Abstract
Background
Microglia, the resident immune cells of the central nervous system (CNS), have two distinct phenotypes in the developing brain: amoeboid form, known to be amoeboid microglial cells (AMC) and ramified form, known to be ramified microglial cells (RMC). The AMC are characterized by being proliferative, phagocytic and migratory whereas the RMC are quiescent and exhibit a slow turnover rate. The AMC transform into RMC with advancing age, and this transformation is indicative of the gradual shift in the microglial functions. Both AMC and RMC respond to CNS inflammation, and they become hypertrophic when activated by trauma, infection or neurodegenerative stimuli. The molecular mechanisms and functional significance of morphological transformation of microglia during normal development and in disease conditions is not clear. It is hypothesized that AMC and RMC are functionally regulated by a specific set of genes encoding various signaling molecules and transcription factors.
Results
To address this, we carried out cDNA microarray analysis using lectin-labeled AMC and RMC isolated from frozen tissue sections of the corpus callosum of 5-day and 4-week old rat brain respectively, by laser capture microdissection. The global gene expression profiles of both microglial phenotypes were compared and the differentially expressed genes in AMC and RMC were clustered based on their functional annotations. This genome wide comparative analysis identified genes that are specific to AMC and RMC.
Conclusions
The novel and specific molecules identified from the trancriptome explains the quiescent state functioning of microglia in its two distinct morphological states.
Background
Microglia are the prime immune effector cells of the central nervous system (CNS). The origin, morphology and role of microglia in health and disease were first elaborately described in 1939 [1]. The amoeboid microglial cells (AMC), which are abundant in the periventricular white matter, namely the corpus callosum (CC) of the brain function as macrophages in the developing brain. Studies have demonstrated that AMC gradually transform into ramified microglial cells (RMC) with advancing age [2].
The time course of development of microglia differs in different regions of the brain [3]. In the CC, AMC preponderate a week before birth in mice and rats [4-6] and actively phagocytose the cellular debris and refine axonal connectivity during the first postnatal week [7-9]. This is followed by the gradual transition into RMC, which survey the brain parenchyma with their fine and non-overlapping ramifications, thereby monitoring chronic and acute insults [10]. Upon activation by trauma, infection or any other neurodegenerative stimuli, microglia retract their ramifications, transform into amoeboid or spherical shape, produce pro-inflammatory cytokines and display phagocytosis [11]. These microglia are known to be activated or reactive.
Studies in chronic, aging associated neuropathologies such as Alzheimer’s disease (AD) [12-14], and Parkinson’s disease (PD) [15] indicate persistent microglial activation as the major causative factor in disease exacerbation. Aging brains are often characterized by the presence of primed microglia, which present an altered cytokine profile in comparison to their counterparts in younger brains [16,17]. These microglia produce an exaggerated inflammatory response when activated [18,19] leading to prolonged cycles of proliferation and production of pro-inflammatory cytokines which eventually render them neurotoxic. Further, chronic microglial activation has been shown to cause the impairment of adult neurogenesis in hippocampus [20] and damage to the periventricular white matter (PWMD) in the early postnatal brain [21]. Hence activated microglia in both postnatal and adult stages can have neurotoxic effects on the CNS by causing excessive inflammation. Identification of ways to attenuate microglia-mediated neuroinflammation, therefore, has been the primary consideration in therapeutic strategy. There is accumulated information on the factors that contribute to the activation, migration, proliferation and immune response of microglia over the years [22,23], but the gene expression and signaling networks that function within these cells are yet to be fully clarified.
Gene expression profiles of microglia from primary cultures are available, but their expression profiles have been found to be altered once isolated from their natural milieu [24]. It is striking that investigation on the expression profiles of functioning genes of AMC and RMC in vivo in their quiescent state have remained elusive. In this connection, we carried out a global gene expression profiling of AMC and RMC in situ by isolating them from the CC of rat brain using laser-capture microdissection (LCM) tool. Overlapping the transcriptome onto several online and commercial databases, our current study aimed to identify molecular candidates that are associated with the morphological transformation and physiological functioning of microglia in the developing brain. Further, we have identified the genes that render ‘stemness’ and ‘monocytic’ functions to AMC and RMC. The transcriptome profiling has also led to the identification of several genes that may be vital in regulating microglial proliferation, differentiation, migration, and ramification.
Methods
Ethics statement
In the handling and care of animals, the International Guiding Principles for Animals Research, as adopted by the Institutional Animal Care and Use Committee (IACUC), National University of Singapore, were followed. All efforts were made to minimize pain and the number of rats used.
Laser-capture microdissection (LCM)
Whole fresh brains were removed from 5-day postnatal Wistar rat pups (n = 3) and 4-week old Wistar rats (n = 3) and placed in liquid nitrogen immediately for a short time and then frozen in a cryostat (Model No. CM 3050 S, Leica Microsystems GmbH, Wetzlar, Germany). The forebrain was sectioned coronally through the CC at 5 μm thickness and mounted on precleaned slides. The sections were fixed in 75% ethanol for 1 min and incubated with peroxidase conjugated isolectin (1: 50, Cat. No. L5391, Sigma-Aldrich Co., MO,USA) for 15 min. The sections were then dehydrated by a graded series of ethanol and cleaned in xylene. The slide was placed on the microscope stage of MMI CellCut (Molecular Machines & Industries, Glattbrugg, Switzerland). The 4 X, 10 X to 40 X objective lenses were used to achieve the proper placement of the cap (for cell collection) above the CC. Lectin stained microglia cells (AMC from 5-day and RMC from 4-week old rat brain CC) were selected and cut by laser and collected into the cap of tube (Cat No. 50202, Molecular Machines &Industries, Glattbrugg, Switzerland). Extra care was taken to minimize the contamination of materials from other cell types while laser dissecting microglia from the CC.
Microarray analysis
Total RNA was extracted from 600 isolated microglia cells per group using RNeasy micro kit (Cat. No. 74004, Qiagen, CA, USA), quantified by Nanodrop 1000 (Thermo Scientific, MA, USA) and hybridized to each microarray chip. RNA (15 ng) was reverse transcripted into the first-strand cDNA using a T7-Oligo (dT) Primer (Two-Cycle Target Labeling and Control Reagent package, Affymetrix, CA, USA). After second-strand cDNA synthesis, the double-stranded cDNA was purified and served as a template in the first cycle of in vitro transcription (IVT) reaction. The unlabeled cRNA was then reverse transcripted into the first-strand cDNA of the second cycle using random primers. Subsequently, the T7-Oligo(dT) Promoter Primer was used in the second-strand cDNA synthesis to generate double-stranded cDNA template containing T7 promoter sequences. Then the double-stranded cDNA was amplified and labeled using a biotinylated nucleotide analog/ribonucleotide mix in the second IVT reaction. The labeled cRNA was then cleaned up, fragmented, and hybridized to Rat Genome 230 2.0 Array (Cat. No.900506, Affymetrix, CA, USA). A total of six arrays (three each for AMC and RMC) were carried out in the present study. The arrays were stained according to the manufacturer’s protocols and then scanned with the Genechip scanner (Affymetrix, CA, USA). Initial analysis of the scanned images was performed by GeneChip Operating Software (GCOS, Affymetrix, CA, USA). For absolute analysis, each chip was normalized to a target intensity of 500, and probe sets were assigned a signal intensity and detection call of Present, Marginal or Absent.
Data analysis and generation of gene lists
The absolute data (signal intensity, detection call and detection p-value) were exported into GeneSpring GX 7.3 software (Agilent Technologies, CA, USA). All the six chips were globally normalized and the genes of over 2-fold differential expression were filtered out and used for functional analysis.
Data normalization and generation of gene lists using MATLAB
Raw CEL files of the six chips were RMA (Robust Multichip Average) normalized using the Affymetrix Expression Console Version 1.1 (Affymetrix, CA, USA). The normalized data was then used to identify differentially expressed genes between AMC and RMC in MATLAB R2009a (MathWorks, MA, USA). For the statistical analysis, we used the ‘Exploring Gene Expression Data’ demo scripts in the Bioinformatics Toolbox™. The data was filtered for removing genes with low expression values and low variance across chips. Further, t-test was performed to retain genes with p-values less than 0.05 and a Volcano Plot was generated to identify the two-fold differentially expressed genes. The microarray data discussed in this publication is MIAME compliant and has been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). It is accessible through GEO Series accession number GSE29885.
Gene expression profile clustering and pathway analysis
Agglomerative average-linkage hierarchical clustering of the different experimental groups was obtained for selected groups of genes with GeneSpring GX 7.3 software (Agilent Technologies, CA, USA) with standard correlation used as the similarity matrix. The gene lists obtained was fed into Pathway Studio 6 software (Ariadne, MD, USA) to generate pathways for identifying interactions between the genes for validation purposes.
Analysis of gene lists
The gene list generated from MATLAB was used to identify functional groups enriched in the AMC and RMC using DAVID Bioinformatics Database [25,26]. To identify the ‘Stemness’ of AMC and RMC, we compared our gene lists to gene lists enriched in embryonic, neural and hematopoietic stem cells [27]. Since the data were accumulated from a different microarray platform, we found orthologs to their genes pertaining to our platform using the online NetAffyx application (Affymetrix, CA, USA). For comparison of our gene expression data to that of peripheral blood monocytes [28], the raw CEL files of monocyte expression data were downloaded from NCBI GEO (Gene Expression Omnibus) and the orthologs pertaining to our platform were identified using the online NetAffyx application. These files were RMA normalized in Affymetrix Expression Console Version 1.1 (Affymetrix, CA, USA) and subsequently the average expression values of the monocyte genes were compared to our microglia gene lists.
Double immunofluorescence staining on postnatal rat brain sections
5-day and 4-week old Wistar rat pups were purchased from the Laboratory Animal Centre, National University of Singapore. The animals were perfused and fixed with 4% paraformaldehyde for further procedure. For double immunofluorescence staining, forebrain sections at 30μm were cut through the corpus callosum using cryostat (Model No. CM 3050 S, Leica Microsystems GmbH, Wetzlar, Germany). The sections were incubated with purified mouse anti-OX-42 Ig (1:50; Cat No. CBL1512, Millipore, MA,USA) along with rabbit anti-ETO (1:100; Cat No. sc-28693, Santa Cruz Biotechnology, Inc. CA, USA) or with rabbit anti-Dcx (1:100, Cat No. ab18723, abcam, Cambridge, UK) or with rabbit anti-Sox4 (1:100; Cat No.sc-20090,Santa Cruz Biotechnology, Inc. CA, USA) or with rabbit anti-Sox11 (1:100; Cat No.sc-20096 ,Santa Cruz Biotechnology, Inc. CA, USA) or with rabbit anti-Sept9 (1:100; Cat No. sc-130263, Santa Cruz Biotechnology, Inc. CA, USA) with rabbit anti-Sept4 (1:100; Cat No. sc-20179, Santa Cruz Biotechnology, Inc. CA, USA) overnight at 4°C. On the following day, the sections were further incubated with either FITC-conjugated goat-anti-mouse IgG (1:100; Cat No. F9137, Sigma-Aldrich Co., MO, USA) or Cy3-conjugated sheep-anti-rabbit IgG secondary antibody (1:100; Cat No. C2306, Sigma-Aldrich Co., MO, USA). The sections were counterstained with DAPI (1 μg/ml, Cat. No. D1306, Invitrogen, CA, USA) and mounted with a fluorescent mounting medium (DakoCytomation, Glostrop, Denmark). Photo-images were captured using a confocal microscope (Olympus FV1000, Tokyo, Japan).
Cell culture
BV-2 cells (a widely used murine microglial cell line) were maintained at 75 cm2 culture flasks in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma, St. Louis, MO, USA; Cat. No. 1152) supplemented with 10% fetal bovine serum (FBS, HyClone, Logan, UT) and cultured in 37°C in a humidified atmosphere of 5% CO2 and 95% air incubator. Cells were seeded on 6-well plates at about a density of 1.0 × 106 per well for RNA isolation.
RNA isolation and real-time RT-PCR for validation of microarray data
Total RNA from laser-captured AMC and RMC was extracted using miRNeasy Mini Kit (Qiagen, Germany, Cat. No.217004) and RNA from BV-2 cells was extracted with RNeasy Mini Kit (Qiagen, Germany, Cat. No. 75161) according to the manufacturer’s instructions and quantified spectrophotometrically. 2 μg of RNA from each sample was added to a total volume of 25 μl reaction mixture containing 2.5 μM of oligo (dT) primer (Promega, Madison, WI USA; Cat. No. C110A), and 200U of Molony Murine Leukemia Virus Reverse Transcriptase (M-MLV, Promega, Madison, WI, USA; Cat. No. M5314). The reaction was initiated by incubating the reaction mixture for 1 h at 42°C for reverse transcription, and stopped by heating for 10 min at 70°C. Aliquot (0.5 μl) of the each reverse transcription product was added to the 10 μl reaction mixture containing QuantiTectRSYBRR Green I, 0.5 μM of each primer corresponding to Runx1t1, Sept9, Sept4, Mbp (rat), Gapdh, Dcx, Mbp (mouse), or β-actin and 4 mM MgCl2 to amplify the genes in ABi 7900HT Fast PCR system (Applied Biosystems, USA). The primer sequences of Runx1t1 are forward: 5′-ACGAACAGCTGCTTCTGGAT-3′and reverse: 5′-TGCTTGGATGTTCTGAGTGC-3′, Sept 9 are forward: 5′-AACCATGTCCCTCGAACTTG-3′ and reverse: 5′-AAGAGAGAGGGGACACGACA-3′, Sept 4 are forward: 5′-CTCATCCGGGAGAAAGATGA-3′ and everse: 5′-GAGCTGATGCAGGGAAG-3′, Mbp are forward: 5′-TACTTGGCCACAGCAAGTACC-3′ and reverse: 5′-GGGTGTACGAGGTGTCACAAT-3′, Gapdh are forward: 5′-ACATGCCGCCTGGAGAAACCTGCCA-3′ and reverse: 5′-TGCCAGCCCCAGCATCAAAGGTGGA-3′. The primer sequences used for the data reported in the supplementary figure are listed in Additional file 1: Sheet S1. After pre-incubation at 95°C for 15 min, the polymerase chain reaction (PCR) was performed as follows: 45 cycles of denaturation at 94°C for 15 s, annealing at 57°C for 25 s, and elongation at 72°C for 15 s.
Results
Laser-capture microdissection of microglial cells from the corpus callosum of 5-day and 4-week old rat brain.
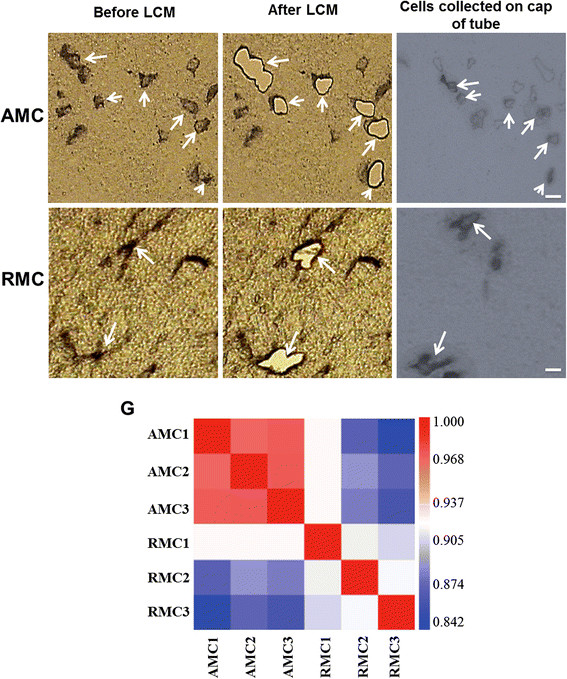
To compare the gene expression profiles of AMC and RMC, we stained both microglial cell types with peroxidase-conjugated lectin and isolated them from the CC of 5-day and 4-week old rat brain respectively. LCM of AMC and RMC from the CC of 5-day old rat brain has been shown in Figure1A-F. Lectin staining has been widely used to selectively stain microglia for study of microglial development in the CNS [29-31]. The cells isolated by LCM were further confirmed to be microglia since the mRNA expression of oligodendrocyte (CNPase), astrocyte (GFAP) and endothelial cell-specific genes (Vimentin) was undetectable (data not shown).
Figure 1 .
A-F. Identification and isolation of amoeboid microglial cells (AMC) and ramified microglial cells (RMC) from the corpus callosum (CC) of 5-day old and 4-weeks old rat brain respectively. Figure A shows AMC and Figure D shows RMC stained with lectin under laser capture microscopy. These cells are laser-cut along their periphery and isolated. Figure B and E show the region of the stained tissue section wherein the cells have been removed, and figure C and F show the isolated cells collected in the cap of vial. Arrows indicate the same cells in all the three images. Scale bars: A-F 50 μm. G. Correlation plots. Correlation plots were generated in Affymetrix Expression Console 1.1 after RMA normalizing raw CEL files of AMC and RMC expression data. The color scale indicates the degree of correlation between two different samples. A value of close to 1 refers to a high correlation.
cDNA microarray and generation of gene lists specific to AMC and RMC
To identify the genes that are differentially expressed between AMC and RMC, we extracted total RNA from AMC and RMC and carried out cDNA microarray using Rat Genome 230 2.0 array (Affymetrix). Each sample contained RNA from six hundred laser-captured microglial cells. To ensure gene expression consistency between samples within the groups, we determined the Pearson’s rank correlation coefficient after normalizing the raw expression data (Figure1G). The gene expression profile from the samples of same group showed a very high correlation of 0.97 ± 0.03 while, a relatively lower correlation value of 0.87 ± 0.03 was observed between samples of different groups. A high correlation coefficient of above 0.8 between the AMC and RMC may be due to the fact that the comparison is between the gene expression profiles of the same cell type., i.e. microglia regardless of the differences in age (5-day and 4-week rat brain) and morphology (amoeboid and ramified).
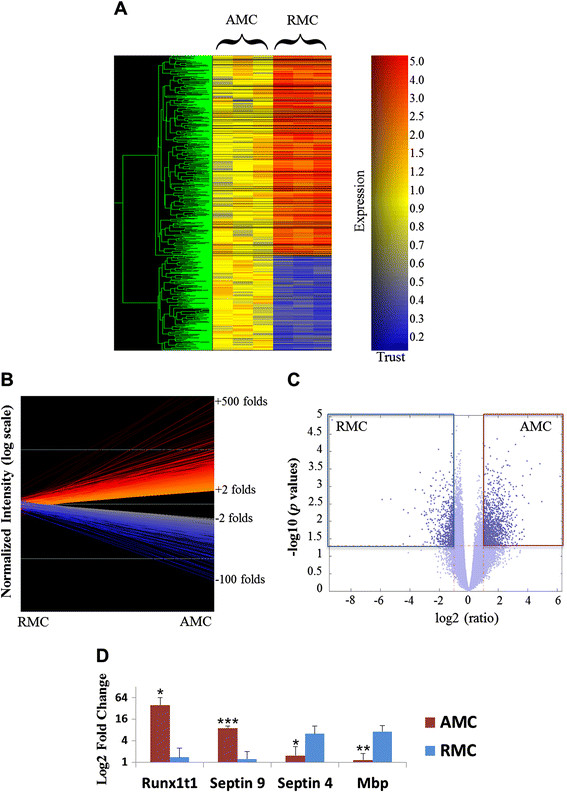
Agglomerative average-linked hierarchical clustering was performed and genes showing over two-fold differential expression between AMC and RMC were identified using GeneSpring 7.3 (Figure2A & B). About 800 genes were found to be differentially regulated in the two sample groups - 537 with upregulation and 258 with downregulation in AMC. A high number of differentially expressed genes identified were either novel or did not have any functional annotation. In view of this, a list of genes with known functional annotations was generated using the statistical functions in the Bioinformatics Toolbox in MATLAB R2009a [32]. This list was generated using a less stringent filtering (p value < 0.05, in contrast to the GeneSpring list which has a p value < 0.01) and contained close to 1400 upregulated genes and 700 downregulated genes in AMC compared to RMC (Figure2C).
Figure 2 .
A. Cluster Analysis. Cluster analysis shows changes in gene expression profiles of AMC and RMC. Agglomerative average-linkage hierarchical clustering of the six independent samples was obtained for selected groups of genes using GeneSpring 7.3. Each colored box represents the normalized expression level of a given gene in each sample and is colored according to the fold change. B. Line Graph. Represents a two-fold differential gene expression between the AMC and RMC. The lines in red represent gens upregulated and those in blue represent genes downregulated in AMC in comparison to RMC. C. Volcano Plot. Within the lateral quadrants (red and blue box) are the genes with two-fold difference and P Value < 0.05. These genes were chosen for generation of functional group lists. D.Validation of Microarray Profile. Histogram shows the qRTPCR validation of two AMC-specific genes (Runx1t1 and Sept9) and two RMC-specific genes (Sept4 and Mbp).
Functional categorization of AMC and RMC
The genes with highest fold change values in AMC and RMC (Table1 and 2) clearly delineate their functions and residing environment. For example, the AMC express genes that are shown to be involved in nervous system development (such as Dpysl3, Crmp1 and Smarca1) [33,34], immune system development (such as Hmgb3 and Sla) [35-38], cell migration during neurodevelopment (Dcx) [39] and the immune response as well in migration of microglia (Cxcr4, a chemokine receptor) [40-42]. The finding of expression of some genes that are known to be neuron-specific (such as Dcx), is interesting and has been further confirmed by immunohistochemical analysis which revealed the expression of Dcx by the AMC in the CC of 5 day old rat brain (Figure3A-C). In addition, mRNA expression of Dcx was detected in the BV-2 microglia (a murine microglia cell line) by RTPCR (Additional file 2: Figure S1).
Table 1.
Top 25 highly-expressed genes in AMC based on fold change. Both MATLAB and GeneSpring analysis are represented
| Genes from MATLAB analysis | Genes from Genespring analysis | ||||||
|---|---|---|---|---|---|---|---|
| Gene Symbol |
Gene Title |
Fold Change |
Function |
Gene Symbol |
Gene Title |
Fold Change |
Function |
| Sla |
Src-like adaptor |
73.34 |
protein binding |
LRRGT00193 |
Unknown |
574.79 |
Unknown |
| Dpysl3 |
dihydropyrimidinase-like 3 |
31.01 |
nervous system development |
Rnf152 |
ring finger protein 152 |
35.04 |
Unknown |
| Sox4 |
SRY (sex determining region Y)-box 4 |
28.33 |
pro-B cell differentiation |
Syncrip |
Synaptotagmin binding, cytoplasmic RNA interacting protein |
26.62 |
mRNA processing |
| Rprm |
reprimo, TP53 dependent G2 arrest mediator candidate |
19.13 |
cell cycle arrest |
Chst8 |
carbohydrate (N- acetylgalactosamine 4–0) sulfotransferase 8 |
26.17 |
sulfur metabolic process |
| Satb2 |
SATB homeobox 2 |
18.05 |
negative regulation of transcription |
Rpl28 |
ribosomal protein L28 |
19.32 |
translation |
| Dcx |
Doublecortin |
16.64 |
neuronmigration |
Slc16a7 |
solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
18.85 |
transport |
| Crmp1 |
collapsin response mediator protein 1 |
15.50 |
neuron development |
Crmp1 |
collapsin response mediator protein 1 |
18.54 |
neuron development |
| Syt16 |
synaptotagmin XVI |
13.53 |
protein binding |
LOC100158225 |
hypothetical protein LOC100158225 |
17.44 |
Unknown |
| Cct8 |
chaperonin containing Tcp1, subunit 8 (theta) |
13.53 |
protein folding |
Bmp2 |
bone morphogenetic protein 2 |
16.43 |
ossification |
| RGD1310352 |
similar to HTGN29 protein; keratinocytes associated transmembrane protein 2 |
13.42 |
Unknown |
Dcx |
doublecortin |
15.26 |
neuron migration |
| Appbp2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
13.28 |
Transport |
Mab21l1 |
mab-21-like 1 (C. elegans) |
14.54 |
positive regulation of cell proliferation |
| Cxcr4 |
chemokine (C-X-C motif) receptor 4 |
12.76 |
ameboidal cell migration |
Hmgb3 |
high mobility group box 3 |
14.14 |
negative regulation of myeloid cell differentiation |
| Ect2 |
epithelial cell transforming sequence 2 oncogene |
12.50 |
cell morphogenesis |
Baz1a |
bromodomain adjacent to zinc finger domain, 1A |
13.89 |
protein binding |
| Hmgb3 |
high mobility group box 3 |
12.47 |
negative regulation ofB cell differentiation |
Sox4 |
SRY (sex determining region Y)-box 4 |
13.43 |
pro-B cell differentiation |
| Hs3st5 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
12.32 |
protein amino acid sulfation |
LOC688455 |
hypothetical protein LOC688455 |
12.93 |
Unknown |
| Cfl2 |
cofilin 2, muscle |
12.21 |
protein binding |
Cxcr4 |
chemokine (C-X-C motif) receptor 4 |
12.92 |
ameboidal cell migration |
| Tnc |
Tenascin C |
11.93 |
negative regulation of cell adhesion |
Spint2 |
serine peptidase inhibitor, Kunitz type, 2 |
12.49 |
serine-type endopeptidase inhibitor activity |
| Nap1l3 |
nucleosome assembly protein 1-like 3 |
10.88 |
nucleosome assembly |
Zfml |
zinc finger, matrin-like |
12.20 |
nucleic acid binding |
| Mex3b |
mex3 homolog B (C. elegans) |
10.78 |
RNA binding |
Nradd |
neurotrophin receptor associated death domain |
11.65 |
signal transduction |
| Smarca1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
10.72 |
brain development |
Bcl7c |
B-cell CLL/lymphoma 7 C |
11.44 |
Unknown |
| Sh3bgrl |
SH3 domain binding glutamic acid-rich protein like |
9.74 |
Unknown |
Tmeff1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
11.43 |
multicellular organismal development |
| Ascc3l1 |
Activating signal cointegrator 1 complex subunit 3-like 1 |
9.58 |
RNA splicing |
Mtus1 |
mitochondrial tumor suppressor 1 |
11.12 |
cell cycle |
| Bex4 |
brain expressed gene 4 |
9.50 |
Nucleus |
Adfp |
Adipose differentiation related protein |
10.85 |
response to organic cyclic substance |
| Fam164a |
family with sequence similarity 164, member A |
9.49 |
Unknown |
Ttk |
Ttk protein kinase |
10.65 |
protein amino acid fig7 phosphorylation |
| LOC294446 | similar to Myristoylated alanine-rich C-kinase substrate (MARCKS) (ACAMP-81) | 9.44 | calmodulin binding | Lrrc20 | leucine rich repeat containing 20 | 10.63 | protein binding |
Table 2.
Top 25 genes in RMC based on fold change. Both MATLAB and GeneSpring analysis are represented
| Genes from MATLAB analysis | Genes from Genespring analysis | ||||||
|---|---|---|---|---|---|---|---|
| Gene Symbol |
Gene Title |
Fold Change |
Function |
Gene Symbol |
Gene Title |
Fold Change |
Function |
| Mobp |
myelin-associated oligodendrocyte basic protein |
623.18 |
nervous system development |
Mog |
myelin oligodendrocyte glycoprotein |
26.34 |
cell adhesion |
| Mog |
myelin oligodendrocyte glycoprotein |
39.14 |
cell adhesion |
Slc5a11 |
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
24.41 |
antigen processing and presentation |
| Mbp |
myelin basic protein |
21.57 |
myelination |
Mbp |
myelin basic protein |
22.86 |
myelination |
| Robo3 |
roundabout homolog 3 (Drosophila) |
17.39 |
neuron migration |
Cyp3a9 |
cytochrome P450, family 3, subfamily a, polypeptide 9 |
19.31 |
sensory perception of smell |
| Mal |
mal, T-celldifferentiation protein |
16.13 |
intracellular protein transport |
Opalin |
oligodendrocytic myelin paranodal and inner loop protein |
15.59 |
Golgi apparatus |
| Dnah12 |
dynein, axonemal, heavy polypeptide 12 |
12.77 |
microtubule-based movement |
Plp1 |
proteolipid protein 1 |
12.00 |
glial cell differentiation |
| Slc5a11 |
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
12.34 |
antigen processing and presentation |
Camk2a |
calcium/calmodulin- dependent protein kinase II alpha |
9.59 |
G1/S transition of mitotic cell cycle |
| Hapln2 |
hyaluronan and proteoglycan link protein 2 |
9.84 |
cell adhesion |
Lgi4 |
leucine-rich repeat LGI family, member 4 |
9.39 |
neuron maturation |
| Plp1 |
proteolipid protein 1 |
9.00 |
glial cell differentiation |
Cpne9 |
copine family member IX |
8.71 |
unknown |
| Ermn |
ermin, ERM-like protein |
8.82 |
morphogenesis of a branching structure |
Znf76 |
zinc finger protein 76 (expressed in testis) |
8.55 |
transcription |
| Tnnc2 |
troponin C type 2 (fast) |
8.60 |
skeletal muscle contraction |
Rhpn1 |
rhophilin, Rho GTPase binding protein 1 |
8.35 |
signal transduction |
| Mag |
myelin-associated glycoprotein |
8.12 |
cell adhesion |
Gng13 |
guanine nucleotide binding protein (G protein), gamma 13 |
8.16 |
G-protein coupled receptor protein signaling pathway |
| Lgi4 |
leucine-rich repeat LGI family, member 4 |
8.10 |
neuron maturation |
Mag |
myelin-associated glycoprotein |
7.99 |
cell adhesion |
| Extl1 |
exostoses (multiple)-like 1 |
8.09 |
protein binding |
Herc6 |
hect domain and RLD 6 |
7.71 |
protein modification process |
| Aldh3b1 |
aldehyde dehydrogenase 3 family, member B1 |
7.81 |
cellular aldehyde metabolic process |
Chn1 |
Chimerin (chimaerin) 1 |
6.31 |
signal transduction |
| Hhatl |
hedgehog acyltransferase-like |
7.79 |
negative regulation of N-terminal protein palmitoylation |
Sept4 |
septin 4 |
5.81 |
cell cycle |
| Gng13 |
guanine nucleotide binding protein (G protein), gamma 13 |
7.44 |
G-protein coupled receptor protein signaling pathway |
Ccdc37 |
coiled-coil domain containing 37 |
5.66 |
unknown |
| Akap8l |
A kinase (PRKA) anchor protein 8-like |
7.30 |
DNA binding |
Cmtm5 |
CKLF-like MARVEL transmembrane domain containing 5 |
5.30 |
membrane |
| Cpne9 |
copine family member IX |
7.24 |
Unknown |
Amhr2 |
anti-Mullerian hormone receptor, type II |
5.08 |
protein amino acid phosphorylation |
| Car12 |
Carbonic anyhydrase 12 |
7.21 |
one-carbon metabolic process |
Sptbn4 |
spectrin, beta, non-erythrocytic 4 |
4.84 |
axonogenesis |
| Srpk3 |
SFRS protein kinase 3 |
7.11 |
protein amino acid phosphorylation |
Gsta3 |
glutathione S-transferase A3 |
4.55 |
glutathione metabolic process |
| Casc1 |
cancer susceptibility candidate 1 |
6.90 |
Unknown |
Grin2c |
glutamate receptor, ionotropic, N-methyl D-aspartate 2 C |
4.53 |
startle response |
| S1pr5 |
sphingosine-1-phosphate receptor 5 |
6.82 |
signal transduction |
Fah |
fumarylacetoacetate hydrolase |
4.50 |
arginine catabolic process |
| Gpd1 |
glycerol-3-phosphate dehydrogenase 1 (soluble) |
6.75 |
carbohydrate metabolic process |
Ggn |
gametogenetin |
4.49 |
multicellular organismal development |
| Itsn2 | intersectin 2 | 6.67 | regulation of Rho protein signal transduction | Ntsr2 | neurotensin receptor 2 | 4.13 | signal transduction |
Figure 3 .
A-F. Differential immunoexpression of Dcx andRunx1t1 in AMC and RMC. Confocal images showing the immunoexpression of Dcx (B; red) and its co-localization (C) in OX42 (A; green) labeled AMC. Immunoexpression of Runx1t1 (E, H; red) and its co-localization (F, I) in OX42 (D, G; green) labeled AMC and RMC in the CC from 5-day (5D) and 4-week (4 W) old rat brain was also observed. Runx1t1 immunoexpression is undetectable in RMC (I) compared to that in the AMC (F). (DAPI – blue). Scale bars: A-C 50 μm, D-I 10 μm.
Interestingly, the RMC express genes involved in myelination (such as Mbp). Mbp-like proteins, also known as Golli proteins have previously been shown to be localized in human microglia at 22 weeks postnatally [43]. MBP mRNA was found to be expressed by laser captured-RMC (Figure2D) and BV-2 microglia (Additional file 2: Figure S1). Other myelin-related genes like Plp1 [44] and Lgi4 [45] have also been found with high expression values in the RMC. Plp1 and Lgi4 were found to be expressed by non-myelinating cells such as the Bergmann glia in cerebellum of the developing mouse brain [46].
On sorting the genes based on p values (Additional file 3: Sheet S2), we found several genes that are specific to AMC such as, genes involved in transcriptional repression (Mbd1 which binds to methylated sites on DNA) [47,48], vesicular trafficking (Snx6, a component of the retromer complex) [49], and microtubule depolymerization (Stmn1) [50]. RMC express genes involved in immune functions such as RT1-A2, which is the MHC of rat and C1ql3, a protein of the complement system [51,52], calcium ion signaling pathway protein, Camk2 [53,54] and sodium dependent glucose transporter gene Slc5a11, known to interact with immune-related genes [55].
Functional clusters of genes specific to AMC and RMC were derived using Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7. The functional groups in AMC involved cell cycle, mRNA processing, ribosome activity, cytoskeleton, and migration (CDC42-RAC pathway) and those in RMC were cellular homeostasis, cell projection, glial cell development, axon ensheathment, and regulation of synaptic transmission and plasticity (Additional file 4: Sheet S3 & Additional file 5: Sheet S4).
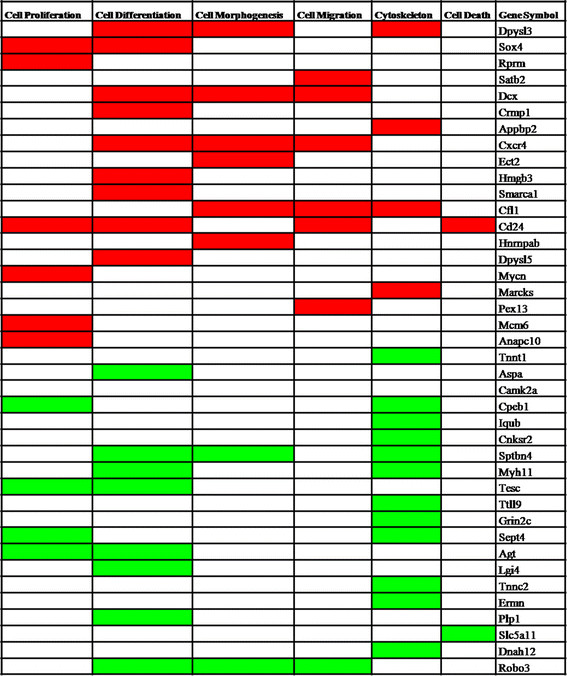
It has been observed that in AMC, genes which are involved in cell proliferation, death, and differentiation are highly expressed, whereas in RMC, genes that are mainly involved in cytoskeletal organization and cell differentiation are highly expressed (Figure4 &5A, Refer Additional file 6: Sheet S5 for entire list of AMC and RMC genes in these functional pathways).
Figure 4 .
Functional clusters to highly-expressed AMC and RMC genes. Heat map shows the top 25 AMC and RMC (arranged according to fold change) and their involvement in major cellular functions. Red shading indicates AMC genes and green shading indicates RMC genes.
Figure 5 .
A. Functional pathways. Graph shows the number of AMC and RMC genes enriched in different functions. It can be noted that the AMC apart from proliferation and differentiation express a high number of genes involved in cell death whereas the RMC express a high number of cytoskeletal genes. B. Stemness of AMC and RMC. Venn diagrams represent the share of Embryonic (ESC), Neural (NSC) and Hematopoietic (HSC) stem cell genes in AMC and RMC. The AMC express a high number of stem cell genes compared to the RMC. The RMC express more HSC-specific genes than NSC- and ESC- specific genes.
Expression of monocyte-specific genes by AMC and RMC
Microglia have been proposed to originate from two distinct sources: the mesenchymal progenitor cells at the yolk sac [56] and bone-marrow derived circulating monocytes [2]. The availability of gene expression profiles for circulating monocytes prompted us to compare and study the functional similarities of AMC and RMC to that of circulating monocytes [28] (Refer Table3 &4 for pathways, Additional file 7: Sheet S6 for full gene list). Monocytic genes highly expressed by AMC (Cxcr4, Csk and Rac1) are known to be involved in several disease pathways such as Parkinson’s disease, Huntington’s disease and HIV infection, phagocytosis and chemokine signaling pathways. Further, the monocytic genes (Hla-c, Cd74, Cd302, Lsp1 and Runx3) expressed by RMC are involved in antigen presentation and lysosome-related functions. The peripheral monocytic genes expressed by the AMC and RMC may be further investigated if they reflect the microglia property in pathological brain. These genes may also provide deeper insights into the similarities between peripheral monocytes and microglia and the re-appearance of these genes during infection or neurodegeneration in the activated microglia may be critical for immune response.
Table 3.
Pathways involving monocytic genes expressed by AMC
| Pathway | Nr. of Genes | P-Value | Benjamini corrected P-value |
|---|---|---|---|
| REACT_17015:Metabolism of proteins |
37 |
2.00E-16 |
1.20E-14 |
| REACT_71:Gene Expression |
42 |
3.30E-13 |
8.60E-12 |
| REACT_1762:3′ -UTR-mediated translational regulation |
24 |
6.20E-13 |
1.10E-11 |
| Ribosome |
17 |
1.30E-10 |
1.40E-08 |
| REACT_6167:Influenza Infection |
24 |
1.90E-09 |
2.50E-08 |
| Proteasome |
10 |
1.20E-06 |
6.90E-05 |
| Pathogenic Escherichia coli infection |
10 |
6.70E-06 |
2.50E-04 |
|
P00029:Huntington disease |
16 |
7.20E-06 |
5.80E-04 |
| REACT_13635:Regulation of activated PAK-2p34 by proteasome mediated degradation |
10 |
1.50E-05 |
1.50E-04 |
|
P00049:Parkinson disease |
12 |
3.40E-05 |
1.40E-03 |
| REACT_9035:APC/C:Cdh1-mediated degradation of Skp2 |
11 |
3.80E-05 |
3.30E-04 |
| REACT_6850:Cdc20:Phospho-APC/C mediated degradation of Cyclin A |
11 |
7.60E-05 |
5.70E-04 |
| REACT_6185:HIV Infection |
18 |
1.70E-04 |
1.10E-03 |
| REACT_11045:Signaling by Wnt |
10 |
2.20E-04 |
1.30E-03 |
|
P00016:Cytoskeletal regulation by Rho GTPase |
10 |
1.40E-03 |
3.70E-02 |
| Fc gamma R-mediated phagocytosis |
9 |
1.90E-03 |
5.10E-02 |
|
P00018:EGF receptor signaling pathway |
11 |
2.40E-03 |
4.80E-02 |
|
P00034:Integrin signalling pathway |
14 |
2.70E-03 |
4.20E-02 |
| Neurotrophin signaling pathway |
10 |
2.70E-03 |
6.00E-02 |
|
P00060:Ubiquitin proteasome pathway |
8 |
4.30E-03 |
5.70E-02 |
| REACT_578:Apoptosis |
12 |
4.90E-03 |
2.50E-02 |
| Chemokine signaling pathway |
12 |
4.90E-03 |
8.80E-02 |
| REACT_1538:Cell Cycle Checkpoints |
11 |
5.30E-03 |
2.50E-02 |
|
P00021:FGF signaling pathway |
10 |
5.80E-03 |
6.50E-02 |
| REACT_383:DNA Replication | 10 | 6.20E-03 | 2.70E-02 |
Table 4.
Pathways involving monocytic genes expressed by RMC
| Pathway | Nr. of Genes | P-Value | Benjamini corrected P-value |
|---|---|---|---|
| Lysosome |
5 |
2.00E-03 |
1.00E-01 |
| Antigen processing and presentation |
4 |
6.60E-03 |
1.60E-01 |
| 70.Signal_peptides_(MHC)_class_I _molecules |
2 |
3.30E-02 |
1.60E-01 |
| Other glycan degradation |
2 |
7.30E-02 |
7.40E-01 |
| 114.Genomic_reformatting_Brain _Ischemia | 2 | 9.70E-02 | 2.30E-01 |
Expression of neural, embryonic and hematopoietic stem cell specific genes by AMC and RMC
Both AMC and RMC express a number of stem cell specific genes [27] (Table5 &6, Additional file 8: Sheet S7). However, the RMC express a lesser number of neural stem cell (NSC) and embryonic stem cell (ESC) specific genes, compared with the AMC. In spite of this, higher percentage of hematopoietic stem cell specific genes (HSC) was found to be expressed in RMC (Figure5B). Our finding that microglia retain HSC specific properties even in the adult brain is suggestive of their hematopoietic lineage.
Table 5.
Stem-cell genes enriched in AMC
| Gene Symbol | Gene Title | Fold Changes | Function |
|---|---|---|---|
| Rprm |
reprimo, TP53 dependent G2 arrest mediator candidate |
19.125 |
cell cycle arrest |
| Cct8 |
chaperonin containing Tcp1, subunit 8 (theta) |
13.525 |
protein folding |
| RGD1310352 |
similar to HTGN29 protein; keratinocytes associated transmembrane protein 2 |
13.419 |
Unknown |
| Ect2 |
epithelial cell transforming sequence 2 oncogene |
12.498 |
cell morphogenesis |
| Cfl2 |
cofilin 2, muscle |
12.211 |
protein binding |
| Sh3bgrl |
SH3 domain binding glutamic acid-rich protein like |
9.7435 |
Unknown |
| Bex4 |
brain expressed gene 4 |
9.4991 |
Unknown |
| Cfl1 |
cofilin 1, non-muscle |
9.3235 |
cytokinesis |
| Cd24 |
CD24 molecule |
9.2544 |
response to hypoxia |
| Ns5atp9 |
NS5A (hepatitis C virus) transactivated protein 9 |
9.1968 |
Unknown |
| Hnrnpab |
heterogeneous nuclear ribonucleoprotein A/B |
8.8712 |
epithelial to mesenchymal transition |
| Mycn |
v-mycmyelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
8.6884 |
regulation of transcription, DNA-dependent |
| LOC294446 |
similar to Myristoylated alanine-rich C-kinase substrate (MARCKS) (ACAMP-81) |
8.5949 |
actin binding |
| Pex13 |
peroxisomal biogenesis factor 13 |
8.4726 |
fatty acid alpha-oxidation |
| Maoa |
monoamine oxidase A |
7.8728 |
catecholamine metabolic process |
| Tmeff1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
7.7994 |
multicellular organismal development |
| Ube2e3 |
ubiquitin-conjugating enzyme E2E 3, UBC4/5 homolog (yeast) |
7.5905 |
modification-dependent protein catabolic process |
| Rab10 |
RAB10, member RAS oncogene family |
7.5637 |
regulation of transcription, DNA-dependent |
| Arf4 |
ADP-ribosylation factor 4 |
7.3522 |
transport |
| Mapre1 |
microtubule-associated protein, RP/EB family, member 1 |
7.0496 |
cell cycle |
| Psme3 |
proteasome (prosome, macropain) activator subunit 3 |
7.0045 |
cell adhesion |
| Tmem43 |
transmembrane protein 43 |
6.9166 |
Unknown |
| March5 |
membrane-associated ring finger (C3HC4) 5 |
6.9148 |
zinc ion binding |
| Sqle |
Squaleneepoxidase |
6.7699 |
cellular aromatic compound metabolic process |
| Apaf1 | apoptotic peptidase activating factor 1 | 6.6501 | neural tube closure |
Table 6.
Stem-cell genes enriched in RMC
| Gene Symbol | Gene Title | Fold Changes | Function |
|---|---|---|---|
| RGD1307882 |
similar to CG9346-PA |
2.9758 |
RNA processing |
| Mll1 |
myeloid/lymphoid or mixed-lineage leukemia 1 |
2.9906 |
DNA repair |
| Fermt3 |
fermitin family homolog 3 (Drosophila) |
3.0629 |
protein binding |
| Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
3.2355 |
DNA integration |
| Mtf1 |
metal-regulatory transcription factor 1 |
3.2416 |
regulation of transcription, DNA-dependent |
| Egr1 |
early growth response 1 |
3.2473 |
negative regulation of transcription from RNA polymerase II promoter |
| Map7 |
microtubule-associated protein 7 |
3.263 |
cell morphogenesis |
| Sfrs11 |
splicing factor, arginine/serine-rich 11 |
3.3325 |
nuclear mRNA splicing, via spliceosome |
| Cd302 |
CD302 molecule |
3.3742 |
binding |
| Ctnnal1 |
catenin (cadherin associated protein), alpha-like 1 |
3.4127 |
unknown |
| Cyp4f6 |
cytochrome P450 4 F6 |
3.4649 |
leukotriene metabolic process |
| Mlec |
Malectin |
3.5937 |
carbohydrate metabolic process |
| Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
3.663 |
negative regulation of cell proliferation |
| Scn1b |
sodium channel, voltage-gated, type I, beta |
3.6699 |
transport |
| Tmbim1 |
transmembrane BAX inhibitor motif containing 1 |
3.7938 |
unknown |
| Pck2 |
phosphoenolpyruvatecarboxykinase 2 (mitochondrial) |
3.8945 |
gluconeogenesis |
| Tnni3 |
troponin I type 3 (cardiac) |
4.0048 |
vasculogenesis |
| Nefh |
neurofilament, heavy polypeptide |
4.2066 |
microtubule cytoskeleton organization |
| Prpf38b |
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
4.3721 |
mRNA processing |
| Tnnt1 |
troponin T type 1 (skeletal, slow) |
4.4062 |
skeletal muscle contraction |
| Zranb2 |
zinc finger, RAN-binding domain containing 2 |
4.598 |
mRNA processing |
| Zrsr1 |
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
5.0348 |
nucleotide binding |
| Lrrc23 |
leucine rich repeat containing 23 |
5.1395 |
protein binding |
| Akap8l |
A kinase (PRKA) anchor protein 8-like |
5.6932 |
DNA binding |
| Tnnc2 | troponin C type 2 (fast) | 8.6024 | skeletal muscle contraction |
Pathway analysis and validation of differentially expressed genes in AMC and RMC
The pathway analysis (Additional file 9: Figure S2) revealed novel molecular networks involving several signaling molecules and pathways within microglia. In order to validate the results obtained from pathway analysis, we randomly selected three transcription factors which are highly expressed in the AMC. They are: Sox4 and Sox11 which are SRY-related HMG-box family of transcription factors [57] and Runt-related transcription factor 1; translocated to, 1 (cyclinD-related) (Runx1t1), a member of the ETO gene family of transcriptional co-repressors [58].
Runx1t1, by forming a fusion protein with Runx1, another member of RUNX family, leads to self-renewal of human monocytic cells thereby impairing differentiation of these cells [59]. Certain genes known to be downregulated by the Runx1-Runx1t1 transcription factor complex such as Socs1, Csf1, and Runx3 are highly expressed by RMC [60] (Additional file 5: Sheet S4). Further, a transcriptional dysregulation caused by this fusion protein was found to cause the over-expression of Sox4 in human progenitor cells [60]. In an earlier study, Sox4 deficient mice exhibited proliferation-defective pro-B cells [61]. Similar networks might function in the AMC and therefore warrants further investigation. Our immunohistochemical analysis revealed that Sox4 (Figure6A-C) is highly expressed in the nucleus and cytoplasm of AMC. Similarly, Sox11 (Figure6D-I) and Runx1t1 (Figure3D-I) are expressed in the AMC, but hardly detectable in the RMC thus validating our microarray results. Further, quantitative real time RTPCR for Runx1t1 using RNA extracted from LCM-captured AMC and RMC showed a very high expression of Runx1t1 in AMC when compared to RMC (Figure2D).
Figure 6 .
A-I. Differential immunoexpression of Sox4 and Sox11 in AMC and RMC. Confocal images showing a high immunoexpression of Sox4 (B; red) and its co-localization (C) in OX42 (A; green) labeled AMC. Immunoexpression of Sox11 (E, H; red) and its co-localization (F, I) in OX42 (D, G; green) labeled AMC and RMC in the CC from 5-day (5D) and 4-week (4 W) old rat brain was also observed. Sox11immuno expression is undetectable in RMC (I) compared to that in the AMC (F). (DAPI – blue).Scale bars: A-I 10 μm.
Differential expression of septin genes in AMC and RMC
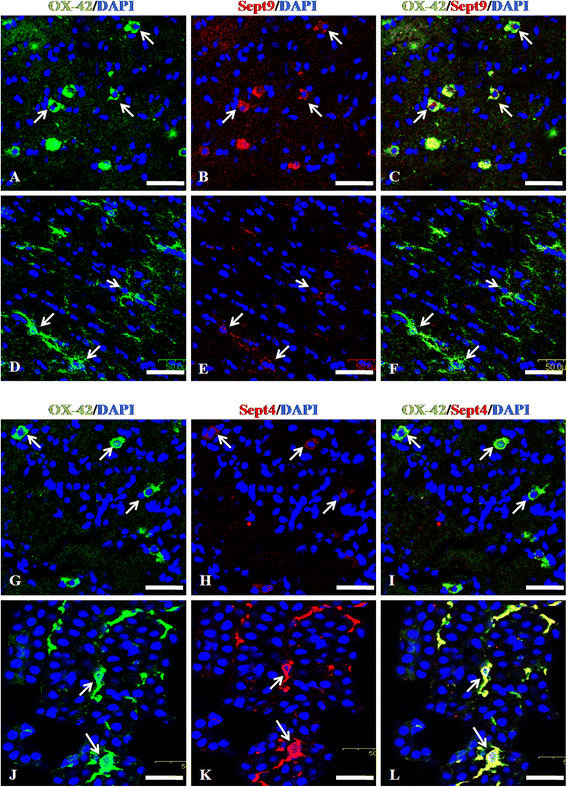
Septins are a family of multifunctional proteins involved in cytoskeletal organisation and cell division [62-64]. They have also been implicated in tumorigenesis and neurodegeneration [65]. In the present study, Septin genes were found to be differentially expressed in AMC and RMC. Sept3, 6, 9 and 11 were expressed in AMC whereas Sept4 and 8 expressed in RMC. The expression of some of these genes was further confirmed by immunohistochemical and quantitative real time RTPCR analysis which showed Sept9 immunoexpression and mRNA expression in the AMC (Figure7A-F and Figure2D) and Sept4 expression in the RMC (Figure7G-L and Figure2D). Since the role of Septins in microglial functioning has not yet been investigated, studies on the Septin family may further our knowledge on the cytoskeletal dynamics involved in proliferation, migration and activation of AMC and RMC.
Figure 7 .
A-L. Immunoexpression of Sept9 and Sept4 in RMC. Confocal images showing the immunoexpression of Sept9 and Sept4 (B, E, H, K; red) and their co-localization (C, F, I, L) in OX42 (A, D, G, J; green) labeled AMC and RMC in the CC from 5-day (5D) and 4-week (4 W) old rat brain.Sept9 immunoexpression is undetectable in RMC (F) compared to that in the AMC (C) whereas Sept4 is undetectable in AMC (I) compared to that in the RMC (L). (DAPI – blue).Scale bars: A-L 50 μm.
Microglia in response to trauma or neurodegenerative stimuli exhibit upregulation of proinflammatory cytokines and chemokines [66]. In the present study, both AMC and RMC exhibited relatively low expression intensities for most of the cytokines such as TNF-α and interleukins and chemokines such as Cxcl3, Cxcl12 and Ccl2 (Additional file 10: Sheet S8).
Discussion
Microglia in the healthy brain express low levels of cytokines and chemokines
This study is a novel attempt to examine the global gene expression profile of microglia in situ and to functionally distinguish the two distinct microglial phenotypes, namely, AMC and RMC. A noteworthy feature of this transcriptome profile was that the expression of cytokines and chemokines in both AMC and RMC was hardly detectable which is in agreement with previous studies [24]. It has been widely shown that the untreated microglia in culture produce some amount of proinflammatory cytokines and chemokines, indicating that culture media stimulate the microglial cells. Significantly, the low expression of cytokines and chemokines in both types of microglia in the present study appears to mimic the transcriptome status of normal microglia in healthy brain in vivo.
AMC express genes involved in cell cycle process and migration whereas RMC express genes involved in synaptic integrity and neuronal maturation
AMC from the first week of postnatal rat brains have a high proliferative capacity [2]. During development, about two-thirds of AMC undergo apoptosis and the rest transform into RMC [67,68]. In accordance with this, microarray analysis in the present study revealed a high expression of cell proliferation/cell cycle-related genes such as Myc and CyclinA2, CyclinB2 and CyclinD1 (Additional file 6: Sheet S5) [69,70] and genes involved in cell death (Figure5A & Additional file 6: Sheet S5) namely, Casp2,Casp3 [71,72] and Apaf1(Glial apoptosis related gene) in the AMC [73]. It is striking that the AMC express Dcx, a protein known to be a marker for migrating neurons [74]. It may be worth investigating the role of Dcx in migration of AMC in the early postnatal brain.
On the other hand, the RMC, apart from cell homeostasis and glial development, appear to contribute to synaptic transmission as they express genes such as Grin2c, S100b and Camk2a (Additional file 5: Sheet S4) [75-77]. This is interesting and supports the recent experimental studies showing the role of microglia in the maintenance and modifications of synaptic integrity in the healthy brain [78,79]. Further, Grin2c, a subunit of NMDA receptor complex is expressed by the microglial cells in the CC and has been shown to be functionally important in microglia-mediated neuroinflammation [80,81]. S100b, a calcium ion binding protein, is also expressed by microglia and relocates around phagosomes during microglial activation and phagocytosis [82,83].
RMC express myelin basic protein (Mbp) which encodes two families of proteins i.e., classic Mbps and Golli-Mbps. Classic Mbps serve as the major protein constituent of myelin in the central and peripheral nervous system whereas, the Golli proteins are known to be broadly distributed in the brain, particularly in the microglia during normal development and inflammation [84] and involved in the interaction between microglia and oligodendrocyte precursor cells during multiple sclerosis [85]. Taken together, these results reveal that microglia are not only involved in immune response and phagocytosis but also play diverse roles in healthy brain.
Both AMC and RMC express cytoskeleton-related genes
Regulation of cytoskeletal dynamics is important to both microglial migration and ramification [86-88]. Apart from cytoskeletal structural proteins such as tubulins and actin, we found that the AMC express cytoskeleton-associated Crmp family proteins [89] such as Crmp-1, Dpysl3 and Dpysl5 and Septin family proteins such as Sept9 and 11 [90,91]. Septins are implicated in cytoskeletal processes such as vesicular trafficking [92]. These cytoskeleton-associated proteins may therefore explain the migration and phagocytosis of AMC during normal development and pathology. In the present study, AMC express Sept9 but not Sept4 whereas, RMC express Sept4, but not Sept9, indicating differential roles of Septin family genes in AMC and RMC. Sept4 has been recently shown to be involved in cortical neuron migration [93]. Absence of Sept4 immunoexpression in the AMC and its high expression in the RMC is suggestive of an important role for this protein in microglial transformation during development.
Expression of monocyte- and stem cell-specific genes by AMC and RMC indicates their stemness and origin
Recent studies have proven that microglia originate from the mesenchymal progenitor cells at the yolk sac [56]. However, microarray studies of various hematopoetic and non-hematopoetic cell types revealed a close relationship between the gene expression profiles of microglia and bone-marrow derived macrophages [94] which are known to differentiate from circulating monocytes. Therefore, we sought to identify the monocyte-specific genes expressed by AMC and RMC. AMC express several monocyte-specific genes including Mcl1 and Id2. Mcl1 is associated with cell viability and differentiation of myeloid cells which include monocytes and macrophages [95,96] and Id2, a negative regulator of basic helix loop helix transcription factors, is involved in the differentiation of myeloid cells [97]. A recent study demonstrated that Id2 is required for bone morphogenic protein (Bmp)-mediated differentiation of microglia into Map2+ neurons and Gfap+ astrocytes [98] suggesting that this gene may promote microglial trans-differentiation. Both Mcl1 and Id2 have been shown to be involved in cell differentiation and their high expression in AMC explains the role of these genes in promoting the maturation of AMC and its transformation into RMC
On the other hand, RMC exhibited increased expression of Lsp1, which binds to the cytoskeleton and is known to be a marker for leucocytes [99]. Overexpression of Lsp1 in neutrophils was associated to defective actin polymerization which render these cells immotile [100]. Further, overexpression of Lsp1 in a highly motile melanoma cell line led to formation of hair-like projections. Thus, upregulation of Lsp1 in the RMC, compared to the AMC may explain the role of this gene in motility and ramification of RMC [101] which are the resident population in the adult brain parenchyma.
Both AMC and RMC express stem cell-specific (ESC, NSC and HSC) genes, indicating their stemness and suggesting that microglia may undergo trans-differentiation. The RMC expressed a high percentage of HSC specific genes in comparison to ESC and NSC specific genes, and this, reinforces the monocytic nature of microglia. For example Mll1, a highly expressed HSC specific gene in the RMC, is a histone methyl transferase whose functional disruption is implicated in human leukemia [102]. Understanding the functions of these HSC specific genes may be important in comprehending the immune system related-roles of AMC and RMC.
AMC express proliferation- and differentiation-related genes, Sox4, Sox11 and Runx1t1
In order to validate the microarray data, we have analyzed the expression patterns of SOX genes (Sox4 and Sox11) which are known to be involved in differentiation and Runx1t1, which is involved in the proliferation of hematopoietic lineage cells. These genes were highly expressed by the AMC and their expression and role have not been studied in microglia, so far. Initially, nuclear expression of the transcription factor Sox11 was shown to be associated with embryonic neurogenesis and lymphopoiesis [103,104]. However, there are no data on the role of SOX genes in microglia in which Sox11 is expressed in the cytoplasm as reported in plasma myeloma cells and other B-cell lymphomas [104]. According to previous studies, the overexpression of the fusion protein, Runx1-Runx1t1 causes the downregulation of Csf-1 (a hematopoietic cytokine known to cause activation of microglia) [105] and Runx3 (a tumour suppressor) [106]. Our expression profile showed the increased expression of Runx1t1 in the AMC and downregulation of Csf-1 and Runx3 in AMC compared to RMC. Functional analysis of these transcription factors may help in understanding microglial proliferation and differentiation.
Conclusions
Overall, the transcriptome profiling has identified several genes, which help in elucidating morphological transformation and functions of AMC and RMC. These genes not only represent the physiological role of microglia in the developing brain but may also be useful therapeutic targets in neuropathologies in which microglia are implicated.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RP performed majority of the experiments and wrote the manuscript. BJ, BN, MJ1 and MJ2 performed some experiments and participated in discussion. JL, SST and EAL participated actively in discussion of the project and editorial work of the manuscript. STD is the Principal Investigator and was instrumental to the execution of the entire project. All of the authors have read and approved the final version of the manuscript.
Supplementary Material
Sheet S1. Primer sequences .
Figure S1. Expression of Dcx and Mbp in BV-2 microglia. PCR was done using cDNA obtained from BV-2 microglia for Dcx and Mbp. Both Dcx and Mbp were found to be expressed by BV-2 microglia as identified by the PCR products in agarose gel electrophoresis.
Sheet S2. AMC and RMC genes with P-value.
Sheet S3. AMC and RMC gene clusters.
Sheet S4. Genes involved in CDC42-RAC pathway for migration, Synaptic transmission and genes downregulated by the Runx1-Runx1t1 complex.
Sheet S5. Functions of AMC and RMC.
Sheet S6. Monocytic genes enriched in AMC and RMC.
Sheet S7. ESC, NSC and HSC genes enriched in AMC and RMC.
Figure S2. Pathway analysis. Novel molecular networks identified by inputting AMC and RMC expression data into Adriane Pathway Studio. Red colored shapes specify AMC genes and violet colored shapes specify RMC genes. Some AMC genes - Sox4, Sox11, Runx1t1 are highlighted in the figure.
Sheet S8. Expression of cytokines in AMC and RMC.
Contributor Information
Rangarajan Parakalan, Email: g0900014@nus.edu.sg.
Boran Jiang, Email: bo.ran.jiang@gmail.com.
Baby Nimmi, Email: a0075544@nus.edu.sg.
Manivannan Janani, Email: g0900038@nus.edu.sg.
Manikandan Jayapal, Email: mjayapal@kau.edu.sa.
Jia Lu, Email: ljia@dso.org.sg.
Samuel SW Tay, Email: samuel_tay@nuhs.edu.sg.
Eng-Ang Ling, Email: eng_ang_ling@nuhs.edu.sg.
S Thameem. Dheen, Email: thameem_dheen@nuhs.edu.sg.
Acknowledgements
This research was supported by research grants from National Medical Research Council, Singapore (NMRC/1113/2007 and EDG10nov039).
References
- PioDel R-H. THE MICROGLIA. Lancet. 1939;233(6036):1023–1026. doi: 10.1016/S0140-6736(00)60571-8. [DOI] [Google Scholar]
- Ling EA, Wong WC. The origin and nature of ramified and amoeboid microglia: a historical review and current concepts. Glia. 1993;7(1):9–18. doi: 10.1002/glia.440070105. [DOI] [PubMed] [Google Scholar]
- Orłowski D, Sołtys Z, Janeczko K. Morphological development of microglia in the postnatal rat brain: A quantitative study. Int J Dev Neurosci. 2003;21(8):445–450. doi: 10.1016/j.ijdevneu.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Brockhaus J, Möller T, Kettenmann H. Phagocytozing ameboid microglial cells studied in a mouse corpus callosum slice preparation. Glia. 1996;16(1):81–90. doi: 10.1002/(SICI)1098-1136(199601)16:1<81::AID-GLIA9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Hirasawa T, Kohsaka S. Visualization of microgilia in living tissues using Iba1-EGFP transgenic mice. Brain Nerve. 2007;59(7):763–772. [PubMed] [Google Scholar]
- Leong S-K, Ling E-A. Amoeboid and ramified microglia: Their interrelationship and response to brain injury. Glia. 1992;6(1):39–47. doi: 10.1002/glia.440060106. [DOI] [PubMed] [Google Scholar]
- Innocenti GM, Clarke S, Koppel H. Transitory macrophages in the white matter of the developing visual cortex. II. Development and relations with axonal pathways. Dev Brain Res. 1983;11(1):55–66. doi: 10.1016/0165-3806(83)90201-8. [DOI] [PubMed] [Google Scholar]
- O’Leary DD, Stanfield BB, Cowan WM. Evidence that the early postnatal restriction of the cells of origin of the callosal projection is due to the elimination of axonal collaterals rather than to the death of neurons. Brain Res. 1981;227(4):607–617. doi: 10.1016/0165-3806(81)90012-2. [DOI] [PubMed] [Google Scholar]
- Hristova M, Cuthill D, Zbarsky V, Acosta-Saltos A, Wallace A, Blight K, Buckley SMK, Peebles D, Heuer H, Waddington SN. et al. Activation and deactivation of periventricular white matter phagocytes during postnatal mouse development. Glia. 2010;58(1):11–28. doi: 10.1002/glia.20896. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. e-Neuroforum. 2005;11(3):95–96. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Ling EA, Ng YK, Wu CH, Kaur C. In: Progress in Brain Research. Volume 132. Castellano Lopez B, editor. Elsevier, MN-S; 2001. Microglia: its development and role as a neuropathology sensor; pp. 61–79. [DOI] [PubMed] [Google Scholar]
- McGeer PL, Itagaki S, Tago H, McGeer EG. Reactive microglia in patients with senile dementia of the Alzheimer type are positive for the histocompatibility glycoprotein HLA-DR. Neurosci Lett. 1987;79(1–2):195–200. doi: 10.1016/0304-3940(87)90696-3. [DOI] [PubMed] [Google Scholar]
- Combs CK, Johnson DE, Karlo JC, Cannady SB, Landreth GE. Inflammatory mechanisms in Alzheimer's disease: inhibition of beta-amyloid-stimulated proinflammatory responses and neurotoxicity by PPARgamma agonists. J Neurosci. 2000;20(2):558–567. doi: 10.1523/JNEUROSCI.20-02-00558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Liu Y, Cooper C, Liu B, Wilson B, Hong JS. Microglia enhance β-amyloid peptide-induced toxicity in cortical and mesencephalic neurons by producing reactive oxygen species. J Neurochem. 2002;83(4):973–983. doi: 10.1046/j.1471-4159.2002.01210.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang T, Pei Z, Miller DS, Wu X, Block ML, Wilson B, Zhang W, Zhou Y, Hong J-S. et al. Aggregated α-synuclein activates microglia: a process leading to disease progression in Parkinson’s disease. FASEB J. 2005;19(6):533–542. doi: 10.1096/fj.04-2751com. [DOI] [PubMed] [Google Scholar]
- Perry VH, Matyszak MK, Fearn S. Altered antigen expression of microglia in the aged rodent CNS. Glia. 1993;7(1):60–67. doi: 10.1002/glia.440070111. [DOI] [PubMed] [Google Scholar]
- Sierra A, Gottfried-Blackmore AC, McEwen BS, Bulloch K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia. 2007;55(4):412–424. doi: 10.1002/glia.20468. [DOI] [PubMed] [Google Scholar]
- Perry VH, Cunningham C, Holmes C. Systemic infections and inflammation affect chronic neurodegeneration. Nat Rev Immunol. 2007;7(2):161–167. doi: 10.1038/nri2015. [DOI] [PubMed] [Google Scholar]
- Godbout JP, Chen J, Abraham J, Richwine AF, Berg BM, Kelley KW, Johnson RW. Exaggerated neuroinflammation and sickness behavior in aged mice after activation of the peripheral innate immune system. FASEB J. 2005;19(10):1329–1331. doi: 10.1096/fj.05-3776fje. [DOI] [PubMed] [Google Scholar]
- Monje ML, Toda H, Palmer TD. Inflammatory Blockade Restores Adult Hippocampal Neurogenesis. Science. 2003;302(5651):1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- Kaur C, Ling EA. Periventricular white matter damage in the hypoxic neonatal brain: Role of microglial cells. Prog Neurobiol. 2009;87(4):264–280. doi: 10.1016/j.pneurobio.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Aloisi F. Immune function of microglia. Glia. 2001;36(2):165–179. doi: 10.1002/glia.1106. [DOI] [PubMed] [Google Scholar]
- Garden G, Möller T. Microglia Biology in Health and Disease. J Neuroimmune Pharmacol. 2006;1(2):127–137. doi: 10.1007/s11481-006-9015-5. [DOI] [PubMed] [Google Scholar]
- Hurley SD, Walter SA, Semple-Rowland SL, Streit WJ. Cytokine transcripts expressed by microglia in vitro are not expressed by ameboid microglia of the developing rat central nervous system. Glia. 1999;25(3):304–309. doi: 10.1002/(SICI)1098-1136(19990201)25:3<304::AID-GLIA10>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane HC, Lempicki R. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: Transcriptional Profiling of Embryonic and Adult Stem Cells. Science. 2002;298(5593):597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Liu H, Shi B, Huang C-C, Eksarko P, Pope RM. Transcriptional diversity during monocyte to macrophage differentiation. Immunol Lett. 2008;117(1):70–80. doi: 10.1016/j.imlet.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya J, Calvo JL, Carbonell AL, Borregon A. A lectin histochemistry study on the development of rat microglial cells. J Anat. 1991;175:229–236. [PMC free article] [PubMed] [Google Scholar]
- Streit WJ, Kreutzberg GW. Lectin binding by resting and reactive microglia. J Neurocytol. 1987;16(2):249–260. doi: 10.1007/BF01795308. [DOI] [PubMed] [Google Scholar]
- Mannoji H, Yeger H, Becker LE. A specific histochemical marker (lectin < i > Ricinus communis</i > agglutinin-1) for normal human microglia, and application to routine histopathology. Acta Neuropathol. 1986;71(3):341–343. doi: 10.1007/BF00688060. [DOI] [PubMed] [Google Scholar]
- Slonim DK, Yanai I. Getting started in gene expression microarray analysis. PLoS Comput Biol. 2009;5(10):e1000543. doi: 10.1371/journal.pcbi.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Strittmatter SM. A family of rat CRMP genes is differentially expressed in the nervous system. J Neurosci. 1996;16(19):6197–6207. doi: 10.1523/JNEUROSCI.16-19-06197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: A regulator of Engrailed gene expression. EMBO J. 2003;22(22):6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth MJ, Curtis DJ, Kirby MR, Garrett-Beal LJ, Seidel NE, Cline AP, Bodine DM. Hmgb3: An HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102(4):1298–1306. doi: 10.1182/blood-2002-11-3541. [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood. 2005;105(2):627–634. doi: 10.1182/blood-2004-07-2551. [DOI] [PubMed] [Google Scholar]
- Dragone LL, Shaw LA, Myers MD, Weiss A. SLAP, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol Rev. 2009;232(1):218–228. doi: 10.1111/j.1600-065X.2009.00827.x. [DOI] [PubMed] [Google Scholar]
- Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like Adaptor Protein (Slap) Is a Negative Regulator of T Cell Receptor Signaling. J Exp Med. 2000;191(3):463–474. doi: 10.1084/jem.191.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6(12):1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- Lavi E, Strizki J, Ulrich A, Zhang W, Fu L, Wang Q, O’Connor M, Hoxie J, Gonzalez-Scarano F. CXCR-4 (Fusin), a co-receptor for the type 1 human immunodeficiency virus (HIV-1), is expressed in the human brain in a variety of cell types, including microglia and neurons. Am J Pathol. 1997;151(4):1035–1042. [PMC free article] [PubMed] [Google Scholar]
- Lu DY, Tang CH, Yeh WL, Wong KL, Lin CP, Chen YH, Lai CH, Chen YF, Leung YM, Fu WM. SDF-1alpha up-regulates interleukin-6 through CXCR4, PI3K/Akt, ERK, and NF-kappaB-dependent pathway in microglia. Eur J Pharmacol. 2009;613(1–3):146–154. doi: 10.1016/j.ejphar.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Wang X, Li C, Chen Y, Hao Y, Zhou W, Chen C, Yu Z. Hypoxia enhances CXCR4 expression favoring microglia migration via HIF-1[alpha] activation. Biochem Biophys Res Commun. 2008;371(2):283–288. doi: 10.1016/j.bbrc.2008.04.055. [DOI] [PubMed] [Google Scholar]
- Tosic M, Rakic S, Matthieu JM, Zecevic N. Identification of Golli and myelin basic proteins in human brain during early development. Glia. 2002;37(3):219–228. doi: 10.1002/glia.10028. [DOI] [PubMed] [Google Scholar]
- Mikoshiba K, Okano H, Tamura TA, Ikenaka K. Structure and Function of Myelin Protein Genes. Annu Rev Neurosci. 1991;14(1):201–217. doi: 10.1146/annurev.ne.14.030191.001221. [DOI] [PubMed] [Google Scholar]
- Bermingham JR, Shearin H, Pennington J, O’Moore J, Jaegle M, Driegen S, van Zon A, Darbas A, Ozkaynak E, Ryu EJ. et al. The claw paw mutation reveals a role for Lgi4 in peripheral nerve development. Nat Neurosci. 2006;9(1):76–84. doi: 10.1038/nn1598. [DOI] [PubMed] [Google Scholar]
- Koirala S, Corfas G. Identification of novel glial genes by single-cell transcriptional profiling of Bergmann glial cells from mouse cerebellum. PLoS One. 2010;5(2):e9198. doi: 10.1371/journal.pone.0009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20(4):1394–1406. doi: 10.1128/MCB.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Meehan RR, Nan X, Bird A. A component of the transcriptional represser MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16(3):256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ, Cullen PJ. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J Cell Sci. 2007;120(1):45–54. doi: 10.1242/jcs.03302. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a Protein That Interacts with Tubulin Dimers and Increases the Catastrophe Rate of Microtubules. Cell. 1996;84(4):623–631. doi: 10.1016/S0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Günther E, Walter L. The major histocompatibility complex of the rat (<SMALL > Rattus norvegicus</SMALL>) Immunogenetics. 2001;53(7):520–542. doi: 10.1007/s002510100361. [DOI] [PubMed] [Google Scholar]
- Iijima T, Miura E, Watanabe M, Yuzaki M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur J Neurosci. 2010;31(9):1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- Balla Z, Hoch B, Karczewski P, Blasig IE. Calcium/Calmodulin-dependent Protein Kinase IIŒ¥2 and Œ ≥ Isoforms Regulate Potassium Currents of Rat Brain Capillary Endothelial Cells under Hypoxic Conditions. J Biol Chem. 2002;277(24):21306–21314. doi: 10.1074/jbc.M200553200. [DOI] [PubMed] [Google Scholar]
- House SJ, Ginnan RG, Armstrong SE, Singer HA. Calcium/calmodulin-dependent protein kinase II-Œ¥ isoform regulation of vascular smooth muscle cell proliferation. Am J Physiol Cell Physiol. 2007;292(6):C2276–C2287. doi: 10.1152/ajpcell.00606.2006. [DOI] [PubMed] [Google Scholar]
- Tsai LJ, Hsiao SH, Tsai LM, Lin CY, Tsai JJ, Liou DM, Lan JL. The sodium-dependent glucose cotransporter SLC5A11 as an autoimmune modifier gene in SLE. Tissue Antigens. 2008;71(2):114–126. doi: 10.1111/j.1399-0039.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER. et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science. 2010;330(6005):841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX Family of Developmental Transcription Factors Based on Sequence and Structural Indicators. Dev Biol. 2000;227(2):239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- Tonks A, Tonks AJ, Pearn L, Pearce L, Hoy T, Couzens S, Fisher J, Burnett AK, Darley RL. Expression of AML1-ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia. 2004;18(7):1238–1245. doi: 10.1038/sj.leu.2403396. [DOI] [PubMed] [Google Scholar]
- Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK, Darley RL. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007;21(12):2495–2505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PAJ, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A. et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380(6576):711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- Field CM, Kellogg D. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9(10):387–394. doi: 10.1016/S0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast: IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69(2):265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: Architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9(6):478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Kinoshita M. The septins. Genome Biol. 2003;4(11):236. doi: 10.1186/gb-2003-4-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheen ST, Kaur C, Ling EA. Microglial activation and its implications in the brain diseases. Curr Med Chem. 2007;14(11):1189–1197. doi: 10.2174/092986707780597961. [DOI] [PubMed] [Google Scholar]
- Wu CH, Wen CY, Shieh JY, Ling EA. A quantitative study of the differentiation of microglial cells in the developing cerebral cortex in rats. J Anat. 1993;182(3):403–413. [PMC free article] [PubMed] [Google Scholar]
- Imamoto K, Leblond CP. Radioautographic investigation of gliogenesis in the corpus callosum of young rats II. Origin of microglial cells. J Comp Neurol. 1978;180(1):139–163. doi: 10.1002/cne.901800109. [DOI] [PubMed] [Google Scholar]
- Huang A, Ho CSW, Ponzielli R, Barsyte-Lovejoy D, Bouffet E, Picard D, Hawkins CE, Penn LZ. Identification of a Novel c-Myc Protein Interactor, JPO2, with Transforming Activity in Medulloblastoma Cells. Cancer Res. 2005;65(13):5607–5619. doi: 10.1158/0008-5472.CAN-05-0500. [DOI] [PubMed] [Google Scholar]
- He Y-Y, Council SE, Feng L, Chignell CF. UVA-Induced Cell Cycle Progression Is Mediated by a Disintegrin and Metalloprotease/Epidermal Growth Factor Receptor/AKT/Cyclin D1 Pathways in Keratinocytes. Cancer Res. 2008;68(10):3752–3758. doi: 10.1158/0008-5472.CAN-07-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Jia Y, Wang C, Lue Y-H, Swerdloff RS, Zhang X-S, Hu Z-Y, Li Y-C, Liu Y-X, Hikim APS. Role of Caspase 2 in Apoptotic Signaling in Primate and Murine Germ Cells. Biol Reprod. 2008;79(5):806–814. doi: 10.1095/biolreprod.108.068833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C-Y, Yang D, Karasuyama H, Rakic P, Flavell RA. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384(6607):368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee M, Acharya S, Ghosh A, Sarkar P, Chatterjee S, Kumar P, Chaudhuri S. Bax and Bid act in synergy to bring about T11TS-mediated glioma apoptosis via the release of mitochondrial cytochrome c and subsequent caspase activation. Int Immunol. 2008;20(12):1489–1505. doi: 10.1093/intimm/dxn109. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin Is a Microtubule-Associated Protein and Is Expressed Widely by Migrating Neurons. Neuron. 1999;23(2):257–271. doi: 10.1016/S0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Rossi P, Sola E, Taglietti V, Borchardt T, Steigerwald F, Utvik JK, Ottersen OP, Kohr G, D’Angelo E. NMDA Receptor 2 (NR2) C-Terminal Control of NR Open Probability Regulates Synaptic Transmission and Plasticity at a Cerebellar Synapse. J Neurosci. 2002;22(22):9687–9697. doi: 10.1523/JNEUROSCI.22-22-09687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds HL, Goussakov I, Nakazawa K, Tonegawa S, Bolshakov VY. Essential function of Œ ± −calcium/calmodulin-dependent protein kinase II in neurotransmitter release at a glutamatergic central synapse. Proc Natl Acad Sci U S A. 2003;100(7):4275–4280. doi: 10.1073/pnas.0530202100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama H, Kn¨pfel T, Endo S, Itohara S. Glial protein S100B modulates long-term neuronal synaptic plasticity. Proc Natl Acad Sci U S A. 2002;99(6):4037–4042. doi: 10.1073/pnas.052020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber MB. Changing Face of Microglia. Science. 2010;330(6005):783–788. doi: 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- Tremblay M-È, Lowery RL, Majewska AK. Microglial Interactions with Synapses Are Modulated by Visual Experience. PLoS Biol. 2010;8(11):e1000527. doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B-S, Braud S, Badger JD, Isaac JTR, Roche KW. Regulation of NR1/NR2C N-Methyl-D-aspartate (NMDA) Receptors by Phosphorylation. J Biol Chem. 2006;281(24):16583–16590. doi: 10.1074/jbc.M513029200. [DOI] [PubMed] [Google Scholar]
- Murugan M, Sivakumar V, Lu J, Ling E-A, Kaur C. Expression of N-methyl D-aspartate receptor subunits in amoeboid microglia mediates production of nitric oxide via NF-κB signaling pathway and oligodendrocyte cell death in hypoxic postnatal rats. Glia. 2011; :n/a–n/a. doi: 10.1002/glia.21121. [DOI] [PubMed] [Google Scholar]
- Smith SP, Shaw GS. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure. 1998;6(2):211–222. doi: 10.1016/S0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Adami C, Sorci G, Blasi E, Agneletti AL, Bistoni F, Donato R. S100b expression in and effects on microglia. Glia. 2001;33(2):131–142. doi: 10.1002/1098-1136(200102)33:2<131::AID-GLIA1012>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Filipovic R, Rakic S, Zecevic N. Expression of Golli proteins in adult human brain and multiple sclerosis lesions. J Neuroimmunol. 2002;127(1–2):1–12. doi: 10.1016/s0165-5728(02)00070-x. [DOI] [PubMed] [Google Scholar]
- Filipoviƒá R, Zeƒçeviƒá N. Interaction between microglia and oligodendrocyte cell progenitors involves Golli proteins. In. 2005;1048:166–174. doi: 10.1196/annals.1342.015. [DOI] [PubMed] [Google Scholar]
- Rezaie P, Male D. Differentiation, Ramification and Distribution of Microglia within the Central Nervous System Examined. Neuroembryology and Aging. 2002;1(1):29–43. [Google Scholar]
- Stuart LM, Bell SA, Stewart CR, Silver JM, Richard J, Goss JL, Tseng AA, Zhang A, Khoury JBE, Moore KJ. CD36 Signals to the Actin Cytoskeleton and Regulates Microglial Migration via a p130Cas Complex. J Biol Chem. 2007;282(37):27392–27401. doi: 10.1074/jbc.M702887200. [DOI] [PubMed] [Google Scholar]
- Ilschner S, Brandt R. The transition of microglia to a ramified phenotype is associated with the formation of stable acetylated and detyrosinated microtubules. Glia. 1996;18(2):129–140. doi: 10.1002/(SICI)1098-1136(199610)18:2<129::AID-GLIA5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Rosslenbroich V, Dai L, Baader SL, Noegel AA, Gieselmann V, Kappler J. Collapsin response mediator protein-4 regulates F-actin bundling. Exp Cell Res. 2005;310(2):434–444. doi: 10.1016/j.yexcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- K-i N, Asano T, Nozawa Y, Inagaki M. Biochemical and Cell Biological Analyses of a Mammalian Septin Complex, Sept7/9b/11. J Biol Chem. 2004;279(53):55895–55904. doi: 10.1074/jbc.M406153200. [DOI] [PubMed] [Google Scholar]
- Hanai N, Nagata K-i, Kawajiri A, Shiromizu T, Saitoh N, Hasegawa Y, Murakami S, Inagaki M. Biochemical and cell biological characterization of a mammalian septin, Sept11. FEBS Lett. 2004;568(1–3):83–88. doi: 10.1016/j.febslet.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Kartmann B, Roth D. Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J Cell Sci. 2001;114(5):839–844. doi: 10.1242/jcs.114.5.839. [DOI] [PubMed] [Google Scholar]
- Shinoda T, Ito H, Sudo K, Iwamoto I, Morishita R, Nagata K-i. Septin 14 Is Involved in Cortical Neuronal Migration via Interaction with Septin 4. Mol Biol Cell. 2010;21(8):1324–1334. doi: 10.1091/mbc.E09-10-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11(11):775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- Zhou P, Qian L, Bieszczad CK, Noelle R, Binder M, Levy NB, Craig RW. Mcl-1 in Transgenic Mice Promotes Survival in a Spectrum of Hematopoietic Cell Types and Immortalization in the Myeloid Lineage. Blood. 1998;92(9):3226–3239. [PubMed] [Google Scholar]
- Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A. 1993;90(8):3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro A, Spirin KS, Shiohara M, Tobler A, Gombart AF, Israel MA, Norton JD, Koeffler HP. Id2 expression increases with differentiation of human myeloid cells. Blood. 1996;87(12):5225–5231. [PubMed] [Google Scholar]
- Niidome T, Matsuda S, Nonaka H, Akaike A, Kihara T, Sugimoto H. A molecular pathway involved in the generation of microtubule-associated protein 2-positive cells from microglia. Biochem Biophys Res Commun. 2008;370(1):184–188. doi: 10.1016/j.bbrc.2008.03.068. [DOI] [PubMed] [Google Scholar]
- Pulford K, Jones M, Banham AH, Haralambieva E, Mason DY. Lymphocyte-specific protein 1: A specific marker of human leucocytes. Immunology. 1999;96(2):262–271. doi: 10.1046/j.1365-2567.1999.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard T, Li Y, Torres M, Guerrero A, Coates T. The 47-kD protein increased in neutrophil actin dysfunction with 47- and 89-kD protein abnormalities is lymphocyte-specific protein. Blood. 1994;83(1):231–241. [PubMed] [Google Scholar]
- Howard TH, Hartwig J, Cunningham C. Lymphocyte-Specific Protein 1†Expression in Eukaryotic Cells Reproduces the Morphologic and Motile Abnormality of NAD 47/89 Neutrophils. Blood. 1998;91(12):4786–4795. [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102(24):8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave M, Wright E, Kun J, Emery J, Cooper L, Koopman P. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev Dyn. 1997;210(2):79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Chen Y-H, Gao J, Fan G, Peterson LC. Nuclear expression of sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol. 2009;23(1):105–112. doi: 10.1038/modpathol.2009.140. [DOI] [PubMed] [Google Scholar]
- Sawada M, Suzumura A, Yamamoto H, Marunouchi T. Activation and proliferation of the isolated microglia by colony stimulating factor-1 and possible involvement of protein kinase C. Brain Res. 1990;509(1):119–124. doi: 10.1016/0006-8993(90)90317-5. [DOI] [PubMed] [Google Scholar]
- Guo WH, Weng LQ, Ito K, Chen LF, Nakanishi H, Tatematsu M, Ito Y. Inhibition of growth of mouse gastric cancer cells by Runx3, a novel tumor suppressor. Oncogene. 2002;21(54):8351–8355. doi: 10.1038/sj.onc.1206037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet S1. Primer sequences .
Figure S1. Expression of Dcx and Mbp in BV-2 microglia. PCR was done using cDNA obtained from BV-2 microglia for Dcx and Mbp. Both Dcx and Mbp were found to be expressed by BV-2 microglia as identified by the PCR products in agarose gel electrophoresis.
Sheet S2. AMC and RMC genes with P-value.
Sheet S3. AMC and RMC gene clusters.
Sheet S4. Genes involved in CDC42-RAC pathway for migration, Synaptic transmission and genes downregulated by the Runx1-Runx1t1 complex.
Sheet S5. Functions of AMC and RMC.
Sheet S6. Monocytic genes enriched in AMC and RMC.
Sheet S7. ESC, NSC and HSC genes enriched in AMC and RMC.
Figure S2. Pathway analysis. Novel molecular networks identified by inputting AMC and RMC expression data into Adriane Pathway Studio. Red colored shapes specify AMC genes and violet colored shapes specify RMC genes. Some AMC genes - Sox4, Sox11, Runx1t1 are highlighted in the figure.
Sheet S8. Expression of cytokines in AMC and RMC.