Abstract
Background
Potential xeroderma pigmentosum group D (XPD), also called excision repair cross-complimentary group two (ERCC2), Lys751Gln and Asp312Asn polymorphisms have been implicated in gastric cancer risk among different ethnicities.
Methods
We aimed to explore the effect of XPD Lys751Gln and Asp312Asn polymorphisms on the susceptibility to gastric cancer among different ethnicities through a systematic review and meta-analysis. Each initially included article was scored for quality appraisal. Desirable data were extracted and registered into databases. 13 studies were ultimately eligible for the meta-analysis of Lys751Gln polymorphism and 9 studies for the meta-analysis of Asp312Asn polymorphism. We adopted the most probably appropriate genetic model (recessive model) for both Lys751Gln and Asp312Asn polymorphisms. Potential sources of heterogeneity were sought out via subgroup and sensitivity analyses, and publication biases were estimated.
Results
Statistically significant findings were apparently noted in Asians but not in Caucasians for both XPD Lys751Gln and XPD Asp312Asn polymorphisms. A statistically significant finding could be seen in noncardia-type gastric cancer for XPD Lys751Gln polymorphism. A statistically significant finding could also be seen in high quality subgroup, small-and-moderate sample size subgroup, articles published after 2007, or PCR-RFLP genotyping subgroup for XPD Asp312Asn polymorphism.
Conclusions
Our meta-analysis indicates that XPD Gln751Gln (CC) genotype and Asn312Asn (AA) genotype may seem to be more susceptible to gastric cancer in Asian populations but not in Caucasian populations, suggesting that the two genotypes may be important biomarkers of gastric cancer susceptibility for Asian populations, the assumption that needs to be further confirmed in well-designed studies among different ethnicities. Gln751Gln (CC) genotype may also be associated with noncardia-type gastric cancer risk, which should also be confirmed among different ethnicities in the future.
Introduction
Although worldwide gastric cancer incidence has decreased, its mortality still ranks second [1]. In China, gastric cancer even constitutes one of the most lethal malignancies [2]. As is widely known, infectious, dietary, environmental, and genetic factors are implicated in gastric carcinogenesis, but those exposed to risk factors who ultimately develop gastric cancer comprises a minor proportion [3], suggesting that host genetic susceptibility plays an important role in gastric cancer risk among different ethnicities. Such various susceptibilities could be explained, in part, by single nucleotide polymorphisms (SNPs) of susceptible genes among different ethnicities [4], [5]. Our previously published meta-analysis papers have provided additional evidence for such ethnically susceptible differences [5], [6].
It is widely acknowledged that DNA must remain stable to undertake its crucial physiological functions, but it is persistently vulnerable to various endogenous and/or exogenous damages and thus its probable mutations could accumulate and carcinogenesis may occur due to the damaged DNA. DNA repair system, however, plays a vital role in maintaining the functions of normal cells and genome integrity through the reversal of the damaged DNA [7]. Inherited functional polymorphisms or accumulated mutations of DNA repair genes may influence the host capacity to repair the damaged DNA and thus modulate cancer risk [8]. SNPs of common DNA repair genes have been identified [9] and demonstrated to be linked to sporadic carcinogenesis [10], [11].
Nucleotide excision repair (NER), one of the major DNA repair pathways in humans, is capable of removing helix-distorting base lesions produced by ultraviolet light (UV) and an array of chemical agents [12]. XPD is believed to participate in DNA unwinding during NER and transcription because it possesses single-strand DNA-dependent ATPase and 5′–3′ DNA helicase activities [13], [14]. XPD (ERCC2) gene, located at chromosome 19q13.3, comprises 23 exons and its polymorphisms are thought to engender structural alterations of NER pathway and influence cancer susceptibility. The most widely investigated XPD polymorphisms in associations with cancer susceptibility comprise a nonsynonymous A>C substitution in exon 23 causing a lysine (Lys) to glutamine (Gln) substitution in codon 751 (Lys751Gln, rs1052559), a nonsynonymous G>A substitution in exon 10 leading to an aspartic acid (Asp) to asparagine (Asn) substitution in codon 312 (Asp312Asn, rs1799793), and a synonymous C>A substitution in exon 6 while conserving the arginine (R) residue in codon 156 (Arg156Arg, rs238406) [15].
In 2005, Huang WY et al. published the first study involved in XPD Lys751Gln polymorphism in relation to gastric cancer risk [16]. Since then, researchers have reported associations of XPD Lys751Gln, Asp312Asn, and/or Arg156Arg with the susceptibility to gastric cancer among different ethnicities, but with mixed or conflicting results [17]–[30]. There is only one published article concerning Arg156Arg polymorphism in relation to gastric cancer risk [28]. To date, there have been three relevant published meta-analysis papers focusing on XPD polymorphisms [31]–[33]. Two articles were mainly concerned with overall cancer susceptibilities rather than gastric cancer susceptibility in depth [31], [32], thus providing less information on its association with gastric cancer risk. More importantly, those three meta-analyses [31]–[33] all failed to adopt the most likely appropriate genetic model, and thus the authentic values of those statistical results could be compromised.
Accordingly, the aim of our meta-analysis was to explore, using the most appropriate genetic model, the effect of XPD polymorphisms on gastric cancer risk among different ethnicities and to identify possible sources of heterogeneity among the eligible studies.
Materials and Methods
Search Strategy
A systematic literature search was performed for articles regarding XPD/ERCC2 SNPs associated with gastric cancer risk. The MEDLINE, EMBASE databases, Chinese National Knowledge Infrastructure (CNKI), Web of Science, and BIOSIS databases were used simultaneously with the combination of terms “XPD”, “ERCC2”, “DNA repair”, “NER”, “Lys751Gln”, “Asp312Asn”, or “Arg156Arg”; “gene”; “polymorphism”, “variant”, or “SNP”; and “gastric cancer”, “gastric carcinoma”, or “stomach cancer” up to December 2011. The search was performed without any restriction on language. The scope of computerized literature search was expanded according to the reference lists of retrieved articles. The relevant original articles were also sought manually.
Study Selection
Studies concerning the association of XPD/ERCC2 SNPs (Lys751Gln, Asp312Asn, and/or Arg156Arg) with gastric cancer risk were included if the following conditions were met: (i) any study described the association of at least one of XPD/ERCC2 SNPs with gastric cancer; (ii) any study reported the numbers of both controls and gastric cancer cases; (iii) results were expressed as odds ratio (OR) with 95% confidence intervals (CI); and (iv) studies were case-control or nested case-control ones.
Methodological Quality Appraisal
To identify high-quality studies, we mainly adopted predefined criteria for Quality Appraisal initially proposed by Thakkinstian et al. [34], adapted by Camargo et al. [35], and refined by Xue et al. [5]–[7]. The criteria (seen in Table S1 online) cover credibility of controls, representativeness of cases, consolidation of gastric cancer, genotyping examination, and association assessment. Methodological quality was independently assessed by two investigators (Lin B and Lu Y). Disagreements were resolved through discussion. Scores ranged from the lowest zero to the highest ten. Articles with the score lower than 6.5 were considered “low-or-moderate quality” ones, whereas those no lower than 6.5 were thought of as “high quality” ones.
Data Extraction
The following data from each article were extracted: authors, year of publication, country, ethnicity of participants (categorized as Caucasians, Asians, etc.), study design, source of controls, number of controls and of cases, genotyping method, distribution of age and gender, Lauren's classification (intestinal, diffuse, or mixed), anatomical classification (cardia or non-cardia cancer), smoking habit, drinking habit and Helicobacter Pylori infection status.
The data were extracted and registered into two databases independently by two investigators (Lin B and Lu Y) who were blind to journal names, institutions or fund grants. Any discrepancy between these two investigators was resolved by the investigator (Xue H), who participated in the discussion with them and made an ultimate decision.
Statistical Analysis
All statistical analyses were performed using STATA statistical software (Version 10.1, STATA Corp, College Station, TX). Two-sided Ps<0.05 were considered statistically significant. Hardy-Weinberg equilibrium (HWE) in controls was calculated again in our meta-analysis. The chi-square goodness of fit was used to test deviation from HWE (significant at the 0.05 level).
Odds ratios (OR) and 95% confidence intervals (95% CI) were used to assess the strength of associations between XPD/ERCC2 SNPs and gastric cancer risk. OR1, OR2, and OR3 were calculated for genotypes CC versus AA, CA versus AA, and CC versus CA for XPD Lys751Gln polymorphism; AA versus GG, GA versus GG, and AA versus GA for XPD Asp312Asn polymorphism; AA versus CC, CA versus CC, and AA versus CA for XPD Arg156Arg polymorphism, respectively. The pairwise differences were used to determine the most appropriate genetic model. If OR1 = OR3≠1 and OR2 = 1, a recessive model is suggested. If OR1 = OR2≠1 and OR3 = 1, a dominant model is implied. If OR2 = 1/OR3 ≠1 and OR1 = 1, a complete overdominant model is suggested. If OR1>OR2>1 and OR1>OR3>1, or OR1<OR2<1 and OR1<OR3<1, a codominant model is indicated [34]. Take XPD Lys751Gln polymorphism as an example to illustrate it. If a dominant model was indicated, the original grouping was collapsed and the new group of C carriers (CC plus CA) was compared with AA genotype; if a recessive model was suggested, CC was compared to the group of AA plus CA; if a complete overdominant model was implied, the group of CC plus AA was compared with CA; or if a codominant model was insinuated, CC was compared with CA and with AA, respectively.
The Q statistic was used to assess heterogeneity across studies included in the meta-analysis. I-squared (I 2) value, representing variation in OR attributable to heterogeneity was then used to quantify the degree of such between-study heterogeneity [36]. According to recently published Venice criteria [37], “I 2<25% represents no heterogeneity, I 2 = 25–50% represents moderate heterogeneity, I 2 = 50–75% represents large heterogeneity, and I 2>75% represents extreme heterogeneity”. Between-study variance Tau-squared (τ2) value was also used to evaluate between-study variance. A fixed-effects model, using Mantel–Haenszel (M–H) method, was employed to calculate the pooled ORs when homogeneity existed on the basis of Q-test p value no less than 0.1.By contrast, a random-effects model, using DerSimonian and Laird method (D+L), was utilized if there was heterogeneity based on Q-test p value less than 0.1. Even for the homogeneity among-studies, D+L method was also used. Meta-regression analyses and subgroup analyses were utilized to explore and control potential sources of heterogeneity across studies. The significance of pooled ORs was tested by Z test (P<0.05 was considered significant).
Sensitivity analysis was performed, in which the meta-analysis estimates were computed after every one study being omitted in each turn.
Finally, publication bias was assessed by performing funnel plots qualitatively, and estimated by Begg's and Egger's tests quantitatively.
Results
Literature Search and Study Selection
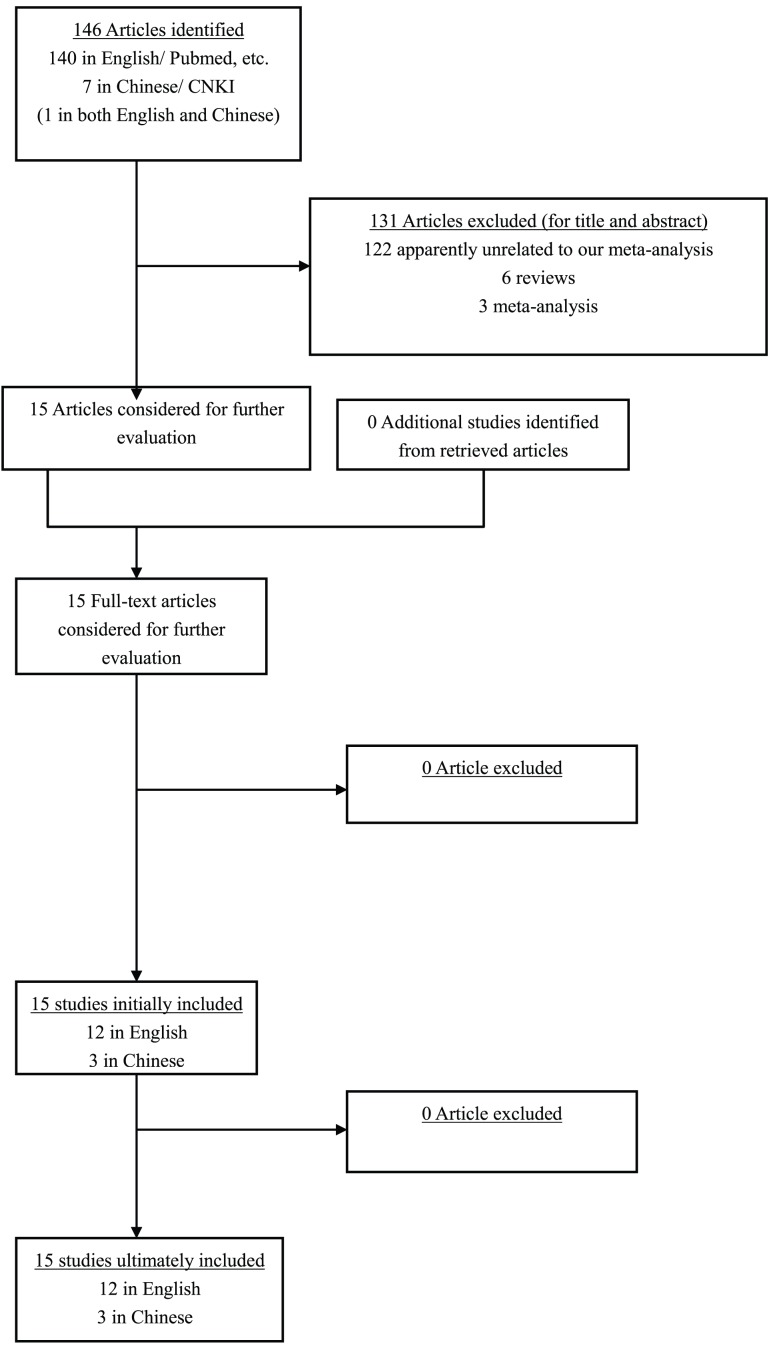
After comprehensive searching, a total of 140 articles in English and 7 in Chinese were retrieved. In our meta-analysis were initially included altogether 15 studies [16]–[30] which met the inclusion criteria. Those 15 studies were preliminarily appropriate to the meta-analysis of the associations with gastric cancer regarding XPD SNPs, among which 13 studies concerned XPD Lys751Gln polymorphism [16]–[26], [28], [29], 9 studies concerned XPD Asp312Asn polymorphism [17]–[21], [23], [27], [28], [30], and only 1 study concerned XPD Arg156Arg polymorphism [28]. 1 article [23] was indexed in both English and Chinese searching engines.
Traditionally speaking, any study that deviated from HWE should have been removed; however, Minelli C et al. recently pointed out that studies that appear to deviate from HWE should be investigated further rather than just excluded unless there are other grounds for doubting the quality of the study [38]. To date, it is still inconclusive whether studies deviated from HWE should be included or excluded in conducting meta-analysis. In our meta-analysis, 3 studies concerned XPD Lys751Gln polymorphism [16], [25], [29] were deviated from HWE, and 3 studies concerned XPD Asp312Asn polymorphism [17], [27], [30] were also deviated from HWE; however, considering that the numbers of participants in those studies were large and given that sensitivity analyses would be conducted, we finally remained those studies in our meta-analysis.
Thus, 13 studies [16]–[26], [28], [29] with a total of 6344 controls and 2750 cases for XPD Lys751Gln polymorphism, and 9 studies [17]–[21], [23], [27], [28], [30] with 3429 controls and 1715 cases for XPD Asp312Asn polymorphism were ultimately eligible for the meta-analysis of XPD polymorphisms. The corresponding characteristics were seen in Table 1 and Table 2. The flow chart of literature search and study selection was illuminated in Figure 1.
Table 1. Study Characteristics of genotypes in gastric cancer cases and controls in the analysis of XPD Lys751Gln polymorphism.
| First author | Year of publication | Quality assessment scores | Genotyping method | Total sample size | Number of controls | Number of cases | Study location | Ethnic group | P values for HWE | Controls, genotypes(n) Lys/Gln | All Cases, genotypes(n) Lys/Gln | ||||
| Lys/Lys | Lys/Gln | Gln/Gln | Lys/Lys | Lys/Gln | Gln/Gln | ||||||||||
| Huang WY | 2005 | 8.5 | MALDI-TOF/hME | 660 | 381 | 279 | Poland | Caucasian | 0.027863595 | 145 | 163 | 73 | 107 | 126 | 46 |
| Ye W# | 2006 | 8 | PCR–RFLP | 598 | 472 | 126 | Sweden | Caucasian | 0.114253032 | 198 | 203 | 71 | 49 | 61 | 16 |
| Lou Y | 2006 | 6 | PCR–RFLP | 438 | 200 | 238 | China | Asian | 0.377267735 | 164 | 33 | 3 | 205 | 30 | 3 |
| Ruzzo A* | 2007 | 7 | PCR–RFLP | 183 | 94 | 89 | Italy | Caucasian | 0.180617442 | 25 | 53 | 16 | 29 | 44 | 16 |
| Zhou RM# | 2007 | 7 | PCR–RFLP | 865 | 612 | 253 | China | Asian | 0.82380608 | 522 | 86 | 4 | 224 | 26 | 3 |
| Capella G# ∧ ¶ ★ | 2008 | 9 | Lightcycler | 1417 | 1172 | 245 | 10 European countries | Caucasian | 0.914544475 | 447 | 555 | 170 | 99 | 105 | 41 |
| Doecke J# | 2008 | 9 | Sequenom iPLEX | 1640 | 1337 | 303 | Australia | Caucasian | 0.220947678 | 575 | 588 | 174 | 127 | 140 | 36 |
| Zhang CZ | 2009 | 6.5 | PCR–RFLP | 419 | 212 | 207 | China | Asian | 0.439890159 | 172 | 39 | 1 | 166 | 39 | 2 |
| Canbay E | 2010 | 7 | PCR–RFLP | 287 | 247 | 40 | Turkey | Turkish population□ | 0.922153834 | 102 | 114 | 31 | 14 | 18 | 8 |
| Long XD∧ + | 2010 | 7.5 | TaqMan-PCR | 977 | 616 | 361 | China | Asian | 6.33741E-08 | 400 | 164 | 52 | 139 | 151 | 71 |
| Palli D | 2010 | 8 | TaqMan-PCR | 841 | 546 | 295 | Italy | Caucasian | 0.098569581 | 177 | 284 | 85 | 90 | 157 | 48 |
| Chen Z | 2011 | 6.5 | PCR–RFLP | 547 | 339 | 208 | China | Asian | 0.698246164 | 282 | 55 | 2 | 166 | 40 | 2 |
| Engin AB | 2011 | 4 | PCR–RFLP | 222 | 116 | 106 | Turkey | Turkish□ population | 0.005846887 | 40 | 43 | 33 | 30 | 56 | 20 |
Data of cardia type of gastric cancer were accessible;
Data of noncardia type of gastric cancer were accessible;
Data of sporadic diffuse-type of gastric cancer were accessible;
esophago-gastric junction adenocarcinoma (EGJAC) was treated as cardia type of gastric cancer in our meta-analysis.
Data of the status of Helicobacter pylori of gastric cancer were accessible.
Three kinds of controls (controls with severe chronic atrophic gastritis, controls without severe chronic atrophic gastritis, and total controls regardless of severe chronic atrophic gastritis) were presented in the study and total controls regardless of severe chronic atrophic gastritis were finally extracted into our database.
Here participants should be better considered as separate Turkish population conducted in our subgroup analysis due to their unknown ethnic backgrounds. RFLP: Restriction fragment length polymorphisms; TaqMan: TaqMan polymerase chain reaction method; MALDI-TOF/hME: SNPs were analyzed using Assisted Laser Desorption Ionization-Time of Flight mass spectrometry (MALDI-TOF MS) and homogeneous MassExtend (hME) chemistry (Sequenom Inc., San Diego, CA); Lightcycler: Polymorphisms were analysed in a LightCycler instrument by melting curve analysis of a fluorescently labelled sensor probe specific for each analysed variant, following manufacturer instructions (Roche Diagnostics, Mannheim, Germany), the results that were confirmed by a second genotyping method, such as restriction analysis, SSCP analysis or direct sequencing; Sequenom iPLEX : SNP typing was conducted using the Sequenom iPLEX protocol (Sequenom, San Diego, CA).
Table 2. Study Characteristics of genotypes in gastric cancer cases and controls in the analysis of XPD Asp312Asn polymorphism.
| First author | Year of publication | Quality assessment scores | Genotyping method | Total sample size | Number of controls | Number of cases | Study location | Ethnic group | P values for HWE | Controls, genotypes(n) G/A (Asp/Asn) | All Cases,genotypes(n) G/A (Asp/Asn) | ||||
| Asp/Asp | Asp/Asn | Asn/Asn | Asp/Asp | Asp/Asn | Asn/Asn | ||||||||||
| Ye W# | 2006 | 8 | PCR–RFLP | 596 | 470 | 126 | Sweden | Caucasian | 0.092544857 | 176 | 237 | 57 | 41 | 69 | 16 |
| Lou Y | 2006 | 6 | PCR–RFLP | 438 | 200 | 238 | China | Asian | 0.018951137 | 176 | 21 | 3 | 189 | 39 | 10 |
| Ruzzo A* | 2007 | 7 | PCR–RFLP | 210 | 121 | 89 | Italy | Caucasian | 0.061339561 | 41 | 67 | 13 | 23 | 46 | 20 |
| Zhou RM# + | 2007 | 7 | PCR–RFLP | 865 | 612 | 253 | China | Asian | 0.527426883 | 528 | 82 | 2 | 221 | 32 | 0 |
| Capella G# ∧ | 2008 | 9 | Lightcycler | 1379 | 1135 | 244 | 10 European countries | Caucasian | 0.985747613 | 444 | 532 | 159 | 110 | 96 | 38 |
| Zhang CZ | 2009 | 6.5 | ARMS-PCR | 419 | 212 | 207 | China | Asian | 0.636402675 | 132 | 72 | 8 | 75 | 117 | 15 |
| Deng SL | 2010 | 3.75 | Direct sequencing | 320 | 160 | 160 | China | Asian | 0.000155604 | 118 | 31 | 11 | 132 | 15 | 13 |
| Chen Z | 2011 | 6.5 | PCR–RFLP | 547 | 339 | 208 | China | Asian | 0.164678753 | 220 | 111 | 8 | 75 | 118 | 15 |
| Yuan T | 2011 | 6 | Direct sequencing | 370 | 180 | 190 | China | Asian | 9.72011E-05 | 133 | 35 | 12 | 156 | 18 | 16 |
Data of cardia type of gastric cancer were accessible;
Data of noncardia type of gastric cancer were accessible;
Data of sporadic diffuse-type of gastric cancer were accessible;
Data of the smoking habits of gastric cancer were accessible. RFLP: Restriction fragment length polymorphisms; Lightcycler: Polymorphisms were analysed in a LightCycler instrument by melting curve analysis of a fluorescently labelled sensor probe specific for each analysed variant, following manufacturer instructions (Roche Diagnostics, Mannheim, Germany), the results that were confirmed by a second genotyping method, such as restriction analysis, SSCP analysis or direct sequencing; ARMS: Amplification refractory mutation system.
Figure 1. The flow chart of literature search and study selection.
Overall Meta-analysis, Meta-regression Analyses and Subgroup Analyses
For XPD Lys751Gln polymorphism, OR1 (p value), OR2 (p value), and OR3 (p value) were 1.22 (p = 0.266), 1.11 (p = 0.369), and 1.02 (p = 0.811), respectively, hardly insinuating a particular model effect of putative susceptible C allele. Heterogeneity chi-squared was 29.83 (d.f. = 12), p value was 0.003, and I 2 was 59.8%. After meta-regression analysis using single covariate (ethnicity composed of Caucasians, Asians, or Turkish population), p values of coefficient t value for Asians, Caucasians, or Turkish population were 0.001, 0.139, and 0.585, respectively; strongly indicating that Asians single covariate could mostly constitute the source of heterogeneity across studies. τ2 for Asians, Caucasians, or Turkish population single covariate were 0, 0.09178, and 0.1347, respectively. For Asians single covariate, τ2 decreased from 0.1233 to 0, indicating Asians single covariate could account for 100% of the source of heterogeneity across studies. When stratified by ethnicity subgroup analysis, OR1 (p value), OR2 (p value), and OR3 (p value) among Asian population were 2.63 (p = 0.002), 1.14 (p = 0.653), and 1.51 (p = 0.034), respectively, highly indicating a recessive model effect of putative susceptible C allele (OR1 = OR3≠1 and OR2 = 1).
For XPD Asp312Asn polymorphism, OR1 (p value), OR2 (p value), and OR3 (p value) were 1.75 (p = 0.015), 1.15 (p = 0.549), and 1.47 (p = 0.003), respectively, highly indicating a recessive model effect of putative susceptible A allele (OR1 = OR3≠1 and OR2 = 1). Heterogeneity chi-squared was 9.91 (d.f. = 8), p value was 0.272, and I 2 was 19.2%, indicating no heterogeneity across studies. After meta-regression analysis using single covariate (ethnicity composed of Caucasians or Asians), p value of coefficient t value for ethnicity single covariate was 0.277, indicating that ethnicity could constitute one of the sources of little heterogeneity across studies. τ2 decreased from 0.0354 to 0.02075, indicating ethnicity could account for 41.4% of the source of little heterogeneity across studies. Similarly, when stratified by ethnicity subgroup analysis, OR1 (p value), OR2 (p value), and OR3 (p value) among Asian population were 2.10 (p = 0.026), 1.22 (p = 0.574), and 1.80 (p = 0.008), respectively, further indicating a recessive model effect of putative susceptible A allele (OR1 = OR3≠1 and OR2 = 1).
Taken together, a recessive genetic model was ultimately chosen for both XPD Lys751Gln and XPD Asp312Asn polymorphisms in our meta-analysis.
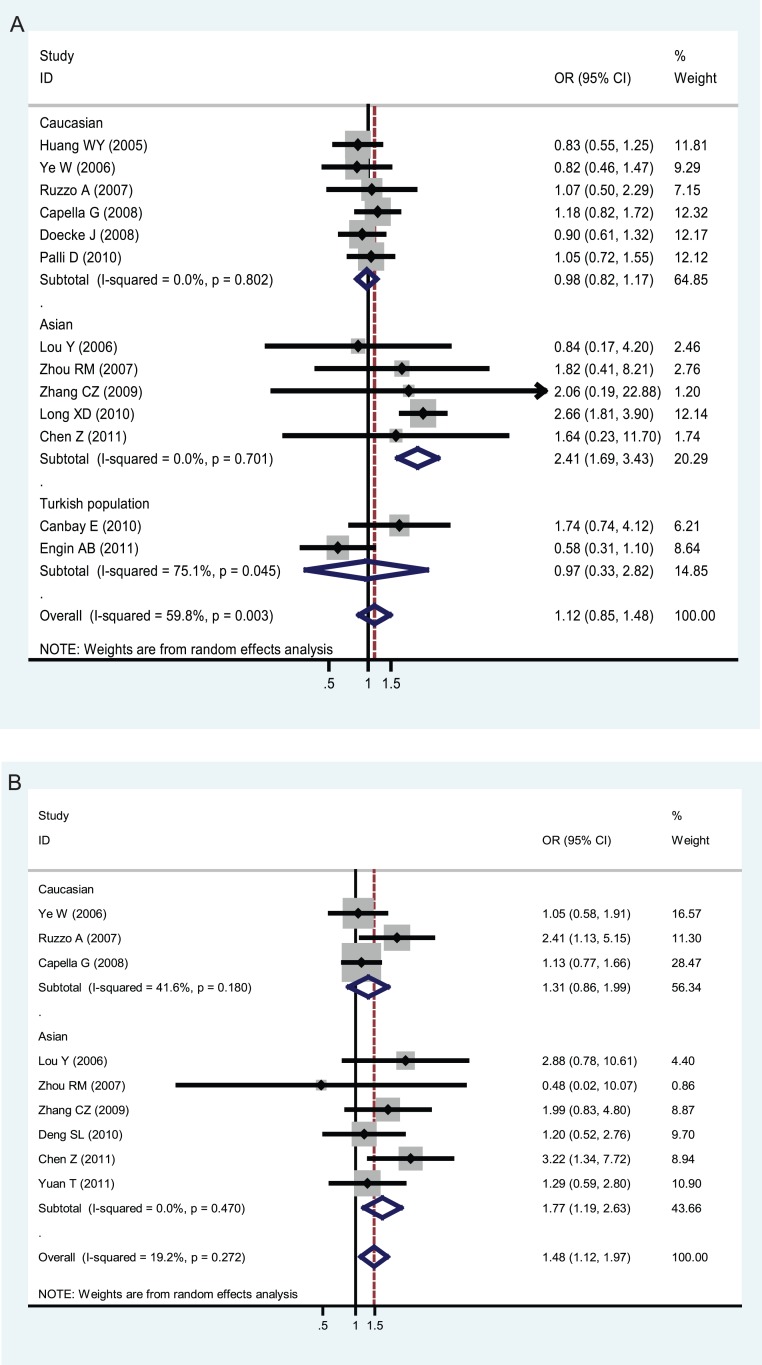
In our meta-analysis, the available data were stratified, in the light of ethnic participants, into Caucasians, Asians, and Turkish population. In Figure 2, statistically significant findings were noted in Asians but not in Caucasians or Turkish population (both XPD Lys751Gln and XPD Asp312Asn polymorphisms). The pooled ORs (95% CIs, p value) were 2.41 (1.69–3.43, p = 0.000), 0.98 (0.82–1.17, p = 0.803), and 0.97 (0.33–2.82, p = 0.954) in Asians, Caucasians, and Turkish population (XPD Lys751Gln polymorphism in Part A) or 1.77 (1.19–2.63, p = 0.005) and 1.31 (0.86–1.99, p = 0.211) in Asians and Caucasians (XPD Asp312Asn polymorphism in Part B), respectively. As for XPD Lys751Gln polymorphism, P values of heterogeneity Q-statistic in Caucasians, Asians, and Turkish population were 0.802, 0.701, and 0.045; I 2 were 0.0%, 0.0%, and 75.1%; and τ2 were 0.0000, 0.0000, and 0.4489, respectively, demonstrating no heterogeneity within Caucasians or Asians, but extreme heterogeneity within Turkish population (shown in Table 3). As for XPD Asp312Asn polymorphism, P values of heterogeneity Q-statistic in Caucasians and Asians were 0.180 and 0.470; I 2 were 41.6% and 0.0%; andτ2 were 0.0587 and 0.0000, respectively, demonstrating no heterogeneity within Asians, but moderate heterogeneity within Caucasians (shown in Table 4).
Figure 2. Odds ratios (ORs) for associations between XPD Lys751Gln and XPD Asp312Asn polymorphisms and gastric cancer risk (based on a recessive genetic model) among different ethnicities. in order of increasing publication year, 2005–2011.
Studies were entered into the meta-analysis sequentially by year of publication. The sizes of the squares indicate the relative weight of each study. Weights were derived from random-effects analysis. Bars, 95% confidence interval (CI). A) The XPD Lys751Gln polymorphism in association with gastric cancer; B) XPD Asp312Asn polymorphism in association with gastric cancer.
Table 3. Stratification for the test of heterogeneity on XPD Lys751Gln based on a recessive model.
| Q-test | I2, % | τ2 | OR(95%CI) | P value | |||
| chi-squared | d.f. | p | |||||
| Overall | 29.83 | 12 | 0.003 | 59.8 | 0.1233 | 1.12(0.85–1.48) | 0.410 |
| Asians | 2.19 | 4 | 0.701 | 0.0 | 0.0000 | 2.41 (1.69–3.43) | 0.000 |
| Caucasians | 2.33 | 5 | 0.802 | 0.0 | 0.0000 | 0.98 (0.82–1.17) | 0.803 |
| Turkish population | 4.02 | 1 | 0.045 | 75.1 | 0.4489 | 0.97 (0.33–2.82) | 0.954 |
| Large sample | 24.05 | 7 | 0.001 | 70.9 | 0.1437 | 1.17 (0.84–1.64) | 0.351 |
| Small-and-moderate sample | 4.68 | 4 | 0.322 | 14.4 | 0.0409 | 0.96 (0.61–1.51) | 0.858 |
| High quality | 25.08 | 10 | 0.005 | 60.1 | 0.1138 | 1.20 (0.91–1.60) | 0.199 |
| Low-and-moderate quality | 0.17 | 1 | 0.684 | 0.0 | 0.0000 | 0.61 (0.34–1.11) | 0.104 |
| High quality Asians | 0.46 | 3 | 0.928 | 0.0 | 0.0000 | 2.54 (1.77–3.65) | 0.000 |
| High quality Caucasians | 2.33 | 5 | 0.802 | 0.0 | 0.0000 | 0.98 (0.82–1.17) | 0803 |
| Publication before or in 2007 | 1.27 | 4 | 0.866 | 0.0 | 0.0000 | 0.89 (0.66–1.19) | 0.428 |
| Publication after 2007 | 24.73 | 7 | 0.001 | 71.7 | 0.1805 | 1.23 (0.84–1.81) | 0.285 |
| Non-cardia type | 3.47 | 1 | 0.063 | 71.2 | 0.1155 | 2.03 (1.16–3.54) | 0.013 |
| Cardia type | 1.08 | 3 | 0.781 | 0.0 | 0.0000 | 0.88 (0.66–1.18) | 0.403 |
| PCR-RFLP genotyping | 5.93 | 7 | 0.547 | 0.0 | 0.0000 | 0.94 (0.68–1.29) | 0.701 |
| TagMan PCR genotyping | 11.04 | 1 | 0.001 | 90.9 | 0.3883 | 1.67 (0.68–4.14) | 0.265 |
Only D+L ORs (95% CI) and P values of D+L estimates provided.
Table 4. Stratification for the test of heterogeneity on XPD Asp312Asn based on a recessive model.
| Q-test | I2, % | τ2 | OR(95%CI) | P value | |||
| chi-squared | d.f. | p | |||||
| Overall | 9.91 | 8 | 0.272 | 19.2 | 0.0354 | 1.48 (1.12–1.97) | 0.007 |
| Asians | 4.58 | 5 | 0.470 | 0.0 | 0.0000 | 1.77 (1.19–2.63) | 0.005 |
| Caucasians | 3.43 | 2 | 0.180 | 41.6 | 0.0587 | 1.31 (0.86–1.99) | 0.211 |
| Large sample | 5.42 | 3 | 0.144 | 44.6 | 0.1053 | 1.35 (0.82–2.21) | 0.239 |
| Small-and-moderate sample | 2.71 | 4 | 0.607 | 0.0 | 0.0000 | 1.74 (1.18–2.56) | 0.005 |
| High quality | 8.55 | 5 | 0.128 | 41.5 | 0.0948 | 1.56 (1.05–2.33) | 0.028 |
| Low-and-moderate quality | 1.36 | 2 | 0.508 | 0.0 | 0.0000 | 1.42 (0.85–2.40) | 0.184 |
| High quality Asians | 1.68 | 2 | 0.433 | 0.0 | 0.0000 | 2.37 (1.29–4.36) | 0.005 |
| High quality Caucasians | 3.43 | 2 | 0.180 | 41.6 | 0.0587 | 1.31 (0.86–1.99) | 0.211 |
| Publication before or in 2007 | 4.34 | 3 | 0.227 | 30.9 | 0.1128 | 1.61(0.89–2.92) | 0.115 |
| Publication after 2007 | 5.41 | 4 | 0.248 | 26.0 | 0.0452 | 1.46 (1.02–2.09) | 0.041 |
| Cardia type | 0.85 | 2 | 0.654 | 0.0 | 0.0000 | 0.89 (0.56–1.43) | 0.642 |
| PCR-RFLP genotyping | 6.59 | 4 | 0.159 | 39.3 | 0.1474 | 1.90 (1.09–3.31) | 0.023 |
| Direct sequencing | 0.02 | 1 | 0.902 | 0.0 | 0.0000 | 1.24 (0.70–2.20) | 0.450 |
Only D+L ORs (95% CI) and P values of D+L estimates provided.
As shown in Table 3 and Table 4, specific data for XPD Lys751Gln and XPD Asp312Asn polymorphisms were stratified, respectively, on the basis of sample size, into two subgroups: large sample (the total number of controls and cases no less than 500) and small-and-moderate sample (the total number of controls and cases less than 500) subgroups. No statistically significant finding was noted in either small-and-moderate sample subgroup or large sample counterpart for XPD Lys751Gln polymorphism, given that the pooled ORs (95% CIs, p value) were 0.96 (0.61–1.51, p = 0.858) for the former and 1.17 (0.84–1.64, p = 0.351) for the latter, respectively; however a statistically significant finding was noted in small-and-moderate sample subgroup but not in large sample counterpart for Asp312Asn polymorphism, given that the pooled ORs (95% CIs, p value) were 1.74 (1.18–2.56, p = 0.005) for the former and 1.35 (0.82–2.21, p = 0.239) for the latter, respectively.
The data were also stratified, in accordance with the quality appraisal scores, into high quality (scores no less than 6.5) and low-and-moderate quality (scores less than 6.5) subgroups. As for XPD Lys751Gln polymorphism, no statistically significant finding was witnessed in either high quality subgroup or low-and-moderate quality counterpart, given that the pooled ORs (95% CIs, p value) were 1.20 (0.91–1.60, p = 0.199) for the former and 0.61 (0.34–1.11, p = 0.104) for the latter. When ethnicity sub-stratification was performed for high quality subgroup, a statistically significant finding was much apparently witnessed among Asians but no statistically significant finding was noted among Caucasians because the pooled ORs (95% CIs, p value) were 2.54 (1.77–3.65, p = 0.000) for the former and 0.98 (0.82–1.17, p = 0.803) for the latter, respectively. As for Asp312Asn polymorphism, a statistically significant finding was witnessed in high quality subgroup but not in low-and-moderate quality counterpart, given that the pooled ORs (95% CIs, p value) were 1.56 (1.05–2.33, p = 0.028) for the former and 1.42 (0.85–2.40, p = 0.184) for the latter. Likewise, when ethnicity sub-stratification was performed for high quality subgroup, a statistically significant finding was much apparently witnessed among Asians but no statistically significant finding was noted among Caucasians because the pooled ORs (95% CIs, p value) were 2.37 (1.29–4.36, p = 0.005) for the former and 1.31 (0.86–1.99, p = 0.211) for the latter, respectively.
The data were additionally stratified, in line with publication time, into the earlier publication subgroup (articles published before or in 2007) and the later publication subgroup (articles published after 2007). As for XPD Lys751Gln polymorphism, no statistically significant findings were observed on the grounds that the pooled ORs (95% CIs, p value) were 0.89 (0.66–1.19, p = 0.428) in the former and 1.23 (0.84–1.81, p = 0.285) in the latter, respectively. As for Asp312Asn polymorphism, a statistically significant finding was observed in the later publication subgroup but not in the earlier publication subgroup on the grounds that the pooled ORs (95% CIs, p value) were 1.46 (1.02–2.09, p = 0.041) in the former and 1.61(0.89–2.92, p = 0.115) in the latter, respectively.
When gastric cancer was classified into non-cardia (or distal) and cardia subtypes, a statistically significant finding was found among non-cardia type but not among cardia type for XPD Lys751Gln polymorphism on the grounds that the pooled ORs (95% CIs, p value) were 2.03 (1.16–3.54, p = 0.013) among non-cardia type and 0.88 (0.66–1.18, p = 0.403) among cardia type. As for Asp312Asn polymorphism, no statistically significant finding was observed among cardia type on the grounds that the pooled OR (95% CIs, p value) was 0.89 (0.56–1.43, p = 0.642), but OR (95% CIs, p value) regarding non-cardia type could not be calculated because only one study [21] clearly mentioned the numbers of genotypes of non-cardia type.
In terms of pathology, gastric cancer could be classified into intestinal, diffuse, or mixed subtypes, but only 1 study [19] clearly dealt with and mentioned the numbers of genotypes of diffuse subtype cancer; thus pathologic subtype stratification could not be done in our meta-analysis.
Confounding factors such as Helicobacter pylori infection or smoking status could not be analyzed in our meta-analysis because necessary relevant data of at least two studies could not be accessible in our meta-analysis.
And when genotyping techniques were considered, no statistically significant finding was noted in each genotyping technique subgroup for XPD Lys751Gln polymorphism because pooled ORs (95% CIs, p value) were 0.94 (0.68–1.29, p = 0.701) and 1.67 (0.68–4.14, p = 0.265) in RFLP and TagMan subgroups, respectively; however a statistically significant finding was noted in RFLP genotyping subgroup but not in Direct sequencing subgroup for Asp312Asn polymorphism because pooled ORs (95% CIs, p value) were 1.90 (1.09–3.31, p = 0.023) and 1.24 (0.70–2.20, p = 0.450), respectively.
Sensitivity Analysis
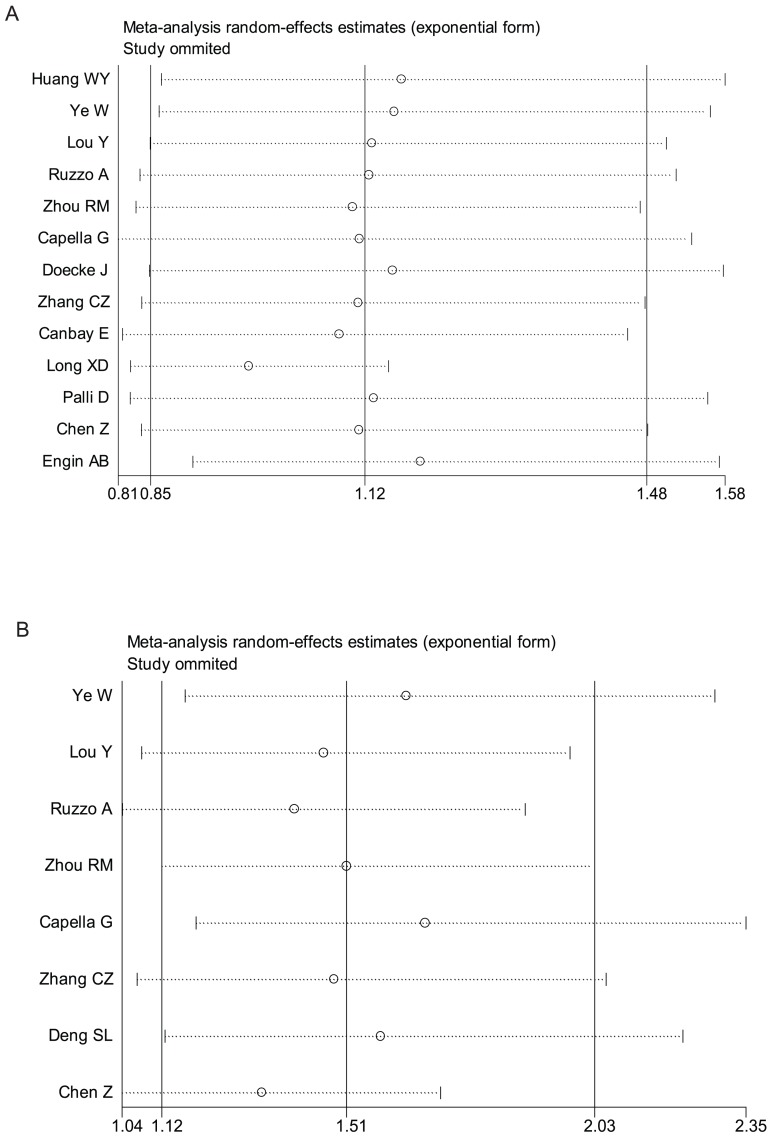
Meta-analyses were conducted repeatedly when each particular study had been removed. The results indicated that random-effects estimates before and after the deletion of each study were similar at large, suggesting moderately high stability of the meta-analysis results. For XPD Lys751Gln polymorphism, as shown in Figure 3 Part A, the most influencing single study on the overall pooled estimates seemed to be the study conducted by Long XD et al. [25], coincidently deviated from HWE, the sensitivity analysis, however, indicated moderately high stability of the results from the facts that the ORs (95% CI, p value) were 1.12(0.85–1.48, p = 0.410) before the removal of that study and 0.98 (0.83–1.15, p = 0.781) after the removal of that study. In view of the study [16] conducted by Huang WY et al. which is deviated from HWE, the ORs (95% CI, p value) were 1.12 (0.85–1.48, p = 0.410) before the removal of that study and 1.17 (0.87–1.58, p = 0.309) after the removal of that study for the all ethnicity, indicating high stability of the results. Similarly, after the removal of the study [29] conducted by Engin AB et al., also deviated from HWE, the OR (95% CI, p value) became 1.19 (0.91–1.57, p = 0.209) for the all ethnicity, indicating high stability of the results. If all the three deviated-HWE studies [16], [25], [29] were removed, the OR (95% CI, p value) became 1.06 (0.88–1.28, p = 0.555) for the all ethnicity, indicating moderately high stability of the results (The illustrating figures were omitted due to the length of paper).
Figure 3. Influence analysis of the summary odds ratio coefficients on the association for XPD Lys751Gln and XPD Asp312Asn polymorphisms with gastric cancer risk (based on a recessive genetic model).
Results were computed by omitting each study (on the left) in turn. Bars, 95% confidence interval. Meta-analysis random-effects estimates (exponential form) were used. A) Influence analysis of the summary odds ratio coefficients on the association for XPD Lys751Gln polymorphism with gastric cancer risk; B) Influence analysis of the summary odds ratio coefficients on the association for XPD Asp312Asn polymorphism with gastric cancer risk.
For Asp312Asn polymorphism, as shown in Figure 3 Part B, the most influencing single study on the overall pooled estimates seemed to be the study conducted by Chen Z et al. [28], the sensitivity analysis, however, indicated moderately high stability of the results from the facts that the ORs (95% CI, p value) were 1.48 (1.12–1.97, p = 0.007) before the removal of that study and 1.32 (1.04–1.70, p = 0.025) after the removal of that study. If all the three deviated-HWE studies [17], [27], [30] were removed, the OR (95% CI, p value) became 1.56 (1.05–2.33, p = 0.028) for the all ethnicity, also indicating moderately high stability of the results (The illustrating figures were omitted due to the length of paper).
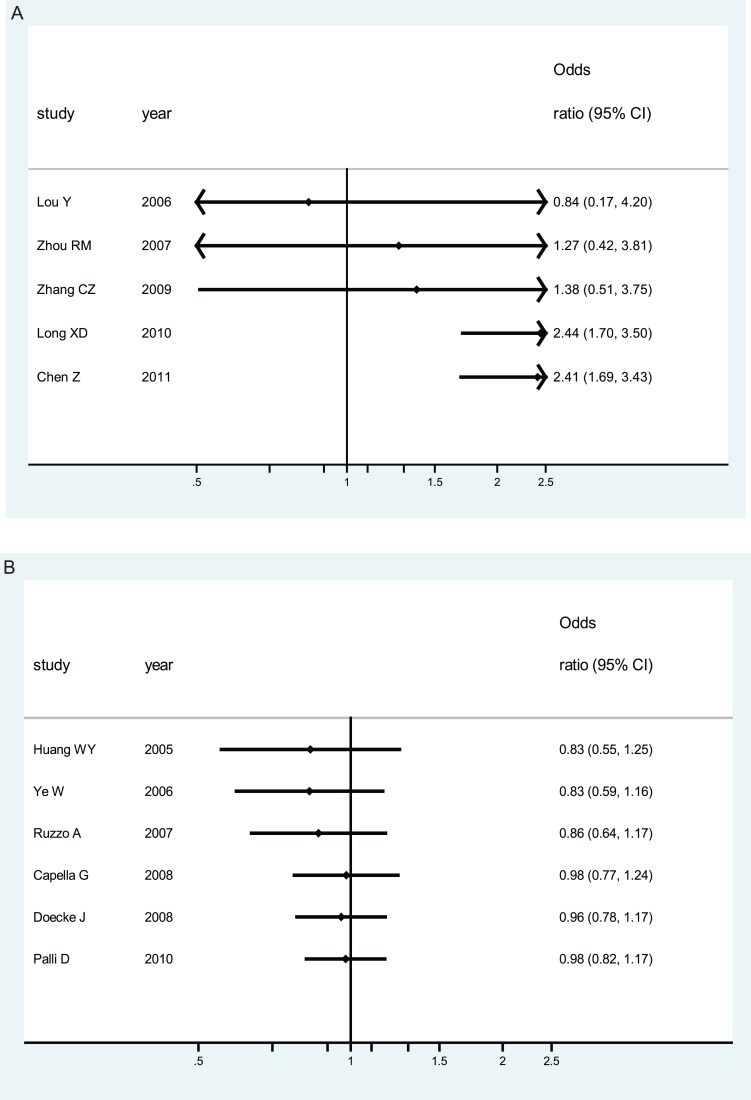
Cumulative Meta-analysis
Cumulative meta-analyses of XPD Lys751Gln polymorphism association were also conducted among Asians and Caucasians via the assortment of publication time and total number of sample size. As shown in Figure 4 part A, the inclinations toward significant associations for XPD Lys751Gln polymorphism could be seen among Asians. In Figure 4 part B the inclinations toward null associations for XPD Lys751Gln polymorphism could be noted among Caucasians in chronological order. Similar inclinations could be observed for Asp312Asn polymorphism among different ethnicity populations (Figures not shown).
Figure 4. Cumulative meta-analysis of associations between XPD Lys751Gln polymorphism and gastric cancer risk among different ethnicities (based on a recessive genetic model). sorted by y publication time and total number of sample size; Horizontal line, the accumulation of estimates as each study was added rather than the estimate of a single study.
A) among Asians; B) among Caucasians.
Publication Bias Analysis
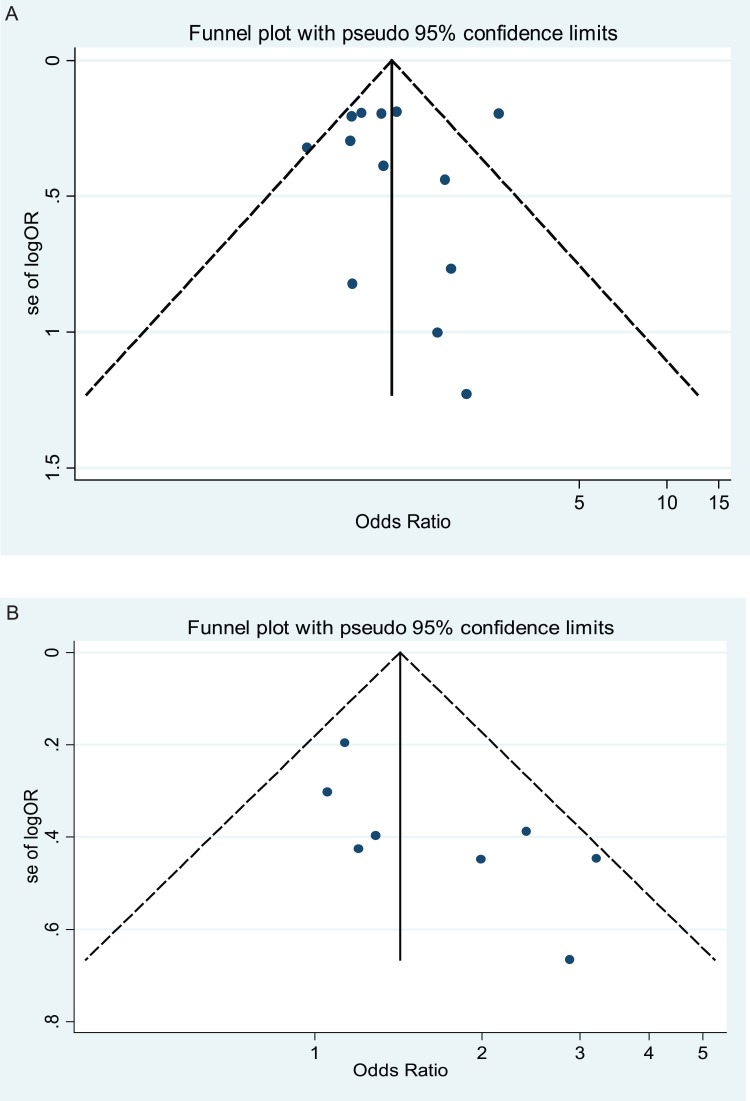
Publication bias was preliminarily examined by funnel plots qualitatively and estimated by Begg's and Egger's tests quantitatively. For XPD Lys751Gln polymorphism, its funnel plot (Figure 5A) showed that dots nearly symmetrically distributed, predominantly within pseudo 95% confidence limits. P values were 0.428 and 0.989 in Begg's test and Egger's test, respectively, insinuating no publication bias. For Asp312Asn polymorphism, its funnel plot (Figure 5B) showed that dots nearly symmetrically distributed, predominantly within pseudo 95% confidence limits. P values were 0.108 and 0.045 in Begg's test and Egger's test, respectively, insinuating no or a little publication bias.
Figure 5. Funnel plot of publication bias for XPD Lys751Gln and XPD Asp312Asn polymorphisms with gastric cancer risk (based on a recessive genetic model).
Note: Funnel plot with pseudo 95% confidence limits was used. A) Funnel plot of publication bias for XPD Lys751Gln polymorphism; B) Funnel plot of publication bias for XPD Asp312Asn polymorphism.
Discussion
In our meta-analysis statistically significant findings were apparently noted in Asians but not in Caucasians for both XPD Lys751Gln and XPD Asp312Asn polymorphisms. Also based on the findings of cumulative meta-analyses, the inclinations toward significant associations in Asians for both XPD Lys751Gln and XPD Asp312Asn polymorphisms could be obviously seen when sorted by publication time and total sample size. XPD Gln751Gln (CC) and Asn312Asn (AA) genotypes may seem to be more susceptible to gastric cancer in Asians. Our meta-analyses suggest that Gln751Gln (CC) and Asn312Asn (AA) genotypes may be important biomarkers of gastric cancer susceptibility for Asians, the assumption that needs to be further confirmed in future well-designed studies in Asian populations. Also the different or even conflicting risk associations, if so, among different ethnicities should be further meticulously investigated and reconfirmed in the future.
For XPD Asp312Asn SNP, although I 2 (%) was 19.2 and τ2 was 0.0354, as shown in Table 4, theoretically representing little or no heterogeneity; practically, after ethnicity subgroup analysis, the values of I 2 (%) and τ2 became 0.0 and 0.0000 for Asians and 41.6 and 0.0587 for Caucasians, respectively, further indicating that, actually, no heterogeneity existed among Asians studies rather than among Caucasians counterparts. Given this, we think that stratification or subgroup analyses are still needed in our meta-analysis. For XPD Asp312Asn polymorphism, our subgroup analyses also indicate that significant associations could be found in the small-and-moderate sample subgroup but not in the large sample counterpart. In large sample subgroup the ORs in the studies conducted by Capellá G et al. [21] and Ye W et al. [18] were both around 1.0, with the highest weight percentage to make the overall OR being statistically insignificant, whereas in small-and-moderate sample subgroup the influences of ORs in the studies conducted by Ruzzo A et al., Zhang CZ et al. and Lou Y et al. [19], [23], [17] were strong enough to make the overall OR to reach the significant value.
For XPD Asp312Asn polymorphism, a statistically significant finding was witnessed in high quality subgroup but not in low-and-moderate quality counterpart. When ethnicity sub-stratification was performed for such a high quality subgroup, a statistically significant finding was even more apparently witnessed among Asians but still no statistically significant finding was noted among Caucasians. For XPD Lys751Gln polymorphism, no statistically significant finding originally noted in high quality subgroup interestingly changed into a statistically significant finding among Asians but still not among Caucasians when ethnicity sub-stratification was performed for such a high quality subgroup. Those findings observed in different ethnic high quality subgroups further indicate the point that XPD Gln751Gln (CC) and Asn312Asn (AA) genotypes may be more susceptible to gastric cancer in Asians rather than in Caucasians. Certainly, high-quality studies should be more advocated to be designed in the future so as to accurately explore the real associations between XPD polymorphisms and gastric cancer risk among different ethnicities. Besides Asians and Caucasians, other ethnic participants, if possible, should be more appraised in the future.
Moreover, as for XPD Lys751Gln polymorphism, 4 [18], [20]–[22] out of 13 eligible studies were dealt with cardia gastric cancer and 2 [21], [25] with noncardia gastric cancer. A statistically significant finding could be noted with noncardia subgroup but not in cardia subgroup. Only 1 included study [19] in our meta-analysis mentioned pathologically sporadic diffuse-type gastric cancer and all the other studies did not mention pathologically Laurence's classification. As for XPD Asp312Asn polymorphism, 3 [18], [20], [21] out of 9 eligible studies were dealt with cardia gastric cancer and 1 [21] with noncardia gastric cancer. No statistically significant finding could be noted in cardia subgroup. Similarly, only 1 included study [19] mentioned pathologically sporadic diffuse-type gastric cancer and all the other studies did not mention pathologically Laurence's classification. As is widely known, cardia-type gastric cancer differs from noncardia-type gastric cancer in etiology, pathology, carcinogenesis, and/or prognosis [39]–[41], so is intestinal-type cancer versus diffuse-type cancer. It could be said that the indiscriminate combination of cardia-type and noncardia-type cases or intestinal-type and diffuse-type cases in the majority of eligible studies may mask or at least underestimate the real strength of the associations [5]–[7].
Furthermore, a variety of confounding factors such as Helicobacter pylori infection, alcoholic drinking, and smoking habits may be associated with increased damage to DNA repair [42]. Unfortunately, those factors could not be appraised in our meta-analysis due to the lack of relevant data.
With the advent of novel genotyping technologies like seminested polymerase chain reaction, TaqMan allelic discrimination test, or real-time PCR, we may witness an upsurge of genetic association studies in the future. As for XPD Asp312Asn polymorphism in our meta-analysis, a statistically significant finding could be noted in PCR-RFLP subgroup but not in Direct sequencing subgroup. As for XPD Lys751Gln polymorphism, however, no statistically significant finding was witnessed in either PCR-RFLP subgroup or TagMan PCR genotyping subgroup. Certainly, the difference should be concerned with extreme caution. Unfortunately, no direct sequencing was used among XPD Lys751Gln polymorphism studies. For a novel genotyping technique to be employed for the study of a particular genetic polymorphism, this technology, to our knowledge, should better be confirmed using direct sequencing. In that case, this new technology can be seen as valid as direct sequencing [43]. Or the sensitivity and specificity of those genotyping techniques need to be explored so as to seek out optimal approaches which could minimize the genotyping errors [5]–[7], [43]. And our opinion is that direct sequencing should be more used in future well-designed studies among different ethnicities.
Finally, the strength of our meta-analysis could be summarized as follows. We sought to find as many publications as we could by means of various searching approaches. The study that appeared to deviate from HWE was not excluded mechanically in our meta-analysis unless there are other convincing grounds for doubting the quality of the study [38]. Even so, we still performed the sensitivity analysis for the removal of all the deviated-HWE studies to further know the stability of the results, that is, the general impact of those deviated-HWE studies on the overall results. We laid more emphasis on assessing biases across studies and pinpointing the potential sources of heterogeneity via subgroup and sensitivity analyses. We comprehensively assessed the publication biases using several means like Begg's and Egger's tests as well as funnel plot tests. In view of this, we convince that the results of our meta-analysis, in essence, are sound and reliable.
To be sure, there are some unavoidable limitations in our meta-analysis. Firstly, the offered information from the included studies is inconsistent. Put it another way, the information about overall gastric cancer susceptibility is predominantly provided, while more important information about pathologic subtypes or anatomic subtypes of gastric cancer is less provided. Thus, the specific subtype results should be considered with caution. Secondly, with the merely published studies included in our meta-analysis, publication bias is very likely to occur, though no or a little statistically significant publication bias is indicated in our meta-analysis. Thirdly, moderate to severe heterogeneity could be witnessed among the included studies. So as to minimize the potential bias, we designed a rigorous protocol before conducting meta-analysis, and performed a scrupulous search for published studies using explicit methods for study selection, data extraction, statistical analysis, adoption of the most appropriate genetic model and sensitivity analysis.
In conclusion, XPD Gln751Gln (CC) and Asn312Asn (AA) genotypes may seem to be more susceptible to gastric cancer in Asian population but not in Caucasian population, suggesting that Gln751Gln (CC) and Asn312Asn (AA) genotypes may be important biomarkers of gastric cancer susceptibility for Asian population, the assumption that needs to be further confirmed in future well-designed studies among different ethnicities. Gln751Gln (CC) genotype may also be associated with the noncardia-type gastric cancer risk, the finding that also needs to be further confirmed.
Supporting Information
Scales for Quality Assessment.
(DOC)
Funding Statement
This work has been supported by The National Natural Science Foundation of China (grants 30830038, 30970842, and 81071180); “973” Project (2012CB932604); New Drug Discovery Project (2012ZX09506-001-005); the Key Project of Science and Technology Commission of Shanghai Municipality (grants 10JC1410000); and the Shanghai Leading Academic Discipline Project (grant S30203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55 (2) 74–108. [DOI] [PubMed] [Google Scholar]
- 2. Yang L (2006) Incidence and mortality of gastric cancer in China. World J Gastroenterol 12 (1) 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suerbaum S, Michetti P (2002) Helicobacter pylori infection. N Engl J Med 347 (15) 1175–1186. [DOI] [PubMed] [Google Scholar]
- 4. Hwang IR, Kodama T, Kikuchi S, Sakai K, Peterson LE, et al. (2002) Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1beta production in Helicobacter pylori infection. Gastroenterology 123: 1793–1803. [DOI] [PubMed] [Google Scholar]
- 5. Xue H, Lin B, Ni P, Xu H, Huang G (2010) Interleukin-1B and interleukin-1 RN polymorphisms and gastric carcinoma risk: a meta-analysis. J Gastroenterol Hepatol 25 (10) 1604–1617. [DOI] [PubMed] [Google Scholar]
- 6. Xue H, Liu J, Lin B, Wang Z, Sun J, et al. (2012) A Meta-analysis of interleukin-8 -251 promoter polymorphism associated with gastric cancer risk. PLoS One 7 (1) e28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xue H, Ni P, Lin B, Xu H, Huang G (2011) X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and gastric cancer risk: A HuGE review and meta-analysis. Am J Epidemiol 173 (4) 363–375. [DOI] [PubMed] [Google Scholar]
- 8. Goode EL, Ulrich CM, Potter JD (2002) Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 11 (12) 1513–1530. [PubMed] [Google Scholar]
- 9. Miller MC 3rd, Mohrenweiser HW, Bell DA (2001) Genetic variability in susceptibility and response to toxicants. Toxicol Lett 120 (1–3) 269–280. [DOI] [PubMed] [Google Scholar]
- 10. Küry S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, et al. (2008) Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer 8: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paz-Elizur T, Sevilya Z, Leitner-Dagan Y, Elinger D, Roisman LC, et al. (2008) DNA repair of oxidative DNA damage in human carcinogenesis: potential application for cancer risk assessment and prevention. Cancer Lett 266 (1) 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillet LC, Schärer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 106 (2) 253–276. [DOI] [PubMed] [Google Scholar]
- 13. Sung P, Bailly V, Weber C, Thompson LH, Prakash L, et al. (1993) Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365 (6449) 852–855. [DOI] [PubMed] [Google Scholar]
- 14. de Boer J, Hoeijmakers JH (2000) Nucleotide excision repair and human syndromes. Carcinogenesis 21: 453–460. [DOI] [PubMed] [Google Scholar]
- 15. Benhamou S, Sarasin A (2002) ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis 17 (6) 463–469. [DOI] [PubMed] [Google Scholar]
- 16. Huang WY, Chow WH, Rothman N, Lissowska J, Llaca V, et al. (2005) Selected DNA repair polymorphisms and gastric cancer in Poland. Carcinogenesis 26 (8) 1354–1359. [DOI] [PubMed] [Google Scholar]
- 17. Lou Y, Song QB, He XM (2006) Association of single nucleotide polymorphism in DNA repair gene XPD with gastric cancer in Han population from northeast region of China. Shi Jie Hua Ren Xiao Hua Za Zhi 14 (32) 3143–3146 [Chinese]. [Google Scholar]
- 18. Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, et al. (2006) The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis 27 (9) 1835–1841. [DOI] [PubMed] [Google Scholar]
- 19. Ruzzo A, Canestrari E, Maltese P, Pizzagalli F, Graziano F, et al. (2007) Polymorphisms in genes involved in DNA repair and metabolism of xenobiotics in individual susceptibility to sporadic diffuse gastric cancer. Clin Chem Lab Med 45 (7) 822–828. [DOI] [PubMed] [Google Scholar]
- 20. Zhou RM, Li Y, Wang N, Dong XJ, Zhang XJ, et al. (2007) Correlation between single nucleotide polymorphism of DNA repair gene XPD and the risks of esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. Zhong Liu 27 (2) 118–122 [Chinese]. [Google Scholar]
- 21. Capellá G, Pera G, Sala N, Agudo A, Rico F, et al. (2008) DNA repair polymorphisms and the risk of stomach adenocarcinoma and severe chronic gastritis in the EPIC-EURGAST study. Int J Epidemiol 37 (6) 1316–1325. [DOI] [PubMed] [Google Scholar]
- 22. Doecke J, Zhao ZZ, Pandeya N, Sadeghi S, Stark M, et al. (2008) Australian Cancer Study. Polymorphisms in MGMT and DNA repair genes and the risk of esophageal adenocarcinoma. Int J Cancer 123 (1) 174–180. [DOI] [PubMed] [Google Scholar]
- 23. Zhang CZ, Chen ZP, Xu CQ, Ning T, Li DP, et al. (2009) [Correlation of XPD gene with susceptibility to gastric cancer]. Ai Zheng 28 (11) 1163–1167 [Chinese]. [DOI] [PubMed] [Google Scholar]
- 24. Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, et al. (2010) Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res 30 (4) 1359–1364. [PubMed] [Google Scholar]
- 25. Long XD, Ma Y, Huang YZ, Yi Y, Liang QX, et al. (2010) Genetic polymorphisms in DNA repair genes XPC, XPD, and XRCC4, and susceptibility to Helicobacter pylori infection-related gastric antrum adenocarcinoma in Guangxi population, China. Mol Carcinog 49 (6) 611–618. [DOI] [PubMed] [Google Scholar]
- 26. Palli D, Polidoro S, D'Errico M, Saieva C, Guarrera S, et al. (2010) Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis 25 (6) 569–575. [DOI] [PubMed] [Google Scholar]
- 27. Deng SL, Chen M, Chen W, Lu W, Huang HL, et al. (2010) Association of DNA repair gene polymorphisms with genetic susceptibility to gastric cancer. 2 (6) 371–374. [DOI] [PubMed] [Google Scholar]
- 28. Chen Z, Zhang C, Xu C, Li K, Hou R, et al. (2011) Effects of selected genetic polymorphisms in xeroderma pigmentosum complementary group D on gastric cancer. Mol Biol Rep 38 (3) 1507–1513. [DOI] [PubMed] [Google Scholar]
- 29. Engin AB, Karahalil B, Engin A, Karakaya AE (2011) DNA repair enzyme polymorphisms and oxidative stress in a Turkish population with gastric carcinoma. Mol Biol Rep 38 (8) 5379–5386. [DOI] [PubMed] [Google Scholar]
- 30. Yuan T, Deng S, Chen M, Chen W, Lu W, et al. (2011) Association of DNA repair gene XRCC1 and XPD polymorphisms with genetic susceptibility to gastric cancer in a Chinese population. Cancer Epidemiol 35 (2) 170–174. [DOI] [PubMed] [Google Scholar]
- 31. Manuguerra M, Saletta F, Karagas MR, Berwick M, Veglia F, et al. (2006) XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol 164 (4) 297–302. [DOI] [PubMed] [Google Scholar]
- 32. Wang F, Chang D, Hu FL, Sui H, Han B, et al. (2008) DNA repair gene XPD polymorphisms and cancer risk: a meta-analysis based on 56 case-control studies. Cancer Epidemiol Biomarkers Prev 17 (3) 507–517. [DOI] [PubMed] [Google Scholar]
- 33. Chen B, Zhou Y, Yang P, Wu XT (2011) ERCC2 Lys751Gln and Asp312Asn polymorphisms and gastric cancer risk: a meta-analysis. J Cancer Res Clin Oncol 137 (6) 939–946. [DOI] [PubMed] [Google Scholar]
- 34. Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, et al. (2005) Systematic review and metaanalysis of the association between{h}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol 162 (3) 201–211. [DOI] [PubMed] [Google Scholar]
- 35. Camargo MC, Mera R, Correa P, Peek RM Jr, Fontham ET, et al. (2006) Interleukin-1beta and interleukin-1 receptor antagonist gene polymorphisms and gastric cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev 15 (9) 1674–1687. [DOI] [PubMed] [Google Scholar]
- 36. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. Br Med J 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ioannidis JP, Boffetta P, Little J, O,Brien TR, Uitterlinden AG, et al. (2008) Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol 37: 120–132. [DOI] [PubMed] [Google Scholar]
- 38. Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J (2008) How should we use information about HWE in the meta-analyses of genetic association studies? Int J Epidemiol 37 (1) 136–146. [DOI] [PubMed] [Google Scholar]
- 39. Heidl G, Langhans P, Mellin W, Bunte H, Grundmann E (1993) Adeno-carcinomas of esophagus and cardia in comparison with gastric carcinoma. J Cancer Res Clin Oncol 120 (1–2) 95–99. [DOI] [PubMed] [Google Scholar]
- 40. Kim MA, Lee HS, Yang HK, Kim W (2005) Clinicopathologic and protein expression differences betweencardia carcinoma and noncardia carcinoma of the stomach. Cancer 103 (7) 1439–1446. [DOI] [PubMed] [Google Scholar]
- 41. Saito H, Fukumoto Y, Osaki T, Fukuda K, Tatebe S, et al. (2006) Distinct recurrence pattern and outcome of adenocarcinoma of the gastric cardia in comparison with carcinoma of other regions of the stomach. World J Surg 30 (10) 1864–1869. [DOI] [PubMed] [Google Scholar]
- 42. Crabtree JE, Covacci A, Farmery SM, Xiang Z, Tompkins DS, et al. (1995) Helicobacter pylori induced interleukin-8 expression in gastric epithelial cells is associated with CagA positive phenotype. J Clin Pathol 48 (1) 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue H, Lin B, An J, Zhu Y, Huang G (2012) Interleukin-10-819 promoter polymorphism in association with gastric cancer risk. BMC Cancer 12: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scales for Quality Assessment.
(DOC)