Abstract
Cells often mount transcriptional responses and activate specific sets of genes in response to stress-inducing signals such as heat or reactive oxygen species. Transcription factors in the RpoH family of bacterial alternative σ factors usually control gene expression during a heat shock response. Interestingly, several α-proteobacteria possess two or more paralogs of RpoH, suggesting some functional distinction. We investigated the target promoters of Rhodobacter sphaeroides RpoHI and RpoHII using genome-scale data derived from gene expression profiling and the direct interactions of each protein with DNA in vivo. We found that the RpoHI and RpoHII regulons have both distinct and overlapping gene sets. We predicted DNA sequence elements that dictate promoter recognition specificity by each RpoH paralog. We found that several bases in the highly conserved TTG in the −35 element are important for activity with both RpoH homologs; that the T-9 position, which is over-represented in the RpoHI promoter sequence logo, is critical for RpoHI–dependent transcription; and that several bases in the predicted −10 element were important for activity with either RpoHII or both RpoH homologs. Genes that are transcribed by both RpoHI and RpoHII are predicted to encode for functions involved in general cell maintenance. The functions specific to the RpoHI regulon are associated with a classic heat shock response, while those specific to RpoHII are associated with the response to the reactive oxygen species, singlet oxygen. We propose that a gene duplication event followed by changes in promoter recognition by RpoHI and RpoHII allowed convergence of the transcriptional responses to heat and singlet oxygen stress in R. sphaeroides and possibly other bacteria.
Author Summary
An important property of living systems is their ability to survive under conditions of stress such as increased temperature or the presence of reactive oxygen species. Central to the function of these stress responses are transcription factors that activate specific sets of genes needed for this response. Despite the central role of stress responses across all forms of life, the processes driving their organization and evolution across organisms are poorly understood. This paper uses genomic, computational, and mutational analyses to dissect stress responses controlled by two proteins that are each members of the RpoH family of alternative σ factors. RpoH family members usually control gene expression during a heat shock response. However, the photosynthetic bacterium Rhodobacter sphaeroides and several other α-proteobacteria possess two or more paralogs of RpoH, suggesting some functional distinction. Our findings predict that a gene duplication event followed by changes in DNA recognition by RpoHI and RpoHII allowed convergence of the transcriptional responses to heat and singlet oxygen stress in R. sphaeroides and possibly other bacteria. Our approach and findings should interest those studying the evolution of transcription factors or the signal transduction pathways that control stress responses.
Introduction
Transcriptional responses to stress are critical to cell growth and survival. In bacteria, stress responses are often controlled by alternative σ factors that direct RNA polymerase to transcribe promoters different from those recognized by the primary σ factor [1], [2]. Therefore, identifying the target genes for a particular alternative σ factor can help identify the functions necessary to respond to a given stress. For example, the transcriptional response to heat shock in Escherichia coli uses the alternative σ factor σ32 to increase synthesis of gene products involved in protein homeostasis or membrane integrity [3]. From available genome sequences, proteins related to E. coli σ32 are conserved across virtually all proteobacteria. This so-called RpoH family of alternative σ factors is characterized by a conserved amino acid sequence (the “RpoH box”) that is involved in RNA polymerase interactions [4], [5]. RpoH family members also possess conserved amino acid sequences in σ factor regions 2.4 and 4.2 that interact with promoter sequences situated approximately −10 and −35 base pairs upstream of the transcriptional start sites, respectively [6]. However, the definition of functional promoters for this family of alternative σ factor using only the presence or the extent of sequence identity for the predicted −10 and −35 binding regions is not a sufficient predictor of transcription activity [7].
While bacteria often possess many alternative σ factors, they usually possess only one member of the RpoH family. However, several α-proteobacteria, including Brucella melitensis [8], Sinorhizobium meliloti [9], [10], Bradyrhizobium japonicum [11], [12], Rhizobium elti [13] and Rhodobacter sphaeroides [14], possess two or more RpoH homologs. In some cases, one or more of these RpoH homologs completely or partially complement the phenotypes of E. coli ΔrpoH mutants, suggesting that these proteins can functionally interact with RNA polymerase and recognize similar promoter elements [8]–[11], [14], [15]. However, in the nitrogen-fixing plant symbiont Rhizobium elti, the ΔrpoH1 mutant was sensitive to heat and oxidative stress while the ΔrpoH2 mutant was sensitive to osmotic stress [13]. Therefore, the additional members of the RpoH family in α-proteobacteria may have roles in other stress responses.
Previous work demonstrated that either R. sphaeroides RpoHI or RpoHII can complement the temperature sensitive phenotype of an E. coli ΔrpoH mutant; that singly mutant R. sphaeroides strains lacking either rpoHI or rpoHII are able to mount a heat shock response; and that RNA polymerase containing either RpoHI or RpoHII can initiate transcription from a common set of promoters in vitro [14]–[16]. Combined, these observations suggest that RpoHI and RpoHII have some overlapping functions in R. sphaeroides. On the other hand, in vitro transcription assays identified promoters that were selectively transcribed by either RpoHI or RpoHII [14], [15]. Moreover, rpoHII is under direct transcriptional control of RpoE, a Group IV alternative σ factor that acts as the master regulator of the response of R. sphaeroides to singlet oxygen stress [17]–[19]. These later results and the recent observation that a ΔrpoHII mutant is more sensitive to singlet oxygen stress than the wild-type strain [15], [17] suggest that RpoHI and RpoHII also have distinct functions in R. sphaeroides. Finally, global protein profiles of R. sphaeroides mutants lacking rpoHI, rpoHII, or both genes, suggested that RpoHI and RpoHII have distinct and overlapping regulons [15], [17], [20]. However, the extent of genes that are direct targets for RpoHI and RpoHII is still unknown because past studies have been unable to distinguish direct from indirect effects on gene expression or identify all the direct targets for either of these σ factors.
In this study, we characterized the RpoHI and RpoHII regulons using a combination of expression microarrays, chromatin immunoprecipitation and computational methods which have been previously been shown to predict correctly direct targets for other alternative σ factors or DNA binding proteins [19], [21]. We found that the genes predicted to be common to the RpoHI and RpoHII regulons function in protein repair or turnover, membrane maintenance, and DNA repair. Genes specific to the RpoHI regulon encode other proteins involved in protein maintenance and DNA repair, whereas genes specific to the RpoHII regulon include proteins involved in maintaining the oxidation-reduction state of the cytoplasmic thiol pool. We used information on the members of each regulon to generate and test hypotheses about DNA sequences that determine promoter specificity of these two RpoH homologs. The observed properties of these two R. sphaeroides RpoH homologs illustrate how duplication of an alternative σ factor and subsequent changes in promoter recognition could have allowed convergence of transcriptional responses to separate signals. In the case of R. sphaeroides, we predict that these events allowed convergence of the transcriptional responses to heat shock and singlet oxygen stresses to be under control of these two RpoH paralogs.
Results
Defining the distinct and overlapping regulons of R. sphaeroides RpoHI and RpoHII
To define members of the RpoHI and RpoHII regulons, we monitored transcript levels and protein-DNA interactions in R. sphaeroides strains ectopically expressing either RpoHI or RpoHII. To generate these strains, we constructed low copy plasmids carrying rpoHI or rpoHII under the control of an IPTG-inducible promoter [22] and conjugated them into R. sphaeroides mutant strains lacking rpoHI [16] or rpoHII [15], respectively. To induce target gene expression, we exposed exponentially growing aerobic cultures to IPTG for one generation before cells were either harvested to extract total RNA for analysis of transcript levels or treated with formaldehyde to prepare samples for chromatin immunoprecipitation on a chip (ChIP-chip) assays. The Western blot analysis used to measure levels of these alternative σ factors demonstrates that cells ectopically expressing RpoHI and RpoHII contained each protein at levels comparable to those following either heat shock or singlet oxygen stress (Figure 1). Thus, these strains can be used to characterize members of the RpoHI and RpoHII regulons.
Figure 1. RpoHI and RpoHII accumulation following heat and singlet oxygen stresses.

Western blots illustrating the levels of RpoHI and RpoHII in wild-type R. sphaeroides (WT) at different times following (A) a shift of temperature from 30°C to 42°C (heat shock) or (B) addition of the photosensitizer methylene blue in the presence of oxygen (singlet oxygen stress). On the same western blots, the levels of FLAG-RpoHI and RpoHII obtained from ectopic expression vectors used in the expression profiling and ChIP-chip experiments under normal conditions. Note that because of the addition of the FLAG polypeptide, RpoHI-FLAG migrates slower than the wild-type RpoHI. The abundance of RpoHI and RpoHII in wild-type cells in the absence of added stress are shown in the first lane. As a gel loading control, the membranes were also subsequently treated polyclonal antibodies against the response regulator PrrA, a control transcription factor who's expression is not known to be dependent on either of the RpoH homologs. The experiment was designed to analyze changes in levels of RpoHI, RpoHII and PrrA before and after a stress, so the differences between panels reflect different exposure times used when developing the Western blots.
As controls for this experiment, we measured the abundance of individual RpoH proteins and a control transcription factor (PrrA) [23], which is not known to be dependent on either alternative σ factor for its expression, when wild type cells were exposed to either heat or singlet oxygen stress. This analysis showed that RpoHI is detectable prior to heat stress, but its levels increase 10 and 20 minutes after the shift to increased temperature (Figure 1A). RpoHI levels remain elevated after the temperature shift but they decline within 60 minutes after heat shock, suggesting that as in the case of E. coli σ32, there is an initial rise in RpoHI levels immediately on heat shock before they return to a new steady state level at elevated temperature [24]. RpoHII was also detected prior to exposure to singlet oxygen and within 10 minutes of exposure to this reactive oxygen species, levels of this protein were increased (Figure 1B). Levels of RpoHII found within 20 minutes after exposure to singlet oxygen remained relatively constant over the time course of this experiment, suggesting a continuous requirement for RpoHII during this stress response (Figure 1B). The abundance of the control transcription factor PrrA did not follow these same trends, suggesting that the observed increases in individual RpoH proteins was associated with these stress responses. In addition, the abundance of individual RpoH proteins did not increase significantly to both stress responses, as expected if these increases were not due to a general increase in protein levels in response to different signals.
To identify transcripts that were increased in abundance as a result of RpoHI or RpoHII activity, we compared mRNA levels of cells expressing RpoHI or RpoHII ectopically to those of control cells lacking either rpoHI or rpoHII. We selected differentially expressed genes with a significance level set for a false discovery rate ≤5% and that displayed at least 1.5-fold higher transcript levels in cells expressing either RpoH family member. This analysis revealed that transcripts from 241 and 186 genes were increased by expression of RpoHI and RpoHII, respectively (Figure 2). These two sets of differentially expressed genes have 60 genes in common.
Figure 2. Overlap between the RpoHI and RpoHII regulons.
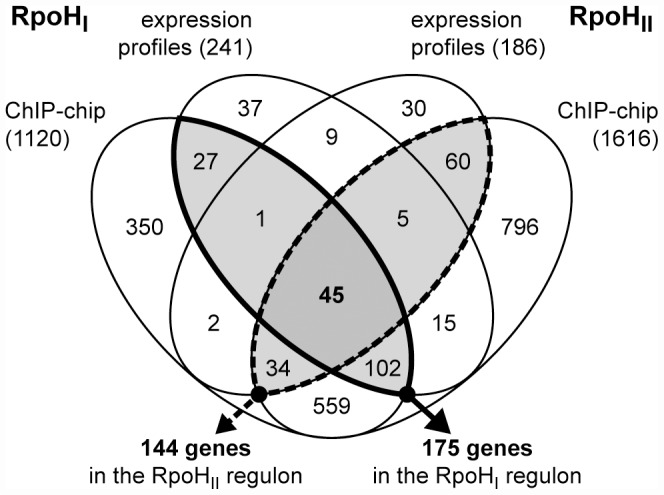
Venn diagram representing the overlaps between genes that were significantly induced by the expression of RpoHI or RpoHII, and genes whose promoters were bound by RpoHI or RpoHII containing RNA polymerase holoenzyme in vivo. The total numbers of genes identified in each study are indicated in the parentheses. The RpoHI (solid outline) and RpoHII (dashed outline) regulons, as defined in this study, are identified by the emphasized outlines. The total numbers of genes contained in each regulon are indicated below the arrows.
We recognize that some of these differentially-expressed transcripts might be not be direct targets for RpoHI and RpoHII. Therefore, to determine which of the above genes were directly transcribed by RNA polymerase holoenzyme containing either RpoHI or RpoHII, we performed ChIP-chip assays from comparable cultures to map direct interactions of RpoHI or RpoHII with genomic DNA. We were able to raise specific antibodies against RpoHII that performed well for the ChIP-chip assay, but repeated attempts to raise suitable antibodies against RpoHI failed. Therefore, we placed a FLAG polypeptide tag [25] at the N-terminus of the RpoHI protein sequence and used anti-FLAG monoclonal antibodies to perform the ChIP-chip assay. As a control we tested and showed that addition of the polypeptide tag did not alter the activity and specificity of RpoHI by comparing the mRNA level profiles of cells expressing the tagged version of RpoHI with cells expressing wild-type RpoHI (Figure S1). In addition, other control experiments showed there was no detectable cross-reaction between FLAG-RpoHI and the antibody used to precipitate RpoHII, and vice versa (data not shown). From the ChIP-chip analysis we identified 812 and 1353 genomic regions enriched after immunoprecipitation with antibodies against RpoHI and RpoHII, respectively, using a significance level set for a false discovery rate ≤5%. Because the signal from a single σ factor binding site extends on average over a 1 kb region, some enriched regions may contain multiple binding sites. To increase the resolution of the putative RpoHI and RpoHII binding sites, we identified the modes of the ChIP-chip signal distributions within each enriched region. This adjustment increased the number of putative binding sites for RpoHI and RpoHII to 1085 and 1765, respectively.
We then identified all the annotated genes that contained a ChIP-chip peak within 300 base pairs upstream of their start codons as a way to define candidate genes or operons in the RpoHI or RpoHII regulons. Included in this list of potential regulon members were genes that are predicted to be co-transcribed using a previous computational analysis of R. sphaeroides operon organization (http://www.microbesonline.org/operons/) [26]. Therefore, by these criteria, the upper limits of the total numbers of genes potentially regulated by RpoHI or RpoHII are 1120 and 1616, respectively (Figure 2). We recognized that a significant number of the putative RpoHI or RpoHII promoters may not be assigned from the ChIP-chip dataset alone, especially because promoter orientation needs to be considered and that because σ factor or RNA polymerase binding events do not always promote transcription. Therefore, we refined the respective RpoHI and RpoHII regulons by intersecting the lists of target genes identified from the ChIP-chip analysis with the lists of candidate genes identified from the expression profiling analysis. After this intersection, we predict that the RpoHI regulon contains 175 genes and the RpoHII regulon contains 144 genes with 45 genes common to both regulons (Figure 2).
Upon examining the annotations of these predicted target genes, the 45 genes that are members of both the RpoHI and RpoHII regulons are predicted to encode mainly for functions related to the electron transport chain, protein homeostasis, and DNA repair (Table 1 and Table S1). The 130 predicted members of the RpoHI regulon also encode functions in these three groups, but with a larger representation for functions associated with protein homeostasis. The 99 predicted members of the RpoHII regulon include fewer proteins predicted to play a role in protein homeostasis and a larger number of proteins predicted to help maintain the oxidation-reduction state of the cytoplasmic thiol pool. However, a large number of genes in both the unique and overlapping RpoHI and RpoHII regulons are annotated as having no predicted functions. Overall, this analysis revealed that RpoHI or RpoHII activate a large set of distinct and overlapping sets of target genes.
Table 1. Compositions of the RpoHI and RpoHII regulons.
| Mainrole§ | Subrole† | Locus‡ |
| RpoHI/II regulon (45 genes) | ||
| Energy metabolism | Electron transport | RSP_0100, RSP_0101, RSP_0102, RSP_0103, RSP_0104, RSP_0105, RSP_0106, RSP_0107, RSP_2805 |
| Other | RSP_0472 | |
| Protein synthesis/fate | Degradation of proteins, peptides, and glycopeptides | RSP_0665, RSP_1076, RSP_1174, RSP_2710, RSP_2806 |
| Other | RSP_1825 | |
| Protein folding and stabilization | RSP_1207 | |
| Ribosomal proteins: synthesis and modification | RSP_0570 | |
| Serine family | RSP_2481 | |
| Fatty acid and cell envelope | Biosynthesis | RSP_0473 |
| Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | RSP_2569 | |
| Other | RSP_3601 | |
| DNA metabolism | DNA replication, recombination, and repair | RSP_2388, RSP_2966 |
| Central intermediary metabolism | Phosphorus compounds | RSP_0013 |
| Cellular processes | Adaptations to atypical conditions | RSP_2617 |
| Signal transduction | Two-component systems | RSP_0847 |
| Unknown function | Unknown function | RSP_2718, RSP_0011, RSP_0370, RSP_1025, RSP_1239, RSP_1760, RSP_2218, RSP_2421, RSP_2625, RSP_2973, RSP_3327, RSP_3599, RSP_0870, RSP_1026, RSP_1421, RSP_1840, RSP_2261, RSP_2265 |
| RpoHI regulon (130 genes) | ||
| Protein synthesis/fate | Amino acid biosynthesis | RSP_0244, RSP_0377, RSP_1475 |
| Degradation of proteins, peptides, and glycopeptides | RSP_0357, RSP_0554, RSP_1408, RSP_1531, RSP_1742, RSP_2412, RSP_2649 | |
| Protein and peptide secretion and trafficking | RSP_1169, RSP_1797, RSP_1798, RSP_1799, RSP_1843, RSP_2540, RSP_2541 | |
| Protein folding and stabilization | RSP_1016, RSP_1173, RSP_1532, RSP_1572, RSP_1805, RSP_4043 | |
| Protein modification and repair | RSP_0559, RSP_0872, RSP_0873, RSP_0874, RSP_0923 | |
| tRNA aminoacylation | RSP_0875 | |
| Energy metabolism | Amino acids and amines | RSP_3957 |
| Electron transport | RSP_0296, RSP_0610, RSP_1194, RSP_1489, RSP_1529, RSP_1576, RSP_2375, RSP_2685, RSP_2945 | |
| Glycolysis/gluconeogenesis | RSP_0361 | |
| Fatty acid and cell envelope | Biosynthesis | RSP_0720, RSP_0929, RSP_2776 |
| Biosynthesis and degradation of murein sacculus and peptidoglycan | RSP_1240 | |
| Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | RSP_0125, RSP_3187 | |
| Degradation | RSP_0409 | |
| Other | RSP_1889 | |
| Biosynthesis of cofactors, prosthetic groups, and carriers | Folic acid | RSP_0930 |
| Lipoate | RSP_2783 | |
| Molybdopterin | RSP_0235, RSP_1071, RSP_1072 | |
| Other | RSP_2658 | |
| Pyridoxine | RSP_1672 | |
| Regulatory functions | DNA interactions | RSP_0014, RSP_2200 |
| Other | RSP_2236 | |
| Protein interactions | RSP_4193 | |
| Transcription factors | RSP_2410 | |
| Transport and binding proteins | Amino acids, peptides and amines | RSP_1564 |
| Cations and iron carrying compounds | RSP_2542, RSP_2891 | |
| Unknown substrate | RSP_2696, RSP_2897 | |
| DNA metabolism | DNA replication, recombination, and repair | RSP_1074, RSP_2815, RSP_4199 |
| Purine ribonucleotide biosynthesis | RSP_2454 | |
| Cellular processes | Adaptations to atypical conditions | RSP_4198 |
| Detoxification | RSP_0890, RSP_1058 | |
| Central intermediary metabolism | Other | RSP_1196, RSP_1949 |
| Sulfur metabolism | RSP_2738 | |
| Signal transduction | Two-component systems | RSP_2130, RSP_3105 |
| Mobile and extra-chromosomal element functions | Transposon functions | RSP_3007 |
| Unknown function | Unknown function | RSP_0126, RSP_0362, RSP_0363, RSP_0408, RSP_0719, RSP_0999, RSP_1104, RSP_1193, RSP_1204, RSP_1238, RSP_1241, RSP_1360, RSP_1406, RSP_1549, RSP_1563, RSP_1573, RSP_1581, RSP_1615, RSP_1671, RSP_1684, RSP_1743, RSP_1852, RSP_2121, RSP_2125, RSP_2214, RSP_2219, RSP_2387, RSP_2638, RSP_2640, RSP_2641, RSP_2739, RSP_2763, RSP_2764, RSP_2816, RSP_2952, RSP_2953, RSP_3067, RSP_3068, RSP_3378, RSP_3426, RSP_3552, RSP_3597, RSP_3598, RSP_3634, RSP_3809, RSP_3810, RSP_4244, RSP_4245, RSP_4248, RSP_4305 |
| RpoHII regulon (99 genes) | ||
| Energy metabolism | Biosynthesis and degradation of polysaccharides | RSP_0482 |
| Electron transport | RSP_0108, RSP_0109, RSP_0110, RSP_0112, RSP_0474, RSP_2785, RSP_3212, RSP_3305, RSP_3537 | |
| Entner-Doudoroff | RSP_2646 | |
| Fermentation | RSP_3164 | |
| Glycolysis/gluconeogenesis | RSP_2736, RSP_4045, RSP_4211 | |
| Other | RSP_0392, RSP_2294 | |
| Pentose phosphate pathway | RSP_2734, RSP_2735 | |
| Sugars | RSP_2937, RSP_3138 | |
| Biosynthesis of cofactors, prosthetic groups, and carriers | Glutathione and analogs | RSP_3272 |
| Heme, porphyrin, and cobalamin | RSP_1197, RSP_1692, RSP_2831 | |
| Menaquinone and ubiquinone | RSP_1175, RSP_1338, RSP_1492, RSP_1869 | |
| Other | RSP_0750, RSP_0898, RSP_2314 | |
| Transport and binding proteins | Amino acids, peptides and amines | RSP_1542, RSP_3274 |
| Carbohydrates, organic alcohols, and acids | RSP_0149, RSP_0150 | |
| Cations and iron carrying compounds | RSP_1546, RSP_2608 | |
| Unknown substrate | RSP_1895, RSP_2802, RSP_3160 | |
| DNA metabolism | DNA replication, recombination, and repair | RSP_1466, RSP_2083, RSP_2414, RSP_2850, RSP_3077, RSP_3423 |
| Pyrimidine ribonucleotide biosynthesis | RSP_3722 | |
| Fatty acid and cell envelope | Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | RSP_1491, RSP_2163, RSP_3721 |
| Degradation | RSP_0119 | |
| Other | RSP_0422, RSP_0595, RSP_0855 | |
| Regulatory functions | DNA interactions | RSP_1083, RSP_4210 |
| Other | RSP_0148, RSP_2631, RSP_3430, RSP_3431 | |
| Transcription factors | RSP_0601 | |
| Cellular processes | Detoxification | RSP_1057, RSP_2389, RSP_2693, RSP_3263 |
| Toxin production and resistance | RSP_2803 | |
| Central intermediary metabolism | Other | RSP_0897, RSP_1258, RSP_1397, RSP_3072 |
| Phosphorus compounds | RSP_0782 | |
| Protein synthesis/fate | Amino acid biosynthesis | RSP_0398 |
| Degradation of proteins, peptides, and glycopeptides | RSP_0686, RSP_1490 | |
| Protein folding and stabilization | RSP_1219 | |
| tRNA and rRNA base modification | RSP_2971 | |
| Unknown function | Unknown function | RSP_0151, RSP_0152, RSP_0269, RSP_0423, RSP_0557, RSP_0799, RSP_0896, RSP_1591, RSP_1956, RSP_1985, RSP_2225, RSP_2268, RSP_3075, RSP_3076, RSP_3089, RSP_3310, RSP_3329, RSP_4144, RSP_4209 |
Summary of the functional annotations of members of the RpoHI and RpoHII regulons in R. sphaeroides defined by the intersections of the results from the expression profiling and chromatin immunoprecipitation experiments.
Classification of the functional main categories according to the JCVI-CRM database (http://cmr.jcvi.org/).
Classification of the functional sub-categories according to the JCVI-CRM database.
Unique locus identifiers for R. sphaeroides 2.4.1.
Predicted differences in promoter sequences recognized by RpoHI or RpoHII
Previous work indicated that RpoHI and RpoHII can recognize and initiate transcription from similar promoter sequences [14], [15], [20]. The characterization of their respective regulons also suggests that some promoters can be transcribed by both σ factors while others are specific to either RpoHI or RpoHII. Therefore, we hypothesized that while the promoter sequences of the two σ factors may be similar, different sequence-specific interactions of RpoHI or RpoHII with promoter elements are the basis of promoter specificity for transcription initiation by RNA polymerase.
To overcome the limited resolution of the ChIP-chip experiment and predict determinants of promoter specificity for RpoHI or RpoHII, we searched the regions upstream of genes in each regulon for conserved sequence elements (137 sequences for RpoHI and 120 sequences for RpoHII). The conserved sequence elements we identified mapped to putative promoter elements that were within 100 bp of the coordinates of the modes of the distributions of the ChIP-chip signal. Thus, the predictions of these searches identified conserved sequence elements that were in agreement with the experimental data. In addition, even though we analyzed the individual RpoHI and RpoHII regulons independently for these motifs, the sequence alignment algorithm converged to the same sequence elements for promoters that were predicted to be recognized by both RpoHI and RpoHII. This result is not surprising given that both σ factors have similar amino acid sequences in their DNA recognition regions and are thus expected to recognize similar promoter sequences. However, this observation supports the hypothesis that RpoHI and RpoHII recognize common promoter sequences in their respective target genes as opposed to distinct promoters.
To predict specificity sequence determinants for each RpoH paralog, the putative distinct and overlapping promoter sequences were sorted into three groups according to the expression profiling and ChIP-chip data sets and converted into sequence logos (Figure 3, Table S2). The sequence logos derived from the three groups include: two groups that are preferentially or selectively bound and transcribed by either RpoHI or RpoHII and one group that is bound and transcribed by both σ factors. As noted above, some promoters appear to be bound by RpoHI or RpoHII without inducing detectable changes in transcript levels. We aligned these promoters separately to determine if they possessed unique characteristics, but no significant differences were detected (data not shown).
Figure 3. Conserved promoter sequences recognized by RpoHI and RpoHII.
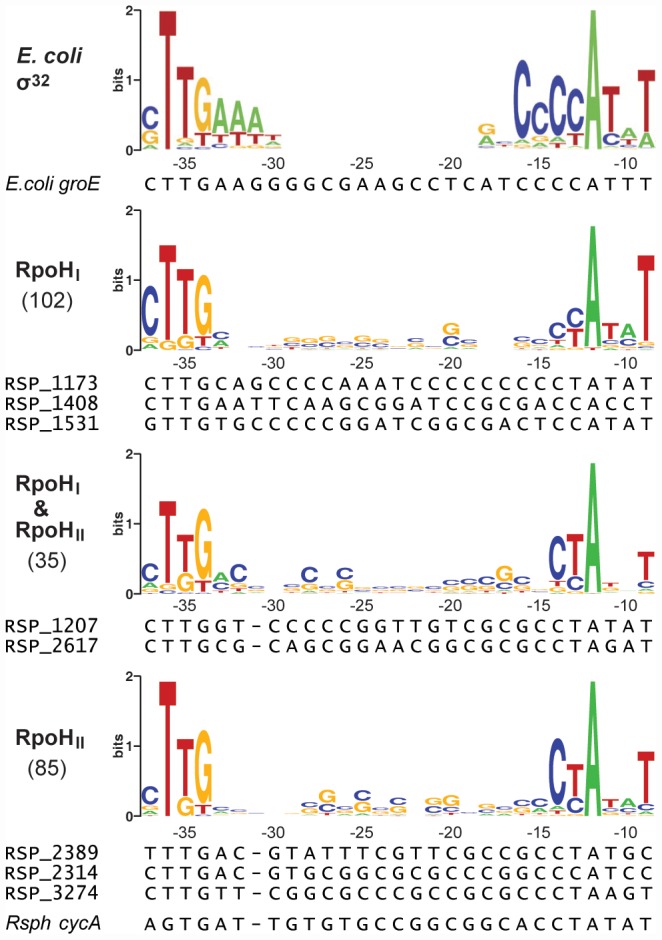
The logos were constructed from promoter sequences alignments sorted into three categories according to their predicted specificity. The consensus sequence for σ32-dependent promoters in E. coli,as determined by Nomaka et al. [37], is shown as a reference. The heights of the letters represent the degree of conservation across sequences (information in bits, logos generated using WebLogo: http://weblogo.berkeley.edu/). The coordinates on the x-axes represent the positions relative to the predicted transcription start site. The numbers of promoter sequences used to create the logos are indicated in parentheses on the left of the logos. Below the logos are the sequence alignments of selected promoters that were used for direct experimental validation.
The conservation of a TTG motif in the −35 region in all three logos is consistent with the importance of this triplet in a previous analysis of at least one promoter known to be recognized by both RpoHI and RpoHII [27]. However, there was also evidence for sequence-specific elements in the logos for each RpoH paralog. In the logo for the RpoHI-dependent promoters, a cytosine is overrepresented at position −37 and a thymine is overrepresented at position −9. In the logo for RpoHII-dependent promoters, cytosine and thymine are overrepresented at positions −14 and −13, respectively.
Overall, the comparison between RpoHI and RpoHII-specific promoter logos allowed us to identify significant differences in the promoter sequences that may be used to adjust promoter selectivity and strength for RpoHI or RpoHII. In addition, the predicted sequence elements for RpoHI or RpoHII promoters are not mutually exclusive. Rather, it appears that promoter specificities for RpoHI or RpoHII are distributed along a gradient using a combination of specific bases at various positions of the −35 or −10 promoter elements.
Degrees of promoter specificity of RpoHI and RpoHII
To test predictions about specificity determinants derived from these logos, we cloned several putative promoters upstream of a lacZ reporter gene and integrated these into the genome of a R. sphaeroides ΔrpoHI ΔrpoHII mutant [15] via homologous recombination. The activity of each promoter was measured by assaying β-galactosidase activity in these R. sphaeroides reporter strains ectopically expressing either RpoHI or RpoHII (Figure 4) at levels comparable to those found during a stress response (see above and Figure 1). The RSP_1173, RSP_1408, and RSP_1531 promoters (which were either predicted to be members of the RpoHI regulon or, in the case of RSP_1173, known to be heat inducible and transcribed by RpoHI [16], had significant activity in the strain expressing RpoHI, but not when the same strain expressed RpoHII (Figure 4). In contrast, the RSP_2314, RSP_2389, and RSP_3274 promoters (which were either predicted to be members of the RpoHII regulon by our analysis or known to be induced by conditions that generate singlet oxygen [17], [18], [20]) showed activity in the presence of RpoHII but not RpoHI (Figure 4). Finally, the RSP_1207 and RSP_2617 promoters (which were predicted to be transcribed by both RpoH proteins and, in the case of RSP_1207, known to be transcribed by RNA polymerase holoenzyme containing either RpoH homolog [15] showed activity in cells containing either RpoHI or RpoHII (Figure 4). Overall, these results support predictions about members of the RpoHI or RpoHII regulons derived by combining the transcription profiling, ChIP-chip and computational analyses.
Figure 4. Relative activities of selected RpoHI- and RpoHII-dependent promoters.
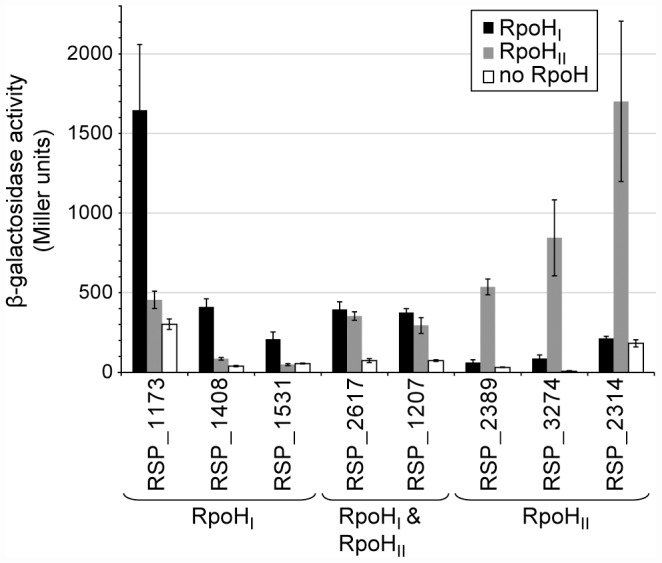
β-galactosidase activity of lacZ operon fusions with selected R. sphaeroides promoter regions monitored in tester strains expressing RpoHI (black), RpoHII (grey), or neither proteins. Genes are grouped according to the gene expression profiles displayed in the gene expression experiments: genes whose expressions were affected only by RpoHI, only by RpoHII, and by both RpoHI and RpoHII. Error bars represents the standard error of the mean from three independent replicates.
To test the predictions about the contributions of individual bases to promoter recognition, we measured the activity of R. sphaeroides RpoHI with an existing library of mutant E. coli groE promoters fused to a lacZ reporter in an E. coli tester strain [7]. The data from this analysis revealed that base substitutions in the TTG motif of the −35 region of this RpoH-dependent promoter (positions −36, −35, and −34) reduced its activity by at least 80% with RpoHI (Figure 5A), as expected from the predictions of promoter logo. We also found a slight increase in promoter activity when position −32 was changed to a cytosine, even though the C-32 is not conserved in RpoHI promoters. This observation is consistent with the results of a previous mutational analysis showing that E. coli σ32 prefers a cytosine at position −32 when the alanine at position 264 of its amino acid sequence is substituted to an arginine (corresponding to R267 of RpoHI) [28], but also suggests that the −32 position is not utilized to distinguish between RpoHI- and RpoHII-specific promoters. In the −10 region of the groE promoter, substitutions of the cytosine at position −14 for an adenine or guanine, the cytosine at position −13 for an adenine, or substitution of the thymine at position −11 for a cytosine, each reduced RpoHI-dependent promoter activity. In addition, a substitution of the adenine at position −12 for a cytosine or changing the thymine at position −9 for any other base reduced RpoHI-dependent activity by >90%. These observations are consistent with the conservation of a thymine at position −9 of the derived RpoHI promoter logo (Figure 3).
Figure 5. Activities of selected mutant promoters when transcribed by RpoHI or RpoHII.
β-galactosidase activity of lacZ operon fusions with selected mutant E. coli (A) or R. sphaeroides (B) promoters monitored in an E. coli tester strain expressing R. sphaeroides RpoHI. The original promoter and specific base substitutions are indicated below the x-axis. (C) β-galatosidase activity of lacZ operon fusions integrated into the genome containing either the wild type of indicated mutant R. sphaeroides cycA P1 promoter in a tester strain expressing the indicated RpoH homolog. Most of the promoter mutations were made in the G-36T cycA P1 background, as this promoter had activity with RpoHI or RpoHII than its wild type (WT) counterpart. Base substitutions are indicated on the x-axis. Error bars represents the standard error of the mean from three independent replicates.
To test the predicted requirement of RpoHI for a thymine at position −9, we also analyzed the properties of two R. sphaeroides promoters in this E. coli tester strain. Activity of the RpoHI-dependent RSP_1531 promoter was reduced by 90% when the thymine at position −9 was changed to a cytosine, whereas the RpoHII-dependent RSP_2314 promoter had higher RpoHI-dependent activity when a thymine was placed at position −9 (Figure 5B). Therefore, this analysis confirmed that position −9 plays a critical role in promoter specificity for RpoHI. In conclusion, the measured effects of mutations in the E. coli groE promoter on RpoHI-dependent transcription confirmed that our models captured elements that are critical for promoter recognition by RpoHI.
We were unable to test activity of R. sphaeroides RpoHII against this groEL promoter library in the same E. coli tester strain (data not shown). Instead, we generated a small set of point mutations in the P1 promoter of the R. sphaeroides cycA promoter (Figure 3) which was previously shown to be transcribed by both RpoHI and RpoHII [27] and measured activity from single-copy fusions of these mutant promoters to lacZ in cells that either lacked both RpoH homologs or that contained a single rpoH gene under control of an IPTG-inducible promoter (Materials and Methods).
By analyzing this promoter library, we found that a G to T mutation at position −36 of cycA P1 (G-36T) increased its transcription by both RpoHI and RpoHII (Figure 5C). This result is consistent with the high predicted information content for T at this position for both RpoHI and HII (Figure 3), as well as the previous observation that the overall increase in activity of cycA P1 is caused by the G-36T mutation [27]. While our RpoHI and RpoHII promoter models (Figure 3) predict that a C could be allowed at position −36, a G-36C mutation lowered activity with RpoHII and had no positive impact on transcription by RpoHI (Figure 5C). Due to the significantly increased in activity from the G-36T mutation in cycA P1, all of the other promoter mutations we tested were generated in this background. Mutations we tested in the −35 region, T-35C and G-34C, resulted in virtually complete loss of cycA P1 activity with either RpoHI and RpoHII when compared to their G-36T parent promoter (Figure 5C), indicating that these bases are essential for transcription initiation by both RpoH homologs. Based on the relatively low information content predicted by our models for other positions in the −35 element (Figure 3), we did not test the effects of other mutations in this region on promoter selectivity by RpoH homologs.
In the predicted −10 region, A-12 has very high information content for both RpoHI and RpoHII, but the sequence logo suggests a T at this position might allow selective recognition by RpoHI (Figure 3). Indeed, a promoter containing a T at position −12 is still active only with RpoHI, suggesting that A-12 is essential for RpoHII activity but not RpoHI activity. The T at position −9 of cycA P1 is also predicted to have significantly higher information content for RpoHI than RpoHII, while a C at this position should have more information content for RpoHII than RpoHI (Figure 3). As predicted, we found RpoHII retained significant activity after placing a T-9C mutation in the context of the G-36T cycA P1 promoter. Furthermore, we found that this mutation completely abolished its activity with RpoHI, illustrating the high information content of a T at this position for transcription by this RpoH homolog. The importance for a T at the analogous position was also observed when testing activity of mutant E. coli groE promoters with RpoHI (T-9C mutation Figure 5A) or assaying function of the R. sphaeroides RSP_1531 promoter (which contains a T, Figure 5B) that is only transcribed to a detectable level by RpoHI (Figure 4). Finally, we also replaced the A at position −10 of the cycA P1 promoter with a G, as the sequence logo suggests there to be little information content at this position for either RpoHI or RpoHII (Figure 3). As predicted, there is little impact of the A-10G mutation on promoter function, though activity with RpoHII is more significantly reduced than that with RpoHI activity (Figure 5C).
Discussion
When organisms encounter environmental or internal stress they often increase the transcription of genes encoding proteins that help mitigate damage to cellular components. Therefore, identifying functions that are involved in transcriptional stress responses is critical to understand both the nature of the damage caused to cellular components and how organisms respond to these challenges. Singlet oxygen and increased temperature are very different phenomena, but in R. sphaeroides the transcriptional responses to these two stresses involve two alternative σ factors, RpoHI and RpoHII, that each belong to the RpoH family [15], [16], [18]. Several other α-proteobacteria contain two or more members of the RpoH family that appear to control different stress responses [13], [29], [30]. However, as it is the case in R. sphaeroides, little is known about the target genes for these multiple RpoH homologs. In this work, we characterized genes that are directly transcribed by R. sphaeroides RpoHI and RpoHII to gain a better understanding of the biological response to heat shock and singlet oxygen stresses. We found that each of these RpoH paralogs control transcription of over 100 genes, suggesting that each of these phenomena lead to large changes in gene expression. However, we also found that there is significant overlap in the RpoHI and RpoHII regulons, creating an unexpectedly extensive connection between the transcriptional responses to these two signals. In addition, we investigated the characteristics of RpoHI- and RpoHII-dependent promoters. This effort allowed us to identify sequence elements that define promoter specificity for each σ factor, thereby allowing cells to selectively partition target genes for each RpoH paralog into different stress responses.
R. sphaeroides RpoHI and RpoHII control the expression of a common set of functions
This work revealed a surprisingly extensive overlap of the RpoHI and RpoHII regulons even though these two homologs activate transcriptional responses to different signals in R. sphaeroides. This suggests that genes activated by these two pathways of the transcriptional regulation network play a role in the physiological response to both these, and even possibly, other stresses. Indeed, the genes regulated by both RpoHI and RpoHII encode known or annotated functions involved in protein homeostasis, DNA repair, and maintenance of cell membrane integrity (Table 1). These types of functions are central to cell viability and may be relevant for the physiological responses to multiple stresses that can have broad primary and secondary effects on cells. Indeed, the predicted functions of the overlapping members of the RpoHI and RpoHII regulons encode functions that are also part of the general stress response regulons for σS in E. coli or σB in Bacillus subtilis [31], [32]. Interestingly, σS homologs are mostly present in β- and γ-proteobacteria, but to date absent from sequenced genomes of α-proteobacteria like R. sphaeroides (http://img.jgi.doe.gov/) [33]. Thus, it is possible that the set of genes controlled by both RpoHI and RpoHII is part of a general stress response that is common to the heat shock, singlet oxygen and possibly other uncharacterized signals in R. sphaeroides [14], [15], [17], [18], [20]. This hypothesis is supported by the observation that R. sphaeroides and R. elti strains lacking both RpoHI and RpoHII are more sensitive to several conditions than strains lacking only one of these proteins [13], [15], [20].
In considering the scope of functions that are regulated by both RpoHI and RpoHII, it is also important to note that this set of genes may be larger than the one we characterized because some promoters known to be transcribed by both σ factors were only marginally affected by ectopic expression of either RpoHI or RpoHII. For example, the RSP_2310 (groES) promoter was shown to be transcribed by both RpoHI and RpoHII in previous in vitro experiments [14] and was detected by our ChIP-chip experiment to be bound by both RpoHI and RpoHII, but did not meet all the criteria of our analysis. Thus, the groES promoter, like other promoters, may be subject to complex regulation in vivo.
RpoHI and RpoHII each control functions specific to heat shock or singlet oxygen stresses, respectively
Our data also significantly extend the number and types of functions that are specifically controlled by RpoHI or RpoHII (Table 1). We expected to find specific sets of target genes because strains lacking either RpoHI or RpoHII displayed different phenotypes [14], [15], [17], [20]. While previous results indicated that accumulation of ∼25 proteins was dependent on RpoHII [17], our data indicate that some 150 genes are directly controlled by each R. sphaeroides RpoH paralog.
Genes in the direct but RpoHI-specific regulon encode functions that are involved in protein homeostasis, maintaining membrane integrity, and DNA repair, as is found for the E. coli σ32 regulon [3] (Table 1) The RpoHI specific regulon is also predicted to encode cation transporters and proteins in the thioredoxin-dependent reduction system (Table 1). Ion transporters can aid the heat shock stress response since exporting cations like iron, which may be released by thermal denaturation of damaged iron-sulfur or other metalloproteins, decreases secondary effects caused by formation of toxic reactive oxygen species [34]. The thioredoxin-dependent reduction system reduces disulfide bonds and peroxides, which are created by protein oxidation, and thereby helps maintain cytoplasmic proteins in a reduced state [35]. Inclusion of these functions in the RpoHI regulon suggests that oxidative damage may be an important secondary effect of heat shock, perhaps caused by protein denaturation or permeabilization of the cell envelope. Overall, these results support the hypothesis that the function of RpoHI in R. sphaeroides is similar to that of σ32 in E. coli for the response to heat shock stress. In addition, it is also possible that RpoHI plays a role in the R. sphaeroides response to other forms of stress. There is precedent for roles of σ32 homologs in other stress responses by other bacteria since the activity of RpoH in Caulobacter crescentus is increased by heavy metal stress [36].
In contrast, rpoHII transcription is under direct control of a Group IV alternative σ factor (RpoE) that serves as the master regulator of the singlet oxygen stress response [18]. In addition, an R. sphaeroides ΔrpoHII mutant is more sensitive to singlet oxygen than a wild-type or ΔrpoHI strain [15], [17]. Therefore, members of the direct RpoHII-specific regulon might be expected to play an important role in the response to singlet oxygen stress. Among the genes in the RpoHII-specific regulon are others predicted to function in maintaining membrane integrity and performing DNA repair, both potential targets for damage by singlet oxygen. However, the RpoHII–specific regulon contains fewer genes encoding functions related to protein homeostasis than found in the RpoHI regulon (Table 1). Other functions apparently unique to the RpoHII regulon include the glutathione-dependent reduction system, which like the thioredoxin-dependent system repair oxidized protein residues and maintain a reduced cytoplasm (Table 1). Even though the thioredoxin- and gluthatione-dependent reduction systems serve similar cellular functions, they are apparently under the control of different RpoH-dependent transcriptional networks in R. sphaeroides. Thus, it is possible that the thioredoxin- and gluthatione-dependent reduction systems preferentially function on different oxidized substrates. Glutathione-dependent reduction systems are known to function on lipids or other types of protein oxidative damage that might be experienced by the cell following singlet oxygen damage [35]. We also found that the RpoHII-specific regulon includes the multi-subunit NADH:quinone oxidoreductase and genes encoding enzymes in heme and quinone biosynthesis (Table 1). Each of these functions are critical for the respiratory and photosynthetic electron transport chains of R. sphaeroides and are known or predicted to contain one or more oxidant-sensitive metal centers. Thus, placement of these genes in the RpoHII-specific regulon suggests that these membrane or bioenergetic functions are damaged by and need to be replaced in the presence of singlet oxygen. Overall, our data indicates that the RpoHII-specific regulon controls expression of functions in the repair of oxidized proteins and replacement or assembly of critical electron transport chain components. Furthermore, the different types of repair functions found in the RpoHII regulon predict that singlet oxygen can damage numerous cellular components.
RpoHI and RpoHII recognize different but compatible promoter sequence elements
Our global gene expression data, results from analysis of gene fusions, as well as previously reported in vitro experiments [14], [15] all indicate that RNA polymerase containing either RpoHI or RpoHII can recognize some promoters in common. This observation is not surprising considering that RpoHI and RpoHII have similar amino acid sequences in their respective promoter recognition regions and are each able to rescue growth of E. coli σ32 mutants [14]–[16]. Likewise, the sequence logos derived here revealed that the promoter sequences recognized by each of the R. sphaeroides RpoH homologs are similar to both each other and to that recognized by E. coli σ32 [37].
Our experiments provide definitive evidence that some promoters are transcribed either exclusively or predominantly by RpoHI or by RpoHII. We were also able to predict and confirm the importance of bases for activity with individual RpoH homologs (particularly those in the −35 element). We have computational and experimental observations that can explain some aspects of promoter selectivity by RpoHI and RpoHII. For example, our experiments identify T-9 and other positions in the −10 element as potential candidates in this discrimination, as one or more substitutions have larger effects on activity with individual RpoHI homologs. Mutation of T-9 to any other base reduced RpoHI-driven expression of GroE promoter by more than 90%, and this same effect was observed using an authentic RpoHI promoter from R. sphaeroides. Importantly, changing the −9 position of an RpoHII R. sphaeroides promoter to T permitted expression by RpoHI. Together, these data suggest that T-9 is either required for or significantly enhances expression of RpoHI promoters, but is likely to be less important for expression of RpoHII promoters, as there is only weak conservation of -9T in RpoHII promoters. Our data also predict that other bases, which are overrepresented in the RpoHII promoters, could be critical for expression by that σ factor. As is the case with E. coli σ32 there are likely to be specificity determinants that lie outside the canonical −35 and −10 elements [7], [37]. Thus, additional in vivo and in vitro experiments with a larger suite of mutant promoters and a library of mutant RpoH proteins are needed to better define the determinants of promoter selectivity by RpoHI and RpoHII.
In conclusion, our results suggest that, at least in R. sphaeroides, RpoHI controls functions that are necessary for maintenance of protein homeostasis and membrane integrity after temperature increase and other cytoplasmic stress, similar to the well-characterized role of E. coli σ32 in the heat shock response [3]. However, we propose that, in R. sphaeroides, some RpoHI-regulated functions are also useful for survival in the presence of other forms of stress because these target genes also contain promoters that are recognized by RpoHII. We propose that the duplication of an ancestral RpoH protein to create a second homolog of this alterative σ factor provided R. sphaeroides the opportunity to connect stress response functions to another stimulus. In this model, rpoHII was placed under the control of the master regulator of the singlet oxygen stress response and the two RpoH proteins evolved to recognize somewhat different but compatible promoter elements to assure the optimal regulation of distinct but overlapping stress regulons. As a result of these events, the transcriptional responses of R. sphaeroides to heat shock and singlet oxygen stress were separable but allowed to converge and contain a common set of functions. It will be interesting to identify and examine other examples of such convergence across bacteria and other organisms that possess multiple homologs of RpoH or other transcription factors.
Materials and Methods
Bacterial strains and growth conditions
E. coli strains were grown in Luria-Bertani medium [38] at 30°C or 37°C. R. sphaeroides strains were grown at 30°C in Sistrom's succinate-based medium [39]. E. coli DH5α was used as a plasmid host, and E. coli S17-1 was used as a donor for plasmid conjugation into R. sphaeroides. The media were supplemented with kanamycin (25 µg/ml), ampicillin (100 mg/ml), chloramphenicol (30 mg/ml), spectinomycin (50 mg/ml), tetracycline (10 mg/ml for E. coli and 1 mg/ml for R. sphaeroides), trimethoprim (30 µg/ml), or 0.1% of L-(+)-arabinose when required. Unless noted, all reagents were used according to the manufacturer's specifications. The list of bacterial strains and plasmids used in this study are summarized in Table S3.
Construction of plasmids for controlled expression of RpoHI and RpoHII in R. sphaeroides
Plasmids for ectopically expressing RpoHI or RpoHII were constructed by separately cloning the rpoHI or rpoHII genes downstream of the IPTG-inducible promoter in pIND4 [22]. DNA fragments containing rpoHI or rpoHII were amplified from R. sphaeroides 2.4.1 genomic DNA using oligonucleotides containing BsrDI and BglII restriction sites (for RpoHII, RSP_0601_BsrDI_F GTAGCAATGCATGGCACTGGACGGATATACCGATC, RSP_0601_BglII_R GTAAGATCTTCATAGGAGGAAGTGATGCACCTCC, and for RpoHI, RSP_2410_BsrDI_F GTAGCAATGCATGAGCACTTACACCAGCCTTC, and RSP_2410_BglII_R GTAAGATCTTCAGGCGGGGATCGTCATGCC). These resulting fragments were digested with BsrDI and BglII and ligated into pIND4 that was digested with BseRI and BglII to create pYSD40 (rpoHI) and pYSD41 (rpoHII), respectively. The pYSD42 plasmid expressing the FLAG-tagged version of RpoHI was constructed following the same procedure but with an oligonucleotide primer containing a sequence encoding for three consecutive copies of the FLAG epitope (DYKDDDDK) at the N-terminus (RSP_2410_3FLAG_BsrDI GTAGCAATGCATGGACTACAAGGACCACGACGGCGACTACAAGGACCACGACATCGACTACAAGGACGACGACGACAAGAGCACTTACACCAGCCTTCCCGCTC). pYSD40, pYSD41, and pYSD42 were conjugated into R. sphaeroides ΔrpoHI [16] and R. sphaeroides ΔrpoHII respectively.
Western blot analysis for the expression of RpoHI and RpoHII
To monitor levels of RpoHI and RpoHII after heat shock, exponential phase aerobic cultures (69% nitrogen, 30% oxygen and 1% carbon dioxide) of wild type R. sphaeroides strain 2.4.1 grown at 30°C, were transferred to a 42°C warm bath with samples collected before heat treatment and at 10 min time intervals after heat shock, up to 60 min. To assess induction resulting from singlet oxygen stress, similarly grown wild type cells were treated with 1 µM methylene blue and exposed to 10 W/m2 incandescent light with samples collected before treatment and at 10 min time intervals after treatment, up to 60 min. Exponentially growing aerobic cultures of R. sphaeroides ΔrpoHI and ΔrpoHII mutants carrying the pYSD40 or pYSD42 plasmids respectively, were treated with 100 µM IPTG for one generation and harvested. All cell samples were resuspended in 3 M urea containing 1× protease inhibitor cocktail (Thermo Scientific, Rockford, IL) and sonicated. Samples were centrifuged to remove debris and total protein concentration of the samples determined with a protein assay kit following the manufacturer protocol (Bio-Rad, Hercules, CA). An equal amount of total protein for each sample was loaded onto a NuPAGE acrylamide gel (Invitrogen, Carlsbad, CA) and run in 1× 4-morpholineethanesulfonic acid running buffer at 150 V for ∼90 min. Proteins were transferred to Invitrolon PVDF membranes (Invitrogen, Carlsbad, CA), which were subsequently incubated for 1 hr in 1× Tris-buffered saline, 0.1% Triton-X, and 5% milk protein. The membranes were incubated with rabbit polyclonal antibodies raised against either RpoHI, RpoHII or PrrA. Horseradish-peroxidase-conjugated goat anti-Rabbit IgG antibody (Thermo Scientific, Rockford, IL) was used as secondary antibody for detection with Super Signal West Dura extended duration substrate (Thermo Scientific, Rockford, IL).
Gene expression microarrays
Triplicate 500 ml cultures were grown aerobically with bubbling (30%O2, 69% N2, 1% CO2) until they reached early exponential phase (OD at 600 nm of 0.15). At this point IPTG (Isopropyl β-D-1-thiogalactopyranoside) was added to a final concentration of 100 µM to induce gene expression from the pIND4 derivatives. After 3 hours incubation (OD at 600 nm of 0.30), 44 ml of cell culture were collected and 6 ml of 5% v/v phenol in ethanol was immediately added. Cells were collected by centrifugation at 6,000 g and frozen at −80°C until sample preparation. RNA extraction, cDNA synthesis, labeling, and hybridization were performed as previously described on Genechip Custom Express microarrays (Affymetrix, Santa Clara, CA) [40]. Processing, normalization, and statistical analysis of the expression profile data were performed in the R statistical software environment (http://www.r-project.org/) [41]. Data were normalized using the affyPLM package with default settings [42]–[44]. The expression microarray data have been deposited in the NCBI's Gene Expression Omnibus [45] and are accessible through GEO Series accession number GSE39806 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE39806).
Chromatin immunoprecipitation on a chip
Cells were harvested at mid-exponential growth (OD at 600 nm of 0.30) from the same cell cultures used for the expression microarray experiment to prepare samples for a ChIP-chip assay [19]. RpoHI-FLAG was immunoprecipitated using commercial monoclonal antibodies against the FLAG polypeptide (DYKDDDDK ) (Sigma Aldrich, St Louis MO). RpoHII was immunoprecipitated with anti-R. sphaeroides RpoHII rabbit serum. Labeled DNA was hybridized on a custom-made tiling microarray, synthesized by NimbleGen (Roche NimbleGen Inc, Madison, WI), covering the genome of R. sphaeroides 2.4.1 [19]. Before data analysis, dye intensity bias and array-to-array absolute intensity variations were corrected using quantile normalization across replicates (limma package in the R environment) [46]. Regions of the genome enriched for occupancy by RpoHI or RpoHII were identified using CMARRT with a false-discovery rate ≤0.05 [47]. The ChIP-chip data have been deposited in the NCBI's Gene Expression Omnibus [45] and are accessible through GEO Series accession number GSE39806 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE39806).
Sequence analyses
DNA sequences were manipulated using custom Python scripts. Operon structure predictions for R. sphaeroides 2.4.1 were obtained from VIMSS MicrobesOnline (http://www.microbesonline.org/operons/) [26]. The promoter sequences predicted to be recognized by RpoHI and RpoHII were discovered using Bioprospector [48] set to search for bi-partite conserved sequence motifs. The promoter sequence alignments were refined using HMMER 1.8.5 [49]. The logo representations of the promoter sequence alignment were generated using WebLogo (http://weblogo.berkeley.edu/) [50], [51].
Construction of plasmid vectors, lacZ reporter promoter fusions, and β-galactosidase assays to assay promoter activity in vivo
To assay the in vivo activity of RpoHI and RpoHII at target promoters, β-galactosidase assays were conducted with R. sphaeroides ΔrpoHI ΔrpoHII mutant strains containing individual reporter gene fusions. To construct this set of reporter strains ∼350 base pair regions upstream of putative target genes: RSP_1173, RSP_1408, RSP_1531, RSP_2314, RSP_2389, RSP_3274, RSP_1207 and RSP_2617, were amplified from genomic DNA using sequence specific primers, with NcoI and XbaI restriction sites at the ends of the upstream and downstream primers respectively. The amplified DNA fragments were purified, digested with NcoI-XbaI and then cloned in a pSUP202 suicide vector containing a promoterless lacZ gene (pYSD51). These Tcr plasmids were then conjugated into an R. sphaeroides ΔrpoHI ΔrpoHII mutant [15], generating single copy promoter-lacZ fusions integrated in the genome. pYSD40, pYSD41 or pIND4 (empty vector) were then conjugated into each of these reporter strains. Exponential phase cultures of these strains, grown by shaking 10 mL in 125 mL conical flasks, were then treated with 100 µM IPTG for one generation and samples analyzed for β-galactosidase activity. β-galactosidase assays were performed as previously described [52]. The data, presented in Miller units, represents the average of three independent replicates.
To test bases that contribute to RpoHI and RpoHII promoter specificity, β-galactosidase assays were conducted in R. sphaeroides tester strains containing reporter gene fusions of the cycA (RSP_0296) P1 promoter with a variety of point mutations (see Results). These reporter strains were constructed as described above, with individual point mutations being generated by overlap extension PCR [53]. β-galactosidase assays were conducted as described above and the data represents the average of three independent replicates. Background LacZ activity from control strains for each promoter fusion containing only the empty pIND4 plasmid (i.e. not expressing either RpoHI or RpoHII) was subtracted from the measured LacZ activity for each mutant promoter.
The construction of the E. coli CAG57102 mutant strain, the promoter library, and the β-galactosidase assay used to test the activity of R. sphaeroides RpoHI in vivo on mutant promoters were described previously [7]. To express R. sphaeroides RpoHI the E. coli rpoH gene of pSAKT32 [7] was replaced with the R. sphaeroides rpoHI gene. At least triplicate assays for β-galactosidase activity were performed on all strains.
Supporting Information
Scatter plot of RpoHI versus FLAG-RpoHI dependent change in gene transcription levels.
(TIF)
RpoHI and RpoHII target genes.
(XLS)
Predicted promoter sequences of RpoHI and RpoHII target genes.
(XLS)
Bacterial strains and plasmids.
(XLS)
Funding Statement
This work was supported by NIGMS grants GM075273 to TJD and GM057755 to Carol A. Gross (University of California at San Francisco). YSD and SI were each fellows on the BACTER Grant from the U.S. Department of Energy Genomics: GTL and SciDAC Programs (DE-FG02-04ER25627). YSD was a recipient of a Wisconsin Distinguished Graduate Fellowship from the University of Wisconsin–Madison College of Agricultural and Life Sciences and of the William H. Peterson Predoctoral Fellowship from the University of Wisconsin–Madison Department of Bacteriology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, et al. (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol [DOI] [PubMed] [Google Scholar]
- 2. Paget M, Helmann J (2003) The σ70 family of sigma factors. Genome Biology 4: 203.201–203.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guisbert E, Yura T, Rhodius VA, Gross CA (2008) Convergence of Molecular, Modeling, and Systems Approaches for an Understanding of the Escherichia coli Heat Shock Response. Microbiol Mol Biol Rev 72: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arsene F, Tomoyasu T, Mogk A, Schirra C, Schulze-Specking A, et al. (1999) Role of region C in regulation of the heat shock gene-specific sigma factor of Escherichia coli, σ32 . J Bacteriol 181: 3552–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joo DM, Nolte A, Calendar R, Zhou YN, Jin DJ (1998) Multiple regions on the Escherichia coli heat shock transcription factor σ32 determine core RNA polymerase binding specificity. J Bacteriol 180: 1095–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wosten M (1998) Eubacterial sigma-factors. FEMS Microbiology Reviews 22: 127–150. [DOI] [PubMed] [Google Scholar]
- 7. Koo BM, Rhodius VA, Campbell EA, Gross CA (2009) Dissection of recognition determinants of Escherichia coli σ32 suggests a composite −10 region with an ‘extended −10’ motif and a core −10 element. Mol Microbiol 72: 815–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delory M, Hallez R, Letesson JJ, De Bolle X (2006) An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16 M. J Bacteriol 188: 7707–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ono Y, Mitsui H, Sato T, Minamisawa K (2001) Two RpoH homologs responsible for the expression of heat shock protein genes in Sinorhizobium meliloti . Molecular and General Genetics 264: 902–912. [DOI] [PubMed] [Google Scholar]
- 10. Oke V, Rushing BG, Fisher EJ, Moghadam-Tabrizi M, Long SR (2001) Identification of the heat-shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti . Microbiology-Sgm 147: 2399–2408. [DOI] [PubMed] [Google Scholar]
- 11. Narberhaus F, Krummenacher P, Fischer HM, Hennecke H (1997) Three disparately regulated genes for σ32-like transcription factors in Bradyrhizobium japonicum . Molecular Microbiology 24: 93–104. [DOI] [PubMed] [Google Scholar]
- 12. Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, et al. (2002) Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9: 189–197. [DOI] [PubMed] [Google Scholar]
- 13. Martinez-Salazar JM, Sandoval-Calderon M, Guo XW, Castillo-Ramirez S, Reyes A, et al. (2009) The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology-Sgm 155: 386–397. [DOI] [PubMed] [Google Scholar]
- 14. Green HA, Donohue TJ (2006) Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J Bacteriol 188: 5712–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green H (2007) The function of two alternate sigma factors, RpoHI and RpoHII, in Rhodobacter sphaeroides [3278859]. United States – Wisconsin: The University of Wisconsin - Madison. 157 p.
- 16. Karls RK, Brooks J, Rossmeissl P, Luedke J, Donohue TJ (1998) Metabolic roles of a Rhodobacter sphaeroides member of the σ2 family. J Bacteriol 180: 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nuss AM, Glaeser J, Klug G (2009) RpoHII activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides . J Bacteriol 191: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anthony JR, Warczak KL, Donohue TJ (2005) A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A 102: 6502–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dufour YS, Landick R, Donohue TJ (2008) Organization and evolution of the biological response to singlet oxygen stress. J Mol Biol 383: 713–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nuss AM, Glaeser J, Berghoff BA, Klug G (2010) Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides . J Bacteriol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dufour YS, Kiley PJ, Donohue TJ (2010) Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet 6: e1001027 doi:10.1371/journal.pgen.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ind AC, Porter SL, Brown MT, Byles ED, de Beyer JA, et al. (2009) Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans . Appl Environ Microbiol 75: 6613–6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Comolli J, Carl A, Hall C, Donohue TJ (2002) Transcriptional activation of the Rhodobacter sphaeroides cytochrome c 2 gene P2 promoter by the response regulator PrrA. J Bacteriol 184: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El-Samad H, Kurata H, Doyle JC, Gross CA, Khammash M (2005) Surviving heat shock: control strategies for robustness and performance. Proc Natl Acad Sci U S A 102: 2736–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Einhauer A, Jungbauer A (2001) The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J Biochem Biophys Methods 49: 455–465. [DOI] [PubMed] [Google Scholar]
- 26. Price MN, Huang KH, Alm EJ, Arkin AP (2005) A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res 33: 880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacGregor BJ, Karls RK, Donohue TJ (1998) Transcription of the Rhodobacter sphaeroides cycA P1 promoter by alternate RNA polymerase holoenzymes. J Bacteriol 180: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kourennaia OV, Tsujikawa L, Dehaseth PL (2005) Mutational analysis of Escherichia coli heat shock transcription factor σ32 reveals similarities with sigma 70 in recognition of the −35 promoter element and differences in promoter DNA melting and −10 recognition. J Bacteriol 187: 6762–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tittabutr P, Payakapong W, Teaumroong N, Boonkerd N, Singleton PW, et al. (2006) The alternative sigma factor RpoH2 is required for salt tolerance in Sinorhizobium sp. strain BL3. Res Microbiol 157: 811–818. [DOI] [PubMed] [Google Scholar]
- 30. Kaufusi PH, Forsberg LS, Tittabutr P, Borthakur D (2004) Regulation of exopolysaccharide synthesis in Rhizobium sp. strain TAL1145 involves an alternative sigma factor gene, rpoH2 . Microbiology 150: 3473–3482. [DOI] [PubMed] [Google Scholar]
- 31. Hecker M, Pane-Farre J, Volker U (2007) SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annual Review of Microbiology 61: 215–236. [DOI] [PubMed] [Google Scholar]
- 32. Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R (2005) Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. Journal of Bacteriology 187: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, et al. (2009) The integrated microbial genomes system: an expanding comparative analysis resource. Nucl Acids Res gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Touati D (2000) Iron and oxidative stress in bacteria. Arch Biochem Biophys 373: 1–6. [DOI] [PubMed] [Google Scholar]
- 35. Carmel-Harel O, Storz G (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annual Review of Microbiology 54: 439–461. [DOI] [PubMed] [Google Scholar]
- 36. McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, et al. (2007) High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol 25: 584–592. [DOI] [PubMed] [Google Scholar]
- 37. Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA (2006) Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev 20: 1776–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell DW (2006) The condensed protocols from Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. v, 800 p. p.
- 39. Sistrom WR (1960) A requirement for sodium in the growth of Rhodopseudomonas sphaeroides . J Gen Microbiol 22: 778–785. [DOI] [PubMed] [Google Scholar]
- 40. Tavano CL, Podevels AM, Donohue TJ (2005) Identification of genes required for recycling reducing power during photosynthetic growth. J Bacteriol 187: 5249–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team (2010) R: A Language and Environment for Statistical Computing. Vienna, Austria.
- 42.Bolstad BM (2004) Low Level Analysis of High-density Oligonucleotide Array Data: Background, Normalization and Summarization. Berkeley: University of California - Berkeley.
- 43. Brettschneider J, Collin F, Bolstad BM, Speed TP (2008) Quality Assessment for Short Oligonucleotide Microarray Data. Technometrics 50: 241–264. [Google Scholar]
- 44.Gentleman R (2005) Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer. xix, 473 p. p.
- 45. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth GK (2005) Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer. pp. 397–420.
- 47. Kuan PF, Chun H, Keles S (2008) CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput 515–526. [PMC free article] [PubMed] [Google Scholar]
- 48. Liu X, Brutlag DL, Liu JS (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138. [PubMed] [Google Scholar]
- 49. Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- 50. Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schilke BA, Donohue TJ (1995) ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c 2 gene. J Bacteriol 177: 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plot of RpoHI versus FLAG-RpoHI dependent change in gene transcription levels.
(TIF)
RpoHI and RpoHII target genes.
(XLS)
Predicted promoter sequences of RpoHI and RpoHII target genes.
(XLS)
Bacterial strains and plasmids.
(XLS)



