Structurally diverse CpG oligonucleotides have a shared ability to activate IFN-dependent ‘core genes’ with antiviral activity while maintaining sequence-specific effects on human pDC.
Keywords: human, pDC, CAL-1, TLR9, gene regulation, oligonucleotide
Abstract
Synthetic ODNs expressing CpG motifs trigger an innate immune response via TLR9. pDCs are major effectors of this response. Two structurally distinct classes of CpG ODNs have been identified that differentially activate pDCs. “K” ODNs trigger the production of TNF-α and IL-6, whereas “D” ODNs preferentially induce the secretion of IFN-α. As K and D ODNs have distinct therapeutic effects, knowledge of their shared and sequence-specific activity is of considerable importance. This work uses the CAL-1 human pDC line to analyze the effect of CpG stimulation on gene expression. Genes up-regulated by both K and D ODNs (n=92) were largely dependent on type I IFN signaling and characterized functionally by antiviral activity. K ODNs induced a short-term increase in IFN-α/β production and uniquely up-regulated genes that supported antibacterial responses. In contrast, D ODNs triggered a persistent increase in IFN-α/β production and uniquely up-regulated genes associated with metabolic functions. Thus, the core functionality of human pDCs mediated by TLR9 ligation rests on a type I IFN response that differs from the response induced by the structural elements unique to specific classes of ODNs.
Introduction
Pathogens stimulate the mammalian immune system to mount a rapid inflammatory response. Supporting this response is TLR9-mediated recognition of the unmethylated CpG motifs present at high frequency in viral and bacterial genomes [1, 2]. The regulation of TLR9-mediated immunity is commonly probed using synthetic ODNs that express CpG motifs. Several discrete classes of CpG ODNs have been identified that differentially activate human pDC, a cell lineage critical to the immunoprotective response [3, 4]. K-type ODNs (also referred to as CpG-B) are linear, single-stranded polynucleotides that express multiple unmethylated CpG motifs [5]. K ODNs trigger a NF-κB-dependent proinflammatory response characterized by the production of IL-6 and TNF-α by pDCs [3]. In contrast, D-type ODNs (also referred to as CpG-A) form complex stem-loop structures and have a poly-G tail that leads them to form G-tetrads [6, 7]. D ODNs stimulate pDCs to produce IFN-α/β rather than TNF-α, an effect that is amplified though an autocrine feedback loop [8, 9].
Although K and D ODNs signal via TLR9, the intracellular trafficking of K differs from that of D ODNs in pDCs [10–12]. Internalized K ODNs enter lysosomal vesicles where they trigger a NF-κB-dependent cascade via TRAF6 [11–13]. In contrast, D ODNs traffic into specialized endolysosomes, where they engage adaptor protein-3 complexes and signal via TRAF3 and IRF7 [10].
Microarray studies can provide insight into the global changes in gene expression elicited by CpG stimulation. Experiments using murine splenocytes, macrophages, and DCs, as well as unfractionated human PBMCs document that inductive and suppressive regulatory networks are triggered during CpG-driven immune responses [14–18]. Those reports showed that CpG stimulation up-regulated the expression of several hundred genes [14–18]. Whereas individual studies described the effect of specific classes of ODNs on the activation of genes and regulatory networks, no general conclusions were reached concerning common mechanisms underlying CpG-mediated immune responses. Indeed, it is unclear whether different classes of ODNs activate similar or distinct gene sets and whether a conserved set of genes is activated by multiple forms of TLR9 ligation.
To assess these issues, microarray studies were performed in which the CAL-1 human pDC line was stimulated for up to 48 h with K or D ODNs. CAL-1 cells are unique in displaying typical plasmacytoid morphology and surface markers characteristic of human pDCs (e.g., CD123+, blood DC antigen-2+, CD11c−) combined with an ability to mirror the response of primary human pDCs to stimulation [19, 20]. Central findings from microarray experiments of CAL-1 cells were verified by selective RT-PCR studies of CpG-activated, primary human pDCs {the large number of pDCs required for microarray studies was unavailable as a result of their low frequency in human peripheral blood (<0.5%) and their susceptibility to activation during the isolation process [21, 22]}. Results identified a set of 92 genes that was reproducibly activated by both K and D ODNs. These common genes differed from those up-regulated by only K or only D ODN in that they included a large number of ISGs associated with antiviral activity. These findings help define the conserved and sequence-specific patterns of gene activation triggered via TLR9 and improve our understanding of the immunomodulatory effects elicited by CpG ODNs.
MATERIALS AND METHODS
ODNs
Endotoxin-free ODNs were synthesized at the Center for Biologics Evaluation and Research core facility (U.S. Food and Drug Administration, Bethesda, MD, USA). An equimolar mixture of three K or D ODN was used in all experiments, as previous studies showed that such mixtures more consistently stimulated cells from multiple donors than did individual ODNs [23]. The K mixture was composed of K3 (5′-ATCGACTCTCGAGCGTTCTC-3′), K23 (5′-TCGAGCGTTCTC-3′), and K123 (5′-TCGTTCGTTCTC-3′), whereas the D mixture contained D19 (5′-GGtgcatcgatgcagGGGGG-3′), D29 (5′- GGtgcaccggtgcagGGGGG-3′), and D35 (5′-GgtgcatcgatgcaggggGG-3′) [6]. ODN 1612 (5′-GCTAGAGCTTAGGCT-3′) served as a control for K ODN, whereas ODN D122 (5′-GgtgcattgatgcagGGGGG) acted as the control for D ODN. In both control ODNs, the immunostimulatory CpG motif was inverted to a nonstimulatory GpC. Bases shown in capital letters are phosphorothioate, and those in lowercase letters are phosphodiester.
Culture and maintenance of CAL-1 cells
The CAL-1 pDC line was grown in RPMI 1640 (Lonza, Walkersville, MD, USA), supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, and 1× minimum essential medium–nonessential amino acid (all from Gibco, Grand Island, NY, USA), plus 10% heat-inactivated FBS (Lonza; hereafter, referred to as complete medium). Cells were maintained in 150 cm2 flasks at a density of 1–2.4 × 106 cells/ml in a total volume of 25 ml and split every 2 days.
Isolation and culture of human pDCs
Human PBMCs were prepared from buffy coats of healthy donors provided by the Department of Transfusion Medicine, U.S. National Institutes of Health (Bethesda, MD, USA), using an Institutional Review Board-approved protocol. PBMCs were obtained by density gradient centrifugation over Histopaque-1077 (Sigma-Aldrich, St. Louis, MO, USA). pDCs were separated using the Diamond pDC Isolation Kit from Miltenyi Biotec (Auburn, CA, USA). Briefly, pDCs were enriched by magnetic labeling of non-pDCs using a biotin-antibody cocktail plus anti-biotin microbeads, followed by autoMACS sorting (program “Depletes”). The resulting population contained 83–87% pDCs and <1% B cells. The remaining cells were TRL9-negative monocytes, T cells, and NK cells. Cell viability exceeded 95%, as determined by trypan blue exclusion. pDCs were cultured in RPMI 1640 containing 2% FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin (Gibco).
ELISA and RT-PCR studies to detect cytokine production
CAL-1 cells (106) in 1 ml medium (enough to cover 80% of the well surface) were plated in 24-well plates and rested overnight in 1% FBS before being stimulated in complete media. Freshly isolated pDCs (5×105) in 0.5 ml medium were plated in 48-well plates and rested for 2 h before stimulation. Based on previous studies, the cells were stimulated with an optimal concentration of 1 μM K ODN or 3 μM D ODN for the times indicated [6]. In some experiments, 5 μg/ml anti-IFNR antibody (mouse mAb against human IFN-α/βR chain 2, PBL Biomedical Laboratories, Piscataway, NJ, USA) was added 30 min prior to CpG stimulation. Human rIFN-β1a (100 IU/ml; PBL Biomedical Laboratories) was used as a positive control to document the induction of type I IFN-dependent genes.
Culture supernatants were analyzed by ELISA for cytokine content. DuoSet ELISA kits (R&D Systems, Minneapolis, MN, USA) were used to detect IL-6 and TNF-α. An IFN-α human multisubtype ELISA kit (PBL Biomedical Laboratories) was used to detect IFN-α, whereas the VeriKine human IFN-β ELISA kit (PBL Biomedical Laboratories) was used to detect IFN-β. All ELISA assays were performed according to the manufacturer's instructions.
RT-PCR studies were conducted by isolating total RNA from CAL-1 cells or primary pDCs using an RNeasy kit (Qiagen, Hilden, Germany). RNA (0.5 μg) was reverse-transcribed into cDNA using the QuantiTect RT kit (Qiagen), according to the manufacturer's instructions. Gene expression levels (normalized to GAPDH) were analyzed using the StepOnePlus RT-PCR system (Applied Biosystems, Foster City, CA, USA). All reagents and probes used in these studies were purchased from Applied Biosystems. The following TaqMan assays were used: MX1 (Hs00895598_m1), OAS2 (Hs00942643_m1), ISG15 (Hs00192713_m1), TRAF4 (Hs01030624_m1), IFNA1 (Hs00256882_s1), IFNB1 (Hs02621180_s1), IL6 (Hs00174131_m1), and TNF (Hs00174128_m1).
Microarray studies to detect changes in gene expression
CAL-1 cells in 15 ml complete RPMI medium were transferred into 75 cm2 flasks and grown to cover 80% of the culture area. They were rested overnight in RPMI plus 1% FBS before being stimulated with 3 μM K or D ODN. Blocking experiments were performed by adding 5 μg/ml neutralizing mAb against the type I IFNR, 30′ prior to CpG administration. All experiments at all time-points were repeated a minimum of three times.
Total RNA was extracted from CAL-1 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), as specified by the manufacturer. The RNA was quantified using a NanoDrop 1000 (NanoDrop Technologies, Thermo Fisher Scientific, Wilmington, DE, USA) and assessed for the absence of degradation by electrophoresis or use of a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Total RNA (20 μg) was reversed-transcribed using 3 μl 10× first-strand buffer (Stratagene, La Jolla, CA, USA), 2 μl (1 μg) oligo(dT)12–18 (Invitrogen), 3 μl (150 U) AffinityScript RT, 2 μl 20× aminoallyl-dUTP/dNTP mix, 1 μl (40 U) RNAse inhibitor (Promega, Madison, WI, USA), and 3 μl 0.1 M DTT in a final volume of 30 μl at 37°C for 1 h, as described previously [24]. For all samples, a universal reference RNA (Stratagene) was processed in parallel. Both cDNAs were purified using a MinElute PCR purification kit (Qiagen) and labeled with Cy5 (sample DNA) or Cy3 (reference RNA) as described previously [14]. The probes were mixed, diluted in 5 μl DMSO plus 1.7 μl 1 M NaHCO3, and hybridized to 36 K human 60-mer ODN array slides (NCI Human Array Set Hs-Operon V 3.0) at 42°C for 18 h in a Micro Array User Interface hybridization system (BioMicro Systems, Salt Lake City, UT, USA), followed by washing, centrifugation, and air-drying.
Analysis of gene expression
Arrays were scanned using a GenePix 4000B scanner (Axon Instruments, Union City, CA, USA) and analyzed using the GenePix Pro 6.0 software tool (Axon Instruments), using Gene Array List files provided by the manufacturer at http://madb.nci.nih.gov/. Data were uploaded to the Center for Information Technology/BioInformatics and Molecular Analysis Section–NCI/Center for Cancer Research Microarray Database and formatted via export function to BRB ArrayTools. Gene expression analysis was performed using BRB ArrayTools Version 3.8.1 developed by the NCI BRB (Bethesda, MD, USA). Data were background-corrected, flagged values were removed, spots in which both signals were <110 were filtered, ratios were log base 2-transformed, and lowest intensity-dependent normalization was used to adjust for differences in labeling intensities of the Cy3 and Cy5 dyes. Analysis was restricted to genes present in >70% of the arrays after filtering. In toto, 2.9 × 104 features were reproducibly tracked in all microarrays. The gene expression profile of all treated groups was compared with that of untreated controls.
A P value cutoff of 10−5 was used to identify genes whose expression was up-regulated by CpG compared with untreated controls. All results were evaluated using IPA (Ingenuity Systems, Redwood City, CA, USA) and BIOBASE (BIOBASE Biological Databases, Germany). The GO Analysis was performed using the GO database AmiGO (http://amigo.geneontology.org).
Statistical analysis
Genes that were expressed differentially in the treatment groups were identified using a random variance t test. The random variance t test is an improvement over the standard, separate t test, as it permits sharing information among genes about within-class variation without assuming that all genes have the same variance [25]. Differences between gene sets (e.g., in terms of affiliation to GO groups) were assessed using two-tailed Fisher exact tests. Differences of fold changes between genes or treatment groups were established by two-sided t tests.
Accession code
Microarray data were deposited in the National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series Accession Number GSE 30,849.
RESULTS
CAL-1 cells mimic the response of human pDCs to CpG stimulation
Human pDCs mount TNF-α- and IL-6-dominated responses when stimulated with K ODN yet generate IFN-α-dominated responses when stimulated with D ODN [3, 8, 9] (Fig. 1 and Supplemental Figs. 1 and 2). These differences have been attributed to K ODN interacting with TLR9 in endosomal vesicles and signaling via TRAF6 and NF-κB while D ODN engage TLR9 in specialized endolysosomes and signal via IRF7 and TRAF3 [10–12]. Previous studies showed that CAL-1 cells share many characteristics of human pDCs, including their expression of TLR9 [19]. To determine whether this cell line mimics the response of human pDCs to CpG stimulation, their patterns of cytokine production were compared after stimulation with K and D ODN.
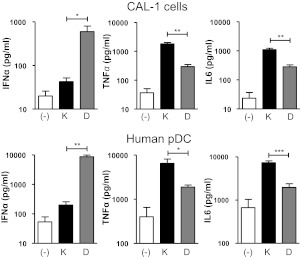
Figure 1. Cytokine production by CAL-1 cells and purified human pDCs treated with K or D ODN.
CAL-1 cells and freshly isolated pDCs were cultured for 24 h in medium (−) supplemented with 1 μM K or 3 μM D ODN (these concentrations were previously found to be optimal [6, 24]). The amount of IFN-α, TNF-α, and IL-6 in culture supernatants was determined by ELISA. Results represent the mean ± sem of three to four independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Consistent with previous reports involving human pDCs [3, 8, 9], CAL-1 cells stimulated for 24 h with K ODN mounted a significantly stronger TNF-α and IL-6 response than those incubated with D ODN, whereas CAL-1 cells stimulated with D ODN preferentially produced IFN-α (P<0.01; Fig. 1). The same pattern and kinetics of responsiveness were observed when freshly isolated human pDCs were stimulated with K versus D ODN, although the absolute magnitude of the responses varied (Fig. 1 and Supplemental Figs. 1 and 2). This difference in magnitude may reflect the presence of preactivated cells in the pDC pool or the twofold higher level of expression of TLR9 by pDCs versus CAL-1 cells (Supplemental Fig. 2B). Based on these and previous findings, we concluded that CAL-1 cells provided a relevant model for evaluating the effect of CpG ODNs on gene expression in human pDCs.
Effect of CpG ODNs on gene expression by CAL-1 cells
Previous studies using murine splenocytes and macrophages showed that the changes in gene expression induced by K ODN peaked within 12 h and declined to near background by 24 h [15, 24]. Thus, initial studies evaluated the response of CAL-1 cells to K and D ODN stimulation after 1, 3, 9, and 27 h. Each time-point was evaluated in at least three independent experiments, and results represent the mean of all responses. Those genes whose level of expression differed significantly from untreated controls were identified using a stringency cutoff of P < 10−5. CAL-1 cells stimulated with control K ODN (structurally similar to K but lacking the CpG motif) failed to up-regulate any genes when compared with untreated controls. Control D ODN (which lack the CpG motif but retain the stem-loop structure and poly G tail characteristic of D ODN) induced limited gene up-regulation. Only those genes activated by D but not control D ODN were included in subsequent analyses.
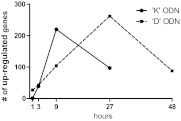
The kinetics of gene up-regulation elicited by K versus D ODN differed (Fig. 2). Consistent with previous studies involving murine cells, gene activation peaked after 9 h of stimulation with K ODN (n=220 genes) and then fell toward baseline. In contrast, gene activation continued to increase through 27 h of D ODN stimulation (n=262; Fig. 2). The effect of D ODN was therefore evaluated at a later time-point (48 h), by which time gene expression returned toward baseline. Combining data from all time-points, a total of 249 genes was up-regulated by K and 305 genes by D ODNs. Ninety-two of these genes were activated by both classes of CpG ODNs (Fig. 3).
Figure 2. Kinetics of gene up-regulation in CAL-1 cells stimulated with CpG ODNs.
CAL-1 cells were incubated with 3 μM K or D ODN and gene expression monitored by microarray. Results represent the average of at least three independent experiments at each time-point. Two additional experiments were performed at the peak of gene up-regulation (9 h for K and 27 h for D ODN), so those time-points reflect the average of five independent studies. Up-regulated genes were identified by comparison with unstimulated controls (evaluated in six independent studies) using a stringency cutoff of P < 10−5. Note that control K ODN (lacking CpG motifs) failed to up-regulate any genes over this period, whereas control D ODN induced limited gene up-regulation (evaluated in four independent experiments). These non-CpG-activated genes were not included in further analyses.
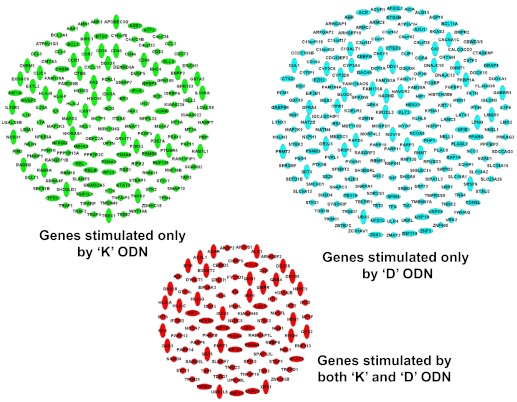
Figure 3. Genes up-regulated by K and/or D ODNs.
CAL-1 cells were stimulated with K or D ODN, as described in Fig. 2. All genes up-regulated significantly at any time-point are shown (P<10−5). Ninety-two genes were stimulated by both K and D ODNs (red), 157 genes were stimulated by only K ODN (green), and 213 by only D ODN (blue).
Functional analysis of genes activated by K and/or D ODNs
All of the genes activated by K and/or D ODNs were categorized functionally using the GO database coupled with IPA. The GO database classifies gene products based on their associated biological processes, cellular components, and molecular functions, whereas IPA characterizes gene products based on their function, expression in disease states, and role in regulatory pathways. Combining the output from both databases facilitates gene function categorization [18, 24].
Consistent with previous studies, genes associated with immune function were preferentially up-regulated by K and D ODN (P<10−8). Antiviral activity (GO Category 0009615) was the most conserved, functional characteristic of the genes up-regulated by both classes of ODNs (P<10−15; Table 1), constituting 23 of these 92 genes. Consistent with antiviral genes being of considerable relevance to CpG-mediated immunity, 18 of the 20 genes most strongly up-regulated by K and D ODNs fell into this category (Table 2).
Table 1. Functional Grouping of Genes Up-Regulated by K and D ODNs.
| Significantly up-regulated genes | Responding to |
||
|---|---|---|---|
| K and D | K only | D only | |
| Total number | 92 | 157 | 213 |
| Functional grouping | |||
| Immune system | 32 | 54 | 25 |
| Innate [n (%)] | 12 (38%) | 13a (24%) | 1 (4%) |
| Adaptive/B and T cell activation [n (%)] | 6 (19%) | 25 (46%) | 7 (28%) |
| Inflammatory [n (%)] | 7 (22%) | 28b (52%) | 5 (20%) |
| Antiviral but not antibacterial [n (%)] | 23c (72%) | 7 (15%) | 5 (20%) |
| Antibacterial but not antiviral [n (%)] | 0 | 19a (35%) | 3 (12%) |
| Metabolic process | 56 | 102 | 116 |
CAL-1 cells were stimulated with 3 μM K or D ODN and changes in gene expression monitored for 1–48 h by microarray, as described in the legend to Fig. 2. All genes up-regulated (P<10−5) significantly at any time-point were identified. These genes were then grouped on the basis of being regulated by only K ODN, only D ODN, or both classes of ODNs. Each group of genes was then functionally categorized using GO and IPA. The functional subgroups of activated genes contributing to host immunity are shown and their fractional distribution as a percentage of the number of immune genes induced by K and/or D ODNs calculated. Fisher exact tests were performed to identify GO groups that were overexpressed significantly. Note that genes in Column 1 were compared with those in Column 2 or 3, whereas those in Columns 2 and 3 were compared with each other.
P < 0.05;
P < 0.01;
P < 0.001.
Table 2. List of All Genes Significantly Up-Regulated (P<10−5) by K and D ODNs.
| Gene symbol | K ODN |
D ODN |
||||||
|---|---|---|---|---|---|---|---|---|
| 1 h | 3 h | 9 h | 27 h | 1 h | 3 h | 9 h | 27 h | |
| ACSL1 | 0.9 | 2.3 | 6.8 | 2.7 | 1.3 | 0.9 | 1.6 | 3.5 |
| ADAR | 1.1 | 1.2 | 2.4 | 2.3 | 1.3 | 1.5 | 2.5 | 3.8 |
| AKAP2 | 1.3 | 1.3 | 2.1 | 1.3 | 1.7 | 1.8 | 2.3 | 2.5 |
| AKT1S1 | 1.0 | 1.3 | 4.0 | 2.6 | 1.8 | 2.6 | 2.1 | 2.4 |
| ALG2 | 1.3 | 1.9 | 3.2 | 2.1 | 1.7 | 1.9 | 2.3 | 3.4 |
| ARHGEF3 | 1.7 | 2.8 | 4.3 | 3.3 | 1.3 | 2.3 | 3.1 | 5.4 |
| AZI2 | 1.1 | 1.5 | 2.7 | 1.5 | 1.0 | 1.2 | 1.6 | 3.3 |
| B2M | 1.0 | 1.4 | 3.3 | 2.9 | 1.2 | 1.1 | 1.6 | 3.0 |
| B3GNT2 | 1.1 | 3.6 | 6.0 | 2.6 | 1.2 | 1.7 | 2.7 | 5.0 |
| CBWD5 | 1.1 | 1.3 | 2.7 | 1.8 | 1.3 | 1.3 | 1.9 | 3.3 |
| CHMP5 | 1.3 | 1.7 | 5.6 | 4.4 | 1.3 | 1.6 | 3.2 | 6.9 |
| CYLD | 1.1 | 1.9 | 3.2 | 2.2 | 1.0 | 1.3 | 1.6 | 3.2 |
| DCK | 1.3 | 1.1 | 2.4 | 1.4 | 1.2 | 1.3 | 1.5 | 3.3 |
| DDX58 | 1.1 | 3.1 | 20.7 | 6.8 | 1.0 | 2.2 | 3.9 | 11.2 |
| DDX60 | 0.9 | 0.9 | 7.0 | 7.6 | 0.8 | 0.8 | 2.6 | 11.8 |
| DHX58 | 0.8 | 2.5 | 5.9 | 2.8 | 0.9 | 0.9 | 2.3 | 6.0 |
| DTX3L | 1.2 | 1.8 | 3.2 | 2.6 | 1.2 | 1.8 | 2.1 | 2.9 |
| DYNLT1 | 0.9 | 1.4 | 3.9 | 1.8 | 1.0 | 0.8 | 1.5 | 3.3 |
| EIF2AK2 | 1.4 | 2.0 | 4.3 | 4.2 | 0.9 | 1.0 | 2.0 | 3.2 |
| EPSTI1 | 1.0 | 2.2 | 35.3 | 34.3 | 0.8 | 1.3 | 8.3 | 37.8 |
| FYTTD1 | 1.1 | 1.3 | 2.8 | 1.7 | 1.1 | 1.2 | 1.5 | 2.5 |
| GCH1 | 1.1 | 1.6 | 4.6 | 1.9 | 1.2 | 1.6 | 1.8 | 3.2 |
| GMPR | 1.2 | 1.8 | 6.8 | 5.1 | 1.2 | 1.5 | 3.4 | 7.2 |
| GNB4 | 0.9 | 1.2 | 2.4 | 1.5 | 1.4 | 1.1 | 1.9 | 3.6 |
| H3F3A/B | 1.1 | 3.0 | 8.1 | 4.9 | 1.1 | 1.4 | 2.6 | 6.9 |
| HERC5 | 1.1 | 2.6 | 16.9 | 10.1 | 0.9 | 1.1 | 3.4 | 14.3 |
| HLA-A | 0.9 | 1.4 | 3.2 | 3.5 | 1.3 | 1.7 | 1.7 | 4.4 |
| HLA-C | 0.8 | 1.4 | 3.5 | 3.5 | 1.4 | 1.8 | 2.1 | 6.1 |
| HLA-G | 0.9 | 1.5 | 3.7 | 3.4 | 1.1 | 1.2 | 1.8 | 3.7 |
| IFI6 | 0.9 | 1.2 | 10.2 | 10.2 | 0.7 | 0.8 | 1.8 | 10.1 |
| IFI44 | 0.9 | 2.5 | 15.5 | 37.7 | 0.9 | 0.9 | 8.2 | 36.1 |
| IFI44L | 0.9 | 2.4 | 7.4 | 11.5 | 0.8 | 1.2 | 4.2 | 8.2 |
| IFIH1 | 1.1 | 5.2 | 30.8 | 12.2 | 0.9 | 1.6 | 4.4 | 17.4 |
| IFIT1 | 1.2 | 13.7 | 23.4 | 9.5 | 0.1 | 5.7 | 9.8 | 19.9 |
| IFIT2 | 1.2 | 39.6 | 42.0 | 23.6 | 0.9 | 6.6 | 5.2 | 16.2 |
| IFIT3 | 0.8 | 3.5 | 5.1 | 6.3 | 0.9 | 2.7 | 5.3 | 7.6 |
| IFIT5 | 0.8 | 5.6 | 8.2 | 3.9 | 0.9 | 1.4 | 2.5 | 7.2 |
| IFITM2 | 1.0 | 1.9 | 8.9 | 11.4 | 1.1 | 1.7 | 5.0 | 21.3 |
| IRF7 | 1.2 | 2.2 | 4.6 | 5.4 | 1.4 | 1.6 | 3.3 | 4.5 |
| ISG15 | 1.2 | 6.0 | 24.8 | 35.8 | 1.1 | 2.2 | 9.9 | 30.2 |
| ISG20 | 0.9 | 2.3 | 15.6 | 12.3 | 0.9 | 1.4 | 4.1 | 14.9 |
| KIAA0040 | 0.9 | 1.5 | 3.3 | 1.6 | 1.0 | 1.1 | 2.1 | 3.5 |
| LAP3 | 1.0 | 1.5 | 4.8 | 3.0 | 0.9 | 1.2 | 2.4 | 5.6 |
| MASTL | 1.0 | 2.5 | 4.1 | 1.9 | 1.3 | 1.0 | 2.5 | 3.3 |
| MFN1 | 1.0 | 1.3 | 2.4 | 1.7 | 1.0 | 0.9 | 1.3 | 2.3 |
| MT1F | 1.4 | 1.5 | 2.4 | 2.1 | 1.6 | 1.5 | 1.3 | 2.0 |
| MX1 | 1.1 | 4.9 | 27.1 | 31.6 | 0.9 | 1.4 | 11.7 | 29.3 |
| NCOA7 | 1.4 | 2.3 | 5.0 | 2.4 | 1.1 | 1.4 | 2.1 | 4.4 |
| NFKB1 | 0.9 | 2.4 | 4.6 | 2.3 | 1.0 | 1.2 | 1.6 | 3.1 |
| NFYB | 1.4 | 1.3 | 2.8 | 1.4 | 1.1 | 1.5 | 1.6 | 3.3 |
| NLRC5 | 1.0 | 1.0 | 2.6 | 1.9 | 0.8 | 0.9 | 2.1 | 2.7 |
| NMI | 1.4 | 1.3 | 3.5 | 2.6 | 1.2 | 1.5 | 2.2 | 4.4 |
| NT5C3 | 1.1 | 3.2 | 34.1 | 20.0 | 0.6 | 1.0 | 2.7 | 15.6 |
| NUB1 | 0.9 | 1.3 | 2.1 | 1.9 | 1.0 | 1.1 | 1.6 | 2.2 |
| OAS2 | 1.0 | 6.4 | 30.9 | 46.2 | 1.3 | 2.4 | 18.9 | 56.8 |
| OAS3 | 0.9 | 1.3 | 7.7 | 5.7 | 0.8 | 0.8 | 2.6 | 8.2 |
| PARP9 | 0.8 | 3.0 | 13.5 | 6.1 | 0.8 | 1.4 | 4.3 | 10.9 |
| PARP12 | 0.9 | 2.6 | 7.2 | 4.7 | 0.7 | 0.9 | 1.4 | 3.3 |
| PARP14 | 1.2 | 3.6 | 13.9 | 16.3 | 1.3 | 2.0 | 8.7 | 21.0 |
| PI4K2B | 1.0 | 2.1 | 4.5 | 2.3 | 0.8 | 1.1 | 1.4 | 2.9 |
| PNPT1 | 1.0 | 1.8 | 7.8 | 3.8 | 1.0 | 1.2 | 2.3 | 6.3 |
| PSMA2 | 1.2 | 1.1 | 2.6 | 2.0 | 1.3 | 1.5 | 1.7 | 3.3 |
| PSMA4 | 1.0 | 0.9 | 3.2 | 1.7 | 1.1 | 1.3 | 1.6 | 2.9 |
| RABGAP1L | 1.1 | 1.4 | 4.7 | 3.3 | 1.1 | 1.2 | 2.4 | 6.5 |
| RBBP6 | 0.8 | 1.4 | 2.4 | 1.2 | 0.9 | 1.0 | 1.2 | 2.0 |
| RGL1 | 1.4 | 4.0 | 8.9 | 5.8 | 1.2 | 2.1 | 2.8 | 5.8 |
| RHOH | 1.2 | 1.6 | 2.1 | 1.0 | 1.2 | 1.8 | 2.0 | 2.7 |
| RNF213 | 1.0 | 1.6 | 7.8 | 5.2 | 0.9 | 1.1 | 2.8 | 7.4 |
| SAMD9 | 0.9 | 5.7 | 16.5 | 6.6 | 1.0 | 1.2 | 2.6 | 8.9 |
| SAMD9L | 1.0 | 11.1 | 33.5 | 17.2 | 0.5 | 1.2 | 3.8 | 15.9 |
| SAT1 | 1.1 | 1.7 | 6.2 | 3.5 | 1.0 | 1.3 | 1.8 | 5.2 |
| SLAMF7 | 1.1 | 2.8 | 5.0 | 4.4 | 1.2 | 1.8 | 2.3 | 3.3 |
| SP100 | 1.0 | 2.0 | 5.6 | 3.0 | 1.1 | 1.4 | 2.4 | 4.8 |
| SP110 | 1.5 | 2.9 | 7.0 | 7.4 | 1.4 | 2.4 | 5.9 | 10.0 |
| SPATS2L | 1.1 | 0.9 | 3.2 | 2.3 | 0.9 | 0.9 | 2.0 | 5.7 |
| STAP1 | 1.2 | 2.1 | 5.6 | 4.4 | 1.3 | 1.4 | 3.1 | 5.5 |
| STAT1 | 1.0 | 6.4 | 24.1 | 14.9 | 0.9 | 1.6 | 7.2 | 22.5 |
| STX11 | 1.1 | 1.6 | 2.7 | 1.8 | 1.3 | 1.4 | 1.7 | 2.0 |
| TAP1 | 0.8 | 2.0 | 4.8 | 3.5 | 1.2 | 0.8 | 2.4 | 4.3 |
| TDRD7 | 1.2 | 2.2 | 8.4 | 4.5 | 1.0 | 1.3 | 2.7 | 9.0 |
| TMCC3 | 1.2 | 1.3 | 4.2 | 4.8 | 0.7 | 0.7 | 1.8 | 4.8 |
| TNFSF10 | 1.4 | 5.6 | 7.3 | 9.7 | 1.0 | 3.8 | 5.1 | 11.8 |
| TNK2 | 1.0 | 1.4 | 2.5 | 2.2 | 1.2 | 1.4 | 1.9 | 2.6 |
| TRAFD1 | 1.1 | 1.6 | 3.2 | 2.1 | 1.3 | 1.4 | 2.2 | 3.0 |
| TRIM21 | 1.3 | 3.5 | 4.6 | 3.8 | 1.3 | 2.2 | 3.5 | 5.2 |
| TRIM22 | 0.9 | 1.2 | 6.6 | 5.4 | 0.9 | 1.3 | 2.9 | 11.4 |
| UBE2L6 | 1.0 | 2.6 | 17.6 | 13.5 | 0.9 | 1.3 | 4.9 | 28.4 |
| UPK3BL | 1.0 | 1.2 | 2.5 | 2.2 | 1.2 | 1.1 | 1.6 | 2.8 |
| USP25 | 1.1 | 1.1 | 2.9 | 2.1 | 1.2 | 1.2 | 1.8 | 2.9 |
| XRN1 | 1.2 | 1.4 | 4.0 | 2.0 | 1.1 | 1.0 | 1.9 | 4.0 |
| ZNF385B | 0.8 | 1.2 | 2.3 | 1.3 | 1.7 | 2.5 | 2.9 | 1.6 |
| ZNFX1 | 1.1 | 2.3 | 3.5 | 2.5 | 1.1 | 1.5 | 2.3 | 2.7 |
CAL-1 cells were stimulated as described in Fig. 2. All genes up-regulated significantly by K and D ODNs (n=92) at any time-point are shown. Shown in bold are those genes that contribute to antiviral immunity (n=23; GO analysis). The level of gene induction is expressed as the fold increase compared with untreated cells.
The function of genes stimulated by a single class of ODN (i.e., K but not D or D but not K) was also examined. Genes up-regulated by only K ODN were functionally associated with the control of bacterial infection and the generation of innate and inflammatory immune responses (P<0.05 for each category when compared with the unique D subset). The genes up-regulated only by D ODN did not fall into any distinct functional groups, as identified by GO or IPA analysis. Rather, most of these genes were associated with metabolic processes, whereas 12% were immune-related (Table 1). Of interest, genes with antiviral activity were up-regulated significantly, more frequently by K and D ODNs than by only K or only D ODN (P<0.001).
Role of type I IFN in the up-regulation of genes activated by CpG ODNs
ISGs were the second group of genes most frequently up-regulated by K and D ODNs (Table 2). This was unexpected, as type I IFN is generally considered to be a component of D- but not K-induced responses (see Fig. 1 and refs. [26–28]). Analysis of the time course over which ISGs were up-regulated provided a clue to this puzzle, as expression of many ISGs rose within 3 h of CpG stimulation. This finding suggested that K ODN might trigger pDCs to produce type I IFN rapidly but for only a brief period.
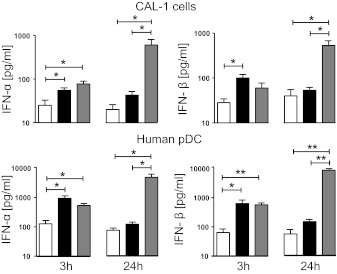
CAL-1 cells and freshly isolated human pDCs were therefore stimulated with both classes of ODNs and supernatants probed for IFN-α/β content at 3 and 24 h. As seen in Fig. 4, both cell types responded to stimulation by K or D ODN, by significantly increasing IFN-α and IFN-β production at 3 h (P<0.05). The magnitude of this type I IFN response continued to rise through 24 h in cells cultured with D ODN but fell to near background levels when pDC or CAL-1 cells were incubated with K ODN for 24 h. This pattern was replicated in studies of IFN-α/β mRNA levels (Supplemental Fig. 1).
Figure 4. Effect of K and D ODN on type I IFN production.
Freshly isolated pDCs and CAL-1 cells were cultured for 3 or 24 h in medium (white) supplemented with 1 μM K (black) or 3 μM D ODN (gray). The concentration of IFN-α and IFN-β in culture supernatants was determined by ELISA. Results represent the mean ± sem of three to four independent experiments. *P < 0.05; **P < 0.01.
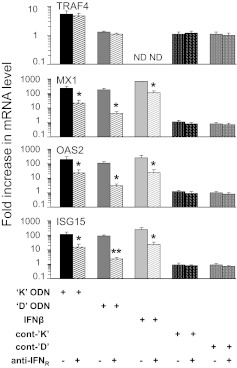
To further examine the role of type I IFN in CpG-driven gene expression, neutralizing antibody against the type I IFNR was added to CpG-stimulated CAL-1 cells. Expression of mRNAs encoding three ISGs (MX1, ISG15, and OAS2) and the non-IFN-related TRAF4 gene (serving as negative control) was evaluated by RT-PCR. Results show that blocking the IFNR reduced expression of all three ISGs by greater than tenfold (P<0.05; Fig. 5). In contrast, blocking the IFNR had no effect on TRAF4 (Fig. 5) nor did treatment with control ODN (±anti-IFNR antibody).
Figure 5. Effect of blocking the type I IFNR of CAL-1 cells on ISGs induced by K and D ODNs.
CAL-1 cells were stimulated with 1 μM K or 3 μM D ODN or their respective control ODN for 12 h in the presence of anti-IFNR antibody. mRNA levels for MX1, OAS2, ISG15, and TRAF4 were monitored by TaqMan RT-PCR. Results show the mean ± sem fold increase in mRNA levels compared with untreated controls (normalized to GAPDH) from three independent experiments. As a positive control, CAL-1 cells were stimulated for 6 h with IFN-β. *P < 0.05; **P < 0.01 versus identically treated cells in the absence of anti-IFNR.
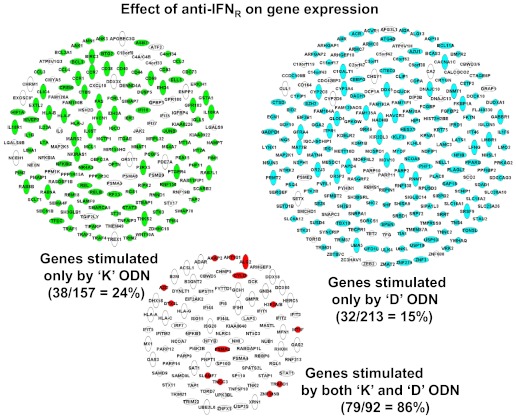
Extending this analysis, the impact of blocking the IFNR on global gene expression induced by K and D ODN stimulation was examined by microarray. To gain the most information from a single analysis, this experiment focused on the time-point of maximal gene up-regulation (9 h for K and 27 h for D ODN; Fig. 3), as this captured >87% of all activated genes. Results show that blocking the IFNR led to a significant decrease in the expression of 86% (79/92) of the genes up-regulated by K and D ODNs (P<0.01; Fig. 6). By comparison, blocking the IFNR reduced the expression of <20% of the genes up-regulated by only K or only D ODN (P<10−32 vs. genes up-regulated by K and D; Fig. 6). These observations support the conclusion that type I IFN is a critical mediator of the activation of genes triggered by K and D ODNs.
Figure 6. Effect of blocking the type I IFNR on gene up-regulation by K and D ODNs in CAL-1 cells.
CAL-1 cells were stimulated with K or D ODN, as described in Fig. 2. Anti-IFNR antibody was added to block autocrine type I IFN signaling. Genes whose expression fell significantly (P<0.01) are shown in white, as is the percent reduction in each group.
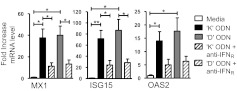
CpG ODNs up-regulate ISGs and STAT1 in primary human pDCs
To verify these key findings, the effect of stimulating human pDCs with CpG ODNs was evaluated. mRNA levels of the ISGs MX1, OAS2, and ISG15 were elevated significantly after 12 h of stimulation with K or D ODN (Fig. 7). Consistent with results from studies of CAL-1 cells (Fig. 5), the addition of anti-IFNR to primary pDCs uniformly reduced ISG expression (Fig. 7). Similarly, K and D ODNs activated phospho-STAT1 (a transcription factor that regulates ISGs; Supplemental Fig. 3) in CAL-1 cells and CD123+ human pDCs. It should be noted that K ODNs activated STAT1 more rapidly than D ODNs, a finding consistent with the more rapid up-regulation of ISGs by K versus D ODN.
Figure 7. Effect of blocking the type I IFNR of primary human pDCs on ISGs induced by K and D ODNs.
Purified human pDCs were stimulated with 1 μM K or 3 μM D ODN for 12 h in the presence of anti-IFNR antibody. mRNA levels for MX1, OAS2, and ISG15 were monitored by TaqMan RT-PCR. Results show the mean ± sem fold increase in mRNA levels compared with untreated controls (normalized to GAPDH) from three independent experiments. *P < 0.05; **P < 0.01.
DISCUSSION
Synthetic ODNs containing unmethylated CpG motifs mimic the immunostimulatory activity of bacterial and viral DNA [1, 2, 4]. Structurally distinct classes of ODNs with unique immunomodulatory properties have been identified, and the nature of the immune response they induce was found to vary as a function of species and cell type [3–9]. Current work sought to differentiate between genes activated by K and D ODNs versus those responding to a single class of ODNs, with the aim of gaining insight into the characteristics of each gene subset.
K ODNs activate human pDCs via a NF-κB-dependent pathway that culminates in the up-regulation of proinflammatory cytokines (e.g., IL-6 and TNF-α; Fig. 1 and Supplemental Fig. 2), whereas D ODNs preferentially (but not solely) induce the IRF7-dependent production of type I IFNs [3, 10–12, 29]. The same pattern of responsiveness was observed in studies of the human CAL-1 pDC line (Figs. 1 and 4 and Supplemental Fig. 1). Although the absolute magnitude of cytokine production differed between CAL-1 cells and freshly isolated pDCs, their pattern of responsiveness upon stimulation with K or D ODN was very similar. As purifying large numbers of resting pDCs from human peripheral blood for microarray studies was not technically feasible (pDCs constitute only 0.2–0.5% of the PBMC pool and are commonly activated during the purification process), CAL-1 cells provided an accessible model for analyzing changes in gene expression induced by CpG ODNs [21, 22]. Key findings from the microarray experiments were then verified by RT-PCR analysis of human pDCs.
Considerable care was taken to insure the quality of the experimental protocols, microarray data, and subsequent analyses. First, all studies were performed on immature CAL-1 cells (maturation was prevented by resting the cells in FBS-deprived medium and studying them before reaching confluence), as mature pDCs and CAL-1 cells lose their ability to produce IFN-α [19, 30, 31]. Second, multiple independent replicates of each experiment were performed, and consistent changes in gene expression observed (R2=0.85±0.06). Third, a stringency cutoff of P < 10−5 was used to select genes whose level of expression differed significantly from unstimulated cells. With the use of these criteria, a total of 462 genes was up-regulated significantly by K or D ODN, whereas 92 were stimulated by both classes (hereafter, referred to as “common” genes). When signaling through the IFNR was blocked, expression levels of a majority (87%) of common genes decreased significantly (P<0.01; Fig. 6). In contrast, IFNR blockade impacted the expression <20% of those genes up-regulated by K or D ODN alone. Thus, type I IFN was the primary inducer of a majority of genes up-regulated by both ligands.
Blocking the type I IFNR was not expected to suppress the expression of most common genes, as earlier reports suggested that K-type ODN had little effect on type I IFN production. Those earlier studies typically evaluated IFN-α/β levels, 1–3 days after CpG stimulation [26–28, 32]. Current findings confirm that D but not K ODN induces high levels of type I IFN at 24 h but further demonstrate that K and D ODNs up-regulate IFN-α/β production by primary pDCs and CAL-1 cells at 3 h (Fig. 4 and Supplemental Fig. 1). This rapid induction of type I IFN was also observed by Kerkmann et al. [9], who concluded that two distinct regulatory pathways mediate IFN-α/β synthesis in human pDCs. The first was deemed to be short-lived, IFNR-independent, and triggered by K and D ODNs. The second was delayed, dependent on IFNR feedback, and mediated solely by D ODNs, which unlike K ODNs, interacted with TLR9 in endosomal vesicles and thus, activated a signaling cascade via IRF7 [3, 9–12, 29]. However, those conclusions were controversial, as Liu et al. [33] claimed that K-type ODNs failed to induce early IFN-α/β in pDCs, mDCs, or human PBMCs. Current findings clarify this issue by showing that K ODNs induce a brief pulse of type IFN production of sufficient magnitude to support ISG up-regulation (Figs. 4–7 and Table 2). Thus, although the duration of type I IFN induction by K and D ODNs differs, their effect on the up-regulation of ISGs after CpG stimulation is similar.
The common genes up-regulated by K and D ODNs included many ISGs. These included MX1, OAS2, ISG15, and eukaryotic initiation factor 2-α kinase 2, which are known to block viral transcription, degrade viral RNA, inhibit translation, and/or modify protein function to inhibit viral replication [34, 35]. The role of ISGs in protecting the host from viral infection is supported by evidence that ISG knockout mice are more susceptible to viral infection (including by influenza and HSV-1) [36–38]. Similarly, human population studies indicate that a single nucleotide polymorphism in ISGs correlates with increased susceptibility to infection by hepatitis C virus, West Nile virus, and HIV [39–41]. The observation that the 92 genes up-regulated by K and D ODNs contain many ISGs (Table 2) leads us to postulate that the core function of TLR9 engagement is the control of viral infection. Consistent with that hypothesis, GO analysis shows that antiviral activity is the dominant functional feature of those common genes (P<10−15; Table 1). Based on these findings, we propose that CpG ODNs may be particularly effective in protecting against viral infection. Supporting such a conclusion are results showing that K and D ODNs prevent lethal vaccinia and HSV-2 infection of mice and that such protection involved type I IFN signaling [42–45]. In this context, it is noteworthy that a Phase I clinical study of K ODNs detected increased expression of ISGs (but not IFN-α/β) in all participants at 24 h [46]. As expression of these ISGs remained elevated for several days, we propose that CpG ODNs may provide durable and better tolerated protection from virus infection than could be achieved by the direct administration of IFN.
Earlier studies reported that CpG-induced innate immunity primarily mediates resistance to bacterial infection [47–49]. Those reports generally examined the effect of K ODNs. In that context, current microarray data demonstrate that a significant subset of genes up-regulated by K ODNs was functionally associated with resistance to bacterial infection (P<10−15), including many proinflammatory genes, such as IL-1β, IL-6, and IL-23, whereas an insignificant number of genes up-regulated by D ODNs had such activity (Table 1 and Supplemental Table 1). Thus, in addition to the conserved antiviral activity shared by ODNs expressing unmethylated CpG motifs, K ODNs manifest the more specialized ability to mediate antibacterial responses. Interestingly, genes uniquely up-regulated by D ODNs were not linked to specific immunological functions or regulatory networks (aside from metabolic functions; Table 1), suggesting that the prolonged induction of type I IFN is their dominant immunological effect.
This work is the first to examine the effect of different classes of CpG ODNs on gene expression in human pDCs. Results show that antiviral immunity mediated via type I IFN represents the core functionality of human pDCs stimulated via TLR9, regardless of CpG ODN structure, backbone, or other nucleotide elements. This finding suggests that the immune response mediated by TLR9 must have broadened over evolutionary periods to enable the recognition of motifs expressed by additional (bacterial) pathogens, consistent with K ODNs preferentially triggering an antibacterial response. This broadening was achieved by diversifying the expression of TLR9 receptors in lysosomal vesicles, as well as endolysosomes, where their activation could trigger additional signaling pathways mediated by different transcription factors (such as IRFs). As clinical trials of CpG ODNs for the treatment of cancer, allergy, and infectious diseases proceed, knowledge of their shared versus sequence-specific effects should aid in the selection of disease-appropriate ODNs.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the U.S. National Institutes of Health, NCI. Analyses were performed using BRB ArrayTools, developed by Richard Simon and Amy Peng Lam.
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
- BRB
- Biometric Research Branch
- Cy3/5
- cyanine 3/5
- GEO
- Gene Expression Omnibus
- GO
- Gene Ontology
- HSV
- herpes simplex virus
- IPA
- Ingenuity Pathway Analysis
- IRF
- IFN-regulatory factor
- ISG
- IFN-stimulated gene
- MX1
- myxovirus resistance-1
- NCI
- National Cancer Institute
- OAS2
- 2′-5′-oligoadenylate synthetase 2
- ODN
- oligonucleotide
- pDC
- plasmacytoid DC
AUTHORSHIP
F.S. and D.M.K. contributed to the conception and design of the research. F.S., C.M., D.T., M.G., and S.K. contributed to acquisition of data. T.M. provided the CAL-1 cell line on which these studies relied. F.S. and D.M.K. contributed to the interpretation of the data and wrote the manuscript. All authors reviewed the manuscript and approved the final version.
DISCLOSURES
Members of the lab of D.M.K. have patents related to the use of CpG ODN. All rights to such patents have been assigned to the federal government. The assertions herein are the private ones of the authors and are not to be construed as official or as reflecting the views of the NCI at large.
REFERENCES
- 1. Hemmi H., Takeuchi O., Kawai T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 2. Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. (2003) Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilliet M., Cao W., Liu Y. J. (2008) Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8, 594–606 [DOI] [PubMed] [Google Scholar]
- 4. Klinman D. M. (2004) Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 4, 249–258 [DOI] [PubMed] [Google Scholar]
- 5. Gursel M., Verthelyi D., Gursel I., Ishii K. J., Klinman D. M. (2002) Differential and competitive activation of human immune cells by distinct classes of CpG oligodeoxynucleotides. J. Leukoc. Biol. 71, 813–820 [PubMed] [Google Scholar]
- 6. Verthelyi D., Ishii K. J., Gursel M., Takeshita F., Klinman D. M. (2001) Human peripheral blood cells differentially recognize and respond to two distinct CpG motifs. J. Immunol. 166, 2372–2377 [DOI] [PubMed] [Google Scholar]
- 7. Yi A. K., Yoon J. G., Yeo S. J., Hong S. C., English B. K., Krieg A. M. (2002) Role of mitogen-activated protein kinases in CpG DNA-mediated IL-10 and IL-12 production: central role of extracellular signal-regulated kinase in the negative feedback loop of the CpG DNA-mediated Th1 response. J. Immunol. 168, 4711–4720 [DOI] [PubMed] [Google Scholar]
- 8. Takauji R., Iho S., Takatsuka H., Yamamoto S., Takahashi T., Kitagawa H., Iwasaki H., Iida R., Yokochi T., Matsuki T. (2002) CpG-DNA-induced IFN-α production involves p38 MAPK-dependent STAT1 phosphorylation in human plasmacytoid dendritic cell precursors. J. Leukoc. Biol. 72, 1011–1019 [PubMed] [Google Scholar]
- 9. Kerkmann M., Rothenfusser S., Hornung V., Towarowski A., Wagner M., Sarris A., Giese T., Endres S., Hartmann G. (2003) Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J. Immunol. 170, 4465–4474 [DOI] [PubMed] [Google Scholar]
- 10. Sasai M., Linehan M. M., Iwasaki A. (2010) Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329, 1530–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guiducci C., Ott G., Chan J. H., Damon E., Calacsan C., Matray T., Lee K. D., Coffman R. L., Barrat F. J. (2006) Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J. Exp. Med. 203, 1999–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., Taniguchi T. (2005) Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434, 1035–1040 [DOI] [PubMed] [Google Scholar]
- 13. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 14. Klaschik S., Tross D., Klinman D. M. (2009) Inductive and suppressive networks regulate TLR9-dependent gene expression in vivo. J. Leukoc. Biol. 85, 788–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tross D., Petrenko L., Klaschik S., Zhu Q., Klinman D. M. (2009) Global changes in gene expression and synergistic interactions induced by TLR9 and TLR3. Mol. Immunol. 46, 2557–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kato A., Homma T., Batchelor J., Hashimoto N., Imai S., Wakiguchi H., Saito H., Matsumoto K. (2003) Interferon-α/β receptor-mediated selective induction of a gene cluster by CpG oligodeoxynucleotide 2006. BMC Immunol. 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amit I., Garber M., Chevrier N., Leite A. P., Donner Y., Eisenhaure T., Guttman M., Grenier J. K., Li W., Zuk O., Schubert L. A., Birditt B., Shay T., Goren A., Zhang X., Smith Z., Deering R., McDonald R. C., Cabili M., Bernstein B. E., Rinn J. L., Meissner A., Root D. E., Hacohen N., Regev A. (2009) Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klaschik S., Tross D., Shirota H., Klinman D. M. (2010) Short- and long-term changes in gene expression mediated by the activation of TLR9. Mol. Immunol. 47, 1317–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maeda T., Murata K., Fukushima T., Sugahara K., Tsuruda K., Anami M., Onimaru Y., Tsukasaki K., Tomonaga M., Moriuchi R., Hasegawa H., Yamada Y., Kamihira S. (2005) A novel plasmacytoid dendritic cell line, CAL-1, established from a patient with blastic natural killer cell lymphoma. Int. J. Hematol. 81, 148–154 [DOI] [PubMed] [Google Scholar]
- 20. Cisse B., Caton M. L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N. S., Kant S. G., Holter W., Rauch A., Zhuang Y., Reizis B. (2008) Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siegal F. P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P. A., Shah K., Ho S., Antonenko S., Liu Y. J. (1999) The nature of the principal type 1 interferon-producing cells in human blood. Science 284, 1835–1837 [DOI] [PubMed] [Google Scholar]
- 22. Gallucci S., Lolkema M., Matzinger P. (1999) Natural adjuvants: endogenous activators of dendritic cells. Nat. Med. 11, 1249–1255 [DOI] [PubMed] [Google Scholar]
- 23. Leifer C. A., Verthelyi D., Klinman D. M. (Jan. 22–27, 2001) CpG ODN mixtures optimally activate human PBMC: interfaces between innate and adaptive immunity. In Keystone Symposia, Keystone, CO, USA (abs. 22). [Google Scholar]
- 24. Klaschik S., Gursel I., Klinman D. M. (2007) CpG-mediated changes in gene expression in murine spleen cells identified by microarray analysis. Mol. Immunol. 44, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 25. Wright G. W., Simon R. M. (2003) A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19, 2448–2455 [DOI] [PubMed] [Google Scholar]
- 26. Vollmer J., Weeratna R., Payette P., Jurk M., Schetter C., Laucht M., Wader T., Tluk S., Liu M., Davis H. L., Krieg A. M. (2004) Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur. J. Immunol. 34, 251–262 [DOI] [PubMed] [Google Scholar]
- 27. Haas T., Metzger J., Schmitz F., Heit A., Muller T., Latz E., Wagner H. (2008) The DNA sugar backbone 2′ deoxyribose determines Toll-like receptor 9 activation. Immunity 28, 315–323 [DOI] [PubMed] [Google Scholar]
- 28. Cooper C. L., Ahluwalia N. K., Efler S. M., Vollmer J., Krieg A. M., Davis H. L. (2008) Immunostimulatory effects of three classes of CpG oligodeoxynucleotides on PBMC from HCV chronic carriers. J. Immune Based Ther. Vaccines 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gursel M., Gursel I., Mostowski H. S., Klinman D. M. (2006) CXCL16 influences the nature and specificity of CpG-induced immune activation. J. Immunol. 177, 1575–1580 [DOI] [PubMed] [Google Scholar]
- 30. Colonna M., Trinchieri G., Liu Y. J. (2004) Plasmacytoid dendritic cells in immunity. Nat. Immunol. 5, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 31. Reizis B., Colonna M., Trinchieri G., Barrat F., Gilliet M. (2011) Plasmacytoid dendritic cells: one-trick ponies or workhorses of the immune system? Nat. Rev. Immunol. 11, 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krug A., Rothenfusser S., Hornung V., Jahrsdorfer B., Blackwell S., Ballas Z. K., Endres S., Krieg A. M., Hartmann G. (2001) Identification of CpG oligonucleotide sequences with high induction of IFN-α/β in plasmacytoid dendritic cells. Eur. J. Immunol. 31, 2154–2163 [DOI] [PubMed] [Google Scholar]
- 33. Liu Y. C., Gray R. C., Hardy G. A., Kuchtey J., Abbott D. W., Emancipator S. N., Harding C. V. (2010) CpG-B oligodeoxynucleotides inhibit TLR-dependent and -independent induction of type I IFN in dendritic cells. J. Immunol. 184, 3367–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadler A. J., Williams B. R. (2008) Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schoggins J. W., Nociari M., Philpott N., Falck-Pedersen E. (2005) Influence of fiber detargeting on adenovirus-mediated innate and adaptive immune activation. J. Virol. 79, 11627–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lenschow D. J., Giannakopoulos N. V., Gunn L. J., Johnston C., O'Guin A. K., Schmidt R. E., Levine B., Virgin H. W. (2005) Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79, 13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arnheiter H., Skuntz S., Noteborn M., Chang S., Meier E. (1990) Transgenic mice with intracellular immunity to influenza virus. Cell 62, 51–61 [DOI] [PubMed] [Google Scholar]
- 38. Zhou A., Paranjape J., Brown T. L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., Silverman R. H. (1997) Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16, 6355–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yakub I., Lillibridge K. M., Moran A., Gonzalez O. Y., Belmont J., Gibbs R. A., Tweardy D. J. (2005) Single nucleotide polymorphisms in genes for 2′-5′-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J. Infect. Dis. 192, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 40. Knapp S., Yee L. J., Frodsham A. J., Hennig B. J., Hellier S., Zhang L., Wright M., Chiaramonte M., Graves M., Thomas H. C., Hill A. V., Thursz M. R. (2003) Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 4, 411–419 [DOI] [PubMed] [Google Scholar]
- 41. Hijikata M., Ohta Y., Mishiro S. (2000) Identification of a single nucleotide polymorphism in the MxA gene promoter (G/T at nt −88) correlated with the response of hepatitis C patients to interferon. Intervirology 43, 124–127 [DOI] [PubMed] [Google Scholar]
- 42. Ashkar A. A., Bauer S., Mitchell W. J., Vieira J., Rosenthal K. L. (2003) Local delivery of CpG oligodeoxynucleotides induces rapid changes in the genital mucosa and inhibits replication, but not entry, of herpes simplex virus type 2. J. Virol. 77, 8948–8956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Svensson A., Bellner L., Magnusson M., Eriksson K. (2007) Role of IFN-α/β signaling in the prevention of genital herpes virus type 2 infection. J. Reprod. Immunol. 74, 114–123 [DOI] [PubMed] [Google Scholar]
- 44. Shen H., Iwasaki A. (2006) A crucial role for plasmacytoid dendritic cells in antiviral protection by CpG ODN-based vaginal microbicide. J. Clin. Invest. 116, 2237–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rees D. G., Gates A. J., Green M., Eastaugh L., Lukaszewski R. A., Griffin K. F., Krieg A. M., Titball R. W. (2005) CpG-DNA protects against a lethal orthopoxvirus infection in a murine model. Antiviral Res. 65, 87–95 [DOI] [PubMed] [Google Scholar]
- 46. Krieg A. M., Efler S. M., Wittpoth M., Al Adhami M. J., Davis H. L. (2004) Induction of systemic TH1-like innate immunity in normal volunteers following subcutaneous but not intravenous administration of CPG 7909, a synthetic B-class CpG oligodeoxynucleotide TLR9 agonist. J Immunother. 27, 460–471 [DOI] [PubMed] [Google Scholar]
- 47. Krieg A. M., Homan L. L., Yi A. K., Harty J. T. (1998) CpG DNA induces sustained IL-12 expression in vivo and resistance to Listeria monocytogenes challenge. J. Immunol. 161, 2428–2434 [PubMed] [Google Scholar]
- 48. Elkins K. L., Rhinehart-Jones T. R., Stibitz S., Conover J. S., Klinman D. M. (1999) Bacterial DNA containing CpG motifs stimulates lymphocyte-dependent protection of mice against lethal infection with intracellular bacteria. J. Immunol. 162, 2291–2298 [PubMed] [Google Scholar]
- 49. Weighardt H., Feterowski C., Veit M., Rump M., Wagner H., Holzmann B. (2000) Increased resistance against acute polymicrobial sepsis in mice challenged with immunostimulatory CpG oligodeoxynucleotides is related to an enhanced innate effector cell response. J. Immunol. 165, 4537–4543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.