:3830–3838. doi: 10.1167/iovs.07-1483. [DOI] [PubMed] [Google Scholar]
- 23.Mordes D., Yuan L., Xu L., Kawada M., Molday R.S., Wu J.Y. Identification of photoreceptor genes affected by PRPF31 mutations associated with autosomal dominant retinitis pigmentosa. Neurobiol. Dis. 2007;26:291–300. doi: 10.1016/j.nbd.2006.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song E.J., Werner S.L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J.W., Elledge S.J., Kirschner M.W., Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niehof M., Borlak J. EPS15R, TASP1, and PRPF3 are novel disease candidate genes targeted by HNF4alpha splice variants in hepatocellular carcinomas. Gastroenterology. 2008;134:1191–1202. doi: 10.1053/j.gastro.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Venkatraj V.S., Fischer S.G., Warburton D., Chellappan S.P. Genomic cloning and chromosomal assignment of the E2F dimerization partner TFDP gene family. Genomics. 1997;39:95–98. doi: 10.1006/geno.1996.4473. [DOI] [PubMed] [Google Scholar]
- 27.Lapouge G., Millon R., Muller D., Abecassis J., Eber M., Bergerat J.P., Klein-Soyer C. Cisplatin-induced genes as potential markers for thyroid cancer. Cell. Mol. Life Sci. 2005;62:53–64. doi: 10.1007/s00018-004-4329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abell A.N., Granger D.A., Johnson G.L. MEKK4 stimulation of p38 and JNK activity is negatively regulated by GSK3beta. J. Biol. Chem. 2007;282:30476–30484. doi: 10.1074/jbc.M705783200. [DOI] [PubMed] [Google Scholar]
- 29.Wen Y., Gorsic L.K., Wheeler H.E., Ziliak D.M., Stephanie Huang R., Eileen Dolan M. Chemotherapeutic-induced apoptosis: a phenotype for pharmacogenomics studies. Pharmacogenet. Genomics. 2011;21:471–488. doi: 10.1097/FPC.0b013e3283481967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark A.L., Zhang W., Zhou T., O'Donnell P.H., Beiswanger C.M., Huang R.S., Cox N.J., Dolan M.E. Population differences in the rate of proliferation of international HapMap cell lines. Am. J. Hum. Genet. 2010;87:829–833. doi: 10.1016/j.ajhg.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang R.S., Kistner E.O., Bleibel W.K., Shukla S.J., Dolan M.E. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol. Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan S., Huang R.S., Zhang W., Bleibel W.K., Roe C.A., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. http://www.R-project.org/ [Google Scholar]
- 36.Pinheiro J., Bates D., DebRoby S., Sarkar D. R Development Core Team. nlme: Linear and Nonlinear Mixed Effects Models. 2011. R Package Version 3.1–98 http://cran.r-project.org/web/packages/nlme/citation.html . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.