Abstract
Phosphorylation of STAT3 (signal transducer and activator of transcription 3) is critical for its nuclear import and transcriptional activity. Although a shorter STAT3β spliceform was initially described as a negative regulator of STAT3α, gene knockout studies have revealed that both forms play critical roles. We have expressed STAT3α and STAT3β at comparable levels to facilitate a direct comparison of their functional effects, and have shown their different cytokine-stimulated kinetics of phosphorylation and nuclear translocation. Notably, the sustained nuclear translocation and phosphorylation of STAT3β following cytokine exposure contrasted with a transient nuclear translocation and phosphorylation of STAT3α. Importantly, co-expression of the spliceforms revealed that STAT3β enhanced and prolonged the phosphorylation and nuclear retention of STAT3α, but a STAT3β R609L mutant, with a disrupted SH2 (Src homology 2) domain, was not tyrosine phosphorylated following cytokine stimulation and could not cross-regulate STAT3α. The physiological importance of prolonged phosphorylation and nuclear retention was indicated by transcriptome profiling of STAT3−/− cells expressing either STAT3α or STAT3β, revealing the complexity of genes that are up- and down-regulated by the STAT3 spliceforms, including a distinct set of STAT3β-specific genes regulated under basal conditions and after cytokine stimulation. These results highlight STAT3β as a significant transcriptional regulator in its own right, with additional actions to cross-regulate STAT3α phosphorylation and nuclear retention after cytokine stimulation.
Keywords: cytokine, interleukin-6 (IL-6), nucleocytoplasmic trafficking, signal transducer and activator of transcription 3 (STAT3), transcription factor, transcriptome analysis
Abbreviations: CLSM, confocal laser-scanning microscopy; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; ERt, substrate-binding portion of the oestrogen receptor; Fc, cytoplasmic fluorescence; FBS, fetal bovine serum; Fn, nuclear fluorescence; Fn/Fc, ratio of nuclear to cytoplasmic fluorescence; GO, gene ontology; gp130, glycoprotein 130; HEK, human embryonic kidney; Hsp90, heat-shock protein of 90 kDa; 4-HT, 4-hydroxytamoxifen; IL, interleukin; iSTAT3, inducible specific STAT3 spliceform; JAK, Janus kinase; MAPK, mitogen-activated protein kinase; MEF, murine embryonic fibroblast; OSM, oncostatin M; SH2, Src homology 2; SHP, SH2 domain-containing protein tyrosine phosphatase; STAT3, signal transducer and activator of transcription 3; VP16, viral protein 16; WT, wild-type
INTRODUCTION
STAT3 (signal transducer and activator of transcription 3), initially identified as an acute-phase response factor binding to the acute-phase response element in IL (interleukin)-6-stimulated hepatocytes, is a pleiotropic transcription factor capable of mediating rapid changes in gene expression following cytokine, hormone or growth factor stimulation [1–3]. The IL-6 family of cytokines, which includes OSM (oncostatin M) and LIF (leukaemia inhibitory factor), signals through the common gp130 (glycoprotein 130) receptor chain to activate STAT3 [4]. This activation of STAT3 requires the phosphorylation of Tyr705 and Ser727. In the most widely accepted paradigm of signalling via STAT3, the phosphorylation of STAT3 Tyr705 by JAKs (Janus kinases) is critical for STAT3 dimerization and subsequent cytokine-stimulated nuclear translocation, whereas the phosphorylation of Ser727 by serine/threonine kinases such as the MAPKs (mitogen-activated protein kinases) enhances STAT3 transcriptional activity [5,6]. Thus phosphorylation of STAT3 provides a key regulatory mechanism communicating extracellular events to cytokine-induced gene expression changes.
The functional importance of STAT3 has been shown by the early embryonic lethality of Stat3−/− mice [7]. Subsequent tissue-specific deletion studies have revealed important roles of STAT3 in inflammatory responses in the liver, proliferation and differentiation in monocytes and neutrophils in response to granulocyte colony-stimulating factor, protection from apoptosis in the mammary epithelium, neuronal cell survival and keratinocyte migration [5,8]. In addition, a persistent activation of STAT3 in a wide variety of cancers and diseases, such as multiple myeloma, head and neck cancer, breast cancer and other solid tumours, leukaemias and lymphomas [9] has further intensified interest in understanding regulators of STAT3 activation.
Two distinct STAT3 isoforms originating from alternative splicing have been described. STAT3α (92 kDa) is 770 amino acids in length, whereas STAT3β (84 kDa) is identical in sequence with the exception of 55 amino acids at the C-terminal tail that are replaced with a unique seven-amino-acid sequence (Figure 1A) [10,11]. As a consequence, the transactivation domain of STAT3β is truncated relative to this domain in STAT3α. This has led to suggestions of impaired transcriptional activity and a role as a dominant-negative regulator of STAT3α [10]. Although the generally lower expression levels of STAT3β compared with STAT3α imply that STAT3α plays a more significant functional role in vivo, there are clear exceptions, such as the levels of STAT3β exceeding STAT3α during myeloid differentiation, pointing to a requirement for high STAT3β levels to act as a mediator during these differentiation events [12–14]. A previous study demonstrating rescue of STAT3−/− embryonic lethality with STAT3β spliceform expression (i.e. in the absence of STAT3α) highlight key STAT3β-specific roles in development [15]. In addition, spliceform-specific functions have been indicated by various in vivo studies showing a requirement for STAT3β during endotoxic assault [16], but a requirement for STAT3α in IL-8 synthesis [17], as well as differential roles for STAT3α and STAT3β in anti-inflammatory responses [15]. Importantly, a recent advance with an oligonucleotide-mediated enforced switching to preferential splicing of STAT3β (rather than STAT3α) has emphasized the anti-tumorigenic activity of STAT3β [18]. This has also validated reprogramming of endogenous splicing, and specifically that of enhancing STAT3β levels significantly over STAT3α levels, as an exciting new therapeutic approach [18]. Clearly, the biochemical mechanisms underlying the distinct functions of STAT3 spliceforms, and in particular that of STAT3β, warrant more in-depth analyses.
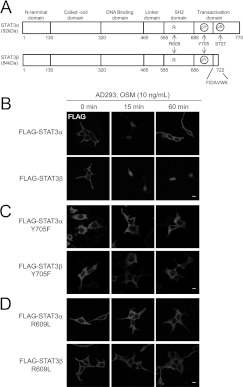
Figure 1. STAT3α and STAT3β are STAT3 spliceforms with different cytokine-stimulated nucleocytoplasmic trafficking.
(A) Schematic diagram of STAT3α (92 kDa) and STAT3β (84 kDa) spliceforms. The arrangement of the various STAT3 subdomains, together with the amino acid numbers at the domain boundaries, is indicated. Amino acids 1–715 are identical in these spliceforms, and the sequence of the shorter unique seven-amino-acid C-terminal tail for STAT3β is shown. Arg609, a key residue for SH2 domain function, and Tyr705 (of both spliceforms) as well as Ser727 (for STAT3α only), which are phosphorylated in active STAT3 forms, are indicated. (B–D) CLSM analysis using anti-FLAG antibody of transiently transfected AD293 cells stimulated with OSM (10 ng/ml) for 0, 15 or 60 min; (B) WT FLAG–STAT3α or FLAG–STAT3β, (C) FLAG–STAT3α/β Y705F mutants and (D) SH2 domain defective FLAG–STAT3α/β R609L mutant. Scale bars represent 10 μm.
To address these distinct functions of the STAT3 spliceforms, we have evaluated the kinetics of nucleocytoplasmic trafficking and phosphorylation of STAT3α and STAT3β in response to cytokine stimulation, particularly focusing on the use of Stat3−/− MEFs (murine embryonic fibroblasts) with inducible expression of either STAT3 spliceform. Our expression of each STAT3 spliceform at a comparable level thus allowed our direct comparison of their functional effects without the confounding effects of different levels of expression. STAT3β exhibited markedly prolonged nuclear translocation and phosphorylation following OSM exposure when compared with STAT3α, which showed more transient responses. Furthermore, a striking cross-regulation of STAT3α by STAT3β was observed upon the co-expression of STAT3β, which enhanced and prolonged STAT3α phosphorylation. Our transcriptome profiling of Stat3−/− MEFs re-expressing either STAT3α or STAT3β showed that the expression of either STAT3 spliceform could reconstitute many of the immediate transcriptional effects of short-term cytokine stimulation noted for WT (wild-type) MEFs. Importantly, analysis after longer cytokine stimulation revealed the large number of genes both up- and down-regulated by either of these STAT3 spliceforms; the physiological significance of prolonged phosphorylation was highlighted with a greater number of genes regulated by STAT3β than regulated by STAT3α. The present study thus highlights STAT3β as a regulator of transcription with an intriguing ability to modulate STAT3α phosphorylation and nuclear retention after cytokine stimulation.
EXPERIMENTAL
Plasmid constructs
Mammalian expression vectors for FLAG epitope-tagged versions of STAT3α and STAT3β were constructed by amplifying the coding region of human STAT3α and STAT3β genes by PCR to create the desired restriction enzyme sites (HindIII and XhoI) for subcloning into the pXJ40-FLAG vector.
The primer pairs used were: FLAG–STAT3α, 5′-GCAAGCTTATGGCCCAATGGAATCAGCTACAG-3′ and 5′-GCCTCGAGTCACATGGGGGAGGTAGCGCACTC-3′; and FLAG–STAT3β, 5′-GCAAGCTTATGGCCCAATGGAATCAGCTACAG-3′ and 5′-GCCTCGAGTTATTTCCAAACTGCATCAATGAA-3′. The STAT3 genes were inserted, in frame, immediately 3′ of the FLAG epitope sequence, thus allowing the expression of N-terminal FLAG-tagged STAT3 proteins in mammalian cells. PCR mutagenesis was used to change Tyr705 to phenylalanine (Y705F) and the critical Arg609 of the SH2 (Src homology 2) domain to leucine (R609L) for both FLAG-tagged STAT3α and STAT3β. The primer pairs used were: STAT3 Y705F, 5′GACCCAGGTAGCGCTGCCCCAGCCCTGAAGACCAAGTTTATC-3′ and 5′-GATAAACTTGGTCTTCAGGGCTGGGGCAGCGCTACCTGGGTC-3′; STAT3 R609L, 5′-TCCAGGCACCTTCCTGCTACTATTCAGTGAAAGCAGCAAA-3′ and 5′-TTTGCTGCTTTCACTGAATAGTAGCAGGAAGGTGCCTGGA-3′.
Lentiviral system and selection of cells with stable STAT3 construct expression
The WT and mutant FLAG-tagged STAT3α and STAT3β constructs were subcloned into a 4-HT (4-hydroxytamoxifen)inducible lentiviral system vector, pF-5UAS-SV40-puroGEV16 [19] via AgeI/NheI restriction enzyme sites created by PCR using the primer pairs 5′-GCACCGGTACCATGGACTACAAGGACGACGAT-3′ and 5′-GCGCTAGCTCACATGGGGGAGGTAGCGCACTC-3′ or 5′-GCACCGGTACCATGGACTACAAGGACGACGAT-3′ and 5′-GCGCTAGCTTATTTCCAAACTGCATCAATGAA-3′.
FLAG-tagged STAT3 expression constructs together with plasmids encoding lentiviral structural components (pCMV-δR8.2 and pCMV-VSV-G) were transfected into HEK (human embryonic kidney)-293FT cells. Lentiviruses were harvested 72 h post-transfection and purified via sterile-filtration. Stat3−/− MEFs were infected with virus for 24 h in the presence of 1 μg of Polybrene (Sigma) and maintained in growth medium for a further 24 h before selection with 10 μg/ml of puromycin (Calbiochem). Puromycin-resistant MEFs were then analysed for STAT3 expression following 4-HT (1 nM) induction.
Cell culture and transfection
AD293 cells and HEK-293FT cells, both variants of HEK-293 cells, COS1, WT MEFs and Stat3−/− MEFs [15] were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS (fetal bovine serum), and penicillin/streptomycin (100 units/ml). Inducible specific STAT3 spliceform (iSTAT3α and iSTAT3β) MEFs were maintained in this same medium but additionally supplemented with 10 μg/ml puromycin (Calbiochem). Transient transfections were carried out using Lipofectamine™ 2000 or Lipofectamine™ LTX with Plus™ according to the manufacturer's instructions (Invitrogen). Cells were cultured in serum-free medium (DMEM supplemented with penicillin/streptomycin) for 16 h prior to treatment with OSM (10 ng/ml, Calbiochem).
Lysate preparation and immunoblot analysis
Cells were lysed in RIPA buffer [50 mM Tris/HCl, pH 7.3, 150 mM NaCl, 0.1 mM EDTA, 1% (v/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% NaF and 100 μM Na3VO4] supplemented with Complete™ protease inhibitors (Roche Diagnostic). Protein samples were resolved by SDS/PAGE and transferred on to a PVDF membrane for immunoblot analysis. The anti-STAT3 antibody (#610189) recognising the shared N-terminal residues of both STAT3α and STAT3β was from BD Biosciences and the anti-phospho-STAT3 (Tyr705) (#9145) antibody was from Cell Signaling Technology. Anti-α-tubulin and -FLAG M2 antibodies were from Sigma. Anti-gp130 and -c-Myc antibodies were from Santa Cruz Biotechnology. Protein bands were visualized by enhanced chemiluminescence and quantified with ImageJ (NIH).
Co-immunoprecipitation
Cells were lysed in Nonidet P40 buffer [1% (v/v) Nonidet P40, 50 mM Tris/HCl, pH 8.0, and 150 mM NaCl] supplemented with Complete™ protease inhibitors. Either mouse anti-FLAG M2 antibodies (Sigma) or rabbit anti-Myc antibodies (Santa Cruz Biotechnology) were added to the extracts and incubated for 1 h at 4°C before the addition of Protein A–agarose (Roche Diagnostic). Immunocomplex pellets were washed extensively and boiled in protein sample buffer before immunoblot analysis.
Immunofluorescence, CLSM (confocal laser-scanning microscopy) and image analysis
Samples were prepared and analysed as described previously [20]. Briefly, OSM-stimulated cells on coverslips were washed three times with ice-cold PBS before fixation using 4% (w/v) paraformaldehyde and permeabilization in 0.2% Triton X-100/PBS or fixation using ice-cold methanol. Non-specific binding was blocked by incubation in 10% (v/v) FBS/PBS. Cells were incubated with primary antibodies [1:400 dilution in 1% (w/v) BSA/PBS] and washed with PBS before incubation with Cy2 (carbocyanine)/Cy3 (indocarbocyanine)-conjugated secondary antibodies (Millipore). Nuclei were stained using DAPI (4′,6-diamidino-2-phenylindole; 1:15000 in PBS) for 5 min. Coverslips were mounted (GelMount, Biomeda) on to glass slides and CLSM was performed using a Leica TCS SP2 imaging system with a ×100 1.35 NA (numerical aperture) objective. Image analysis from digitized confocal images was carried out using ImageJ as described previously [21]. Briefly, an area was measured in the nucleus and cytoplasm of cells stained with antibodies from ten different fields from three individual experiments (n=3) to determine the fluorescence of the nuclear (Fn) and cytoplasmic (Fc) STAT3 proteins. The nuclear to cytoplasmic fluorescence ratio (Fn/Fc) was calculated after the subtraction of values for background fluorescence.
RNA preparation and microarray analysis
Total RNA was extracted from Stat3−/− and iSTAT3α and iSTAT3β MEFs using a Purelink RNA mini-kit (Invitrogen) according to the manufacturer's protocols and stored at −80°C. Total RNA (1 μg) was analysed using Affymetrix GeneChip mouse gene 1.0 ST arrays at the Molecular Genomics Facility (Peter MacCallum Cancer Centre, Melbourne, Australia). Data for Stat3−/−, iSTAT3α and iSTAT3β MEFs were obtained with biological replicates (n=3) and combined for statistical analysis. The data were imported and normalized using the R-package aroma.affymetrix [22]. RMA background correction and quantile normalization was applied. Statistical significance of differential expression was determined using LIMMA [23]. The P-values were adjusted using the Benjamini–Hochberg method to reduce false discovery rates. An adjusted P-value cut-off of 0.05 and log fold-change cut-off (LOGFC)≥1 or ≤−1 were used to derive the complete gene lists for all conditions. Further analysis to determine the genes regulated by STAT3 spliceform expression in iSTAT3α and/or iSTAT3β MEFs, but not regulated as a consequence of parallel signalling events (e.g. MAPK activation), was performed by comparing gene sets with that derived from Stat3−/− MEFs. Thus genes also recorded in the Stat3−/− MEFs were removed to create the gene lists presented. GO (gene ontology) analysis on these lists was then carried out by performing functional annotations of genes using DAVID Bioinformatics [24,25] and further grouped into their parent GO term using CateGOrizer [26].
Validation of microarray results with quantitative real-time PCR
Total RNA was reverse transcribed to cDNA using RT High Capacity kit (Applied Biosystems) according to the manufacturer's protocols. Quantitative real-time TaqMan® PCR was performed using 50 ng of cDNA in a 20 μl reaction volume containing TaqMan® Gene Expression Master Mix and a specific TaqMan® Gene Expression Assay (AssayIDs: Aim2, Mm01295719_m1; Cxcl10, Mm00445231_m1; Ifi44, Mm00505670_m1; Crip1, Mm01740674_g1; Plce1, Mm00457691_m1; Il18, Mm00434225_m1; Adamts9, Mm00614433_m1; Cdh11, Mm00515466_m1; Ilk, Mm00439671_g1) by Applied Biosystems. Amplification of cDNA was carried out in a 48-well Step One real-time PCR system (Applied Biosystems) using the PCR conditions as follows: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The data were normalized to β-actin (AssayID: Actb, Mm00607939_s1) in the respective samples and data quantification was carried out using the 2−ΔΔCT method and expressed as a log2 fold change which is equivalent to the microarray LOGFC. Quantification was performed on three independent occasions.
Statistical analysis
Statistical analysis was carried out using Graphpad Prism 5 software. Data comparisons between WT MEFs and iSTAT3α or iSTAT3β MEFs under OSM stimulation for the corresponding timepoints were performed using an unpaired Student's t test. All values are shown as means±S.E.M., with P<0.05 considered statistically significant.
RESULTS
Different nuclear retention of the STAT3 spliceforms STAT3α and STAT3β following cytokine stimulation
Two STAT3 proteins, STAT3α and the shorter STAT3β isoform that differ only in the C-terminal sequence of their transactivation domains, arise from alternative splicing during the transcription of the STAT3 gene (Figure 1A). To extend the studies addressing the isoform-specific roles of these proteins [12–17], we initially assessed their nucleocytoplasmic trafficking in the absence and presence of cytokine stimulation. AD293 cells were transiently transfected to express N-terminal FLAG-tagged STAT3α and STAT3β and then stimulated with OSM, a member of the IL-6 cytokine family. Immunostaining using the anti-FLAG antibody (Figure 1B) together with routine staining of cell nuclei with DAPI (results not shown), followed by CLSM, showed that FLAG–STAT3α was largely cytosolic under basal conditions, but predominantly nuclear following 15 min of OSM stimulation (Figure 1B, upper panels). A comparable increase in nuclear localization of FLAG–STAT3β was observed following 15 min of OSM treatment, but strikingly FLAG–STAT3α showed cytoplasmic localization following 60 min of OSM treatment, whereas FLAG–STAT3β remained predominantly nuclear (Figure 1B, lower panels). Subcellular fractionation has also been used to evaluate nuclear retention of active STAT3, although the proportions of nuclear STAT3 observed in this approach can be somewhat lower than observed in CLSM/immunostaining experiments [27]. Our fractionation studies, with nuclear/cytosolic separation confirmed by detection of PARP [poly(ADP-ribose) polymerase; nucleus] and α-tubulin (cytosol) showed the sustained nuclear retention of FLAG–STAT3β over the 60 min of OSM treatment (Supplementary Figure S1A at http://www.BiochemJ.org/bj/447/bj4470125add.htm). Furthermore, using CLSM visualization of FLAG–STAT3 proteins in transfected COS1 cells, we observed greater nuclear retention of FLAG–STAT3β than of FLAG–STAT3α over 120 min of OSM stimulation as shown by the co-localization with DAPI staining (Supplementary Figures S1B and S1C). Thus, despite sharing 93% identity (100% identity within the N-terminal 715 amino acids), STAT3α and STAT3β show markedly different nuclear retention times following cytokine stimulation.
The kinetics of nuclear translocation and retention were further investigated for STAT3 mutants. Specifically, mutation of STAT3 Tyr705 abolishes the tyrosine phosphorylation considered essential for its nuclear translocation under cytokine-stimulated conditions, whereas mutation of Arg609 disrupts the phosphotyrosine binding of the SH2 domain of STAT3 [28,29]. Analysis of the Y705F or R609L mutants of FLAG-tagged STAT3α and STAT3β showed no changes in subcellular localization upon OSM stimulation, consistent with the requirement for Tyr705 phosphorylation and a functional SH2 domain for cytokine-stimulated changes of either spliceform (Figures 1C and 1D).
Enhanced Tyr705 phosphorylation and nuclear retention of STAT3β following cytokine stimulation
To assess the Tyr705 phosphorylation of the different STAT3 spliceforms, we used lentiviral transduction [19] to produce stable cell lines in a Stat3−/− MEF [15] background with 4-HT-inducible expression of either STAT3α or STAT3β. The key elements of the viral constructs are shown in Figure 2(A). Of importance, under basal conditions, the transcription activator VP16 (viral protein 16) fused to the substrate-binding portion of the oestrogen receptor (GAL4–ERt2–VP16) would be sequestered by cytosolic Hsp90 (heat-shock protein of 90 kDa) in cells expressing these constructs and thus unable to activate the expression of specific STAT3 proteins. However, upon incubation with the oestrogen receptor ligand 4-HT, competitive binding of 4-HT to the GAL4–ERt2–VP16 protein dissociates Hsp90 to allow the expression of either STAT3α or STAT3β by these constructs. 4-HT-inducible expression of either FLAG–STAT3α or FLAG–STAT3β in these cell lines (iSTAT3α and iSTAT3β respectively) was confirmed by immunoblotting alongside the detection of endogenous STAT3 in WT MEFs (Figure 2B). Our expression of each STAT3 spliceform was at a comparable level, thus allowing our direct comparison of their functional effects and biochemical actions attributable to their different C-terminal sequences without confounding effects of different levels of expression.
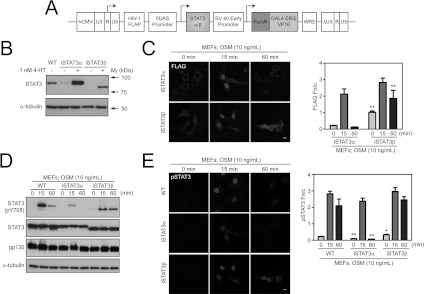
Figure 2. Absence of STAT3β leads to a lower and more transient STAT3α Tyr705 phosphorylation and nucleocytoplasmic trafficking.
(A) Schematic diagram of the lentiviral system used to drive the 4-HT-inducible expression of either STAT3α or STAT3β in Stat3−/− MEFs, thus creating iSTAT3α and iSTAT3β MEF lines. (B) Lysates of WT, iSTAT3α and iSTAT3β MEFs treated with or without 4-HT (1 nM) were immunoblotted for STAT3 proteins using anti-STAT3 antibody. Immunoblotting with an α-tubulin antibody was used to indicate equivalent level of protein loading. (C) WT and 4-HT (1 nM)-treated iSTAT3α and iSTAT3β MEFs were stimulated with OSM (10 ng/ml) for 0, 15 or 60 min before immunofluorescence analysis using the anti-FLAG antibody (left-hand panels). Image analysis and quantification was carried out using ImageJ software to determine the nuclear fluorescence (Fn) and cytoplasmic fluorescence (Fc) corrected for background for cells taken from ten different fields and averaged for each time point for three independent experiments (right-hand panel). Results were calculated as the Fn/c ratio. The histogram shows the mean±S.E.M. Asterisks indicate values that are statistically significant when compared with the WT cells (**P≤0.001). (D) WT and 4-HT (1 nM) treated iSTAT3 MEFs (iSTAT3α and iSTAT3β) were stimulated with OSM (10 ng/ml) for 0, 15 and 60 min. Cell lysates were collected and immunoblot analysis was carried out using anti-phospho-STAT3 Tyr705 (pY705) antibody for activated STAT3 proteins, anti-STAT3 antibody to indicate total STAT3 protein levels, as well as anti-gp130 and anti-α-tubulin antibodies to indicate equivalent protein loading. (E) WT and 4-HT (1 nM)-treated iSTAT3α and iSTAT3β MEFs were stimulated with OSM (10 ng/ml) for 0, 15 or 60 min and stained with anti-phospho-STAT3 Tyr705 antibody (left-hand panels). The phospho-STAT3 Fn/Fc ratio was calculated as above (right-hand panel), and the histogram shows the mean±S.E.M. Asterisks indicate values that are statistically significant when compared with the WT cells (*P≤0.05; **P≤0.001).
Analysis of iSTAT3α and iSTAT3β MEFs by CLSM after immunostaining for the FLAG epitope and DAPI staining of cell nuclei showed the dominance of nuclear STAT3 following 15 min of OSM treatment, with an ensuing rapid loss of FLAG–STAT3α from the nucleus but nuclear retention of FLAG–STAT3β by 60 min of OSM treatment (Figure 2C, left-hand panels, and Supplementary Figure S2A at http://www.BiochemJ.org/bj/447/bj4470125add.htm for the DAPI, FLAG and overlay images). This is consistent with the observations in transiently transfected AD293 and COS1 cells (Figure 1B and Supplementary Figures S1B and S1C respectively). Quantitative analysis of the relative levels of FLAG–STAT3 protein in the nucleus and in the cytosol, expressed in terms of the nuclear to cytoplasmic ratio (Fn/Fc) for FLAG staining, confirmed the transient nuclear retention of FLAG–STAT3α following OSM treatment together with the sustained retention of FLAG–STAT3β in the nucleus under these conditions (Figure 2C, right-hand panel). In addition, this analysis showed a statistically significantly higher retention of STAT3β under basal non cytokine-stimulated conditions.
In the absence of endogenous STAT3 in this system, the kinetics of Tyr705 phosphorylation of FLAG–STAT3α or FLAG–STAT3β could also be defined. This indicated a higher basal Tyr705 phosphorylation for FLAG–STAT3β (Figure 2D, and Supplementary Figure S2B that shows a longer exposure for the pTyr705 STAT3 immunoblot) consistent with the enhanced basal nuclear retention of FLAG–STAT3β as noted earlier (Figure 2C). Further analysis following cytokine treatment showed prolonged FLAG–STAT3β Tyr705 phosphorylation over the 60 min period examined, in contrast with transient Tyr705 phosphorylation of FLAG–STAT3α (Figure 2D). Furthermore, a strikingly lower level of STAT3α Tyr705 phosphorylation in the absence of endogenous STAT3β was consistently observed across multiple independent experiments, including in MEFs independently virally transduced with inducible Myc-epitope-tagged STAT3α expression constructs (I.H.W. Ng, unpublished work).
We analysed the nuclear retention of phospho-Tyr705 STAT3 in WT MEFs, noting the intense nuclear phospho-STAT3 detected at 15 min of OSM stimulation, but the continued detection of nuclear phospho-STAT3 following 60 min of OSM stimulation (Figure 2E and Supplementary Figure S2C for DAPI and phospho-STAT3 detection). Analysis of the nuclear retention of phospho-STAT3α or -STAT3β in iSTAT3α or iSTAT3β was undertaken in parallel by co-staining the cells examined in Figure 2(C) for phospho-STAT3 localization. This analysis further confirmed a rapid loss of phospho-STAT3α from the nucleus and prolonged nuclear retention of phospho-STAT3β (Figure 2E, left-hand panels, and Supplementary Figure S2A for the DAPI, FLAG, phospho-STAT3 and overlay images).
Quantitative analysis of the fluorescence intensities (Figure 2E, right-hand panel) indicated that the nuclear levels of phospho-STAT3 were highest at 15 min post-activation with OSM in all cases in WT, iSTAT3α and iSTAT3β MEFs, and declined rapidly in the case of iSTAT3α MEFs. In contrast, both WT and iSTAT3β MEFs showed prolonged levels of nuclear retention of phospho-STAT3 at 60 min post-treatment with OSM. Furthermore, under basal non-cytokine-stimulated conditions, the levels of phospho-STAT3 detected in iSTAT3α MEFs were significantly lower than in WT MEFs, whereas the levels of nuclear phospho-STAT3 in iSTAT3β MEFs were significantly higher. These results show the prolonged activation/phosphorylation and nuclear retention of STAT3β when compared with STAT3α, but also suggest a cross-regulation by STAT3β to sustain STAT3α Tyr705 phosphorylation and nuclear retention.
Prolonged STAT3α Tyr705 phosphorylation in the presence of STAT3β is dependent on a functional STAT3β SH2 domain
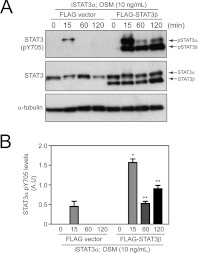
To test further the modulation of STAT3α Tyr705 phosphorylation by STAT3β, iSTAT3α MEFs were transiently transfected to co-express STAT3β in the presence of STAT3α. The reconstitution of STAT3β into iSTAT3α MEFs, albeit with an overexpression of STAT3β to higher levels than usually observed in the WT cells, led to increased STAT3α Tyr705 phosphorylation following OSM stimulation over the 15–120min period examined, as seen by immunoblot analysis (Figure 3A) and confirmed by quantitative analyses over three independent experiments (Figure 3B). Similarly, ectopic expression of high levels of STAT3β in COS1 cells led to a prolonged STAT3α Tyr705 phosphorylation up to 120 min as shown by immunoblotting and subsequent quantification in three independent experiments (Supplementary Figure S3 at http://www.BiochemJ.org/bj/447/bj4470125add.htm). These results confirm an action of these high levels of STAT3β to cross-regulate STAT3α phosphorylation and nuclear retention, and so complement our observations (Figure 2) that an absence of STAT3β decreases phosphorylation and nuclear retention of STAT3α.
Figure 3. Expression of STAT3β up-regulates and prolongs STAT3α Tyr705 phosphorylation.
(A) Protein lysates were prepared from iSTAT3α MEFs transiently transfected with empty FLAG vector or FLAG–STAT3β and stimulated with OSM (10 ng/ml) for 0, 15, 60 or 120 min. Lysates were immunoblotted with anti-STAT3 antibody as an indicator of total STAT3 proteins and anti-phosopho-STAT3 Tyr705 (pY705) antibody for activated STAT3 proteins. α-Tubulin was blotted to indicate an equivalent level of protein loading. (B) Densitometry analysis of Tyr705 phospho-STAT3α bands from immunoblots (n=3) was carried out using ImageJ software. The histogram shows the mean levels of pSTAT3α±S.E.M. Asterisks indicate values that are statistically significant when compared with the control bands of corresponding time point (*P≤0.01; **P≤0.001).
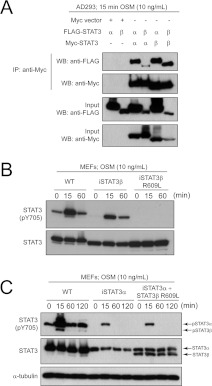
To examine further how STAT3β cross-regulates STAT3α phosphorylation, STAT3 heterodimer formation was demonstrated in co-immunoprecipitation experiments of epitope-tagged STAT3 isoforms ectopically expressed in transfected AD293 cells following 15 min of OSM treatment (Figure 4A). To evaluate a requirement for heterodimerization of the STAT3 isoforms in this novel cross-regulation mechanism, we evaluated a STAT3β R609L SH2 domain mutant that we demonstrated was not phosphorylated on Tyr705 following OSM stimulation (Figure 4B). In comparing the effect of STAT3β R609L with that of WT STAT3β in the iSTAT3α MEF cell system, we demonstrated that the STAT3β R609L mutant derivative could not prolong phosphorylation of STAT3α in the iSTAT3α MEFs (Figure 4C) when compared with the demonstrated actions of WT STAT3β (Figure 3). The results indicate the requirement for a functional SH2 domain of STAT3β in the cross-regulation of phosphorylation of STAT3α, consistent with the effects of STAT3β on STAT3α being dependent on STAT3 dimerization and/or phosphorylation.
Figure 4. A functional SH2 domain is required for Tyr705 phosphorylation and dimerization of STAT3 proteins.
(A) AD293 cells were co-transfected with the following combinations and exposed for 15 min to OSM (10 ng/ml): FLAG–STAT3α with empty Myc vector, FLAG–STAT3β with empty Myc vector, FLAG–STAT3α with Myc–STAT3α, FLAG–STAT3β with Myc–STAT3α, FLAG–STAT3α with Myc–STAT3β or FLAG–STAT3β with Myc–STAT3β. Cell lysates were immunoprecipitated (IP) with anti-Myc antibody and immunoblotted (WB) with anti-FLAG and anti-Myc antibodies to detect homo- and hetero-dimerization of STAT3α and STAT3β. (B) 4-HT (1 nM)-treated WT, iSTAT3β and iSTAT3β R609L mutant MEFs stimulated with OSM (10 ng/ml) for 0, 15, 60 or 120 min were immunoblotted for total STAT3 proteins and phospho-STAT3 Tyr705 (pY705) for activated STAT3 proteins. (C) iSTAT3α MEFs transiently expressing FLAG–STAT3β R609L were stimulated with OSM (10 ng/ml) for 0, 15, 60 or 120 min and immunoblotted for total STAT3 proteins and phospho-STAT3 Tyr705. α-Tubulin was blotted to indicate an equivalent level of protein in each loaded sample.
Transcriptional profiling reveals STAT3β-dependent gene expression changes under basal and cytokine-stimulated conditions
To extend these biochemical analyses to the biological consequences of altered nuclear retention, we conducted transcriptional profiling to define the transcriptional roles for the STAT3 spliceforms. All mRNA samples were prepared on three independent occasions from the different MEF lines, under basal conditions or following cytokine stimulation, as indicated. All samples were subjected to gene microarray analysis using Affymetrix GeneChip mouse gene 1.0 ST arrays. With the analysis of these samples, a statistical significance cut-off was set at P<0.05, then a list of genes with a LOGFC of ≤−1 and ≥1 (i.e. a 2-fold decrease or increase in expression upon STAT3 re-expression) was recorded.
We first examined the impact of re-expression and cytokine-stimulated activation of STAT3 spliceforms in the Stat3−/− background by comparison with the transcriptional changes noted for WT MEFs under these same conditions. This analysis reveals the extent of reconstitution possible in this system in which the STAT3 spliceforms are only re-expressed for 2 days prior to their activation and analysis for their transcriptional roles. Thus, in this analysis, all genes altered in expression in a STAT3-dependent fashion were derived from the comparison with the Stat3−/− cells that had also been stimulated with OSM for 30 min. In this system, 219 genes changed in expression in WT MEFs when compared with Stat3−/− MEFs under the conditions of 30 min of OSM stimulation. Notably, and as presented in Table 1, 46 of these genes were regulated at this level of statistical significance upon re-expression of STAT3α or STAT3β and stimulation with OSM. Furthermore, an additional 37 were regulated by STAT3α re-expression and OSM stimulation and an additional 17 were regulated by STAT3β re-expression and OSM stimulation. Thus a large group of genes regulated in OSM-stimulated WT cells were accounted at this high level of statistical confidence by STAT3α or STAT3β re-expression and cytokine stimulation. These results provide evidence that the reconstitution with STAT3 spliceforms provides a robust and physiologically relevant system to define transcriptional consequences of these STAT3 proteins.
Table 1. Summary of OSM-stimulated gene changes in WT MEFs defined as STAT3-dependent by comparisons with changes in OSM-stimulated (30 min) Stat3−/− MEFs, recapitulated by the OSM stimulation (30 min) of Stat3−/− MEFs re-expressing STAT3α or STAT3β for 48 h.
| Common gene expression changes (shared by OSM-stimulated iSTAT3α and iSTAT3β) | Gene expression changes re-established by STAT3α | Gene expression changes re-established by STAT3β | ||||||
|---|---|---|---|---|---|---|---|---|
| ↑ by STAT3α/β | ↓ by STAT3α/β | ↑ by STAT3α | ↓ by STAT3α | ↑ by STAT3β | ↓ by STAT3β | |||
| 12 | 34 | 25 | 12 | 5 | 12 | |||
| C3 | 0610010O12Rik | Fam180a | Podxl | Abcb1b | Gm8773 | 2810047C21Rik1 | Aspa | Ak3l1 |
| Ccl2 | 2610018G03Rik | Fam184a | Prg4 | Acta2 | H2-K1 | Adamts3 | Cacna2d1 | Atp11c |
| Gstm5 | 2900062L11Rik | Fhl1 | Prss12 | Adamts9 | H2-M2 | Akr1c18 | Gbp4 | Ccdc112 |
| Gyg | 4930506M07Rik | Foxr2 | Rex2 | Casp4 | Osmr | Fgfbp1 | Gm7669 | Chchd7 |
| Ifitm3 | Akr1c13 | Il18 | Sema3d | Ccl9 | Rnd1 | Gpm6a | Phlda1 | Elovl7 |
| Igf1 | Armcx1 | Macc1 | Sorcs1 | Cdh11 | Saa3 | Itih2 | Gja1 | |
| Igfbp7 | Atp8a1 | Mpp7 | Tmem108 | Cxcl12 | Slc43a3 | Pde3b | Gm447 | |
| Il1r1 | Car9 | Muc16 | Trf | Cyp1b1 | Steap1 | Ppargc1a | Peg10 | |
| Myc | Cldn15 | Nt5e | Upk3b | Cyr61 | Tagln2 | Sepp1 | Rbm28 | |
| Sh3kbp1 | Crip1 | Nxt2 | Vmn2r50 | Ddah1 | Tmem176a | Vmn2r43 | Rcan2 | |
| Tmem140 | Cysltr1 | Plce1 | Ecscr | Tmem176b | Zfp772 | Tmod2 | ||
| Tnc | Efemp1 | Plxdc2 | Enpp2 | Tmem88 | Zic1 | Upk1b | ||
| Fn1 | ||||||||
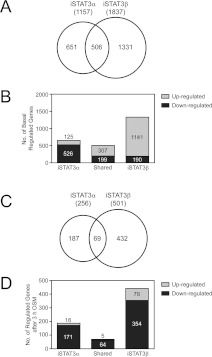
Prompted by the different nuclear levels of STAT3 proteins under basal conditions (Figure 2C and Figure 2E), we next explored how the reconstitution of the Stat3−/− MEFs with either isoform would impact on gene expression as an indication of the basal activities of STAT3α or STAT3β. Validation of gene expression changes was undertaken for selected genes using quantitative real-time PCR, confirming the actions of STAT3 spliceform expression to result in common as well as unique changes in gene expression (Supplementary Figure S4 at http://www.BiochemJ.org/bj/447/bj4470125add.htm). The profiling results, summarized diagrammatically in Figure 5(A), indicate the large number of gene expression changes, either unique for re-expression of STAT3α (651 genes with statistically significant changes), unique for STAT3β (1331 genes with statistically significant changes), or shared between STAT3α and STAT3β (506 genes with statistically significant changes). Notably, grouping of the gene expression changes by up-regulation (LOGFC of ≥1) or down-regulation (LOGFC of ≤−1) emphasized the large number of genes up-regulated specifically by STAT3β (1141 genes) when compared with those up-regulated specifically by STAT3α (125 genes) or shared by STAT3α and STAT3β (307 genes) under these basal conditions (Figure 5B). Thus, for STAT3β re-expression, the number of genes up-regulated (1141 genes in iSTAT3β only) was 6-fold greater than the numbers down-regulated (190 genes in iSTAT3β only).
Figure 5. Different transcriptional changes following reconstitution of STAT3α or STAT3β in Stat3−/− MEF cells.
(A) Analysis of basal-regulated gene expression following re-expression of the STAT3 spliceforms in iSTAT3α and iSTAT3β MEFs. The Venn diagram illustrates the numbers of gene changes recorded to be statistically significant (P≤0.05 and a LOGFC of ≥1 or ≤−1) in iSTAT3α and iSTAT3β MEFs when compared with Stat3−/− MEFs. (B) The histogram shows the number of genes that are up-regulated (grey bars) or down-regulated (black bars) in the respective iSTAT3α and iSTAT3β MEFs. The numbers of regulated genes shared by iSTAT3α and iSTAT3β are indicated. (C) Analysis of OSM-stimulated gene expression following re-expression of the STAT3 spliceforms in iSTAT3α and iSTAT3β MEFs. The Venn diagram illustrates the numbers of gene changes recorded to be statistically significant (P≤0.05 and a LOGFC of ≥1 or ≤−1) in OSM-stimulated iSTAT3α and iSTAT3β MEFs when compared with OSM-stimulated STAT3−/− MEF cells when assessed 48 h after induction of STAT3 spliceform expression and following 3 h of OSM stimulation. (D) The histogram shows the number of genes that are up-regulated (grey bars) or down-regulated (black bars) in the respective OSM-stimulated iSTAT3α and iSTAT3β MEFs. The numbers of regulated genes shared by iSTAT3α and iSTAT3β following OSM stimulation are indicated.
In examining the likely biological significance of these gene expression changes under basal non-cytokine stimulated conditions further, GO analyses using the online tools DAVID Bioinformatics [24,25] and CateGOrizer [26] revealed large numbers of STAT3β-regulated genes involved in metabolism, protein metabolism (including transcription and translation), transport, cell organization and biogenesis (Table 2). To define whether any of these changes in gene expression may also underlie a capacity of STAT3β to cross-regulate STAT3α phosphorylation and nuclear retention, we specifically searched the genes in the GO categories of transport (Supplementary Table S1 at http://www.BiochemJ.org/bj/447/bj4470125add.htm) and signal transduction (Supplementary Table S2 at http://www.BiochemJ.org/bj/447/bj4470125add.htm) for regulators of STAT3 activation; however, these gene lists do not reveal any statistically significant changes in known STAT3 regulators following STAT3β re-expression in this system. Although there may be additional undescribed regulators of STAT3 within the gene lists examined, our results further emphasize the likely direct actions of STAT3β on STAT3α via heterodimer formation rather than indirect actions through downstream transcriptional differences.
Table 2. Summary of the GO terms of genes in MEFs re-expressing either STAT3α or STAT3β spliceforms.
| GO term | Common | iSTAT3α | iSTAT3β |
|---|---|---|---|
| Metabolism | 56 | 140 | 394 |
| Developmental processes | 31 | 3 | 57 |
| Cell organization and biogenesis | 32 | 46 | 113 |
| Transport | 15 | 24 | 124 |
| Protein metabolism | 3 | 65 | 161 |
| Stress response | 36 | 9 | 40 |
| Signal transduction | 25 | 30 | 58 |
| Cell death | 21 | − | 28 |
| Cell proliferation | 24 | 22 | 54 |
| RNA metabolism | 11 | 21 | 37 |
| Cell cycle | 11 | 29 | 63 |
| DNA metabolism | 11 | 10 | 40 |
| Cell adhesion | 6 | − | − |
| Cell–cell signalling | − | − | 3 |
We finally examined the gene expression outcomes following cytokine stimulation (OSM 3 h). This has allowed our assessment of the impact of the different STAT3 spliceforms, with different nuclear retention times after activation, on gene expression profiles following exposure to cytokine. All samples, again prepared in triplicate, were subjected to gene microarray analysis using AffymetrixGeneChip mouse gene 1.0 ST arrays and a statistical significance cut-off was set at P<0.05 and list of genes with a LOGFC of ≤−1 and ≥1 (i.e. a 2-fold decrease or increase in expression upon OSM exposure) were recorded. These results, summarized in Figure 5(C), indicate 501 gene changes (418 down-regulated, i.e. 354 unique plus 64 shared down-regulated changes) in iSTAT3β MEFs, twice as many in iSTAT3α MEFs (256 genes, 235 down-regulated, i.e. 171 unique plus 64 shared down-regulated changes); an implication being that the longer nuclear retention time for activated phospho-STAT3β, when STAT3β is expressed at levels comparable with that of STAT3α, has a greater impact on transcription. Significantly, 69 of these gene changes (i.e. 64 down-regulated plus five up-regulated) were shared between the STAT3 isoforms, with twice as many gene changes (432 genes) unique to iSTAT3β MEFs when compared with iSTAT3α MEFs (187 unique changes). Although many of the genes observed to change were down-regulated, 78 genes were increased in expression in cytokine-stimulated iSTAT3β MEFs. This compared with only 16 up-regulated in cytokine-stimulated iSTAT3α MEFs. These results clearly illustrate that the effects of STAT3β on transcription in response to cytokine exposure are not restricted to transcriptional repression, and conversely that the effects of STAT3α are not dominated by increased gene expression under these conditions of analysis. Taken together, these results emphasize the differences in gene expression profiles that result from activation of STAT3α or STAT3β, when expressed at comparable levels, in response to cytokine stimulation.
Classification by GO terms was undertaken for the genes altered following cytokine exposure (Table 3). An evaluation to parallel those in Supplementary Tables S1 and S2 included evaluation of the GO terms transport (Supplementary Table S3 at http://www.BiochemJ.org/bj/447/bj4470125add.htm) and signal transduction (Supplementary Table S4 at http://www.BiochemJ.org/bj/447/bj4470125add.htm). When comparing the transport gene sets between basal and 3 h of OSM stimulation (Supplementary Table S1 compared with Supplementary Table S3), four genes (Atp5d, Atp6v0b, Ipo9 and Tnpo2) in iSTAT3β MEFs appeared in both sets, whereas the gene sets for iSTAT3α did not overlap. Further consideration of the GO lists showed the dominance of STAT3β to regulate genes classified in the GO term of developmental processes, with 39 genes (Supplementary Table S5 at http://www.BiochemJ.org/bj/447/bj4470125add.htm) altered in expression in iSTAT3β MEFs, but no genes in this GO category altered in iSTAT3α MEFs. Similarly, in the GO term cell adhesion, three genes (Thbs1, Fbln2 and Cyr61) were regulated exclusively in the iSTAT3β MEFs. These groupings contrasted with the genes classified in the GO term protein metabolism (Supplementary Table S6 at http://www.BiochemJ.org/bj/447/bj4470125add.htm) with regulation in iSTAT3α or iSTAT3β or shared in both iSTAT3α and iSTAT3β MEFs. Furthermore, some GO terms showed changes in iSTAT3α and iSTAT3β MEFs only, such as transport (Supplementary Table S3), cell organization and biogenesis (Supplementary Table S7 at http://www.BiochemJ.org/bj/447/bj4470125add.htm) or cell proliferation (Supplementary Table S8 at http://www.BiochemJ.org/bj/447/bj4470125add.htm). Conversely, some GO terms were restricted to iSTAT3α MEFs, such as cell cycle (Supplementary Table S9 at http://www.BiochemJ.org/bj/447/bj4470125add.htm). This analysis clearly highlights numerous genes regulated by both STAT3α and STAT3β during cytokine stimulation, but also changes unique to either iSTAT3α or iSTAT3β. Thus there is both functional overlap as well as unique roles for these two different STAT3 spliceforms.
Table 3. Summary of the GO terms of genes regulated after 3 h of OSM treatment of MEFs re-expressing either STAT3α or STAT3β spliceforms.
| GO term | Common | iSTAT3α | iSTAT3β |
|---|---|---|---|
| Metabolism | 30 | 60 | 110 |
| Developmental processes | − | − | 39 |
| Cell organization and biogenesis | − | 8 | 26 |
| Transport | − | 16 | 11 |
| Protein metabolism | 15 | 21 | 47 |
| Stress response | 5 | − | − |
| Signal transduction | 3 | − | 6 |
| Cell proliferation | − | 6 | 20 |
| RNA metabolism | − | 16 | − |
| Cell cycle | − | 10 | − |
| Cell adhesion | − | − | 3 |
DISCUSSION
STAT3α, the predominant STAT3 spliceform in many cell types, typically shows rapid phosphorylation and nuclear translocation following cytokine stimulation [10]. Although the biological functions of the STAT3 spliceforms have remained a subject of debate since their initial description [10,11], their activation by the same cytokine stimuli and JAK-mediated Tyr705 phosphorylation enhancing their nuclear import is consistent with their identical regulatory regions, including the coiled-coiled and SH2 domains [30,31]. The present study shows that the different C-terminal domain sequences of the STAT3 spliceforms markedly prolong STAT3β Tyr705 phosphorylation and nuclear retention following OSM treatment when compared with STAT3α under the same cytokine-stimulated conditions. This is consistent with an earlier report of nuclear retention of STAT3β following cell exposure to IL-6 [32]. Importantly, the present study documents the actions of STAT3β to influence STAT3α phosphorylation and nuclear retention. Specifically, in the presence of cytokine stimulation, and dependent on heterodimer formation with STAT3β, the phosphorylation and nuclear retention of STAT3α can be prolonged to more closely resemble that of STAT3β. Furthermore, our transcriptional profiling results comparing gene expression changes driven by comparable levels of the different STAT3 spliceforms have revealed a greater number of genes regulated by STAT3β under both basal and cytokine-stimulated conditions when compared with the numbers of genes regulated by STAT3α under the same conditions. Taken together, our results highlight that STAT3β is a potent transcriptional regulator with sustained nuclear retention and is also able to cross-regulate/enhance the transcriptional activity of STAT3α.
The prolonged phosphorylation of STAT3β Tyr705 may be a result of different recognition by tyrosine phosphatases that normally target STAT3α. Thus the dephosphorylation of either spliceform of STAT3 in the nucleus would be expected to be a crucial regulatory step prior to its CRM1-mediated nuclear export [32]. On the basis of experiments using WT and TC45−/− cells, the nuclear tyrosine phosphatase TC45 has been implicated in the dephosphorylation of STAT3 Tyr705 [33]. Supporting this, a combination of results from binding assays using a catalytically inactive TC45 mutant and deletion studies of STAT3α indicated an interaction between the C-terminal domain of STAT3α and TC45 [34]. Thus an absence of this interaction of TC45 with STAT3β due its different STAT3β C-terminal sequence may contribute to the prolonged Tyr705 phosphorylation and nuclear retention of STAT3β. However, in exploring this possible mechanism, we were able to co-immunoprecipitate epitope-tagged TC45 with STAT3α or STAT3β, demonstrating that either isoform can interact with TC45 (I.H.W. Ng, unpublished work). Furthermore, we demonstrated that the overexpression of catalytically inactive TC45 or the use of TC45 siRNA (small interfering RNA) that depleted TC45 protein levels by >70% could not prolong STAT3α Tyr705 phosphorylation after cytokine treatment of iSTAT3α MEFs (I.H.W. Ng, unpublished work). This lack of effect may reflect a redundancy of actions of the tyrosine phosphatases targeting STAT3 as the combined knockdown of TC45 in combination with the cytoplasmic tyrosine phosphatases SHP1 (SH2 domain-containing protein tyrosine phosphatase 1) [35,36] and SHP2 [37,38] has also been shown more recently to be insufficient in prolonging STAT3 phosphorylation [39]. Thus negative regulation of STAT3 is more complex than originally anticipated and further work is needed to define the repertoire of tyrosine phosphatases targeting the STAT3 spliceforms, and in particular to identify phosphatases capable of targeting STAT3α but that are not able to dephosphorylate nuclear phospho-STAT3β.
An unanticipated observation in our present study was the modulation of STAT3α phosphorylation by the presence of STAT3β. In initial studies in Stat3β−/− MEFs, no changes in STAT3α phosphorylation in the absence of STAT3β were observed [15]. Possible reasons for the differences between those results and our studies in the iSTAT3α cells could include the differences in the experimental systems employed, particularly our use of FLAG-tagged STAT3 constructs and the expression levels of the STAT3 isoforms achieved in our lentivirus-based inducible expression system, rather than endogenous levels of the STAT3 isoforms. Indeed, others have also concluded that there were no noticeable changes in STAT3α regulation in the absence of STAT3β (i.e. in Stat3β−/− MEFs) following cytokine stimulation [16]. However, direct side-by-side comparisons of STAT3α phosphorylation in Stat3β−/−, Stat3β+/− or Stat3β+/+ cells were not presented in those studies and the loss of STAT3 DNA binding or STAT3 reporter gene activity appeared to be greater than could be anticipated based solely on the ratios of STAT3α/STAT3β in WT cells [16]. Importantly, the cellular context may also be a critical factor in determining the extent of cross-regulation of the STAT3 spliceforms. For example, although lipopolysaccharide-modulation of hepatic STAT3α in the absence of STAT3β was reported to be unperturbed, levels of STAT3α phospho-Tyr705 were lower at the 1.5 h and 6 h timepoints of treatment in the absence of STAT3β [15]. These observations indicate that the effects of the loss of STAT3β on STAT3α regulation are further supported by a study in liver showing that the adenoviral delivery of STAT3β, followed by cytokine stimulation with IL-6, potentiates phosphorylation of STAT3α [40].
As there has been increasing evidence of basal nucleocytoplasmic shuttling of STAT3 [6,41–43], and that basal STAT3 has been shown to have transcriptional activity under basal conditions [44,45], we explored whether this phenomenon of cross-regulation could be attributed to changes in expression of STAT3α regulators when STAT3β is present. In profiling the transcriptional consequences of re-expression of STAT3α or STAT3β in Stat3−/− MEFs, our analysis revealed the range of gene expression changes shared by these STAT3 isoforms, but also large sets of STAT3β-specific differences under basal conditions. To our knowledge, this is the first transcriptome profile for STAT3β in a Stat3−/− background and highlights the importance of unphosphorylated STAT3β in the regulation of diverse subsets of genes. Importantly, in the context of understanding cross-regulation mechanisms, no known STAT3α regulators were identified in the genes significantly altered in expression by STAT3β, thus suggesting that the action of STAT3β to cross-regulate STAT3α is not dependent on longer-term transcriptional events.
We therefore also explored the possibility of direct cross-regulation mediated by a STAT3β–STAT3α interaction. Dimerization between STAT3 proteins has been a prerequisite for nuclear translocation upon activation and this interaction occurs via its functional SH2 domain [28,29]. Previously, the R609L mutation that disrupts the SH2 domain function of STAT3 has only been made in the context of STAT3α, but its expression in cells with endogenous STAT3α levels precluded its detailed characterization of phosphorylation and nuclear translocation kinetics [29]. As revealed in the present study, STAT3β R609L could no longer up-regulate or prolong STAT3α Tyr705 phosphorylation. Thus the regulation of STAT3α by STAT3β appears to require a functional STAT3β SH2 domain and/or the tyrosine phosphorylation of STAT3β.
In evaluating further the gene expression changes in the Stat3−/− MEFs as driven uniquely by the re-expression of STAT3β, we noted prominent representation of several GO classes: metabolism (394 STAT3β-specific changes), protein metabolism (161 STAT3β-specific changes), transport (124 STAT3β-specific changes) as well as cell organization and biogenesis (113 STAT3β-specific changes). Furthermore, STAT3β re-expression was sufficient to change the expression for genes for the GO class of cell death, but our statistical analyses showed that no genes in this class were uniquely regulated by STAT3α. These results thus highlight the large repertoire of STAT3α- and STAT3β-dependent changes, and that the STAT3β-dependent changes do not simply recapitulate the STAT3α-dependent changes observed.
STAT3 transcriptional activity has been attributed to its transactivation domain that binds transcription co-activators such as p300 [46]. STAT3α and STAT3β would thus activate a common subset of genes via their interaction with co-activators to form enhanceosome complexes [47]. Alternatively, other shared transcription factor partners, such as c-Jun, which can regulate induction of the α2-macroglobulin promoter [48], may underpin the regulation of genes targeted by either STAT3α or STAT3β. However, the truncated transactivation domain of STAT3β has led to the suggestion that STAT3β may lack transcriptional activity and so act as a dominant-negative regulator of STAT3α [10]. Initial support for this came with the repression and/or down-regulation of a number of recognized STAT3α target genes when STAT3β was overexpressed [10,48]. Furthermore, in COS cells, STAT3β was unable to initiate a transcriptional response as determined by luciferase reporter assay in cells expressing the ICAM-1 (intercellular adhesion molecule 1) promoter [10], and in cancer cells STAT3β suppressed the transformation activity of STAT3α by repressing the expression of Bcl-xL, p21WAF/CIP1 and cyclin D1, leading to apoptosis and regression of the cancer cells [49,50]. The ability of STAT3β to rescue STAT3−/− embryonic lethality has clearly indicated that STAT3β can perform at least some of the roles of STAT3α [15]. Furthermore, STAT3β has been shown to initiate transcription of the p27Kip1 gene in myeloblastic cells [51], α1-anti-chymotrypsin and α2-macroglobulin in hepatocytes [52], acute-phase genes in the liver during inflammation [15], and so can act as an up-regulator of transcription of specific gene sets.
The differences in gene expression profiles in the presence of the different STAT3 spliceforms, but also following cell exposure to cytokine, highlight further the remarkable spliceform-dependent differences in gene expression. Thus both STAT3α and STAT3β are transcriptional regulators following cytokine stimulation, and STAT3β should not simply be viewed as a repressor or negative regulator of gene transcription. Indeed, although the established paradigms illustrate STAT3α as a transcriptional activator, it is clear from previous studies that STAT3α can also act as a negative regulator of its target gene expression. Notably, STAT3 has been demonstrated to activate or repress its direct target genes in NIH 3T3 cells, with OSM treatment increasing six of 18 direct target genes specifically tested, but decreasing expression of ten of these 18 direct target genes [53]. Similarly, the STAT3-dependent repression of genes has been shown to be critical for muscle cell differentiation [53]. These results are consistent with the association of STAT3 with both active and inactive promoters in embryonic stem cells [54] and the reported actions of STAT3 to down-regulate expression of specific target genes, such as that recently described for the negative growth regulator Necdin [55]. In addition to direct gene regulatory mechanisms, increased attention should be directed to more complex regulatory mechanisms, such as those requiring STAT3-dependent up-regulation of microRNAs (such as miR-21 and miR-181b-1 [56]) that mediate repression of gene expression.
In conclusion, the present study reinforces the transcriptional functions of STAT3β under basal conditions as well as its direct actions to modulate STAT3α activation following cytokine stimulation. These functions of STAT3β indicate its importance as a modulator of gene expression in its own right, but also now highlight the exciting possibility that an additional important action of STAT3β may be in extending the activation kinetics for STAT3α. Given the striking changes in the levels of STAT3α and STAT3β noted during myeloid differentiation [12–14], and the interest in the directed expression of STAT3β in the place of STAT3α by manipulation of alternative splice regulation, further exploration of these mechanisms of these differences and transcription factor cross-regulation is clearly warranted.
Online data
AUTHOR CONTRIBUTION
Ivan Ng performed and analysed all the experiments. Ivan Ng, Dominic Ng, David Jans and Marie Bogoyevitch designed the experiments, discussed the analyses and results interpretation, and wrote the paper.
ACKNOWLEDGEMENTS
We thank Dr John Silke (Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia) for the inducible lentiviral expression constructs and Professor Valeria Poli (Department of Genetics, Biology and Biochemistry, University of Turin, Italy) for the WT and Stat3−/− MEFs. We thank Jason Li (Bioinformatics Core Facility, Research Division Peter MacCallum Cancer Centre, Melbourne, Australia) for statistical analysis of the microarray gene expression data.
FUNDING
This work was supported by a National Health and Medical Research Council (NHMRC) Project Grant [grant number 353592 (to M.A.B.)] and a National Heart Foundation of Australia Grant-in-aid [grant number G09M4435 (to D.C.H.N.)]. I.H.W.N. is a recipient of a Monash University: Monash Graduate Scholarship and Faculty of Medicine International Postgraduate Research Scholarship, D.C.H.N. is a recipient of a University of Melbourne MDHS CR Roper Fellowship, and D.A.J. is an NHMRC Senior Principal Research Fellow [number #APP1002486].
References
- 1.Akira S., Nishio Y., Inoue M., Wang X. J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 2.Lutticken C., Wegenka U. M., Yuan J., Buschmann J., Schindler C., Ziemiecki A., Harpur A. G., Wilks A. F., Yasukawa K., Taga T., et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 3.Zhong Z., Wen Z., Darnell J. E., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 4.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Muller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy D. E., Lee C. K. What does Stat3 do? J. Clin. Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L., McBride K. M., Reich N. C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poli V. STAT3 function in vivo. In: Sehgal P. B., editor. Signal Transducers and Activators of Transcription (STATs) Dordecht: Kluwer Academic Publishers; 2003. pp. 492–512. [Google Scholar]
- 9.Yu H., Jove R. The STATs of cancer–new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 10.Caldenhoven E., van Dijk T. B., Solari R., Armstrong J., Raaijmakers J. A., Lammers J. W., Koenderman L., de Groot R. P. STAT3β, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 11.Shao H., Quintero A. J., Tweardy D. J. Identification and characterization of cis elements in the STAT3 gene regulating STAT3 α and STAT3 β messenger RNA splicing. Blood. 2001;98:3853–3856. doi: 10.1182/blood.v98.13.3853. [DOI] [PubMed] [Google Scholar]
- 12.Biethahn S., Alves F., Wilde S., Hiddemann W., Spiekermann K. Expression of granulocyte colony-stimulating factor- and granulocyte-macrophage colony-stimulating factor-associated signal transduction proteins of the JAK/STAT pathway in normal granulopoiesis and in blast cells of acute myelogenous leukemia. Exp. Hematol. 1999;27:885–894. doi: 10.1016/s0301-472x(99)00017-x. [DOI] [PubMed] [Google Scholar]
- 13.Dewilde S., Vercelli A., Chiarle R., Poli V. Of alphas and betas: distinct and overlapping functions of STAT3 isoforms. Front. Biosci. 2008;13:6501–6514. doi: 10.2741/3170. [DOI] [PubMed] [Google Scholar]
- 14.Hevehan D. L., Miller W. M., Papoutsakis E. T. Differential expression and phosphorylation of distinct STAT3 proteins during granulocytic differentiation. Blood. 2002;99:1627–1637. doi: 10.1182/blood.v99.5.1627. [DOI] [PubMed] [Google Scholar]
- 15.Maritano D., Sugrue M. L., Tininini S., Dewilde S., Strobl B., Fu X., Murray-Tait V., Chiarle R., Poli V. The STAT3 isoforms α and β have unique and specific functions. Nat. Immunol. 2004;5:401–409. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- 16.Yoo J. Y., Huso D. L., Nathans D., Desiderio S. Specific ablation of Stat3β distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell. 2002;108:331–344. doi: 10.1016/s0092-8674(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 17.Yeh M., Gharavi N. M., Choi J., Hsieh X., Reed E., Mouillesseaux K. P., Cole A. L., Reddy S. T., Berliner J. A. Oxidized phospholipids increase interleukin 8 (IL-8) synthesis by activation of the c-src/signal transducers and activators of transcription (STAT)3 pathway. J. Biol. Chem. 2004;279:30175–30181. doi: 10.1074/jbc.M312198200. [DOI] [PubMed] [Google Scholar]
- 18.Zammarchi F., de Stanchina E., Bournazou E., Supakorndej T., Martires K., Riedel E., Corben A. D., Bromberg J. F., Cartegni L. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17779–17784. doi: 10.1073/pnas.1108482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeap Y. Y., Ng I. H., Badrian B., Nguyen T. V., Yip Y. Y., Dhillon A. S., Mutsaers S. E., Silke J., Bogoyevitch M. A., Ng D. C. c-Jun N-terminal kinase/c-Jun inhibits fibroblast proliferation by negatively regulating the levels of stathmin/oncoprotein 18. Biochem. J. 2010;430:345–354. doi: 10.1042/BJ20100425. [DOI] [PubMed] [Google Scholar]
- 20.Ng D. C., Ng I. H., Yeap Y. Y., Badrian B., Tsoutsman T., McMullen J. R., Semsarian C., Bogoyevitch M. A. Opposing actions of extracellular signal-regulated kinase (ERK) and signal transducer and activator of transcription 3 (STAT3) in regulating microtubule stabilization during cardiac hypertrophy. J. Biol. Chem. 2011;286:1576–1587. doi: 10.1074/jbc.M110.128157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poon I. K., Oro C., Dias M. M., Zhang J., Jans D. A. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 2005;65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- 22.Bengtsson H., Wirapati P., Speed T. P. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics. 2009;25:2149–2156. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Huang da W., Sherman B. T., Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Huang da W., Sherman B. T., Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z. L., Bao J., Reecy J. M. CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J. Bioinform. 2008;9:108–112. [Google Scholar]
- 27.Sehgal P. B. Paradigm shifts in the cell biology of STAT signaling. Semin. Cell. Dev. Biol. 2008;19:329–340. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuai K., Horvath C. M., Huang L. H., Qureshi S. A., Cowburn D., Darnell J. E., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang T., Kee W. H., Seow K. T., Fung W., Cao X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol. Cell. Biol. 2000;20:7132–7139. doi: 10.1128/mcb.20.19.7132-7139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer T. S., Sanders L. K., Park O. K., Nathans D. Functional differences between Stat3α and Stat3β. Mol. Cell. Biol. 1997;17:5307–5316. doi: 10.1128/mcb.17.9.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilks A. F. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., Qiu J., Dong S., Redell M. S., Poli V., Mancini M. A., Tweardy D. J. Stat3 isoforms, α and β, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3β mapping to its unique C-terminal end. J. Biol. Chem. 2007;282:34958–34967. doi: 10.1074/jbc.M704548200. [DOI] [PubMed] [Google Scholar]
- 33.ten Hoeve J., de Jesus Ibarra-Sanchez M., Fu Y., Zhu W., Tremblay M., David M., Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto T., Sekine Y., Kashima K., Kubota A., Sato N., Aoki N., Matsuda T. The nuclear isoform of protein-tyrosine phosphatase TC-PTP regulates interleukin-6-mediated signaling pathway through STAT3 dephosphorylation. Biochem. Biophys. Res. Commun. 2002;297:811–817. doi: 10.1016/s0006-291x(02)02291-x. [DOI] [PubMed] [Google Scholar]
- 35.Han Y., Amin H. M., Frantz C., Franko B., Lee J., Lin Q., Lai R. Restoration of shp1 expression by 5-AZA-2′-deoxycytidine is associated with downregulation of JAK3/STAT3 signaling in ALK-positive anaplastic large cell lymphoma. Leukemia. 2006;20:1602–1609. doi: 10.1038/sj.leu.2404323. [DOI] [PubMed] [Google Scholar]
- 36.Han Y., Amin H. M., Franko B., Frantz C., Shi X., Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- 37.Xu D., Qu C. K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008;13:4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann U., Schmitz J., Weissenbach M., Sobota R. M., Hortner M., Friederichs K., Behrmann I., Tsiaris W., Sasaki A., Schneider-Mergener J., et al. SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130. J. Biol. Chem. 2003;278:661–671. doi: 10.1074/jbc.M210552200. [DOI] [PubMed] [Google Scholar]
- 39.Kim D. J., Tremblay M. L., Digiovanni J. Protein tyrosine phosphatases, TC-PTP, SHP1, and SHP2, cooperate in rapid dephosphorylation of Stat3 in keratinocytes following UVB irradiation. PLoS ONE. 2010;5:e10290. doi: 10.1371/journal.pone.0010290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramadoss P., Unger-Smith N. E., Lam F. S., Hollenberg A. N. STAT3 targets the regulatory regions of gluconeogenic genes in vivo. Mol. Endocrinol. 2009;23:827–837. doi: 10.1210/me.2008-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrmann A., Vogt M., Monnigmann M., Clahsen T., Sommer U., Haan S., Poli V., Heinrich P. C., Muller-Newen G. Nucleocytoplasmic shuttling of persistently activated STAT3. J. Cell Sci. 2007;120:3249–3261. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 42.Pranada A. L., Metz S., Herrmann A., Heinrich P. C., Muller-Newen G. Real time analysis of STAT3 nucleocytoplasmic shuttling. J. Biol. Chem. 2004;279:15114–15123. doi: 10.1074/jbc.M312530200. [DOI] [PubMed] [Google Scholar]
- 43.Vogt M., Domoszlai T., Kleshchanok D., Lehmann S., Schmitt A., Poli V., Richtering W., Muller-Newen G. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J. Cell Sci. 2011;124:900–909. doi: 10.1242/jcs.072520. [DOI] [PubMed] [Google Scholar]
- 44.Yang J., Chatterjee-Kishore M., Staugaitis S. M., Nguyen H., Schlessinger K., Levy D. E., Stark G. R. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 45.Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., Stark G. R. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuringa J. J., Schepers H., Vellenga E., Kruijer W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 2001;495:71–76. doi: 10.1016/s0014-5793(01)02354-7. [DOI] [PubMed] [Google Scholar]
- 47.Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 48.Schaefer T. S., Sanders L. K., Nathans D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niu G., Shain K. H., Huang M., Ravi R., Bedi A., Dalton W. S., Jove R., Yu H. Overexpression of a dominant-negative signal transducer and activator of transcription 3 variant in tumor cells leads to production of soluble factors that induce apoptosis and cell cycle arrest. Cancer Res. 2001;61:3276–3280. [PubMed] [Google Scholar]
- 50.Sinibaldi D., Wharton W., Turkson J., Bowman T., Pledger W. J., Jove R. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene. 2000;19:5419–5427. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 51.de Koning J. P., Soede-Bobok A. A., Ward A. C., Schelen A. M., Antonissen C., van Leeuwen D., Lowenberg B., Touw I. P. STAT3-mediated differentiation and survival and of myeloid cells in response to granulocyte colony-stimulating factor: role for the cyclin-dependent kinase inhibitor p27Kip1. Oncogene. 2000;19:3290–3298. doi: 10.1038/sj.onc.1203627. [DOI] [PubMed] [Google Scholar]
- 52.Sasse J., Hemmann U., Schwartz C., Schniertshauer U., Heesel B., Landgraf C., Schneider-Mergener J., Heinrich P. C., Horn F. Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol. Cell. Biol. 1997;17:4677–4686. doi: 10.1128/mcb.17.8.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyder M., Huang X. Y., Zhang J. J. Identification of novel direct Stat3 target genes for control of growth and differentiation. J. Biol. Chem. 2008;283:3791–3798. doi: 10.1074/jbc.M706976200. [DOI] [PubMed] [Google Scholar]
- 54.Kidder B. L., Yang J., Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haviland R., Eschrich S., Bloom G., Ma Y., Minton S., Jove R., Cress W. D. Necdin, a negative growth regulator, is a novel STAT3 target gene down-regulated in human cancer. PLoS ONE. 2011;6:e24923. doi: 10.1371/journal.pone.0024923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iliopoulos D., Jaeger S. A., Hirsch H. A., Bulyk M. L., Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.