Abstract
Prochlorococcus and Synechococcus, which numerically dominate vast oceanic areas, are the two most abundant oxygenic phototrophs on Earth. Although they require solar energy for photosynthesis, excess light and associated high UV radiations can induce high levels of oxidative stress that may have deleterious effects on their growth and productivity. Here, we compared the photophysiologies of the model strains Prochlorococcus marinus PCC 9511 and Synechococcus sp. WH7803 grown under a bell-shaped light/dark cycle of high visible light supplemented or not with UV. Prochlorococcus exhibited a higher sensitivity to photoinactivation than Synechococcus under both conditions, as shown by a larger drop of photosystem II (PSII) quantum yield at noon and different diel patterns of the D1 protein pool. In the presence of UV, the PSII repair rate was significantly depressed at noon in Prochlorococcus compared to Synechococcus. Additionally, Prochlorococcus was more sensitive than Synechococcus to oxidative stress, as shown by the different degrees of PSII photoinactivation after addition of hydrogen peroxide. A transcriptional analysis also revealed dramatic discrepancies between the two organisms in the diel expression patterns of several genes involved notably in the biosynthesis and/or repair of photosystems, light-harvesting complexes, CO2 fixation as well as protection mechanisms against light, UV, and oxidative stress, which likely translate profound differences in their light-controlled regulation. Altogether our results suggest that while Synechococcus has developed efficient ways to cope with light and UV stress, Prochlorococcus cells seemingly survive stressful hours of the day by launching a minimal set of protection mechanisms and by temporarily bringing down several key metabolic processes. This study provides unprecedented insights into understanding the distinct depth distributions and dynamics of these two picocyanobacteria in the field.
Keywords: marine cyanobacteria, Synechococcus, Prochlorococcus, light/dark cycle, light stress, UV radiations, oxidative stress, photophysiology
Introduction
Phytoplanktonic cells, and in particular cyanobacteria, experience dramatic daily fluctuations of solar radiations, which can become suboptimal for photosynthetic processes around midday. Photosystem II (PSII) is particularly sensitive to these changes in photon fluxes and under unfavorable or stressful conditions its activity can decline more rapidly than most other physiological processes (Berry and Björkman, 1980; Demmig-Adams and Adams, 1992; Aro et al., 1993; Andersson and Aro, 2001). Photodamages to PSII are thought to start by the inactivation of the oxygen-evolving complex, which is caused by the dissociation of the Mn4Ca2+ cluster. This process leads to the production of long-lived P680+, the oxidized form of the reaction center chlorophyll (Chl) pair, a particularly strong oxidant which in turn provokes the destruction of the PSII core protein D1 (Hakala et al., 2006; Nishiyama, 2006). At low irradiances, the rate of photosynthetic electron transport is proportional to the photon flux density and damaged D1 polypeptides can be removed from the PSII reaction center and rapidly replaced by newly synthesized D1 proteins (Park et al., 1995; Tyystjarvi and Aro, 1996; Nixon et al., 2005; Ohnishi et al., 2005). However, at higher irradiances, the rate at which the PSII reaction center is damaged can exceed its repair rate, which results in an increase of inactivated PSII centers and a subsequent decline of the quantum yield of photosynthesis, resulting from photoinhibitory fluorescence quenching (Powles, 1984; Prásil et al., 1992; Aro et al., 1993; Andersson and Aro, 2001).
Although the visible part of the solar spectrum (400–700 nm), also called photosynthetically active radiations (PAR), is responsible for most photoinhibitory effects, the contribution of UV-B (280–315 nm) and, to a least extent, UV-A (315–400 nm) is also notable in the uppermost layer of the ocean (Dring et al., 2001; van de Poll et al., 2001; He and Häder, 2002b). UV-B can indeed damage the photosynthetic apparatus about 100-fold more efficiently than visible light and these radiations might directly affect PSII proteins and the Mn4Ca2+ cluster (Sarvikas et al., 2006; Caldwell et al., 2007). UV and high visible radiations can also cause indirect photoinhibitory effects via the production of reactive oxygen species (ROS; He and Häder, 2002a,b; Rastogi et al., 2010), mainly formed within reaction centers (Asada, 1999) and light-harvesting complexes (Knox and Dodge, 1985; Zolla and Rinalducci, 2002). ROS are powerful oxidizing agents which can react with DNA, lipids, and proteins. Although these compounds are inevitably produced by cell metabolism, even under optimal growth conditions, their production is drastically enhanced when cells are exposed to a variety of stresses, including excess visible light and UV radiations (UVR; Latifi et al., 2005; Ross et al., 2006; Houot et al., 2007; Allakhverdiev and Murata, 2008). The effect of ROS on PSII photoinhibition is thought to act primarily by inhibiting the de novo synthesis of proteins, including those required for the repair of PSII (Nishiyama et al., 2004; Nishiyama, 2006; Takahashi and Murata, 2008). A direct effect of ROS on the inactivation of PSII reaction center has also been suggested through triggering D1 degradation (Vass et al., 1992; Aro et al., 1993; Miyao et al., 1995; Keren et al., 1997; Lupinkova and Komenda, 2004). In any case, ROS clearly have a major role in light-mediated photoinhibition as well as in other environmental stresses (Nishiyama, 2006; Allakhverdiev and Murata, 2008; Latifi et al., 2009). Thus, survival of phototrophic organisms depends upon the amount of ROS produced and their efficiency in scavenging these oxygen species.
In this context, marine picocyanobacteria belonging to the genera Synechococcus and Prochlorococcus constitute two relevant and complementary models to study acclimation processes to high light and UVR and their interrelationships with oxidative stress. In oceanic ecosystems, these two organisms numerically dominate the phytoplanktonic community (Partensky et al., 1999a; Scanlan, 2003) and are considered to be the two most abundant photosynthetic organisms on Earth, with a substantial contribution to Chl biomass and primary production (Liu et al., 1997; Partensky et al., 1999a; Agawin et al., 2000; Garcia-Pichel et al., 2003). Members of the marine Synechococcus genus are ubiquitously distributed and are most abundant in coastal regions and mesotrophic open ocean surface waters (Partensky et al., 1999a; Zwirglmaier et al., 2008), whereas Prochlorococcus preferentially thrives in warm, stratified, oligotrophic tropical, and subtropical marine areas (Partensky et al., 1999b; Zubkov et al., 2000; Johnson et al., 2006). In the field, these organisms experience large variations in irradiance, linked to the combination of the light/dark (L/D) cycle, water mixing, and a variable cloudiness (MacIntyre et al., 2000). Moreover, their tiny size (0.5–0.8 and 0.8–1.2 μm diameter for Prochlorococcus and Synechococcus, respectively) confers them a high surface to volume ratio, optimizing their photon capture, and making them particularly sensitive to UVR (Llabres and Agusti, 2006, 2010).
Like other photosynthetic organisms, marine cyanobacteria have evolved a variety of protection mechanisms to ensure their growth and survival in highly illuminated habitats. These mechanisms include thermal dissipation of excess light excitation, structural changes of the photosynthetic machinery as well as enzymatic and non-enzymatic scavenging systems to eliminate ROS, in particular those produced in photosynthetic membranes (for reviews, see Bailey and Grossman, 2008; Latifi et al., 2009). However, several pieces of evidence suggest that Prochlorococcus and Synechococcus lineages could deal differently with light stress. Indeed, two P. marinus strains (PCC 9511 and SS120, a high light- and a low-light-adapted ecotype, respectively) were found to be more sensitive to a transient exposure to high irradiances than three Synechococcus spp. strains representative of various trophic environments and exhibiting different pigmentation (RS9917, RCC307, and WH8102; Six et al., 2007b). Similarly, measurements of cell abundances and/or mortality rates of field populations of picocyanobacteria exposed to different levels of natural solar radiations showed that Prochlorococcus exhibited a lower resistance to UVR than Synechococcus in surface waters of the central Atlantic Ocean (Llabres and Agusti, 2006; Agusti and Llabres, 2007) and the Mediterranean Sea (Sommaruga et al., 2005; Llabres and Agusti, 2010).
In order to reveal potential differences in circadian metabolic rhythms between these two genera, the photophysiology of the model strains P. marinus PCC 9511 and Synechococcus sp. WH7803 was examined at different times of a modulated L/D cycle of visible light (hereafter VL) with or without UV. Additionally, the diel variability of the sensitivity of Prochlorococcus and Synechococcus to oxidative stress, as triggered by different H2O2 concentrations was investigated. Expression of key genes involved in photosynthesis, light, and oxidative stress response and a number of other processes were also monitored in order to get insights about the molecular bases of the observed physiological differences.
Materials and Methods
Strains and culture conditions
The two model strains P. marinus PCC 9511 [a strain genetically very close, if not identical, to MED4 (Rippka et al., 2000; Rocap et al., 2003)] and Synechococcus sp. WH7803 (Scanlan et al., 2009) used in this study were grown at 22°C in PCR-S11 medium (Rippka et al., 2000), supplemented with 1 mM NaNO3. All experiments were performed under modulated L/D conditions using a computer-controlled illumination device (cyclostat), as detailed elsewhere (Holtzendorff et al., 2001; Kolowrat et al., 2010). This system allows the simulation of a bell-shaped 12/12 h L/D cycle, which induces a good synchronization of cell division, as observed for field populations (Vaulot et al., 1995; Jacquet et al., 1998). The maximal VL irradiance (at virtual noon) was set at 870 μmol photons m−2 s−1, corresponding to the reference light condition. In order to test the specific effects of UVR, the same experiments were repeated but with supplementing the VL condition with modulated UV-A and UV-B radiations (hereafter VL + UV), provided by UV-A-340 fluorescent lamps (Q-Panel Lab products, Cleveland, OH, USA). UVR reached 7.59 W m−2 UV-A (320–340 nm) and 0.57 W m−2 UV-B (280–320 nm) at virtual noon, corresponding to levels representative of natural doses measured in the upper layer of nutrient-poor, oceanic areas (Helbling et al., 1992). Two replicate cultures were acclimated to L/D cycles for at least 2 weeks prior to start monitoring the different parameters. During the experiment, two replicate cultures were grown with a continuous input of fresh medium, in order to maintain cells in exponential growth throughout the whole sampling period. One microliter aliquots were taken every hour, fixed for 10 min with grade I glutaraldehyde (0.25% final concentration; Sigma Aldrich, Saint-Louis, MO, USA), then frozen at −80°C for delayed analyses of cell abundance and cell cycle using a FACS Canto flow cytometer (Becton Dickinson Biosciences, San Jose, CA, USA), as previously described (Marie et al., 1997, 1999). Because of the continuous dilution, growth rate of these cultures was indirectly assessed from cell cycle data (μcc) using the method described by Carpenter and Chang (1988), as detailed earlier (Kolowrat et al., 2010). To study the kinetics of response to light fluctuations, cultures were sampled at 6, 9, 12, 15, 18, 20, 22, and 2 h over 3 days for measuring a variety of parameters described below.
Pigment and fluorescence measurements
Photosynthetic pigments were extracted in 95% methanol and analyzed by HPLC, as previously described (Everroad et al., 2006). Whole cell fluorescence emission spectra with excitation at 530 nm were recorded for Synechococcus using a LS-50B spectrofluorometer (Perkin Elmer, Waltham, MA, USA). The PSII quantum yield (FV/FM) was measured using a Pulse Amplitude Modulated fluorimeter (PhytoPAM, Walz, Effeltrich, Germany) connected to a LabPro chart recorder allowing the direct visualization of fluorescence traces (Vernier, Beaverton, OR, USA). A 2 mL aliquot was dark acclimated for 5 min in a quartz cuvette with a mirrored facet facing the photomultiplier to enhance the signal. The modulated light was then turned on to measure the basal fluorescence level F0 with modulated excitation at 440 nm for Prochlorococcus cells and 520 nm for Synechococcus. The maximal fluorescence FM was determined by applying a light saturating pulse in the presence of 50 μM of the PSII inhibitor 3-(30,4-dichlorophenyl)-1,1-dimethylurea (DCMU) under ca. 2,000 μmol photons m−2 s−1. The PSII quantum yield was calculated as:
| (1) |
where FV is the variable fluorescence.
Photosystem II repair
At each sampling time point, a 20 mL volume of each replicate culture was sampled and split into two 100 mL quartz Erlenmeyer flasks, and one of them was supplemented with 500 μg mL−1 lincomycin, an inhibitor of protein translation (Six et al., 2007a). The two flasks were immediately brought back into the culture system and 2 mL aliquots were collected at 0, 15, 30, and 60 min after the sampling to measure the PSII quantum yield, as described above. The PSII quantum yield was then plotted over time for both the control and lincomycin-treated sub-cultures and plots were fitted with an exponential decay function. The PSII repair rate was estimated as the difference between the exponential decay rates in the absence and presence of lincomycin (Six et al., 2007a, 2009; Campbell and Tyystjarvi, 2012).
Immunoblotting
Cell pellets were resuspended in extraction buffer (140 mM Tris base, 105 mM Tris-HCl, 0.5 mM ethylenediaminetetraacetic acid, 2% lithium dodecyl sulfate, 10% glycerol, and 0.1 mg mL−1 PefaBloc protease inhibitor (Roche, Basel, Switzerland) and total protein concentration was determined using a Lowry protein assay kit (Bio-Rad, Hercules, CA, USA) and bovine serum albumin as protein standards. Samples were then denatured with 50 mM dithiothreitol and heated for 2 min at 80°C and 2 μg total protein was loaded on a 4–12% gradient acrylamide precast NuPAGE Bis-Tris mini-gel (Invitrogen, Carlsbad, CA, USA) along with recombinant standards of D1 or D2 proteins (Agrisera, Vännäs, Sweden) to establish a standard curve. Gels were electrophoresed and the proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane, then immediately immersed into Tris Buffer Saline-Tween (TBS-T) buffer, pH 7.6 (0.1% Tween 20, 350 mM sodium chloride, 20 mM Trizma base) containing 2% (w:v) ECL Advance blocking agent (Amersham Biosciences, Piscataway, NJ, USA). Aliquots of primary antibodies against D1 or D2 proteins (Agrisera) were diluted at 1:50 000 in TBS-T in the presence of 2% blocking agent and membranes were soaked into this solution for 1 h with agitation. After extensive washing of the membrane with TBS-T buffer, anti-rabbit secondary antibodies were applied with the same procedure as for primary antibodies. Membranes were developed by chemoluminescence using the ECL Advance reagent kit (Amersham Biosciences) and visualized with a LAS4000 imager equipped with a CCD camera (GE Healthcare, Waukesha, WI, USA). Signals were quantified using the ImageQuant software and the recombinant protein standard curve. To ease comparisons of the diel trends between strains and conditions, data were normalized at 6:00 am.
Oxidative stress assays
Samples collected from dawn to dusk (6, 9, 12, 15, and 18 h time points) were split into aliquots of 2.5 mL and subjected to a series of H2O2 concentrations ranging from 0.1 to 1,800 μM. These culture aliquots were then incubated in the culturing system for 50 min and the PSII quantum yield was measured as described above. To quantitatively estimate the PSII resistance to such oxidative stress at each time point, the decay of FV/FM as a function of increasing H2O2 concentration was plotted as a percentage of FV/FM of the control cultures. The decay curves were then fitted with a two-parameter hyperbolic decay function:
| (2) |
The b-value proved to be informative because it integrates both the minimal H2O2 concentration needed to cause a decrease of FV/FM and the decrease rate of the PSII quantum yield. Thus, this parameter can be used as a proxy to assess the global resistance of PSII to oxidative stress induced by H2O2.
RNA extraction and real time quantitative PCR
Samples for RNA extraction were harvested and extracted as previously described in Kolowrat et al. (2010) for eight data points per L/D cycle and for two different days, corresponding to biological replicates. Briefly, cell pellets, resuspended in Trizol (Invitrogen, Carlsbad, CA, USA), were extracted using the miRNeasy kit as recommended by the manufacturers (Qiagen, Valencia, CA, USA), followed by two successive DNase treatments performed on the miRNeasy columns using the Qiagen RNase-free DNase Set (Qiagen).
Real time quantitative PCR (hereafter qPCR) was performed on a set of P. marinus PCC 9511 and Synechococcus sp. WH7803 genes, representative of key metabolic processes (Table S1 in Supplementary Material). Design and optimization of gene specific primers were performed as previously described (Six et al., 2007b) using PrimerExpress™ software v2.0 (Applied Biosystems) and by checking for every set of primers, the linearity of the CT (cycle at threshold) vs. cDNA content within a dilution range of cDNA. Reverse transcription was carried out on 100 ng RNA using SuperScriptII reverse transcriptase (Gibco-BRL, Gaithersburg, MD, USA). qPCR was performed in triplicate on the cDNA obtained after dilution, using the DNA Engine/Chromo4 Real Time PCR-Detector (Bio-Rad, Hercules, CA, USA) and the absolute SYBR Green ROX Mix (Abgene, Epsom, UK), as previously described (Garczarek et al., 2008). Gene expression profiles monitored during L/D cycles were expressed as the ratio of gene expression vs. expression of the aperiodic gene rnpB (Mary and Vaulot, 2003; Zinser et al., 2009; Kolowrat et al., 2010) and normalized to the 6:00 time point sampled under VL conditions, using the 2−ΔΔCT method (Schmittgen and Livak, 2008).
Results
Cell cycle
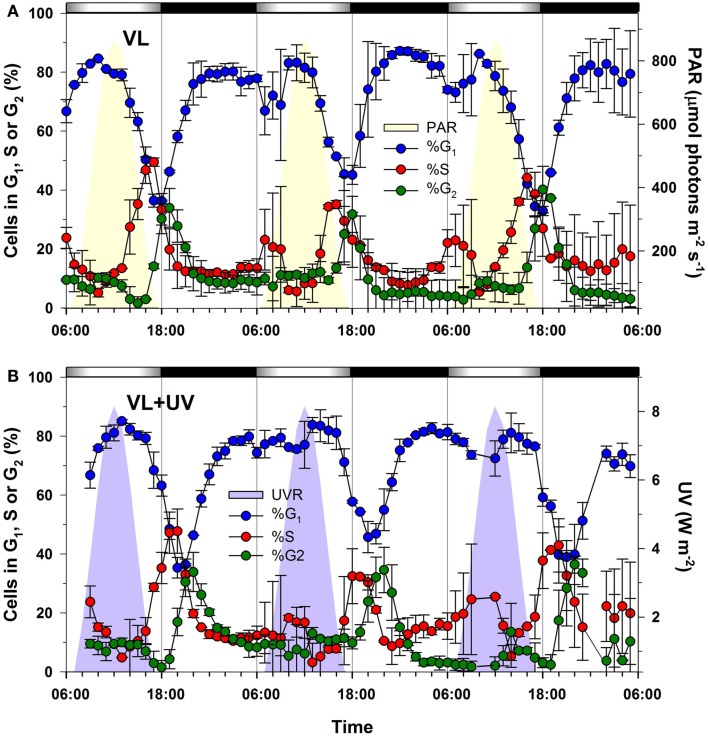
The cell cycle of Synechococcus sp. WH7803 was strongly synchronized by the alternation of light and darkness, with a peak of DNA replicating cells (S cells) occurring at ca. 16–17:00, i.e., 1–2 h before the light-to-dark transition (LDT; 18:00), in modulated VL only and about 3 h later for cells acclimated to modulated VL + UV (Figure 1). A comparable delay of the S phase was previously reported for P. marinus PCC 9511 in the presence of UVR, but the peak of S cells occurred at the LDT in VL and about at the same time as for Synechococcus in VL + UV (Kolowrat et al., 2010). Another notable difference with Prochlorococcus is the occurrence of a second minor S peak in the early morning followed by a small bump of G2 cell abundance around noontime (mainly visible during the first and third cycle), suggesting some ultradian growth (Shalapyonok et al., 1998). Mean growth rates of Synechococcus cultures were assessed from the percentages of cells in S and G2 (μcc) using the method described by Carpenter and Chang (1988). They were not statistically distinct between the two conditions (average over 3 days and two replicates: μcc = 1.00 ± 0.21 day−1 in VL; μcc = 1.23 ± 0.11 day−1 in VL + UV) but were higher than in Prochlorococcus (μcc = 0.67 ± 0.05 day−1 in VL; μcc = 0.68 ± 0.03 day−1 in VL + UV; Kolowrat et al., 2010). This difference is attributable in part to the shorter delay observed between the maxima of S and G2 cells in Synechococcus (tG2 − tS ∼ 2.5 h) than in Prochlorococcus (tG2 − tS ∼ 4 h), as this parameter is used for the calculation of growth rates in the μcc method (Carpenter and Chang, 1988).
Figure 1.
Effect of UV exposure on the timing of the cell cycle phases of Synechococcus sp. WH7803 cells grown over a modulated 12/12 h L/D cycle with or without UV radiations. (A) Distribution of G1 (blue), S (red), and G2 (green) phases for cells acclimated to VL. (B) Same for VL + UV conditions. Error bars indicate mean deviation for two biological replicates. Note that only the total UVR (UV-A + UV-B) plot is shown in graph (B) since PAR was the same as in graph (A). White and black bars above graphs indicate light and dark periods, also delineated by gray vertical bars and areas filled in yellow (PAR) or purple (UV). Abbreviations: L/D, light/dark; PAR, photosynthetically available radiations; VL, visible light; UV, ultraviolet.
Pigment ratios
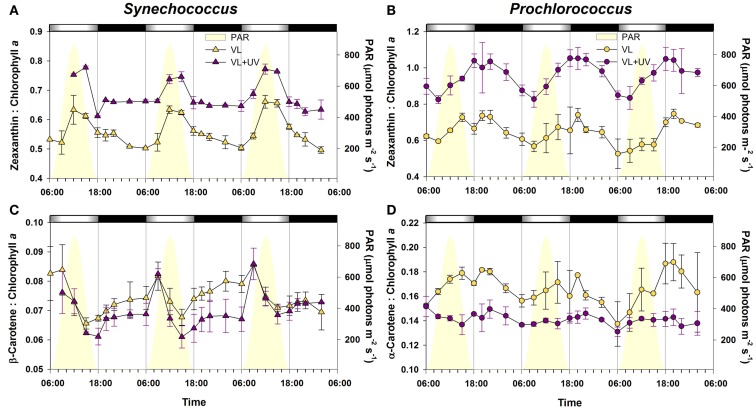
Comparative diel variations of molar ratios of the main pigments in the two marine picocyanobacteria under both light conditions are reported in Figure 2. For Synechococcus grown under VL, the zeaxanthin (Zea) to Chl a ratio (Figure 2A) was maximal between noon and the early afternoon, and then decreased sharply till the LDT and at a lower rate during the night. In VL + UV, the pattern was globally similar, except that (i) the values were higher in VL + UV than in VL, (ii) the midday peak had a lower average amplitude (15.4 vs. 25.4% increase under VL + UV and VL, respectively), and (iii) the ratio remained stable during the night. By comparison, the β-carotene (β-Car) to Chl a ratio of Synechococcus grown in VL systematically peaked at 9:00, decreased till 15:00 then increased again for the rest of the diel cycle (Figure 2C). The pattern was very similar under UV, except that night values were slightly lower during the first two L/D cycles. The pattern of pigment ratios observed in Prochlorococcus exhibited a number of differences compared to Synechococcus. In VL, the Zea to divinyl-(DV-) Chl a ratio started to increase at mid-morning and was maximal around 20:00, then decreased during the rest of the night (Figure 2B), corresponding to the cell division phase. The same diel pattern was observed under VL + UV. Although the variation range was moderate for the α-Car to DV-Chl a ratio in VL, its diel pattern was comparable to that of the Zea to DV-Chl a, except that it increased immediately after dawn (Figure 2D). There were almost no diel oscillations of this ratio under VL + UV. The DV-Chl b to a ratio exhibited no clear diel pattern in either light condition (data not shown).
Figure 2.
Daily variations of the ratios of the two major carotenoids (zeaxanthin and carotene) to chlorophyll a for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of VL with or without UV radiations. (A,C) Synechococcus sp. WH7803. (B,D) Prochlorococcus marinus PCC 9511. White and black bars above graphs indicate light and dark periods, also delineated by vertical bars and areas filled in yellow. Error bars indicate mean deviation for two biological replicates. Abbreviations as in Figure 1.
Photosystem II function and repair
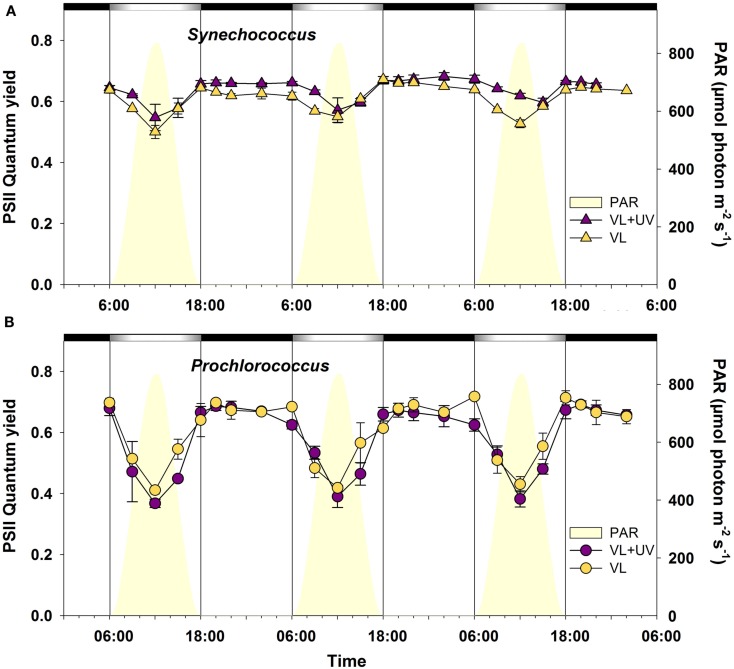
In order to investigate how the daily variations of VL and UVR could affect the PSII activity of cells, we followed the PSII quantum yield in Synechococcus sp. WH7803 and P. marinus PCC 9511 cultures during three consecutive L/D cycles (Figure 3). For both strains and under both light conditions, the FV/FM ratio showed a cyclic evolution, reaching a maximum at night and a minimum at virtual noon. This midday drop, originating either from PSII photoinactivation or from dissipative non-photochemical quenching (NPQ) of fluorescence (or both), was larger for Prochlorococcus than for Synechococcus under both light conditions. Indeed, there was a relative decrease of the PSII quantum yield of about 40% for Prochlorococcus, while in Synechococcus it never exceeded 20% (Figure 3). The diel patterns of PSII activities showed also some differences between VL and VL + UV, with slightly higher late night and morning yields for Synechococcus and lower yields at 6:00, 12:00, and 15:00 for Prochlorococcus cells in the latter condition.
Figure 3.
Daily variations of the photosystem II maximal quantum yield (FV/FM) for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of VL with or without UV radiations. (A) Synechococcus sp. WH7803, (B) Prochlorococcus marinus PCC 9511. White and black bars above graphs indicate light and dark periods, also delineated by vertical bars and areas filled in yellow. Error bars indicate mean deviation for two biological replicates. Abbreviations as in Figure 1.
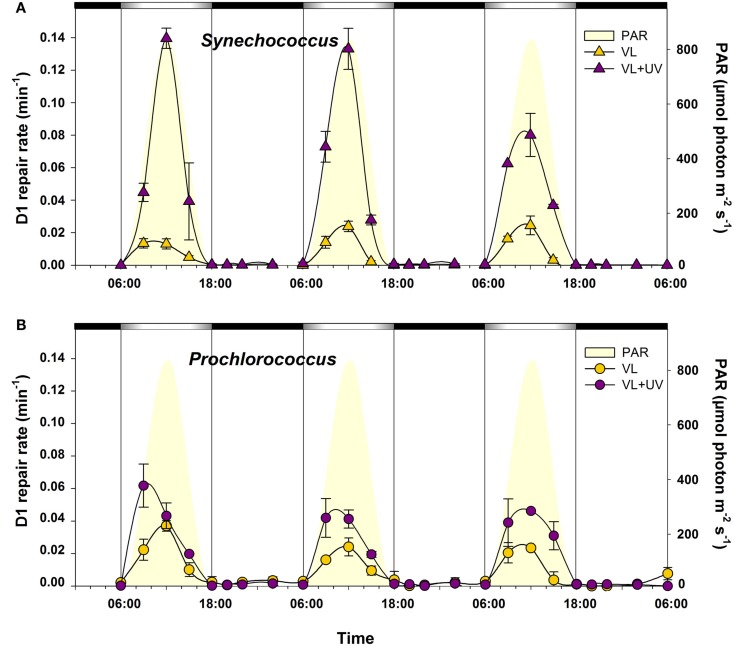
Variations in the PSII repair activity of the cells, as assessed by comparing the PSII quantum yield of sub-cultures incubated with or without lincomycin, an inhibitor of protein synthesis, also showed a cyclic pattern, with no measurable PSII repair occurring during the night (Figure 4). Moreover, cells acclimated to VL and VL + UV conditions exhibited a very different repair rate, especially in Synechococcus. While VL exposure only led to a moderate induction of the PSII repair rate with regard to night levels in Synechococcus, a sharp increase of this rate (from three- to six-fold) was observed for cells acclimated to UV-supplemented light (Figure 4A). Such an UV-induced increase of the daily PSII repair rate was also observed in Prochlorococcus cells, although much more limited (less than twofold), and the maximal rate were already reached at 9:00 (Figure 4B).
Figure 4.
Daily variations of the photosystem II repair activity for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of VL with or without UV radiations. (A) Synechococcus sp. WH7803, (B) Prochlorococcus marinus PCC 9511. White and black bars above graphs indicate light and dark periods, also delineated by vertical bars and areas filled in yellow. Error bars indicate mean deviation for two biological replicates. Abbreviations as in Figure 1.
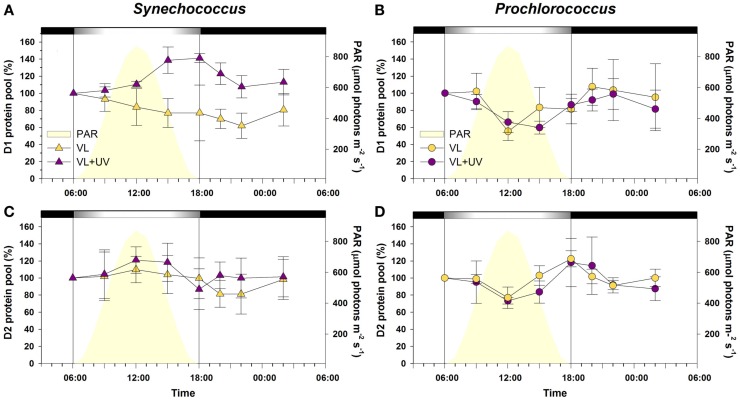
Photosystem II core protein pools
In Synechococcus cells grown under VL, both the D1 and D2 protein pools showed no significant diel oscillations during the L/D cycle (Figure 5). Under VL + UV, while D2 also remained stable, cells progressively accumulated D1 proteins during daytime and the pool reached a maximum at the LDT (40% increase compared to the value at 6:00). Prochlorococcus cells showed a quite different diel pattern of D1 and D2 protein pools. Under VL, both proteins showed minimal contents during the day (30–40% decrease), when irradiance was maximal, with a subsequent full recovery during the night. A similar pattern was observed under VL + UV, but with an extended period of low PSII core protein content.
Figure 5.
Daily variations of the total pools of photosystem II core proteins D1 and D2 for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of high visible light with or without UV radiations. (A,C) Synechococcus sp. WH7803, (B,D) Prochlorococcus marinus PCC 9511. White and black bars above graphs indicate light and dark periods, also delineated by vertical bars and areas filled in yellow. Data represent the mean ± standard deviation (n = 3–4) of two biological replicates and two consecutive days. Abbreviations as in Figure 1.
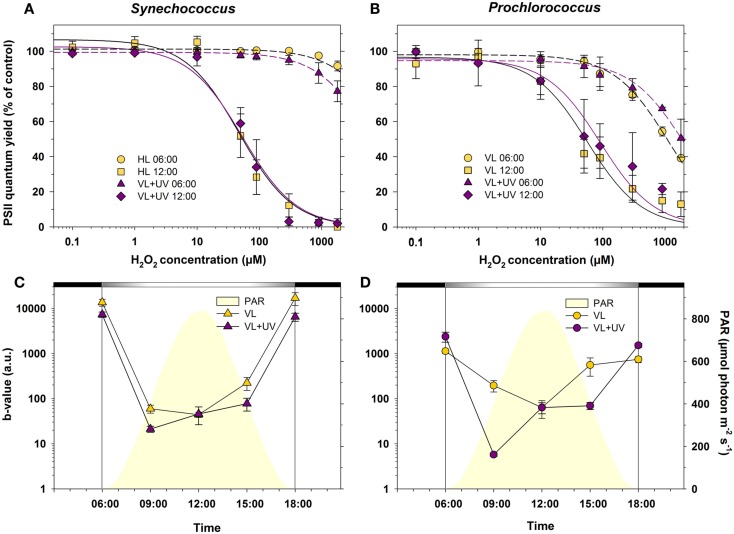
Resistance of PSII to oxidative stress
Synechococcus and Prochlorococcus grown under VL and VL + UV conditions exhibited quite different PSII responses to oxidative stress (Figure 6). Synechococcus sub-cultures sampled at 6:00 (i.e., at the dark-to-light transition) showed no significant change in their PSII quantum yield until H2O2 concentrations as high as 300 and 1,800 μM in VL + UV and VL, respectively. Sub-cultures sampled at 12:00 were affected by concentrations as low as 10 and 50 μM H2O2, the photoinhibitory effect increasing with the concentration of oxidizing agent until a total shutdown of PSII activity at 300 and 900 μM H2O2 for VL + UV and VL, respectively (Figure 6A). PSII inactivation occurred at lower doses in Prochlorococcus at both time points, starting at about 100 μM H2O2 for cultures collected at 6:00 and reaching 60 and 50% of photoinhibition at 1,800 μM H2O2 in VL and VL + UV, respectively (Figure 6B), compared to less than 20% for Synechococcus cells at the same time point. Furthermore, as for Synechococcus, Prochlorococcus sub-cultures sampled at noon were sensitive to lower H2O2 concentrations than cultures sampled at 6:00.
Figure 6.
Daily variations of photosystem II (PSII) resistance to oxidative stress for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of high visible light with or without UV radiations. (A,B) Maximal PSII quantum yield (FV/FM) measured at 06:00 and at noon, after 50 min exposure to a range of H2O2 concentrations of Synechococcus sp. WH7803 (A) or Prochlorococcus marinus PCC 9511 cells (B). Data were fitted with a two-parameter hyperbolic decay function. (C,D) PSII resistance to oxidative stress in Synechococcus (C) and Prochlorococcus (D) cells, as estimated by the b-value (see Materials and Methods). White and black bars above graphs (C,D) indicate light and dark periods, also delineated by vertical bars and areas filled in yellow. Data represent the mean ± standard deviation (n = 4) of two biological replicates and two consecutive days. Abbreviations as in Figure 1.
In order to compare the rates of decay of the PSII quantum yield obtained at various times of the day, FV/FM vs. H2O2 concentration curves for all time points (only those obtained for 6:00 and 12:00 are shown in Figures 6A,B) were fitted with a two-parameter hyperbolic decay function. The daily variations of the b-value, characterizing the global resistance of PSII to oxidative stress induced by H2O2 (see Materials and Methods), are shown in Figures 6C,D. As expected from results at 6:00 and 12:00 (Figures 6A,B), PSII proved to be much more sensitive to an artificially induced oxidative stress during the day than during the night in both picocyanobacteria. Still, Synechococcus PSII was seemingly much more resistant than Prochlorococcus to this stress, when applied in the dark. It is also worth noting that the sensitivity to oxidative stress was enhanced for UV-acclimated cells, especially in Prochlorococcus, which displayed a dramatic drop of the b-value at 09:00 compared to those grown in VL only. However, after this sharp mid-morning drop, the b-value rose again during the day in both strains.
Transcriptomic response
Photosynthesis
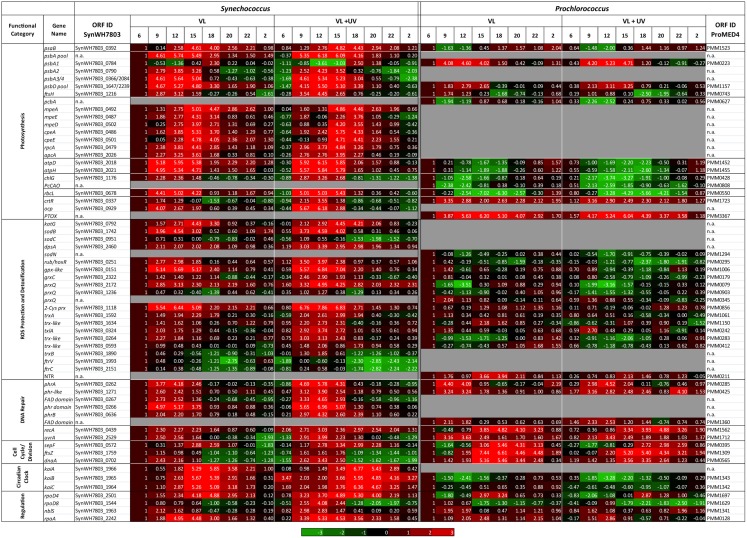
The diel expression pattern of a selection of genes involved in photosynthesis and a number of other processes related to (or affected by) light or UV stress were analyzed by qPCR in P. marinus PCC 9511 and Synechococcus sp. WH7803 strains, grown under both VL and VL + UV conditions (Figure 7). As expected, photosynthetic genes were among those showing the strongest diel oscillations. In both strains, the daily variations of the total pools of psbA and psbD transcripts, encoding the two major subunits of the reaction center II (D1 and D2, respectively), closely matched the modulated fluctuations of growth irradiance, with a slight increase in the relative amplitude of the peak of psbA transcripts under UVR. It is worth noting that in the case of Synechococcus, which possesses four psbA genes (compared to only one in P. marinus MED4/PCC9511; Garczarek et al., 2008), this global expression pattern translates the temporal succession of the single copy gene encoding the D1.1 isoform (SynWH7803_0784) and the three copies encoding the D1.2 isoforms (SynWH7803_0790, 0366, and 2084), the former being repressed at noon and maximum at the LDT, while the three latter exhibited an opposite behavior (note that the expression levels of SynWH7803_0366 and 2084 cannot be measured separately). Accordingly, the expression pattern of PMM0743 and SynWH7803_1216, encoding the FtsH2 protease that is involved in the clearance of damaged D1 proteins from inactivated PSII (Komenda et al., 2006, 2010), showed a broad maximum during the period of highest photon fluxes (9:00–15:00) in both species. However, in Prochlorococcus, this gene showed a minimum at dusk, then a continuous increase during the dark period and the early morning, while in Synechococcus its relative expression levels were minimal all over the dark period from 18:00 to 6:00.
Figure 7.
Daily variations of the relative expression of selected genes, as measured by quantitative real time PCR, for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of high visible light with or without UV radiations. All data, expressed as log2 (Fold Change), were normalized to the value observed at 06:00 in the VL condition for each strain and are shown as a range of red (relative upregulation) or green (relative downregulation) colors, with intensity depending on gene expression levels, as indicated in the scale bar below the data. Genes are classified by functional categories, as indicated in the first column. A more detailed listing of these data showing mean deviation for two biological replicates as well as gene products is provided as Table S2 in Supplementary Material.
In contrast to reaction center II genes, the diel expression pattern of psaB, encoding one of the two major subunits of the PSI core, was very different between the two cyanobacteria. While in Synechococcus its expression level peaked between 15:00 and 18:00 and remained low for the rest of the L/D cycle, Prochlorococcus psaB transcript level was at its lowest around noontime and was maximal during the night period. Genes encoding light-harvesting systems also behaved quite differently between Prochlorococcus and Synechococcus. In the former organism, pcbA transcripts displayed a comparable pattern during the day to that of psaB transcripts with a significant downregulation in both light conditions, though the relative level of expression was even lower at noon for UV-irradiated cells. In contrast, for Synechococcus in VL, all genes encoding the different phycobiliprotein α-subunits (apcA, rpcA, cpeA, and mpeA) and phycoerythrin (PE) linkers (cpeE, mpeD, mpeE) were highly upregulated during the light period, with maximal expression at 15:00. While the expression patterns of genes coding for allophycocyanin, phycocyanin, and PEI subunits and their linkers were little affected by UVR, the expression of PEII genes showed a drop at noon. Interestingly, the mpeE gene, encoding the linker binding the distal PEII disk in this strain (Six et al., 2007c), even exhibited a 3-h delay of its maximal expression level in VL + UV compared to VL only.
Pigment biosynthesis genes also showed large variations of relative expression over the day under both light conditions. The chlG gene, encoding the chlorophyll a synthase, also behaved in an opposite way between Prochlorococcus and Synechococcus, with minimal and maximal expression levels at noon, respectively, in both cases more marked for UV-acclimated cells. Accordingly, the diel expression pattern of PcCao, a Prochlorococcus-specific gene encoding the chlorophyll b synthase (Satoh and Tanaka, 2006), also somewhat mirrored diel variations of growth irradiances. In contrast, the ctrR gene, encoding the β-Car hydroxylase that catalyzes the last biosynthesis step of zeaxanthin, usually considered as a photoprotective pigment, was maximally expressed during daytime in both organisms, but with a different global pattern, since in Prochlorococcus there was a strong surge in expression levels at 9:00 am followed by a progressive decline for the rest of the day, whereas in Synechococcus the diel pattern again closely matched the growth irradiance oscillations.
Most marine Synechococcus strains, including WH7803, contain the ocp gene, which encodes the orange carotenoid protein (OCP), a pigment-protein complex likely located between the phycobilisomes (PBS) and the PSII reaction center and involved in the dissipation of excess energy as heat (Boulay et al., 2008; Kirilovsky and Kerfeld, 2012). In VL, WH7803 ocp was maximally expressed during the day with a peak at 9:00, whereas the expression maximum was considerably higher and broadened toward noontime in UV-acclimated cells, suggesting a key role of this gene in the protection against UVR. Although all Prochlorococcus strains lack ocp, due to the absence of functional PBS, high light-adapted Prochlorococcus strains (including MED4/PCC9511), low-light-adapted ecotype LLI (Scanlan et al., 2009) as well as a few marine Synechococcus strains (though not WH7803) possess another potentially important gene for photoprotection, ptox, which encodes a plastid terminal oxidase. This enzyme was suggested to extract electrons from the electron transport chain between PSII and PSI and to re-oxide the plastoquinone pool reduced by PSII (Bailey et al., 2008; Berg et al., 2011). Interestingly, ptox exhibited very large variations in relative expression with a maximum at 15:00 (Figure 7), though contrary to ocp, its diel expression pattern was virtually identical between VL and VL + UV.
The ATPase genes atpD et atpH showed similar expression patterns in Synechococcus with high values from 9:00 to 15:00 in VL and even higher under VL + UV, whereas expression of these genes decreased all over the day in Prochlorococcus followed by a night recovery (Figure 7; see also Figure S3 in Kolowrat et al., 2010). An opposite behavior between the two strains was also observed for the rbcL gene, coding for the large subunit of RuBisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase), the major enzyme of the carbon fixation process, except that in Prochlorococcus, there was a much sharper drop of expression levels than for atp genes at 15:00 in VL, and at 18:00 under VL + UV (Figure 7).
The strikingly different diel expression patterns observed between Prochlorococcus and Synechococcus for genes involved in light-harvesting, ATP formation, as well as the light-independent reactions of photosynthesis (Calvin–Benson cycle) suggest that most photosynthetic processes are probably controlled by distinct regulation networks in these two cyanobacteria, despite their close phylogenetic relatedness.
Redox and ROS detoxification
Most genes involved in the regulation of redox state and ROS detoxification pathways that are shared by Synechococcus sp. WH7803 and P. marinus PCC 9511 were differentially regulated during the L/D cycle in both picocyanobacteria and their diel expression patterns were comparable in VL and VL + UV conditions. However, the daily amplitudes of variations of ROS genes were often much larger in Synechococcus than in Prochlorococcus. Three of the most differentially expressed genes in the former organism, i.e., those coding for rubredoxin (rub), glutathione peroxidase (SynWH7803_0151), and a 2-cys peroxiredoxin (2-Cys prx), had their maximal daily expression around noontime, again suggesting a tight regulation by light, and their relative expression level significantly increased under UV. In contrast, in Prochlorococcus, orthologs of these three genes (PMM0295, PMM1006, and PMM0856, respectively) showed no clear diel pattern. Interestingly, for each cyanobacterium, the different peroxiredoxin gene copies exhibited diel patterns very distinct from one another.
Synechococcus sp. WH7803 and P. marinus PCC 9511 also possess specific sets of genes involved in ROS-scavenging systems. Among these strain-specific genes, it is worth noting that the catalase-peroxidase gene katG of Synechococcus was notably upregulated during the day, with a peak at 15:00, while Prochlorococcus PMM0211, a NADPH-dependent thioredoxin-disulfide reductase (NTR system) was by far the most differentially expressed ROS-scavenging gene of this organism, with a maximal expression at 18:00 in VL. At last, while the only sod gene present in Prochlorococcus, sodN, encoding a Ni-binding superoxide dismutase (SOD), was slightly downregulated at noon relative to nighttime in VL and VL + UV, Synechococcus sodB, encoding a Fe-binding SOD, was in contrast strongly upregulated during the light period under both conditions. The second sod gene of Synechococcus, sodC that encodes a Cu/Zn-binding SOD, showed only faint diel variations of its expression in VL, but was slightly downregulated at 20:00 in VL + UV.
Cell cycle, DNA repair, and circadian clock
In Synechococcus sp. WH7803, the DNA replication initiation factor gene dnaA was upregulated during the day with a broad peak of expression during the 9:00–15:00 time period in both light conditions. Subtle differences can however be noted between the two conditions, since the expression maximum seemingly occurred at 9:00 in VL and at noon in VL + UV (Figure 7). The ftsZ gene, which controls the biosynthesis of the Z-ring formed in the middle of the cell prior to cell division, showed very similar diel expression patterns to dnaA, except for systematically lower amplitudes of variations over the L/D cycle. The last cell cycle gene that was examined, sepF, which codes for a protein interacting with FtsZ during septum formation, showed a bell-shaped diel expression pattern peaking at 15:00 in both light conditions, though relative mRNA levels were again slightly higher in UV-acclimated cells.
In P. marinus PCC 9511 grown in VL, the ftsZ gene expression level peaked at 15:00, whereas dnaA and sepF both reached a maximum 3 h later. The relative expression levels of all three genes were considerably lower during the day when cells were grown under VL + UV, as previously reported (Kolowrat et al., 2010).
Since UVR are known to have deleterious effects on DNA structure, we also examined the diel expression patterns of a few genes involved in DNA repair pathways. The recA gene, which encodes an ATPase involved in the repair of double-strand breaks (DSBs) by homologous recombination (Chen et al., 2008), showed a broad maxima during daytime in Synechococcus cells grown in VL, while the relative expression of this gene was enhanced under VL + UV and remain high until 20:00 (Figure 7). In contrast, in Prochlorococcus, the recA expression peak occurred at the LDT in VL and was delayed by 2 h in the presence of UVR. The uvrA gene, encoding one subunit of the excinuclease UvrABC, an enzyme of the nucleotide excision DNA repair (NER) pathway, exhibited a sharp increase of its expression level during the lit period in both picocyanobacteria, with a peak at noon under both VL and VL + UV conditions. However, the maximal uvrA expression level was higher in UV-acclimated Synechococcus cells than in VL, whereas in Prochlorococcus, patterns were comparable between the two conditions (except at 9:00; Figure 7; Kolowrat et al., 2010).
Both picocyanobacteria also possess a family of genes related to DNA photolyases. In Synechococcus, the pattern of the photolyase gene phrA (Ng et al., 2000) closely matched the L/D cycle, with a peak at noon, which was threefold higher in VL + UV with regard to VL only (Figure 7). On the contrary, in Prochlorococcus, the phrA expression peak heights were similar in the two light conditions, even though the relative phrA mRNA level was higher at mid-morning in VL than VL + UV (Kolowrat et al., 2010). Like other marine Synechococcus but unlike Prochlorococcus, WH7803 possesses a second member of the DNA photolyase family (PhrB), which was recently suggested to be a cryptochrome (Goosen and Moolenaar, 2008). Interestingly, its diel expression pattern was quite similar to phrA, but with a reduced amplitude of variation of the relative mRNA amounts during daytime, even if there was again a very strong relative increase of the expression maximum under UV. The same was observed for a gene encoding an uncharacterized photolyase relative in Synechococcus (SynWH7803_1271), while its ortholog in Prochlorococcus (PMM0425) showed no clear diel pattern. Other, shorter members of the Phr family were also analyzed. Indeed, Synechococcus possesses a cluster of two genes (SynWH7803_0266 and 0267), each of which corresponds to one of the two domains of photolyases (photolyase and FAD-binding domains, respectively). So, once translated, they might potentially form an additional heterodimeric photolyase complex, as suggested by their diel pattern similar to phrA and high relative expression level (Figure 7). Interestingly, Prochlorococcus also possesses a gene (PMM1360) encoding a FAD-binding domain-containing protein, but surprisingly no gene coding for the other half of a photolyase (Scanlan et al., 2009; Partensky and Garczarek, 2010). Again, this gene had a very similar diel pattern as phrA from the same organism but with a much lower expression level.
The diel expression of genes involved into the circadian clock machinery was also studied given their central role in controlling global rhythmic transcriptional activity of the cells. While all marine Synechococcus possess the three kai genes coding for the core circadian oscillator, Prochlorococcus lacks kaiA and its circadian clock rather behaves like an “hourglass” that is reset every morning (Holtzendorff et al., 2008; Axmann et al., 2009). Interestingly, kai genes had a completely different behavior between the two cyanobacteria. In VL, all three kai genes of Synechococcus sp. WH7803 exhibited large diel variations with a broad maximum relative expression level at the end of the light period (15:00–18:00), while in Prochlorococcus, the expression of kaiB was minimal at noon, then increased continuously until dawn and there was no significant diel oscillation of kaiC mRNA. Presence of UVR caused a strong decrease of the level of expression of kaiABC genes at 15:00 in Synechococcus leading to a sharpening of their expression peaks, whereas in Prochlorococcus, the kaiB gene was further downregulated at noon without time shift. We also looked at the expression of rpaA, which codes for a DNA-binding protein acting as the response regulator of the KaiC-interacting kinase SasA, a two-component system that mediates the diel oscillations generated by KaiC phosphorylation to global transcription rhythms (Takai et al., 2006). RpaA is also known to be involved in the regulation of energy transfer from PBS to PSI and to interact with ferredoxin (Ashby and Mullineaux, 1999; Hanke et al., 2011). In both organisms, rpaA was maximally expressed around noontime and UVR had no significant effect on its diel transcription pattern.
Two genes encoding type II σ factors (rpoD4 and rpoD8), that have been shown to modulate gene expression under different conditions and to respond to L/D stimuli (Imamura et al., 2003; Summerfield and Sherman, 2007), were also analyzed to assess their potential role in controlling global gene expression during L/D transitions. In Synechococcus, rpoD4 exhibited a comparable diel expression pattern in VL and VL + UV, with a continuous increase during the day, a peak at the LDT then a progressive night decrease. In contrast, rpoD8 showed only mild variations in VL while its diel pattern was prominent in UV-acclimated cells, with a strong upregulation during the day and a downregulation during the night. In Prochlorococcus, the pattern of these genes has been previously described (Kolowrat et al., 2010). Briefly, rpoD8 has a low maximal expression in the mid-morning to noon period, whereas rpoD4 peaked at the end of the light period (see also Figure 7).
At last, we looked at the expression of nblS, encoding a membrane-bound, sensor histidine kinase involved in the control by light of the expression of several photosynthesis-related genes, including all three psbA genes, the cpcBA operon, and some hli genes (van Waasbergen et al., 2002; Kappell et al., 2006). In VL, this gene was upregulated during the day with a peak in the morning and its maximum expression level increased almost twofold in the presence of UVR (Figure 7).
Discussion
Differences in photosystem activity and regulation between Prochlorococcus and Synechococcus
The alternation of light and darkness is one of the most predictive events that cyanobacteria have to deal with in the field. Strong variations of PAR occur over a daily timescale and are associated, in near surface waters, with concomitant changes of UVR fluxes. Here, we compare PCC9511, a strain representative of the Prochlorococcus HLI clade found in near surface oligotrophic waters, to WH7803, a Synechococcus strain characteristic of mesotrophic areas. Although these model strains do not represent the whole physiological diversity existing within these two genera, our data clearly show that both Prochlorococcus and Synechococcus cells are able to tune their photosynthetic apparatus to diurnal irradiance fluctuations. However, Prochlorococcus proved to be more sensitive than Synechococcus to photoinhibition by high photon fluxes, as suggested by a marked drop of the cellular pool of PSII core proteins (Figure 5) and a larger decrease of the PSII quantum yield (Figure 3) around noontime. It is noteworthy however that the latter phenomenon might also partly be due to NPQ of PSII fluorescence associated with photoprotective dissipation of light energy as heat (Bailey et al., 2005; Boulay et al., 2008). The diel changes of the photosynthetic activity observed here for P. marinus PCC 9511 (Figure 3) are quite comparable with those previously described for this strain grown in similar light conditions (Bruyant et al., 2005), except that our cultures exhibited a higher FV/FM during the night (∼0.7 vs. ∼0.55), likely translating a slightly better physiological status. However, they contrast with those obtained on the closely related MED4 strain by Zinser et al. (2009), who did not observed any significant diel variation of the FV/FM, likely because it was grown at lower irradiance (∼230 vs. 870 μmol photon m−2 s−1 here) and over a different L/D cycle (14/10 vs. 12/12 h). Another noticeable observation from our study was that UVR did not cause any appreciably stronger photoinhibitory effect than VL in both genera, likely due to an increased repair rate under VL + UV compared to VL. However, this UV-induced repair was much more important for Synechococcus than Prochlorococcus (∼twofold vs. ∼fivefold at noon, respectively). Accordingly, in the former organism, the relative D1 content was enhanced in the presence of UVR, while in Prochlorococcus the noontime drop of D1 was somewhat extended for UV-acclimated cells (see also transcriptomic analyses of the psbA genes below). These results are consistent with those obtained by Six et al. (2007a), who observed that in response to a transient high light exposure, P. marinus PCC 9511 exhibited a lower PSII repair rate (0.9 PSII gained per second) than a range of marine Synechococcus strains (1.1–1.6 PSII s−1). In both strains in VL, this rate was more or less proportional to the instantaneous irradiance. However for cells grown under VL + UV, while the repair rate was comparable between the two strains at 9:00 and 15:00, it was significantly depressed at noon in Prochlorococcus compared to Synechococcus. This suggests that the PSII repair capacity of Prochlorococcus was already maximum around 400 μmol photons m−2 s−1 (Figure 4B).
The occurrence of different D1 encoding gene copies in Prochlorococcus and Synechococcus could, at least partially, explain such a distinct behavior. Indeed, while Prochlorococcus strains have one to three identical psbA gene copies (one in PCC 9511), coding for a single D1:1-like isoform (Hess et al., 1995; Partensky and Garczarek, 2003), Synechococcus strains possess three to six psbA genes, with only one copy coding for a D1:1 isoform and two to five copies (three in WH7803), coding for D1:2 isoforms (Garczarek et al., 2008). The respective role of these isoforms has been widely studied in the literature, both in freshwater cyanobacteria (Bustos et al., 1990; Clarke et al., 1993; Campbell et al., 1995, 1998a; Sass et al., 1997; Kos et al., 2008) and in marine picocyanobacteria (Garcia-Fernandez et al., 1998; Garczarek et al., 2008). Although some variations among cyanobacteria have been observed, it is generally accepted that the D1:1 isoform would confer a higher PSII activity (Campbell et al., 1996), while D1:2 would provide a lower quantum yield but a higher PSII resistance to photoinhibition (Krupa et al., 1991;Campbell et al., 1995, 1998a; Tichy et al., 2003). A variety of environmental cues, including UV exposure, can induce the exchange of these isoforms (Sicora et al., 2006, 2008; Garczarek et al., 2008; for a review, see Bouchard et al., 2006). Here, we indeed noticed in Synechococcus sp. WH7803 an opposite expression pattern of D1:1 (SynWH7803_0784) and D1:2 isoforms encoding genes (SynWH7803_0790, 0366, and 2084) during daytime (Figure 7). It is worth noting that while there were only slight discrepancies in the expression levels of the different D1:2 encoding genes between VL and VL + UV, the D1:1 encoding gene was about fivefold more repressed at noon under the latter condition, suggesting a complete replacement of the D1:1 isoform by D1:2 isoform(s) that may contribute to the slight midday decrease in PSII quantum yield (Figure 3). Interestingly, although the single isoform in Prochlorococcus is phylogenetically a D1:1 isoform, as confirmed by the occurrence of a Gln residue at position 130 of the amino acid sequence (instead of Glu in D1:2; Clarke et al., 1993; Giorgi et al., 1996), its transcriptomic pattern was clearly closer from that of D1:2 isoforms, at least in our culture conditions. It is likely however that it presents a lower resistance to PSII photoinactivation compared to a true D1:2, as suggested by the larger drop of FV/FM at noon (Figure 3) and the concomitant lower PSII repair rate (Figure 4), compared to Synechococcus.
Interestingly, the D2 subunit of the PSII core is also encoded by a single psbD gene in all Prochlorococcus genomes, whereas all Synechococcus sequenced so far possess two nearly identical psbD genes, as in most other cyanobacteria (such as e.g., Synechococcus sp. PCC 7942; Golden et al., 1989), with one co-transcribed with psbC that encodes the internal PSII antenna protein CP43 (Garczarek et al., 2001) and the other isolated in the genome. It is likely that as for psbA, the two copies are differently regulated in response to light and/or UV stress as previously reported in freshwater model cyanobacteria (Bustos and Golden, 1992; Kos et al., 2008), although this was not checked in the present study. Alternatively, this may simply contribute to a higher expression level of this key photosynthetic gene, possibly enabling a higher turnover of the corresponding protein.
Another strategy used by cyanobacteria to cope with excess light energy is to decrease the relative amount of PSI reaction center complexes, an adjustment that was shown to decelerate the rate of photosynthetic electron transport (Murakami and Fujita, 1991; Hihara et al., 1998; Muramatsu and Hihara, 2003). Indeed, experiments on Synechocystis sp. PCC 6803 mutants impaired in their ability to modulate photosystem stoichiometry showed that this capacity is indispensable for growth under continuous high irradiance (Hihara et al., 1998; Fujimori et al., 2005). Accordingly, in Synechococcus sp. WH7803 the relative psaB levels were low before dawn (Figure 7), likely reflecting a lower PSI cell content at this time of the day, as previously observed in Crocosphaera watsonii (Saito et al., 2011). In contrast, this was not the case in Prochlorococcus, in which PSI core transcripts were maximal over most of the dark period.
Differential regulation of light-harvesting systems in response to high light and UV radiations
The striking structural differences between the major PSII antenna complexes of Prochlorococcus and Synechococcus may also be partially responsible for the different sensitivity of these picocyanobacteria to UV stress. Indeed, whilst Synechococcus, as most cyanobacteria, possess a large membrane-extrinsic antenna, the PBS (Sidler, 1994; Six et al., 2007c), Prochlorococcus, like the other two green oxyphotobacteria, Prochloron and Prochlorothrix, use a transmembrane Chl a/b-binding Pcb antenna (LaRoche et al., 1996; Garczarek et al., 2003; Partensky and Garczarek, 2003). These dissimilar antenna structures may have important consequences on the way excitation energy is funneled downhill toward the reaction centers, on the regulation of this process as well as on the amount and nature of damages caused by UVR on Prochlorococcus and Synechococcus cells, since both organisms show some absorption capacities in the 300- to 400-nm band, which are likely related to their antenna (see e.g., Ong and Glazer, 1991; Claustre et al., 2002). Comparing and monitoring UV cross-sections for both picocyanobacteria would help answering this question.
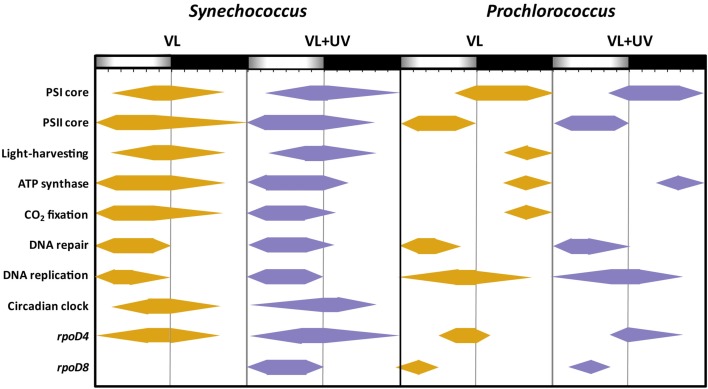
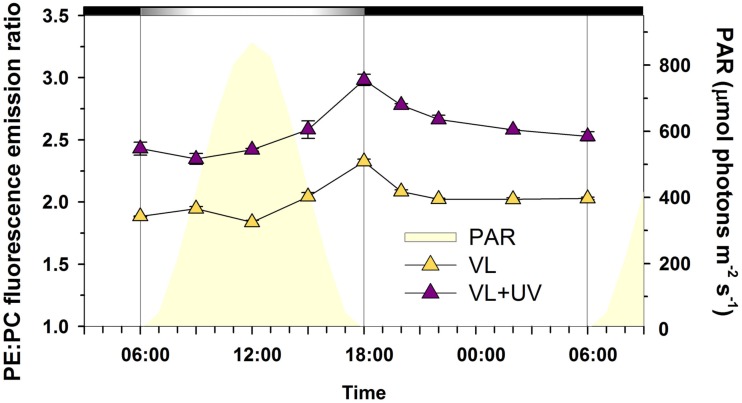
Here, we indeed observed an opposite expression pattern between the genes encoding antenna systems from these two organisms, with a strong downregulation of the pcb gene in Prochlorococcus during day time under VL (see also Garczarek et al., 2001) that was even more dramatic under VL + UV, while all examined PBS genes were upregulated in the afternoon in both light conditions (Figure 8). A similar result was previously obtained in L/D-entrained Cyanothece sp. ATCC 51142 cultures, though in the latter case maximum expression was centered at midday (Toepel et al., 2008). These observations suggest that the biosynthesis of antenna complexes occurs at different times of the day in the two picocyanobacteria and is under different light regulation controls. The fact that PBS genes remained upregulated under VL + UV suggests that the buildup of PBS complexes was only moderately affected by these radiations. This contrasts with a previous study where cultures of Synechococcus sp. WH8102 grown in continuous low-light were subjected to a sudden shift to UV (Six et al., 2007b). This stress provoked a strong decrease in the relative expression of all PBS genes, associated with a disconnection of the PBS complexes from the thylakoid membrane as well as a dissociation of the distal PEII disks of PBS rods. In the present study, where cells were acclimated to either VL or VL + UV for several weeks, the PE to PC fluorescence emission ratio did not exhibit a peak at noon in either light condition, as would be expected if terminal PE subunits (i.e., PEII) were disrupted (Figure A1 in Appendix). The higher values of this ratio in UV-acclimated Synechococcus cells are likely not related to a higher PE content of the PBS, but rather to permanently decoupled PEII subunits, which would dissipate incident light as fluorescence. Consistently, UV also induced a slight drop of the expression of most PEII genes at noon and a 3-h delay in the timing of the expression peak of mpeE, which encodes the linker binding the terminal PEII disk in Synechococcus sp. WH7803 (Six et al., 2007c).
Figure 8.
Scheme of the daily patterns of gene upregulation for several important functional for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of high visible light with or without UV radiations. This figure, derived from Figure 7, shows for each individual the time intervals during which they were significantly upregulated [|log2 (Fold Change)| > 1.0], with regard to the expression level measured at 6:00 in VL.
Differential photoprotection mechanisms
Prochlorococcus and Synechococcus have also developed specific mechanisms to dissipate excess light energy. Most Synechococcus strains possess one gene encoding the orange carotenoid protein (OCP; for a recent review, see Kirilovsky and Kerfeld, 2012), which is thought to mediate energy dissipation as heat through interaction with the PBS core, thus inducing a NPQ of PSII fluorescence. Here, we observed that the ocp gene was upregulated during daytime in VL and that UVR dramatically enhanced its expression in the early light period that may trigger a temporary increase of the OCP cellular pool and/or activity. To our knowledge, this is a first time that UV is shown to control the expression of this gene since it is usually believed that OCP is only blue light sensitive (Wilson et al., 2008; Kirilovsky and Kerfeld, 2012).
While all Prochlorococcus strains lack OCP, all high light-adapted strains (including PCC 9511) and some low-light-adapted strains possess an homolog of PTOX, which is thought to extract electrons between PSII and PSI and to combine them with protons and oxygen to generate water (Bailey et al., 2005, 2008). The ptox gene was (with ftsZ) one of the two most differentially expressed genes among those analyzed here in PCC 9511, with a maximal relative fold change over the day of ∼73-fold [or log2(FC) = 6.2] at 15:00. These high values are consistent with previous results on the closely related strain P. marinus MED4 (Zinser et al., 2009; Berg et al., 2011). So the alternative electron flow to oxygen triggered by PTOX might be an important mechanism used by Prochlorococcus to struggle against excess light energy arising to PSII (Bailey et al., 2008; Berg et al., 2011), although high light induced proteins (HLIPs), which were not analyzed in the present study, are also likely important contributors in this process (He et al., 2001).
Other key actors in photoprotection mechanisms are carotenoids. While the cellular localization and precise action mechanism of Zea is unclear yet, Car are known to be mostly bound to PS reaction centers, two of them in close vicinity of Chls, and help mitigate oxidative damages, mainly by quenching the 1O2 species resulting from the de-excitation of Chl triplets (Telfer, 2005). Both Zea:Chl a and Car:Chl a ratios appeared to be tightly coupled to the L/D cycle in Synechococcus, but less so in Prochlorococcus, for which we observed a continuous increase of both ratios during most of the day and a symmetrical drop during the night concomitant with cell division, as previously noticed by Claustre et al. (2002). In Synechococcus, the sharp drop of the β-Car:Chl a ratio from noon to dusk, which was more pronounced in UV-exposed cells (Figure 2C), likely reflects the progressive destruction of β-Car molecules by oxidative stress (Telfer, 2005). β-Car was then seemingly regenerated at low rate during the night, then much more rapidly during the first hours of the day, suggesting a light-dependency of the β-Car biosynthesis process, as previously reported in other cyanobacteria (Steiger et al., 2005; Ryu et al., 2010). Similarly, the light-modulated variations of the Zea:Chl a ratio during the day in Synechococcus in both light conditions could be due either to a higher degradation rate of Chl a relative to Zea or to a higher synthesis rate of the latter pigment, a likely photoprotective mechanism against high midday photon fluxes. The observed strong upregulation of crtR at high irradiances tend to favor the second hypothesis (Figure 7). A further increase of the relative crtR transcript levels at noon in VL + UV, which was not translated into a significantly higher midday increase in the Zea:Chl a ratio than in VL (Figure 3A), suggests that Zea msolecules might have a particularly high turnover under UV. In both genera, the systematically higher values of Zea:Chl a ratio under VL + UV than VL only suggest that UV exposure induces a comparable response as a long-term acclimation to high irradiance, as previously reported in other marine cyanobacterial strains (Kana and Glibert, 1987; Moore et al., 1995; Six et al., 2004). Altogether, our results suggest the occurrence of a light-controlled anabolism/catabolism cycle of both photoprotectants (Car and Zea) in Synechococcus, while diel changes in the pigment content of Prochlorococcus cells were rather related to the cell division cycle or other factors not analyzed here such as PS stoichiometry and/or antenna size.
Differential protection mechanisms against reactive oxygen species-induced damages
Light-harvesting complexes are not only sources but also major targets of ROS (Sies and Menck, 1992; Krieger-Liszkay, 2005). Because of their localization within the thylakoid membrane around PSII complexes (Bibby et al., 2003), the divinyl-Chl a/b-binding Pcb antennae of P. marinus PCC 9511 likely produce ROS, particularly noxious for this photosystem. Indeed, it has been shown in other Chl b-containing organisms that excited singlet Chls lead to the synthesis of Chl triplet states that can react with 3O2 to produce the very reactive species1O2 (Sies and Menck, 1992; Krieger-Liszkay, 2005). Assays performed here by directly adding H2O2 to sub-cultures at different time points of the modulated L/D cycle strongly suggest that the effect of ROS increased in a light-dependent manner for both organisms. Consistently, a synergistic effect of light and oxidative stress on PSII photoinactivation was previously demonstrated by Blot et al. (2011) who showed in Synechococcus sp. WH7803 that ROS may induce PSII inactivation through both direct damages to the reaction center II and inhibition of the PSII repair cycle. The latter phenomenon resulted in faster PSII inactivation in high light- than in low-light-acclimated cultures, due to their higher D1 turnover. Furthermore, Prochlorococcus was also more sensitive to H2O2-triggered stress than Synechococcus during the night, which might be related to a lower intrinsic resistance of its PSII.
The difference in ROS sensitivity between Prochlorococcus and Synechococcus strains might be explained, at least partially, by a more complete set of genes involved in ROS protection and detoxification in Synechococcus. Indeed, most Prochlorococcus lineages have experienced an extensive genome streamlining during evolution, resulting in the loss of a large number of non-essential but potentially useful genes in this context (Figure 7; Dufresne et al., 2005; Scanlan et al., 2009; Partensky and Garczarek, 2010). For instance, while Synechococcus sp. WH7803 synthesizes one catalase/peroxidase (KatG), Prochlorococcus has neither catalases nor peroxidases. Prochlorococcus also lack the ftrC and ftrV genes, encoding the two subunits of ferredoxin-thioredoxin reductase (FTR), as well as one ferredoxin and one thioredoxin, potentially associated to this complex (Dufresne et al., 2008). Furthermore, while Synechococcus possesses two superoxide dismutase (SOD), one Fe-type and one Cu-Zn-type, Prochlorococcus has only one, Ni-type SOD (SodN; Scanlan et al., 2009). In the present study, the expression levels of most ROS genes showed strikingly higher amplitudes of variation over the day in Synechococcus than in Prochlorococcus. Even though transcriptomic data are not necessarily synchronized with the activity of the corresponding enzymes, the lower resistance of Prochlorococcus to high VL and UVR could at least partially be due to a higher sensitivity to light-driven oxidative stress.
Conclusion
The comparison of Synechococcus and Prochlorococcus cultures acclimated to VL supplemented or not with UVR revealed relatively few physiological responses specific to UV. This notably includes a shift of the DNA synthesis phase (Figure 1; see also Kolowrat et al., 2010), an increase of the Zea:Chl a ratio (Figure 2) and an enhanced PSII repair rate (Figure 4). Accordingly, UVR seemingly also had limited effects at the transcriptomic level, as shown by the globally similar diel expression patterns between VL and VL + UV in both strains (Figures 7 and 8). It is worth noting however that the relative expression of a few genes was either enhanced (e.g., psbD in both strains, D1:2 encoding genes and crtR in Synechococcus only) or reduced (e.g., Synechococcus D1:1 encoding gene or Prochlorococcus pcbA) in UV-acclimated cultures. A handful of genes, including rpoD4/8 in Prochlorococcus and kaiABC in Synechococcus, also exhibited a delayed expression peak by about 3 h. However, this surprisingly did not translate into any conspicuous changes in the diel patterns of most of the other genes examined here, as could have been expected from the known regulatory role of the circadian clock and sigma factors on gene transcription (see e.g., Summerfield and Sherman, 2007; Ito et al., 2009).
The most striking result of the present study is likely the markedly distinct response to diurnal light variations between Prochlorococcus and Synechococcus, despite their close phylogenetic relatedness (Scanlan et al., 2009). These two picocyanobacteria indeed exhibited very different degrees of PSII photoinactivation at noon that can be partly explained by their distinct (i) PSII repair capacity (Figure 4; see also Six et al., 2007a), (ii) ability to modulate photoprotective pigments (Zea and Car, Figure 2), and (iii) resistance capacity against oxidative stress (Figure 6). Comparative transcriptomic analyses also revealed that, in Synechococcus, genes coding for a number of protective systems were maximally expressed during hours of highest irradiance, i.e., when these mechanisms are most critical to cope with transitory stressful conditions. This includes several genes involved in ROS detoxification enzymes, DNA repair genes as well as genes involved in photoprotection and/or dissipation of excess energy, such as crtR and ocp (Figure 7). In contrast, in Prochlorococcus, very few genes of these pathways, including ptox and psbA, were maximally expressed around midday, while others, such as phrA and crtR, had already reached their maximal (saturating) expression at mid-morning. Furthermore, many photosynthetic genes that were upregulated during the day in Synechococcus were in contrast downregulated in Prochlorococcus, including the PSI core gene psaB as well as genes involved in light-harvesting, ATP synthase and CO2 fixation (Figure 8). Similarly, glgA (encoding glycogen synthase) mRNA abundance was recently shown to exhibit a maximum diel expression at midday in Synechococcus sp. WH8103 grown under a 16/8-h L/D cycle, while it peaked in concert with rbcLS in Prochlorococcus marinus MED4 during the LDT (Wyman and Thom, 2012). Thus altogether, it seems that while Synechococcus, as many other cyanobacteria (Kucho et al., 2004, 2005; Stockel et al., 2008; Toepel et al., 2008, 2009; Shi et al., 2010), efficiently copes with the diurnal changes in photon fluxes, Prochlorococcus rather displays a stress-like response at midday. The latter response is most likely related to the high irradiance level used in the present study (reaching 870 μmol photons m−2 s−1 at noon), which are typically found in the upper mixed layer of tropical oligotrophic oceans (Holtzendorff et al., 2001). Indeed, several genes that were found here to be downregulated during the light period (e.g., pcb, rbcL, chlG) were in contrast upregulated in Prochlorococcus cultures grown at lower irradiances, provided as continuous (Berg et al., 2011) or cyclic light (Zinser et al., 2009). Despite the very atypical transcriptomic response observed here, Prochlorococcus was able to recover high PSII quantum yield at night and to maintain an optimal growth rate under these conditions. Thus, we hypothesize that, in contrast to Synechococcus sp. WH7803, P. marinus PCC 9511 cells manage to cope with harmful light conditions by bringing down temporarily some of the main metabolic processes and by launching a minimal set of protection mechanisms during stressful hours. Whether the absence of a true circadian clock in Prochlorococcus (Holtzendorff et al., 2008; Axmann et al., 2009) is involved in this differential management of excess light compared to Synechococcus still remains to be investigated.
Altogether, our study reinforces previous studies depicting Prochlorococcus as a very specialized organism restricted to a narrow environmental niche, while Synechococcus has adopted a generalist strategy enabling it to cope with more variable environmental conditions, a difference consistent with the distinct habitats in which these two organisms predominate (Scanlan, 2003; Kettler et al., 2007; Dufresne et al., 2008; Scanlan et al., 2009).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Aquatic_Microbiology/10.3389/fmicb.2012.00285/abstract
List of primers used for real time PCR reactions. Most primers are targeting a specific gene. Exceptions include the psbD pool and psbA pool primers, which were designed to amplify the two psbD gene copies and all four psbA copies present in Synechococcus sp. WH7803, respectively. Furthermore, one primer set targets two psbA genes (SynWH7803_0366 and 2084), which have nearly identical coding sequence and 5′-UTR. Genes are classified by functional categories, as indicated in the first column. The Cyanorak database of picocyanobacteria protein families is publicly accessible at http://www.sb-roscoff.fr/Phyto/cyanorak/
Daily variations of the expression of selected genes, as measured by real time quantitative PCR, for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of VL with or without UV radiations. All data are expressed as log2 (Fold Change) ± mean deviation for two biological replicates. For each gene, transcript levels are normalized to the reference time point 6:00 in VL. Genes are classified by functional categories, as indicated in the first column. This table is a detailed version of Figure 7 of the main manuscript.
Acknowledgments
This work was supported by the French programs ANR PELICAN (PCS-09-GENM-200) and EMBRC France (INFRA-2010-2.2.5), the European Union programs MicroB3 (UE-contract-287589) MaCuMBA (FP7-KBBE-2012-6-311975). Daniella Mella-Flores was supported by the National Commission of Scientific and Technological Investigation of Chile (CONICYT). We thank Roseline Edern, Fabienne Jalabert, and the Roscoff Culture Collection for maintaining the Synechococcus and Prochlorococcus strains used in this study.
Appendix
Figure A1.
Phycoerythrin to phycocyanin (PE:PC) fluorescence emission ratio as measured by spectrofluorimetry (at 572 and 651 nm, respectively) in Synechococcus sp. WH7803 cells grown under a L/D cycle with and without UV. This ratio showed significantly higher values in VL + UV than in VL-acclimated cells. In both light conditions, it increased during the day by ∼25% compared to the value at dawn then decreased during the night. The amplitude of these oscillations (ca 0.4 unit) was very low compared to those reported for Synechococcus sp. WH8102 acclimated to a large range of irradiance (2.5 units; Six et al., 2004). Further investigation is required to determine whether the diel pattern of PE:PC fluorescence emission ratio rather results from a decrease in the efficiency of energy transfer in the PBS rod and/or a progressive accumulation of free phycoerythrins during the day. These light-induced oscillations might be indirectly related with state transitions (Campbell et al., 1998b; Mullineaux and Emlyn-Jones, 2005), a light-dependent process which usually exhibits large amplitudes in marine Synechococcus (Six et al., personal communication).
References
- Agawin N. S. R., Duarte C. M., Agusti S. (2000). Nutrient and temperature control of the contribution of picoplankton to phytoplankton biomass and production. Limnol. Oceanogr. 45, 591–600 10.4319/lo.2000.45.3.0591 [DOI] [Google Scholar]
- Agusti S., Llabres M. (2007). Solar radiation-induced mortality of marine pico-phytoplankton in the oligotrophic ocean. Photochem. Photobiol. 83, 793–801 10.1111/j.1751-1097.2007.00144.x [DOI] [PubMed] [Google Scholar]
- Allakhverdiev S. I., Murata N. (2008). Salt stress inhibits photosystems II and I in cyanobacteria. Photosyn. Res. 98, 529–539 10.1007/s11120-008-9334-x [DOI] [PubMed] [Google Scholar]
- Andersson B., Aro E. M. (2001). “Photodamage and D1 protein turnover in photosystem II,” in Regulation of Photosynthesis, eds Aro E. M., Andersson B. (Dordrecht: Springer; ), 377–393 [Google Scholar]
- Aro E. M., Virgin I., Andersson B. (1993). Photoinhibition of photosystem 2 – inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134 10.1016/0005-2728(93)90134-2 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- Ashby M. K., Mullineaux C. W. (1999). Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol. Lett. 181, 253–260 10.1111/j.1574-6968.1999.tb08852.x [DOI] [PubMed] [Google Scholar]
- Axmann I. M., Duhring U., Seeliger L., Arnold A., Vanselow J. T., Kramer A., Wilde A. (2009). Biochemical evidence for a timing mechanism in Prochlorococcus. J. Bacteriol. 191, 5342–5347 10.1128/JB.00419-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S., Grossman A. (2008). Photoprotection in cyanobacteria: regulation of light harvesting. Photochem. Photobiol. 84, 1410–1420 10.1111/j.1751-1097.2008.00453.x [DOI] [PubMed] [Google Scholar]
- Bailey S., Mann N. H., Robinson C., Scanlan D. J. (2005). The occurrence of rapidly reversible non-photochemical quenching of chlorophyll a fluorescence in cyanobacteria. FEBS Lett. 579, 275–280 10.1016/j.febslet.2004.11.091 [DOI] [PubMed] [Google Scholar]
- Bailey S., Melis A., MacKey K. R., Cardol P., Finazzi G., Van Dijken G., Berg G. M., Arrigo K., Shrager J., Grossman A. (2008). Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim. Biophys. Acta 1777, 269–276 10.1016/j.bbabio.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Berg G. M., Shrager J., Van Dijken G., Mills M. M., Arrigo K. R., Grossman A. R. (2011). Responses of psbA, hli and ptox genes to changes in irradiance in marine Synechococcus and Prochlorococcus. Aquat. Microb. Ecol. 65, 1–14 10.3354/ame01528 [DOI] [Google Scholar]
- Berry J., Björkman O. (1980). Photosynthetic response and adaptation to temperature in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 31, 491–543 [Google Scholar]
- Bibby T. S., Mary I., Nield J., Partensky F., Barber J. (2003). Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051–1054 10.1038/nature01933 [DOI] [PubMed] [Google Scholar]
- Blot N., Mella-Flores D., Six C., Le Corguille G., Boutte C., Peyrat A., Monnier A., Ratin M., Gourvil P., Campbell D. A., Garczarek L. (2011). Light history influences the response of the marine cyanobacterium Synechococcus sp. WH7803 to oxidative stress. Plant Physiol. 156, 1934–1954 10.1104/pp.111.174714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard J. N., Roy S., Campbell D. A. (2006). UVB effects on the photosystem II D1 protein of phytoplankton and natural phytoplankton communities. Photochem. Photobiol. Sci. 82, 936–951 10.1562/2005-08-31-IR-666 [DOI] [PubMed] [Google Scholar]
- Boulay C., Abasova L., Six C., Vass I., Kirilovsky D. (2008). Occurrence and function of the orange carotenoid protein in photoprotective mechanisms in various cyanobacteria. Biochim. Biophys. Acta 1777, 1344–1354 10.1016/j.bbabio.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Bruyant F., Babin M., Genty B., Prasil O., Behrenfeld M. J., Claustre H., Bricaud A., Garczarek L., Holtzendorff J., Koblizek M., Dousova H., Partensky F. (2005). Diel variations in the photosynthetic parameters of Prochlorococcus strain PCC 9511: combined effects of light and cell cycle. Limnol. Oceanogr. 50, 850–863 10.4319/lo.2005.50.3.0850 [DOI] [Google Scholar]
- Bustos S. A., Golden S. S. (1992). Light-regulated expression of the psbD gene family in Synechococcus sp. strain PCC 7942: evidence for the role of duplicated psbD genes in cyanobacteria. Mol. Gen. Genet. 232, 221–230 [DOI] [PubMed] [Google Scholar]
- Bustos S. A., Schaefer M. R., Golden S. S. (1990). Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J. Bacteriol. 172, 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell M. M., Bornman J. F., Ballaré C. L., Flint S. D., Kulandaivelu G. (2007). Terrestrial ecosystems, increased solar ultraviolet radiation, and interactions with other climate change factors. Photochem. Photobiol. Sci. 6, 252–266 10.1039/b700019g [DOI] [PubMed] [Google Scholar]
- Campbell D., Bruce D., Carpenter C., Gustafsson P., Oquist G. (1996). Two forms of the photosystem II D1 protein alter energy dissipation and state transitions in the cyanobacterium Synechococcus sp. PCC 7942. Photosyn. Res. 47, 131–144 10.1007/BF00016176 [DOI] [PubMed] [Google Scholar]
- Campbell D., Eriksson M. J., Oquist G., Gustafsson P., Clarke A. K. (1998a). The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 364–369 10.1073/pnas.95.1.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D., Hurry V., Clarke A. K., Gustafsson P., Öquist G. (1998b). Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol. Mol. Biol. Rev. 667–683, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D., Zhou G. Q., Gustafsson P., Oquist G., Clarke A. K. (1995). Electron transport regulates exchange of two forms of photosystem II D1 protein in the cyanobacterium Synechococcus. EMBO J. 14, 5457–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. A., Tyystjarvi E. (2012). Parameterization of photosystem II photoinactivation and repair. Biochim. Biophys. Acta 1817, 258–265 10.1016/j.bbabio.2011.04.010 [DOI] [PubMed] [Google Scholar]
- Carpenter E. J., Chang J. (1988). Species-specific phytoplankton growth rates via diel DNA synthesis cycles. I. Concept of the method. Mar. Ecol. Prog. Ser. 43, 105–111 10.3354/meps043105 [DOI] [Google Scholar]
- Chen Z., Yang H., Pavletich N. P. (2008). Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–484 10.1038/nature06971 [DOI] [PubMed] [Google Scholar]
- Clarke A. K., Hurry V. M., Gustafsson P., Öquist G. (1993). Two functionally distinct forms of the photosystem II reaction center D1 protein in the cyanobacterium Synechococcus sp. PCC7942. Proc. Natl. Acad. Sci. U.S.A. 90, 11985–11989 10.1073/pnas.90.24.11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claustre H., Bricaud A., Babin M., Bruyant F., Guillou L., Le Gall F., Marie D., Partensky F. (2002). Diel variations in Prochlorococcus optical properties. Limnol. Oceanogr. 47, 1637–1647 10.4319/lo.2002.47.6.1637 [DOI] [Google Scholar]
- Demmig-Adams B., Adams W. W. (1992). Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 7, 1–116 [Google Scholar]
- Dring M. J., Wagner A., Luning K. (2001). Contribution of the UV component of natural sunlight to photoinhibition of photosynthesis in six species of subtidal brown and red seaweeds. Plant Cell Environ. 24, 1153–1164 10.1046/j.1365-3040.2001.00765.x [DOI] [Google Scholar]
- Dufresne A., Garczarek L., Partensky F. (2005). Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6, R14. 10.1186/gb-2005-6-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A., Ostrowski M., Scanlan D. J., Garczarek L., Mazard S., Palenik B. P., Paulsen I. T., Tandeau De Marsac N., Wincker P., Dossat C., Ferriera S., Johnson J., Post A. F., Hess W. R., Partensky F. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9, R90. 10.1186/gb-2008-9-5-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everroad C., Six C., Partensky F., Thomas J. C., Holtzendorff J., Wood A. M. (2006). Biochemical bases of type IV chromatic adaptation in marine Synechococcus spp. J. Bacteriol. 188, 3345–3356 10.1128/JB.188.9.3345-3356.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T., Higuchi M., Sato H., Aiba H., Muramatsu M., Hihara Y., Sonoike K. (2005). The mutant of sll1961, which encodes a putative transcriptional regulator, has a defect in regulation of photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 139, 408–416 10.1104/pp.105.064782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez J. M., Hess W. R., Houmard J., Partensky F. (1998). Expression of the psbA gene in the marine oxyphotobacteria Prochlorococcus spp. Arch. Biochem. Biophys. 359, 17–23 10.1006/abbi.1998.0862 [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F., Johnson S. L., Youngkin D., Belnap J. (2003). Small-scale vertical distribution of bacterial biomass and diversity in biological soil crusts from arid lands in the Colorado Plateau. Microb. Ecol. 46, 312–321 10.1007/s00248-003-1004-0 [DOI] [PubMed] [Google Scholar]
- Garczarek L., Dufresne A., Blot N., Cockshutt A. M., Peyrat A., Campbell D. A., Joubin L., Six C. (2008). Function and evolution of the psbA gene family in marine Synechococcus: Synechococcus sp. WH7803 as a case study. ISME J. 2, 937–953 10.1038/ismej.2008.46 [DOI] [PubMed] [Google Scholar]
- Garczarek L., Partensky F., Irlbacher H., Holtzendorff J., Babin M., Mary I., Thomas J. C., Hess W. R. (2001). Differential expression of antenna and core genes in Prochlorococcus PCC 9511 (Oxyphotobacteria) grown under a modulated light-dark cycle. Environ. Microbiol. 3, 168–175 10.1046/j.1462-2920.2001.00173.x [DOI] [PubMed] [Google Scholar]
- Garczarek L., Poupon A., Partensky F. (2003). Origin and evolution of transmembrane Chl-binding proteins: hydrophobic cluster analysis suggests a common one-helix ancestor for prokaryotic (Pcb) and eukaryotic (LHC) antenna protein superfamilies. FEMS Microbiol. Lett. 222, 59–68 10.1016/S0378-1097(03)00241-6 [DOI] [PubMed] [Google Scholar]
- Giorgi L. B., Nixon P. J., Merry S. A., Joseph D. M., Durrant J. R., De Las Rivas J., Barber J., Porter G., Klug D. R. (1996). Comparison of primary charge separation in the photosystem II reaction center complex isolated from wild-type and D1-130 mutants of the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 271, 2093–2101 10.1074/jbc.271.4.2093 [DOI] [PubMed] [Google Scholar]
- Golden S. S., Cho D. S., Nalty M. S. (1989). Two functional psbD genes in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 171, 4707–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., Moolenaar G. F. (2008). Repair of UV damage in bacteria. DNA Repair (Amst.) 7, 353–379 10.1016/j.dnarep.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Hakala M., Rantamaki S., Puputti E. M., Tyystjarvi T., Tyystjarvi E. (2006). Photoinhibition of manganese enzymes: insights into the mechanism of photosystem II photoinhibition. J. Exp. Bot. 57, 1809–1816 10.1093/jxb/erj189 [DOI] [PubMed] [Google Scholar]
- Hanke G. T., Satomi Y., Shinmura K., Takao T., Hase T. (2011). A screen for potential ferredoxin electron transfer partners uncovers new, redox dependent interactions. Biochim. Biophys. Acta 1814, 366–374 10.1016/j.bbapap.2010.09.011 [DOI] [PubMed] [Google Scholar]
- He Q. F., Dolganov N., Bjorkman O., Grossman A. R. (2001). The high light-inducible polypeptides in Synechocystis PCC6803 – expression and function in high light. J. Biol. Chem. 276, 306–314 10.1074/jbc.M102000200 [DOI] [PubMed] [Google Scholar]
- He Y.-Y., Häder D.-P. (2002a). Involvement of reactive oxygen species in the UV-B damage to the cyanobacterium Anabaena sp. J. Photochem. Photobiol. B 66, 73–80 10.1016/S1011-1344(02)00231-2 [DOI] [PubMed] [Google Scholar]
- He Y. Y., Häder D. P. (2002b). Reactive oxygen species and UV-B: effect on cyanobacteria. Photochem. Photobiol. Sci. 1, 729–736 10.1039/b110365m [DOI] [PubMed] [Google Scholar]
- Helbling E. W., Villafane V. E., Ferrario M., Holm-Hansen O. (1992). Impact of natural ultraviolet radiation on rates of photosynthesis and on specific marine phytoplankton species. Mar. Ecol. Prog. Ser. 80, 80–89 10.3354/meps080089 [DOI] [Google Scholar]
- Hess W. R., Weihe A., Loiseaux-De Goër S., Partensky F., Vaulot D. (1995). Characterization of the single psbA gene of Prochlorococcus marinus CCMP 1375 (Prochlorophyta). Plant Mol. Biol. 27, 1189–1196 10.1007/BF00020892 [DOI] [PubMed] [Google Scholar]
- Hihara Y., Sonoike K., Ikeuchi M. (1998). A novel gene, pmgA, specifically regulates photosystem stoichiometry in the cyanobacterium Synechocystis species PCC 6803 in response to high light. Plant Physiol. 117, 1205–1216 10.1104/pp.117.4.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzendorff J., Partensky F., Jacquet S., Bruyant F., Marie D., Garczarek L., Mary I., Vaulot D., Hess W. R. (2001). Diel expression of cell cycle-related genes in synchronized cultures of Prochlorococcus sp strain PCC 9511. J. Bacteriol. 183, 915–920 10.1128/JB.183.3.915-920.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzendorff J., Partensky F., Mella D., Lennon J. F., Hess W. R., Garczarek L. (2008). Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms 23, 187–199 10.1177/0748730408316040 [DOI] [PubMed] [Google Scholar]
- Houot L., Floutier M., Marteyn B., Michaut M., Picciocchi A., Legrain P., Aude J.-C., Cassier-Chauvat C., Chauvat F. (2007). Cadmium triggers an integrated reprogramming of the metabolism of Synechocystis PCC6803, under the control of the Slr1738 regulator. BMC Genomics 8, 350. 10.1186/1471-2164-8-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S., Yoshihara S., Nakano S., Shiozaki N., Yamada A., Tanaka K., Takahashi H., Asayama M., Shirai M. (2003). Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J. Mol. Biol. 325, 857–872 10.1016/S0022-2836(02)01242-1 [DOI] [PubMed] [Google Scholar]
- Ito H., Mutsuda M., Murayama Y., Tomita J., Hosokawa N., Terauchi K., Sugita C., Sugita M., Kondo T., Iwasaki H. (2009). Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 106, 14168–14173 10.1073/pnas.0910787106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet S., Lennon J. F., Marie D., Vaulot D. (1998). Picoplankton population dynamics in coastal waters of the northwestern Mediterranean Sea. Limnol. Oceanogr. 43, 1916–1931 [Google Scholar]
- Johnson Z. I., Zinser E. R., Coe A., Mcnulty N. P., Woodward E. M., Chisholm S. W. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311, 1737–1740 10.1126/science.1118052 [DOI] [PubMed] [Google Scholar]
- Kana T. M., Glibert P. M. (1987). Effect of irradiances up to 2000 μE m-2 s-1 on marine Synechococcus WH7803 – I. Growth, pigmentation, and cell composition. Deep Sea Res. 34, 479–485 10.1016/0198-0149(87)90001-X [DOI] [Google Scholar]
- Kappell A. D., Bhaya D., Van Waasbergen L. G. (2006). Negative control of the high light-inducible hliA gene and implications for the activities of the NblS sensor kinase in the cyanobacterium Synechococcus elongatus strain PCC 7942. Arch. Microbiol. 186, 403–413 10.1007/s00203-006-0154-0 [DOI] [PubMed] [Google Scholar]
- Keren N., Berg A., Vankan P. J. M., Levanon H., Ohad I. (1997). Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: the role of back electron flow. Proc. Natl. Acad. Sci. U.S.A. 94, 1579–1584 10.1073/pnas.94.4.1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler G., Martiny A. C., Huang K., Zucker J., Coleman M. L., Rodrigue S., Chen F., Lapidus A., Ferriera S., Johnson J., Steglich C., Church G., Richardson P., Chisholm S. W. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3, e231. 10.1371/journal.pgen.0030231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilovsky D., Kerfeld C. A. (2012). The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta 1817, 158–166 10.1016/j.bbabio.2011.04.013 [DOI] [PubMed] [Google Scholar]
- Knox J. P., Dodge A. D. (1985). Singlet oxygen and plants. Phytochemistry 24, 889–896 10.1016/S0031-9422(00)83147-7 [DOI] [Google Scholar]
- Kolowrat C., Partensky F., Mella-Flores D., Le Corguillé G., Boutte C., Blot N., Ratin M., Ferréol M., Lecomte X., Gourvil P., Lennon J. F., Kehoe D. M., Garczarek L. (2010). Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus PCC9511. BMC Microbiol. 10, 204. 10.1186/1471-2180-10-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuvikova S., De Vries R., Mullineaux C. W., Tichy M., Nixon P. J. (2006). The FtsH protease Slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281, 1145–1151 10.1074/jbc.M503852200 [DOI] [PubMed] [Google Scholar]
- Komenda J., Knoppova J., Krynicka V., Nixon P. J., Tichy M. (2010). Role of FtsH2 in the repair of Photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn(4) cluster. Biochim. Biophys. Acta 1797, 566–575 10.1016/j.bbabio.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Kos P. B., Deak Z., Cheregi O., Vass I. (2008). Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the cyanobacterium Thermosynechococcus elongatus BP-1. Biochim. Biophys. Acta 1777, 74–83 10.1016/j.bbabio.2008.05.292 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A. (2005). Singlet oxygen production in photosynthesis. J. Exp. Bot. 56, 337–346 10.1093/jxb/erh237 [DOI] [PubMed] [Google Scholar]
- Krupa Z., Oquist G., Gustafsson P. (1991). Photoinhibition of photosynthesis and growth responses at different light levels in psbA gene mutants of the cyanobacterium Synechococcus. Physiol. Plant. 82, 1–8 10.1034/j.1399-3054.1991.820101.x [DOI] [Google Scholar]
- Kucho K., Okamoto K., Tsuchiya Y., Nomura S., Nango M., Kanehisa M., Ishiura M. (2005). Global analysis of circadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 187, 2190–2199 10.1128/JB.187.6.2190-2199.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K., Tsuchiya Y., Okumoto Y., Harada M., Yamada M., Ishiura M. (2004). Construction of unmodified oligonucleotide-based microarrays in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: screening of the candidates for circadianly expressed genes. Genes Genet. Syst. 79, 319–329 10.1266/ggs.79.189 [DOI] [PubMed] [Google Scholar]
- LaRoche J., Van Der Staay G. W., Partensky F., Ducret A., Aebersold R., Li R., Golden S. S., Hiller R. G., Wrench P. M., Larkum A. W., Green B. R. (1996). Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 15244–15248 10.1073/pnas.93.26.15244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A., Jeanjean R., Lemeille S., Havaux M., Zhang C. C. (2005). Iron starvation leads to oxidative stres in Anabaena sp. strain PCC 7120s. J. Bacteriol. 187, 6596–6598 10.1128/JB.187.18.6596-6598.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A., Ruiz M., Zhang C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278 10.1111/j.1574-6976.2008.00134.x [DOI] [PubMed] [Google Scholar]
- Liu H. B., Nolla H. A., Campbell L. (1997). Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 12, 39–47 10.3354/ame012039 [DOI] [Google Scholar]
- Llabres M., Agusti S. (2006). Picophytoplankton cell death induced by UV radiation: evidence for oceanic Atlantic communities. Limnol. Oceanogr. 51, 21–29 10.4319/lo.2006.51.1.0021 [DOI] [Google Scholar]
- Llabres M., Agusti S. (2010). Effects of ultraviolet radiation on growth, cell death and the standing stock of Antarctic phytoplankton. Aquat. Microb. Ecol. 59, 151–160 10.3354/ame01392 [DOI] [Google Scholar]
- Lupinkova L., Komenda J. (2004). Oxidative modifications of the photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochem. Photobiol. 79, 152–162 [DOI] [PubMed] [Google Scholar]
- MacIntyre H. L., Kana T. M., Geider R. J. (2000). The effect of water motion on short-term rates of photosynthesis by marine phytoplankton. Trends Plant Sci. 5, 12–17 10.1016/S1360-1385(99)01504-6 [DOI] [PubMed] [Google Scholar]
- Marie D., Partensky F., Jacquet S., Vaulot D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63, 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D., Partensky F., Vaulot D., Brussaard C. (1999). Enumeration of phytoplankton, bacteria, and viruses in marine samples. Curr. Protoc. Cytom. 10, 11.11.11–11.11.15 [DOI] [PubMed] [Google Scholar]
- Mary I., Vaulot D. (2003). Two-component systems in Prochlorococcus MED4: Genomic analysis and differential expression under stress. FEMS Microbiol. Lett. 226, 135–144 10.1016/S0378-1097(03)00587-1 [DOI] [PubMed] [Google Scholar]
- Miyao M., Ikeuchi M., Yamamoto N., Ono T. (1995). Specific degradation of the D1 protein of photosystem-II by treatment with hydrogen-peroxide in darkness – implications for the mechanism of degradation of the D1 protein under illumination. Biochemistry 34, 10019–10026 10.1021/bi00031a025 [DOI] [PubMed] [Google Scholar]
- Moore L. R., Goericke R., Chisholm S. W. (1995). Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116, 259–275 10.3354/meps116259 [DOI] [Google Scholar]
- Mullineaux C. W., Emlyn-Jones D. (2005). State transitions: an example of acclimation to low-light stress. J. Exp. Bot. 56, 389–393 10.1093/jxb/eri064 [DOI] [PubMed] [Google Scholar]
- Murakami A., Fujita Y. (1991). Regulation of photosystem stoichiometry in the photosynthetic system of the cyanophyte Synechocystis PCC 6714 in response to light intensity. Plant Cell Physiol. 32, 223–230 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Hihara Y. (2003). Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216, 446–453 [DOI] [PubMed] [Google Scholar]
- Ng W. O., Zentella R., Wang Y. S., Taylor J. S. A., Pakrasi H. B. (2000). PhrA, the major photoreactivating factor in the cyanobacterium Synechocystis sp strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch. Microbiol. 173, 412–417 10.1007/s002030000164 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y. (2006). A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim. Biophys. Acta 1757, 742–749 10.1016/j.bbabio.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Allakhverdiev S. I., Yamamoto H., Hayashi H., Murata N. (2004). Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry 43, 11321–11330 10.1021/bi036178q [DOI] [PubMed] [Google Scholar]
- Nixon P. J., Sarcina M., Diner B. A. (2005). The D1 and D2 core proteins. Photosystem II 22, 71–93 10.1007/1-4020-4254-X_5 [DOI] [Google Scholar]
- Ohnishi N., Allakhverdiev S. I., Takahashi S., Higashi S., Watanabe M., Nishiyama Y., Murata N. (2005). Two-step mechanism of photodamage to photosystem II: step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry 44, 8494–8499 10.1021/bi047518q [DOI] [PubMed] [Google Scholar]
- Ong L. J., Glazer A. N. (1991). Phycoerythrins of the marine unicellular cyanobacteria. J. Biol. Chem. 266, 9519–9527 [PubMed] [Google Scholar]
- Park Y. I., Chow W. S., Anderson J. M. (1995). The quantum yield of photoinactivation of photosystem II in pea leaves is greater at low than high photon exposure. Plant Cell Physiol. 36, 1163–1167 [Google Scholar]
- Partensky F., Blanchot J., Vaulot D. (1999a). “Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review,” in Marine Cyanobacteria, eds Charpy L., Larkum A. (Monaco: Musée Océanographique; ), 457–475 [Google Scholar]
- Partensky F., Hess W. R., Vaulot D. (1999b). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky F., Garczarek L. (2003). “The photosynthetic apparatus of chlorophyll b- and d-containing oxychlorobacteria,” in Photosynthesis in Algae, eds Larkum A. W. D., Douglas S. E., Raven J. A. (Dordrecht: Kluwer Academic Publishers; ), 29–62 [Google Scholar]
- Partensky F., Garczarek L. (2010). Prochlorococcus: advantages and limits of minimalism. Annu. Rev. Mar. Sci. 2, 305–331 10.1146/annurev-marine-120308-081034 [DOI] [PubMed] [Google Scholar]
- Powles S. B. (1984). Photoinhibition of photosynthesis induced by visible light. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35, 15–44 10.1146/annurev.arplant.35.1.15 [DOI] [Google Scholar]
- Prásil O., Adir A., Ohad I. (1992). “Dynamics of photosystem II: mechanism of photoinhibition and recovery processes,” in Topics in photosynthesis – the photosystems: structure, function and molecular biology, ed. Barber J. (Amsterdam: Elsevier Science Publishers; ), 295–348 [Google Scholar]
- Rastogi R. P., Singh S. P., Hader D. P., Sinha R. P. (2010). Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem. Biophys. Res. Commun. 397, 603–607 10.1016/j.bbrc.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Rippka R., Coursin T., Hess W., Lichtlé C., Scanlan D. J., Palinska K. A., Iteman I., Partensky F., Houmard J., Herdman M. (2000). Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 50, 1833–1847 [DOI] [PubMed] [Google Scholar]
- Rocap G., Larimer F. W., Lamerdin J., Malfatti S., Chain P., Ahlgren N. A., Arellano A., Coleman M., Hauser L., Hess W. R., Johnson Z. I., Land M., Lindell D., Post A. F., Regala W., Shah M., Shaw S. L., Steglich C., Sullivan M. B., Ting C. S., Tolonen A., Webb E. A., Zinser E. R., Chisholm S. W. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047 10.1038/nature01947 [DOI] [PubMed] [Google Scholar]
- Ross C., Santiago-Vazquez L., Paul V. (2006). Toxin release in response to oxidative stress and programmed cell death in the cyanobacterium Microcystis aeruginosa. Aquat. Toxicol. 78, 66–73 10.1016/j.aquatox.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Ryu J. Y., Song J. Y., Chung Y., Park Y. M., Chow W. S., Park Y. I. (2010). Transcript accumulation of carotenoid biosynthesis genes in the cyanobacterium Synechocystis sp. PCC 6803 during the dark-to-light transition is mediated by photosynthetic electron transport. Plant Biotechnol. Rep. 4, 149–155 10.1007/s11816-010-0130-7 [DOI] [Google Scholar]
- Saito M. A., Bertrand E. M., Dutkiewicz S., Bulygin V. V., Moran D. M., Monteiro F. M., Follows M. J., Valois F. W., Waterbury J. B. (2011). Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci. U.S.A. 108, 2184–2189 10.1073/pnas.1006943108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvikas P., Hakala M., Patsikka E., Tyystjarvi T., Tyystjarvi E. (2006). Action spectrum of photoinhibition in leaves of wild type and npq1-2 and npq4-1 mutants of Arabidopsis thaliana. Plant Cell Physiol. 47, 391–400 10.1093/pcp/pcj006 [DOI] [PubMed] [Google Scholar]
- Sass L., Spetea C., Mate Z., Nagy F., Vass I. (1997). Repair of UV-B induced damage of photosystem II via de novo synthesis of the D1 and D2 reaction centre subunits in Synechocystis sp. PCC 6803. Photosyn. Res. 54, 55–62 10.1023/A:1005895924892 [DOI] [Google Scholar]
- Satoh S., Tanaka A. (2006). Identification of chlorophyllide a oxygenase in the Prochlorococcus genome by a comparative genomic approach. Plant Cell Physiol. 47, 1622–1629 10.1093/pcp/pcl026 [DOI] [PubMed] [Google Scholar]
- Scanlan D. J. (2003). Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol. 47, 1–64 10.1016/S0065-2911(03)47001-X [DOI] [PubMed] [Google Scholar]
- Scanlan D. J., Ostrowski M., Mazard S., Dufresne A., Garczarek L., Hess W. R., Post A. F., Hagemann M., Paulsen I., Partensky F. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299 10.1128/MMBR.00035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Shalapyonok A., Olson R. J., Shalapyonok L. S. (1998). Ultradian growth in Prochlorococcus spp. Appl. Environ. Microbiol. 64, 1066–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Ilikchyan I., Rabouille S., Zehr J. P. (2010). Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J. 4, 621–632 10.1038/ismej.2009.148 [DOI] [PubMed] [Google Scholar]
- Sicora C. I., Appleton S. E., Brown C. M., Chung J., Chandler J., Cockshutt A. M., Vass I., Campbell D. A. (2006). Cyanobacterial psbA families in Anabaena and Synechocystis encode trace, constitutive and UVB-induced D1 isoforms. Biochim. Biophys. Acta 1757, 47–56 10.1016/j.bbabio.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Sicora C. I., Brown C. M., Vass I., Cheregi O., Campbell D. A. (2008). The psbA gene family responds differentially to light and UVB stress in Gloeobacter violaceus PCC 7421, a deeply divergent cyanobacterium. Biochim. Biophys. Acta 777, 130–139 [DOI] [PubMed] [Google Scholar]
- Sidler W. A. (1994). “Phycobilisome and phycobiliprotein structure,” in The Molecular Biology of Cyanobacteria, ed. Bryant D. A. (Dordrecht: Kluwer Academic Publishers; ), 139–216 [Google Scholar]
- Sies H., Menck C. F. M. (1992). Singlet oxygen induced DNA damage. Mutat. Res. 275, 367–375 10.1016/0921-8734(92)90039-R [DOI] [PubMed] [Google Scholar]
- Six C., Finkel Z. V., Irwin A. J., Campbell D. A. (2007a). Light variability illuminates niche-partitioning among marine picocyanobacteria. PLoS ONE 2, e1341. 10.1371/journal.pone.0001341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six C., Joubin L., Partensky F., Holtzendorff J., Garczarek L. (2007b). UV-induced phycobilisome dismantling in the marine picocyanobacterium Synechococcus sp. WH8102. Photosyn. Res. 92, 75–86 10.1007/s11120-007-9170-4 [DOI] [PubMed] [Google Scholar]
- Six C., Thomas J. C., Garczarek L., Ostrowski M., Dufresne A., Blot N., Scanlan D. J., Partensky F. (2007c). Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol. 8, R259. 10.1186/gb-2007-8-12-r259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six C., Sherrard R., Lionard M., Roy S., Campbell D. A. (2009). Photosystem II and pigment dynamics among ecotypes of the green alga Ostreococcus. Plant Physiol. 151, 379–390 10.1104/pp.109.140566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six C., Thomas J. C., Brahamsha B., Lemoine Y., Partensky F. (2004). Photophysiology of the marine cyanobacterium Synechococcus sp. WH8102, a new model organism. Aquat. Microb. Ecol. 35, 17–29 10.3354/ame035017 [DOI] [Google Scholar]
- Sommaruga R., Hofer J. S., Alonso-Saez L., Gasol J. A. (2005). Differential sunlight sensitivity of picophytoplankton from surface Mediterranean coastal waters. Appl. Environ. Microbiol. 71, 2154–2157 10.1128/AEM.71.4.2154-2157.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger S., Jackisch Y., Sandmann G. (2005). Carotenoid biosynthesis in Gloeobacter violaceus PCC4721 involves a single crtI-type phytoene desaturase instead of typical cyanobacterial enzymes. Arch. Microbiol. 184, 207–214 10.1007/s00203-005-0004-5 [DOI] [PubMed] [Google Scholar]
- Stockel J., Welsh E. A., Liberton M., Kunnvakkam R., Aurora R., Pakrasi H. B. (2008). Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. U.S.A. 105, 6156–6161 10.1073/pnas.0711068105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield T. C., Sherman L. A. (2007). Role of sigma factors in controlling global gene expression in light/dark transitions in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 189, 7829–7840 10.1128/JB.01036-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Murata N. (2008). How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 13, 178–182 10.1016/j.tplants.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Takai N., Nakajima M., Oyama T., Kito R., Sugita C., Sugita M., Kondo T., Iwasaki H. (2006). A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12109–12114 10.1073/pnas.0602955103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A. (2005). Too much light? How beta-carotene protects the photosystem II reaction centre. Photochem. Photobiol. Sci. 4, 950–956 10.1039/b507888c [DOI] [PubMed] [Google Scholar]
- Tichy M., Lupinkova L., Sicora C., Vass I., Kuvikova S., Prasil O., Komenda J. (2003). Synechocystis 6803 mutants expressing distinct forms of the Photosystem II D1 protein from Synechococcus 7942: relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim. Biophys. Acta 1605, 55–66 10.1016/S0005-2728(03)00064-1 [DOI] [PubMed] [Google Scholar]
- Toepel J., Mcdermott J. E., Summerfield T. C., Sherman L. A. (2009). Transcriptional analysis of the unicellular, diazotrophic cyanobacterium Cyanothece sp. ATCC 51142 grown under short day / night cycles. J. Phycol. 45, 610–620 10.1111/j.1529-8817.2009.00674.x [DOI] [PubMed] [Google Scholar]
- Toepel J., Welsh E., Summerfield T. C., Pakrasi H. B., Sherman L. A. (2008). Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light-dark and continuous-light growth. J. Bacteriol. 190, 3904–3913 10.1128/JB.00206-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjarvi E., Aro E. M. (1996). The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc. Natl. Acad. Sci. U.S.A. 93, 2213–2218 10.1073/pnas.93.5.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Poll W. H., Eggert A., Buma A. G. J., Breeman A. M. (2001). Effects of UV-B-induced DNA damage and photoinhibition on growth of temperate marine red macrophytes: habitat-related differences in UV-B tolerance. J. Phycol. 37, 30–37 10.1046/j.1529-8817.2001.037001030.x [DOI] [Google Scholar]
- van Waasbergen L. G., Dolganov N., Grossman A. R. (2002). nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J. Bacteriol. 184, 2481–2490 10.1128/JB.184.9.2481-2490.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass I., Styring S., Hundal T., Koivuniemi A., Aro E. M., Andersson B. (1992). Reversible and irreversible intermediates during photoinhibition of photosystem II: stable reduced QA species promote chlorophyll triplet formation. Proc. Natl. Acad. Sci. U.S.A. 89, 1408–1412 10.1073/pnas.89.4.1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D., Marie D., Olson R. J., Chisholm S. W. (1995). Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial Pacific Ocean. Science 268, 1480–1482 10.1126/science.268.5216.1480 [DOI] [PubMed] [Google Scholar]
- Wilson A., Punginelli C., Gall A., Bonetti C., Alexandre M., Routaboul J. M., Kerfeld C. A., Van Grondelle R., Robert B., Kennis J. T., Kirilovsky D. (2008). A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl. Acad. Sci. U.S.A. 105, 12075–12080 10.1073/pnas.0804276105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman M., Thom C. (2012). Temporal orchestration of glgA (encoding glycogen synthase) expression and glycogen accumulation in the oceanic picoplanktonic cyanobacterium, Synechococcus sp. strain WH8103. Appl. Environ. Microbiol. 78, 4744–4747 10.1128/AEM.00254-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E. R., Lindell D., Johnson Z. I., Futschik M. E., Steglich C., Coleman M. L., Wright M. A., Rector T., Steen R., Mcnulty N., Thompson L. R., Chisholm S. W. (2009). Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE 4, e5135. 10.1371/journal.pone.0005135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla L., Rinalducci S. (2002). Involvement of active oxygen species in degradation of light-harve sting proteins under light stresses. Biochemistry 41, 14391–14402 10.1021/bi0265776 [DOI] [PubMed] [Google Scholar]
- Zubkov M. V., Sleigh M. A., Burkill P. H. (2000). Assaying picoplankton distribution by flow cytometry of underway samples collected along a meridional transect across the Atlantic Ocean. Aquat. Microb. Ecol. 21, 13–20 10.3354/ame021013 [DOI] [Google Scholar]
- Zwirglmaier K., Jardillier L., Ostrowski M., Mazard S., Garczarek L., Vaulot D., Not F., Massana R., Ulloa O., Scanlan D. J. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used for real time PCR reactions. Most primers are targeting a specific gene. Exceptions include the psbD pool and psbA pool primers, which were designed to amplify the two psbD gene copies and all four psbA copies present in Synechococcus sp. WH7803, respectively. Furthermore, one primer set targets two psbA genes (SynWH7803_0366 and 2084), which have nearly identical coding sequence and 5′-UTR. Genes are classified by functional categories, as indicated in the first column. The Cyanorak database of picocyanobacteria protein families is publicly accessible at http://www.sb-roscoff.fr/Phyto/cyanorak/
Daily variations of the expression of selected genes, as measured by real time quantitative PCR, for picocyanobacterial cells acclimated to a modulated 12/12 h L/D cycle of VL with or without UV radiations. All data are expressed as log2 (Fold Change) ± mean deviation for two biological replicates. For each gene, transcript levels are normalized to the reference time point 6:00 in VL. Genes are classified by functional categories, as indicated in the first column. This table is a detailed version of Figure 7 of the main manuscript.