Abstract
Background
microRNAs (miRNAs) have been found to play an essential role in the modulation of numerous biological processes in eukaryotes. Chlamydomonas reinhardtii is an ideal model organism for the study of many metabolic processes including responses to sulfur-deprivation. We used a deep sequencing platform to extensively profile and identify changes in the miRNAs expression that occurred under sulfur-replete and sulfur-deprived conditions. The aim of our research was to characterize the differential expression of Chlamydomonas miRNAs under sulfur-deprived conditions, and subsequently, the target genes of miRNA involved in sulfur-deprivation were further predicted and analyzed.
Results
By using high-throughput sequencing, we characterized the microRNA transcriptomes under sulphur-replete and sulfur-deprived conditions in Chlamydomonas reinhardtii. We predicted a total of 310 miRNAs which included 85 known miRNAs and 225 novel miRNAs. 13 miRNAs were the specific to the sulfur-deprived conditions. 47 miRNAs showed significantly differential expressions responding to sulfur-deprivation, and most were up-regulated in the small RNA libraries with sulfur-deprivation. Using a web-based integrated system (Web MicroRNAs Designer 3) and combing the former information from a transcriptome of Chlamydomonas reinhardtii, 22 miRNAs and their targets involved in metabolism regulation with sulfur-deprivation were verified.
Conclusions
Our results indicate that sulfur-deprivation may have a significant influence on small RNA expression patterns, and the differential expressions of miRNAs and interactions between miRNA and its targets might further reveal the molecular mechanism responding to sulfur-deprivation in Chlamydomonas reinhardtii.
Background
Sulfur is an essential trace element for all organisms, and is widely used in biochemical processes. Many enzymes and antioxidant molecules such as glutathione contains sulfur. Organically bonded sulfur is a component of all proteins, in the amino acids cysteine and methionine. Generally, sulfate is the most stable form of sulfur. The available pools of sulfate can vary significantly as environmental conditions change. Most organisms have a limited capacity to store sulfur, and thus require different strategies to optimize sulfur use for survival. The ability of microbes to acclimate to periods of nutrient insufficiency is essential to their survival in the natural environment [1]. The unicellular green alga Chlamydomonas reinhardtii is an ideal model organism for the study of many metabolic processes including response to sulfur-deprivation. Sulfur-deprived Chlamydomonas cells have been used for microarray-based RNA abundance studies [2,3], RNA-seq analysis [4], determination of metabolite profiles [5], and sustained production of H2 [6-8]. Chlamydomonas reinhardtii exhibits several responses to sulfur deprivation, including changes of the photosynthetic apparatus, the synthesis of enzymes, cell wall structure, SO42- transport activity, and cell size [4].
Recently, there has been considerable interest in understanding the impacts of sulfur deprivation on miRNA. miRNAs were first found in Caenorhabditis elegans through forward genetic screens of the lin-4 and let-7 mutants [9,10]. Since then, genetic studies on various organisms have revealed that miRNAs are universally present and are key components of various gene regulatory pathways in eukaryotes. Chlamydomonas miRNAs were discovered independently by two groups [11,12], showing that miRNAs exist not only in multicellular systems but also in unicellular eukaryotes. Experimental approaches and bioinformatics-assisted screening have identified approximately 85 Chlamydomonas reinhardtii miRNAs, which are listed in the miRBase version15.0 http://microrna.sanger.ac.uk/s-equences/index.shtml. To investigate the role of Chlamydomonas reinhardtii miRNAs involved in sulfur deprivation, we used a deep sequencing platform to extensively profile and identify changes in the miRNAs expression that occur during sulfur-replete and sulfur-deprived conditions.
Results
The small RNA profile of Chlamydomonas reinhardtii in both sulfur-replete and sulfur-deprived conditions
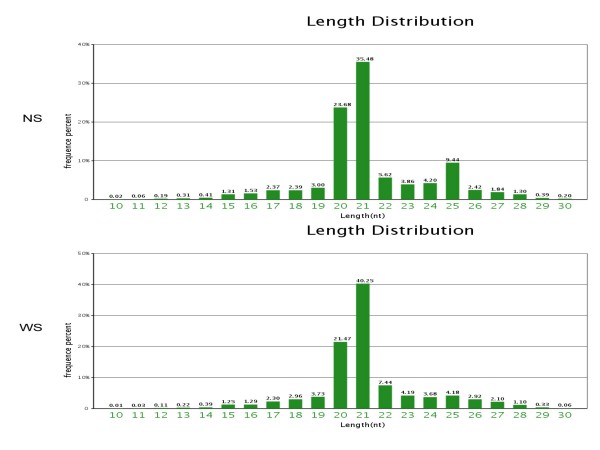
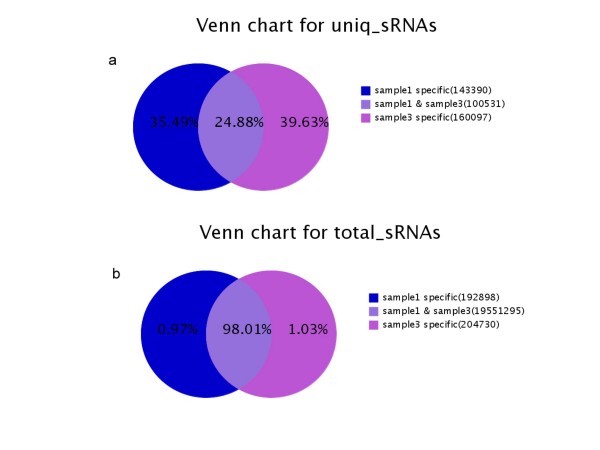
The equal numbers of algal cells were resuspended under continuous illumination for up to 72 h in TAP (with sulfate 40.55 mg/L) or TAP-S (the sulfate concentration was less than 0.42 mg/L). Two small RNA libraries were constructed using the algal cells under sulphur-replete (+S library) and with sulfur-deprived conditions (-S library) respectively. Sequencing of the Chlamydomonas reinhardtii small RNA libraries was performed with Solexa high-throughput sequencing. We obtained 11,080,539 reads from the sulfur-deprived (-S) library and 11,284,767 reads from the sulfur-replete (+S) library. After discarding low quality and shorter than 18 (nt) sequences, 9,918,931(-S library) and 10,029,992 (+S library) clean reads ranging from 18 to 30 nt were collected (Table 1). The majority of the small RNA sequences obtained from the -S and + S libraries were 20-25 nt in size, which was the typical size range for Dicer derived products (Figure 1). Comparing the common small RNAs sequences between -S and + S libraries, although the total small RNAs sequences in both libraries reached 98.01%, the common categories of unique small RNAs were only 24.88%. It indicates that sulfur-deprivation led to a significant influence on the small RNA expression patterns in Chlamydomonas reinhardtii (Figure 2).
Table 1.
Statistics of small RNA sequences from Chlamydomonas reinhardtii + S and -S libraries
| Sequences generated | % | |
|---|---|---|
| +S library | ||
| Total reads | 11284767 | |
| High quality | 10762852 | 100% |
| Adaptor3 null | 3304 | 0.03% |
| Insert null | 2815 | 0.03% |
| Adaptor5 contaminants | 63608 | 0.59% |
| Smaller than 18 nt | 663116 | 6.16% |
| PolyA | 17 | 0.00% |
| Clean reads | 10029992 | 93.19% |
| -S library | ||
| Total reads | 11080539 | |
| High quality | 10572756 | 100% |
| Adaptor3_null | 3499 | 0.03% |
| Insert null | 3575 | 0.03% |
| Adaptor5 contaminants | 58280 | 0.55% |
| Smaller than 18 nt | 588466 | 5.57% |
| PolyA | 6 | 0.00% |
| Clean reads | 9918931 | 93.82% |
Total reads: total sequenced reads; High quality: number of high quality reads with no N, no more than 4 bases whose quality score is lower than 10 and no more than 6 bases whose quality is lower than 13; Adaptor3 null: number of reads with no 3' adaptor; Insert null: number of reads with no insertion; Adaptor5 contaminants: number of 5' contaminants; Smaller than 18 nt: number of reads less than 18 nt. Generally, small RNA tags are between 18 ~ 30 nt long. So too short tags should be removed from data for future analysis; polyA: number of reads wih polyA; Clean reads: number of clean reads after adaptors and contaminants are removed which are used in the following analysis.
Figure 1.
The size distribution of small RNAs in both + S (NS) and -S (WS) libraries.
Figure 2.
Summary of the common and specific tags of small RNAs in + S (sample 1) and -S (sample 3) libraries, including the summary of unique tags (a) and total tags (b). (a) sample1 specific: number of unique sample 1 (+S) specific sRNAs and the percentage; sample3 specific: number of unique sample 3 (-S) specific sRNAs and the percentage; sample1 & sample3: number of unique common sRNAs between two samples and percentage. (b) sample1 specific: number of total sample1 (+S) specific sRNAs and the percentage; sample3 specific: number of total sample3 (-S) specific sRNAs and the percentage; sample1 & sample3: number of total common sRNAs between two samples and percentage.
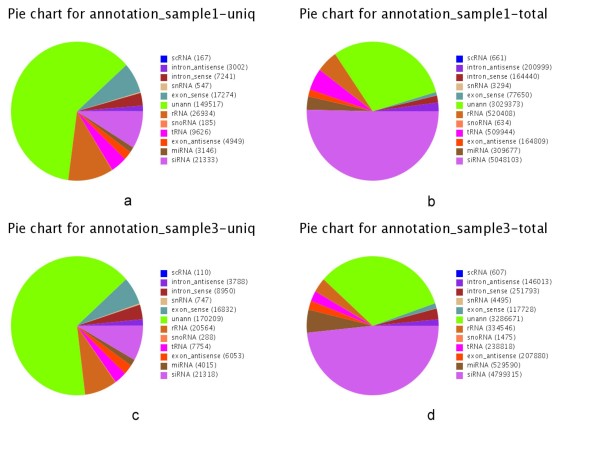
The 20-24 nt sequences from the + S and -S libraries were aligned to the draft Chlamydomonas reinhardtii genome using SOAP [13]. A total of 4,598,243 (+S) and 6,039,480 (-S) sequences were found to match the genome perfectly (see Additional file 1: Figure S1). These small RNAs were used for further analysis. With the alignment to Genbank, Rfam, Exon and Intron, the composition of small RNAs were annotated as siRNA, miRNA, snRNA, snoRNA etc. (Figure 3). Among all the categories of small RNAs, siRNAs expression (small interfering RNA) did not change on sulfur deprivation. However, the microRNAs expression showed more significant up-regulation in response to sulfur-deprivation (Table 2).
Figure 3.
Small RNA annotation of + S (a, b) and -S (c, d) libraries. Charts a and c show the numbers of unique sRNA tags matched to all categories of RNA; charts b and d show the number of total sRNA tags matched to all categories of RNA.
Table 2.
Composition of the small RNAs in the + S and -S libraries
| Unique sRNA (percentage values) | Total sRNA (percentage values) | |||
|---|---|---|---|---|
| +S | -S | +S | -S | |
| Total | 243921 | 260628 | 10029992 | 9918931 |
| Exon antisense | 4949 (2.02) | 6053 (2.3) | 164809 (1.6) | 207880 (2.09) |
| Exon sense | 17274 (7.08) | 16832 (6.4) | 77650 (0.7) | 117728 (1.1) |
| Intron antisense | 3002 (1.2) | 3788 (1.46) | 200999 (2.0) | 146013 (1.4) |
| Intron sense | 7241 (2.9) | 8950 (3.4) | 164440 (1.63) | 251793 (2.5) |
| miRNA | 3146 (1.2) | 4015 (1.5) | 309677 (3.0) | 529590 (5.3) |
| rRNA | 26934 (11.0) | 20564 (7.8) | 520408 (5.1) | 334546 (3.3) |
| scRNA | 167 (0.06) | 110 (0.04) | 661 (0.00) | 607 (0.00) |
| siRNA | 21333 (8.7) | 21318 (8.1) | 5048103 (50.3) | 4799315 (48.3) |
| snRNA | 547 (0.2) | 747 (0.2) | 3294 (0.032) | 4495(0.04) |
| snoRNA | 185 (0.07) | 288 (0.11) | 634 (0.00) | 1475 (0.01) |
| tRNA | 9626 (3.4) | 7754 (2.9) | 509944 (5.08) | 238818 (2.0) |
| unann | 149517 (61.2) | 170209 (65.0) | 3029373 (30.2) | 3286671 (33.0) |
Identifying novel potential miRNAs in Chlamydomonas reinhardtii
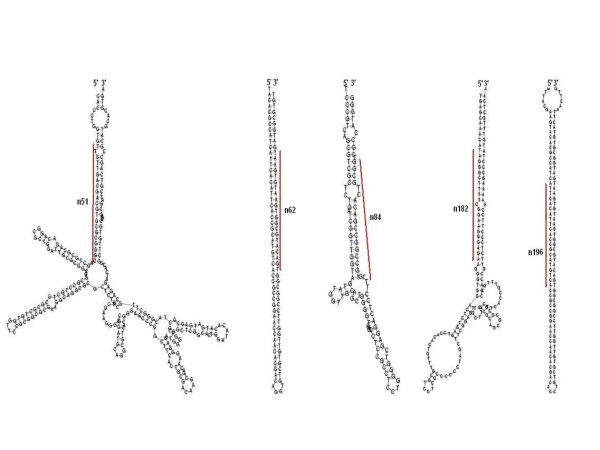
To date, miRBase had a collection of 85 Chlamydomonas miRNA. The characteristic hairpin structure of miRNA precursor can be used to predict novel miRNA. We used the predictive software Mireap to predict novel miRNAs by exploring their secondary structures, and the minimum free energy of the unannotated small RNA tags which could be mapped to the genome. We predicted 225 novel miRNAs by Solexa sequencing and Mireap predictive software in Chlamydomonas reinhardtii. Some of the novel potential miRNAs (n51, n62, n84, n182, n196, Figure 4) have more than ten thousand reads but were not detected by former research [11,12]. These results showed that high-throughput sequencing of small RNAs is also an ideal strategy to analyze small RNAs profiles and identify novel potential miRNAs in Chlamydomonas reinhardtii.
Figure 4.
The hairpin secondary structures of 5 predicted miRNA precursors (n51, n62, n84, n182, n196). The positions of mature miRNAs are highlighted in red.
Differential expression of Chlamydomonas reinhardtii miRNAs in sulfur-deprived and sulphur-replete conditions
In order to detect the effect of sulfur-deprivation on Chlamydomonas reinhardtii miRNAs, the expression abundance of 310 candidate miRNAs (minimum short RNA sequence depth above 1) in both -S and + S libraries was examined. Overall, approximately 15% (47 miRNA) of miRNAs showed significant changes in expression (fold changes > 2) upon sulfur-deprivation. Among these miRNA with altered expression, 24 known miRNAs and 23 predicted miRNAs were up-regulated with 13 miRNAs being -S library specific (Tables 3, 4).
Table 3.
Known Chlamydomonas reinhardtii miRNAs that are responsive to sulfur-deprivation
| pairwise | miR-name | +S-std | -S-std | fold-change (log2 -S/+S) | p-value | sig-lable |
|---|---|---|---|---|---|---|
| +S/-S | miR1144a.1 | 2.7916 | 6.8556 | 1.29619070 | 3.00277572489945e-05 | ** |
| +S/-S | miR1144b | 67.7967 | 174.2123 | 1.36155953 | 4.25092513184362e-107 | ** |
| +S/-S | miR1147.1 | 645.5638 | 2759.7732 | 2.09591812 | 0 | ** |
| +S/-S | miR1148.2 | 5.2842 | 13.2071 | 1.32155674 | 3.68981982177614e-09 | ** |
| +S/-S | miR1149.1 | 0.1994 | 1.4114 | 2.82338960 | 0.00218223161858286 | ** |
| +S/-S | miR1149.2 | 35.3938 | 81.9645 | 1.21150253 | 4.19876393573889e-43 | ** |
| +S/-S | miR1150.3 | 0.3988 | 1.2098 | 1.60103125 | 0.0464712840858496 | * |
| +S/-S | miR1153.1 | 1051.8453 | 2991.5522 | 1.50797170 | 0 | ** |
| +S/-S | miR1153.2* | 44.8654 | 96.4822 | 1.10465953 | 9.53780690450724e-44 | ** |
| +S/-S | miR1155 | 18.9432 | 63.0108 | 1.73391907 | 3.9298538924937e-56 | ** |
| +S/-S | miR1156.1 | 31.6052 | 729.5141 | 4.52870201 | 0 | ** |
| +S/-S | miR1156.2 | 37.1885 | 302.6536 | 3.02473905 | 0 | ** |
| +S/-S | miR1158 | 1.5952 | 4.5368 | 1.50793775 | 0.000148301262915249 | ** |
| +S/-S | miR1159.2 | 0.7976 | 2.1172 | 1.40842024 | 0.0148368997880162 | * |
| +S/-S | miR1160.2 | 0.6979 | 36.3951 | 5.70458009 | 4.07725858511646e-98 | ** |
| +S/-S | miR1160.3 | 0.0997 | 9.1744 | 6.52387649 | 1.14199975919422e-26 | ** |
| +S/-S | miR1164 | 1.5952 | 4.0327 | 1.33800877 | 0.00109191543677247 | ** |
| +S/-S | miR1166.1 | 0.6979 | 2.7221 | 1.96362783 | 0.000450528886605393 | ** |
| +S/-S | miR1172.1 | 232.3033 | 921.3694 | 1.98777004 | 0 | ** |
| +S/-S | miR1172.2 | 308.0760 | 1139.3365 | 1.88683570 | 0 | ** |
| +S/-S | miR906-3p | 11.8644 | 42.5449 | 1.84234706 | 1.13973265129334e-41 | ** |
| +S/-S | miR909.1 | 0.3988 | 1.4114 | 1.82338960 | 0.0180099225615881 | * |
| +S/-S | miR910 | 260.9175 | 924.7972 | 1.82554332 | 0 | ** |
| +S/-S | miR912 | 2830.5107 | 6891.9725 | 1.28385457 | 0 | ** |
pairwise: pair of samples in differentially expression analysis; miR-name: miRNA name; -std: normalized expression level of miRNA in a sample; fold change (log2 -S/+S): fold change of miRNAs in the pair of samples; p-value: p value which reflects the significance of miRNA differential expression between samples. Less p value shows more significance of difference of miRNA between samples; sig-label: significance label, **: fold change (log2) > 1 or fold change (log2) < -1, and p value < 0.01. *: fold change (log2) > 1 or fold change (log2) < -1, and 0.01 < = p value < 0.05
Table 4.
Predicted Chlamydomonas reinhardtii miRNAs that are responsive to sulfur-deprivation
| Name | Sequence (5'-3') | L (nt) | Location in the geonome | MFE (kcal mol-1) | Reads +S/-S |
|---|---|---|---|---|---|
| n006 | UCCAGCUGGGCGGCCGUCUCC | 21 | chromosome_10:4733426:4733510:- | -38.6 | 1004/2798 |
| n015 | UUCUACCCAAGAGGCUGUGUA | 21 | chromosome_12:2479175:2479337:+ | -93.9 | 84/264 |
| n030 | UCAAAGCUAGGAGCCAUGAAG | 21 | chromosome_14:139326:139506:+ | -129.4 | 1347/4625 |
| n046 | UGUUCGGAGAUCCUUGUGCAUG | 22 | chromosome_16:3072001:3072107:- | -101.1 | 117/373 |
| n051 | UUGUUGACGACGUGCGCGGGC | 21 | chromosome_17:204043:204314:- | -129.8 | 41137/86593 |
| n052 | UGACACAUGGAACAACACAAC | 21 | chromosome_1:3554627:3554814:+ | -150.9 | 287/505 |
| n062 | UGACAUGCGGUGAAUGUGAAU | 21 | chromosome_3:5564702:5564806:+ | -105.4 | 15185/49088 |
| n077 | UCAUGAAGCGGAUACUGUGAA | 21 | chromosome_7:743887:744064:- | -103.91 | 1160/61 |
| n083 | UGGGCCUGUUGUGCACGUUCC | 21 | scaffold_28:208520:208678:- | -115.9 | 104/662 |
| n084 | UUGUGCCGGCCGACACUGCGG | 21 | scaffold_33:45408:45519:+ | -49.9 | 77092/55878 |
| n152 | UGUACGGCGACCUGCAAAUGG | 21 | chromosome_10:5980485:5980652:+ | -115.7 | 0/215 |
| n168 | AGGAUGACCGUCAUGAUUGCG | 21 | chromosome_13:4630466:4630659:- | -167 | 0/4628 |
| n169 | GUCAUUAAGACCGUCGGCAAU | 21 | chromosome_14:1815533:1815835:+ | -206.26 | 0/170 |
| n182 | UAGGGCUUUUCGGAAGGGAGA | 21 | chromosome_17:4302900:4303054:+ | -83.2 | 0/20463 |
| n196 | AUUCACAUUCACCGCAUGUCA | 21 | chromosome_3:5564693:5564816:+ | -112 | 0/16257 |
| n197 | UCCUCCUCCUUGACGUCGGCG | 21 | chromosome_3:7309395:7309503:+ | -50.1 | 0/321 |
| n198 | AGCAGCUUCCCACUCCCACGACC | 23 | chromosome_3:2104211:2104422:- | -66.5 | 0/415 |
| n200 | UGCGCAGCGGCAUCAUCUGGA | 21 | chromosome_4:2994696:2994894:- | -157.7 | 0/367 |
| n207 | UAGCAGUCUGAACCAAAGUCG | 21 | chromosome_7:2564607:2564734:+ | -92.1 | 0/931 |
| n209 | UAUGGGCAGUUGUACUAAAUC | 21 | chromosome_7:4354369:4354571:+ | -136.8 | 0/564 |
| n212 | UGGGCCUCACGGCGGCGGACC | 21 | chromosome_7:5449979:5450131:+ | -140.8 | 0/270 |
| n214 | AGGGCCAACAGCUUUGACCGG | 21 | chromosome_7:5670376:5670487:+ | -103.9 | 0/557 |
| n222 | AAUGCCAGCAGCUCCACGCCC | 21 | scaffold_18:257:618:+ | -158.6 | 0/4384 |
Name: the name of predicted Chlamydomonas reinhardtii miRNAs that are responsive to sulfur-deprivation; Sequence: sequence cloned in small RNA libraries; L, the length of miRNA; MFE: the adjusted minimum free energy (MFE) representing the MFE of 100 nucleotides; Reads: the effect of sulfur-deprivation on miRNA expression, +S/-S, normalized sequencing frequencies in the +S and -S library.
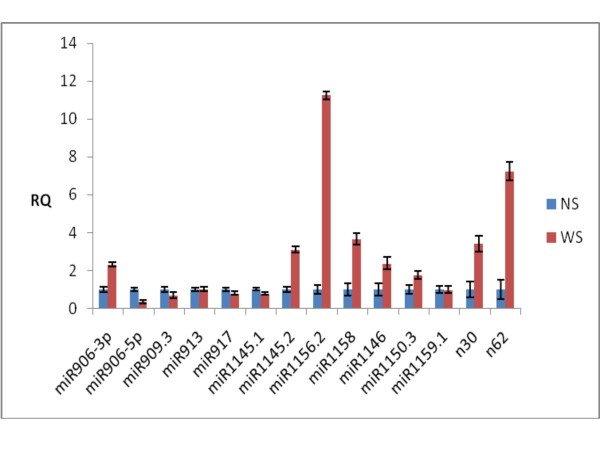
To confirm the expression changes of Chlamydomonas miRNAs and their response to sulfur-deprived stress, we used quantitative RT-PCR analysis to validate the results of the high throughput sequencing. Fourteen miRNAs, which included 12 known miRNAs and 2 novel predicted miRNAs, were selected at random. The quantitative RT-PCR experimental results of 11 miRNAs matched these of high throughput sequencing data (Figure 5 and Tables 3, 4). However, 3 of the chosen miRNAs did not show the matched results. We deduce that this was likely due to the low quality of primers [14] or low abundance of miRNAs, and further research is needed for this problem.
Figure 5.
quantitative RT-PCR analysis of fold changes for the expression of fourteen miRNAs between + S(NS) and -S(WS).
Target predictions of Chlamydomonas reinhardtii miRNAs involved sulfur-deprivation
The target genes of Chlamydomonas. reinhardtii miRNAs were predicted by a web-base integrated system, Web MicroRNAs Designer 3 http://wmd3.weigelworld.org/cgi-bin/webapp.cgi. The 47 miRNA, which showed differential expressions (fold changes > 2) to sulfur-deprivation were screened for target predictions. We divided the predicted target genes into different group according to their putative functions including photosynthesis, carbon metabolism, lipid metabolism and other processes. We analyzed the differential expression correlation between the miRNA and the target genes [3,4] in response to sulfur-deprivation; consequently, we confirmed 17 miRNAs and the target genes involved in response to sulfur-deprivation in Chlamydomonas reinhardtii (Table 5).
Table 5.
General overview of Chlamydomonas reinhardtii miRNAs and their functional targets responded to sulfur-deprivation
| Potential Role | miRNAs ID | Expression of miRNA | Functional Target Gene | Expression of gene [3,4] |
|---|---|---|---|---|
| Lipids metabolism | miR914 | Up | Lipoxygenase | Down |
| n196 | -S only | SC5D, C-5 sterol desaturase | Down | |
| n214 | -S only | Glycerol-3-phosphate dehydrogenase | Unchanged | |
| n222 | -S only | LPB1, lowphosphate bleaching protein | Up | |
| Proteolisis and peptidolisis | miR906.3p | Up | 26S proteasome regulatory subunit | Unchanged |
| miR1147.1 | Up | COP signalosome subunit 5 | Down | |
| n198 | -S only | Peptidase M14, carboxypeptidase A | Unchanged | |
| n222 | -S only | Peptidase M14, carboxypeptidase A | Unchanged | |
| Photosynthesis | miR1150.3 | Up | Cytochrome P450 CYP3/CYP5/CYP6/CYP9 subfamilies | Down |
| miR1166.1 | Up | Phosphatidylglycerophosphate synthase | Down | |
| miR909.1 | Up | pheophorbide a oxygenase | Unchanged | |
| Carbon metabolism | miR1158 | Up | 6-phosphogluconate dehydrogenase, decarboxylating | Up |
| n222 | -S only | NADP malic enzyme | Down | |
| Transporters and translocons | n034 | -S only | PTA3, proton/phosphate symporter | Down |
| n197 | +S only | amino acid transporter | Up | |
| DNA binding, RNA binding, transcription and translation processes | miR1166.1 | Up | ribosome biogenesis pescadillo-like protein | Down |
| n197 | +S only | PRPL1, plastid ribosomal protein | Down | |
| Purine and pyrimidine metabolism | miR1147.1 | Up | Adenylate/guanylate kinase | Down |
| Inositol metabolism | miR1150.3 | Up | phosphatidylinositol 3-kinase-related protein kinase | Unchanged |
| Amino acid metabolism | miR1156.2 | Up | N-acetyltransferase | Down |
| Kinases and phosphatases | n005 | Up | Serine/threonine protein kinase | Up |
| Redox processes | n030 | Up | cytochrome P450, CYP85 clan | Unchanged |
Potential role: potential role of target genes in metabolism; miRNAs ID: name of miRNA; expression of miRNA: the expression level of C. reinhardtii miRNAs after sulfur-deprivation; functional target gene: possible functional target genes by target gene prediction; expression of gene: the expression of target genes reported by published references
Discussion
The reliability of the method to identify Chlamydomonas miRNA
miRNAs are a group of small non-coding RNAs that play an important role in various developmental and stress response processes through negative control of gene expression [15]. miRNA identification had been reported in Chlamydomonas reinhardtii [11,12], but only 85 miRNAs were annotated and collected to miRBase in previous studies. In this study we characterized a total of 310 Chlamydomonas reinhardtii miRNAs which included the 85 known miRNAs and 225 novel miRNAs by using Solexa sequencing technology with prediction software Mireap. We found that most known miRNAs were abundant, and that the 225 novel miRNAs were less abundant or specific to the miRNA of -S library. It further confirmed Solexa sequencing technology as a high-throughput sequencing system, which was able to produce highly accurate, reproducible and quantitative readouts of small RNAs [16,17].
The Chlamydomonas miRNAs expression patterns for responding to sulfur-deprivation
The unicellular green alga Chlamydomonas reinhardtii has been used as a model organism for studying the responses of eukaryotes to sulfur-deprivation [4]. The completion of Chlamydomonas reinhardtii genome sequencing and substantially expressed sequence tag (EST) project has greatly increased its utility as a model system [18]. Although some studies have been carried out on Chlamydomonas miRNAs regulation [11,12], this is the first report on the miRNAs which responded to sulphur-deprivation stress in Chlamydomonas reinhardtii. By comparing the expression levels of two miRNAs libraries(+S library and -S library), we found that approximately 15% of miRNAs expressions showed evident changes (fold changes > 2) with sulfur deprivation (Tables 3, 4). These results further indicate that sulfur-responsive processes are complicated and need a lot of miRNAs to participate in the regulation of gene expression at both transcriptional and post-transcriptional levels [3,4]. Interestingly, although most miRNA which were reported by several groups [10,11] were found in this study, we found that no Chlamydomonas miRNAs have identifiable orthologs with miR395s. miR395s play a very important roles in the sulfur-deprived response in higher plants [19,20] where miR395 regulates sulfate distribution and metabolism in the plant cells. Further, miR395 targets a low-affinity sulfate transporter AST68 (At5g10180) and 3 enzymes in the sulfate assimilation pathway (i.e., ATP sulfurylases [(APS1: At3g22890), (APS3: At4g14680) and (APS4: At5g43780)] [19,20]. The highly conserved miR395 and the presence of multiple copies of miR395 in diverse plant species is necessary for sulfur homeostasis. The lack of a universally conserved miR395s family in Chlamydomonas reinhardtii suggests that green algae miRNAs may have a different pathway for responding to sulfur-deprivation than higher plants.
Target genes of Chlamydomonas miRNAs involved in sulfur-deprivation
miRNAs negatively regulated their targets by cleavage-induced degradation, and the effects of miRNAs over expression were thus often reflected in decreased mRNA levels of the target gene [21]. Although we found the differential expression of multitudinous Chlamydomonas miRNA between sulfur-replete and sulfur-deprived conditions, it is difficult to confirm their target genes because of the complex mechanism of interaction between miRNAs and their target transcripts was not determined. Zhao et al (2009) selected the precursor of miRNA cre-MIR1162 as backbone to design amiRNAs silencing the MAA7 and RBCS2 genes [22]. Attila et al (2009) selected the precursor of miRNA cre-MIR 1157 as a backbone to efficiently produce amiRNAs targeting COX90, PSY and DCL1 genes [23]. James et al (2010) also used amiRNAs silencing HydA1, HydA2, and Hyd3 genes [24]. This far we have not found the specific bioinformatics tools to predict Chlamydomonas miRNAs targets, so we chose the web-base integrated system (Web MicroRNAs Designer 3) to predict Chlamydomonas miRNAs involved in sulfur-deprivation. The predicted results contained false positive rates similar to other reports on miRNA target prediction [14], so it was necessary to verify these predicted targets. However, it was more difficult to screen the miRNAs targets without the 3' UTR sequence database and abundance information of mRNA expression for response to sulfur-deprivation because most miRNA targeted the 3' UTR of mRNA and expression abundances showed the negative correlative between miRNA and their target mRNA. Fortunately, the Chlamydomonas reinhardtii trancriptome was characterized from sulfur-replete and sulfur-depleted conditions [3,4], which provided very important information to analyze the interaction between miRNA and its target mRNA. Theoretically, with the sulfur-deprivation stress, the amount of related mRNAs expression will increased, the transcription levels of its target genes should be decreased by miRNA-mediated gene silencing. In fact, we found that expression abundances of most miRNAs showed a negative correlation with their levels of target mRNA under sulfur-deprivation among 22 miRNAs which regulated the metabolic activities (Table 5). Interestingly, the four target genes which encoded low phosphate bleaching protein (LPB), 6-phosphogluconate dehydrogenase (6-PGDH), decarboxylating amino acid transporter and serine/threonine protein kinase were up-regulated while the expressions of the their miRNA including n222, miR1158, n197 and n005 were up. LPB is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation [25]. 6-PGDH is the first enzyme for pentose phosphate pathway (PPP), which is common for plant responses to abiotic stresses, and serine/threonine protein kinase is required for acclimation of the alga to sulfur deprivation [26]. These four genes are important for Chlamydomonas reinhardtii to survive under sulfur-deprivation, so their miRNA should be down regulated when the algal cell is stressed by sulfur deprivation. However, our results from high-throughput sequencing demonstrate that their regulated miRNAs are increased with sulfur deprivation. These results may indicate that the expression of the 4 genes are regulated by multiple factor, the miRNA regulation may not be the major force. Further experiments are needed to verify this hypothesis.
We were particularly interested in the relative miRNA and its targets for hydrogen bioproduction. The differential expression profile of Chlamydomonas miRNAs led to massive changes in gene expression and metabolism which was closely associated with H2 photo-production. Several miRNAs targeted to genes involved photosynthesis created an anaerobic environment and induced the activity of hydrogenase. The target gene of miR1166.1 encoded the PG phosphate synthase protein. PG is synthesized from cytidyldiphosphate (CDP)-diacylglycerol and glycerol-3-phosphate by the catalytic action of PG-phosphate synthase. Being the only phospholipid in cyanobacteria, PG was required for the accumulation of chlorophyll-protein complexes in the thylakoid membrane and for the normal functioning of photosystem II (PSII) [27]. In the absence of O2, in order to generate ATP, green algae resorted to anaerobic photosynthetic metabolism by miRNA regulation and evolved H2 in the light and consumed endogenous substrates.
Conclusions
We have performed a deep-sequencing analysis of miRNAs in Chlamydomonas reinhardtii and provided a genome-wide, quantitative view of how sulfur-deprivation impacts the expression of small RNAs in Chlamydomonas reinhardtii. Our data confirm 24 known miRNAs and 23 predicted miRNAs with altered expression under sulfur-deprivation, most of which were up-regulated and 13 were -S library specific. Target predictions revealed that a variety of metabolic processes may be affected by changing the expression of miRNAs. Our study has delivered new insights into the role of miRNAs involved in sulfur-deprivation and provided a new approach to understand the biohydrogen production from the small RNA level in Chlamydomonas reinhardtii.
Methods
Growth of the algae
Chlamydomonas reinhardtii CC849 were obtained from Chlamydomonas Genetic Centre (c/o Dr. Elizabeth H. Harris, Department of Botany, Duke University, Durham, NC27706, USA). The algal strain was grown in a Tris-Acetate-Phosphate (TAP) medium at 25°C and under continuous cool-white fluorescent lamps(≈200 μmol photons m-2 s-1). To impose S deprivation [6,8], the liquid cultures were grown into mid-logarithmic phase, algal cells were collected by centrifugation, were washed twice with liquid TAP medium without S (TAP-S, for 1 L of Medium: 2X Filner's Beijernicks Solution 25 ml; 1 M Potassium Phosphate 1 ml; Trace mineral solution 1 ml; Tris-Base 2.42 g; adjust pH to 7.0 by Glacial Acetic Acid. Sulfur-deprivation media (TAP-S) were prepared by replacement of the S-salts by their chloride counterparts). Equal numbers of cells were resuspended in TAP or TAP-S under continuous illumination for up to 72 h, with cell aliquots were collected for RNA isolation, and the sulfate concentration in the supernatant was determined by Dionex ICS-1100 ion chromatogram. Small RNA library construction was carried out as follows: for the + S library, RNA was isolated from the algal cells which were resuspended in sulfur-replete TAP media; for the -S library, RNA was isolated from the algal cells which were resuspended in sulfur-free media TAP-S.
Preparation of total RNA
Total RNA was extracted using Trizol reagent (Invitrogen). The 72 h cells cultured at 25°C in TAP and TAP-S were collected. Total RNA was extracted according to the manufacturer's protocol. The quality of RNA was examined by using an Agilent 2100 Bioanalyzer. The same amount of total RNA was used to construct the two libraries and the samples were prepared in a similar manner. Sequencing of the two libraries was performed on the Illumina's Solexa Sequencer and the samples were run side by side.
Small RNA library construction and high-throughput sequencing
After PAGE purification of small RNA molecules under 30 bases and ligation of a pair of Solexa adaptors to their 5'and 3'ends (Illumina, San Diego, CA. USA), the small RNA molecules were amplified using the adaptor primers for 17 cycles and the fragments around 90 bp (small RNA + adaptors) were isolated with agarose gel. The purified DNA was used directly for cluster generation and sequencing analysis using the Illumina's Solexa Sequencer according to the manufacturer's instructions. The image files generated by the sequencer were processed to produce digital-quality data. The following procedures were performed with Solexa to summarize the data: evaluation of the sequencing quality, calculation of the length distribution of small RNA reads, and filtration of the reads contaminated by rRNA, tRNA, mRNA, snRNA, and snoRNA. Finally, clean reads were compared with a miRBase database (release 15.0).
Bioinformatics analysis
Data quality and length distribution
We eliminated some contaminant reads from the fq file and to obtain the final clean reads and then summarized the length distribution of these clean reads. Normally, length of small RNA is between 18 nt and 30 nt. The length distribution analysis was helpful to see the composition of small RNA samples. For example, miRNA is normally 21 nt or 22 nt, siRNA is 24 nt, and piRNA is 30 nt. The data was processed by the following steps: 1) Elimination of low quality reads (the criteria for this was listed in the explanation of meaning of each row in the result tables); 2) Elimination of reads with 5' primer contaminants; 3) Elimination of reads without 3' primer; 4) Elimination of reads without the insert tag; 5) Elimination of reads with poly A; 6) Elimination of reads shorter than 18 nt; 7) Summarization of the length distribution of the clean reads.
Mapping to genome
We mapped the small RNA tags to the genome by SOAP to analyze their expression and distribution on the genome. Program and Parameters: soap -v 0 -r 2 -s 7 -p 7 -a clean.fa -d ref_genome.fa -o match_genome.soap.
Summary of known miRNA alignment
We aligned the small RNA to the miRNA precursor of corresponding species (using mature miRNA if there was no precursor information of that species in miRBase14.0) to obtain the miRNA count as well as base bias on the first position of identified miRNAs with certain lengths and on each position of all identified miRNAs respectively.
siRNA identification
Small interfering RNA (siRNA) is a 22-24 nt long double-strand RNA, each strand of which is 2 nt longer than the other on the 3' end. According to this structural feature, we aligned tags from clean reads to each other to find sRNAs meeting this criteria. These tags might be potential siRNA candidates. Program and Parameters: Software developed by BGI-tag2siRNA.
Alignment to Genbank
We annotated the small RNA tags with rRNA, scRNA, snoRNA, snRNA and tRNA from Genbank and elimination of matched tags from unannotated tags. Program and Parameters: blastall -p blastn -FF -e 0.01
Alignment to Rfam
Annotation of the small RNA tags with sequences from Rfam and elimination of matched tags from unannotated tags. Program and Parameters: blastall -p blastn -FF -e 0.01
Exon and intron alignment
We aligned small RNA tags to exons and introns of mRNA http://genome.jgi-psf.org/Chlre3/Chlre3.download.ftp.html to find the degraded fragments of mRNA in the small RNA tags. Program and Parameters: Software developed by BGI-overlap
Small RNA annotation
We Summarized all prior alignments and annotation before. In the previous alignment and annotation, some small RNA tags may be mapped to more than one category. To make every unique small RNA map to only one annotation, we followed the following priority rule: rRNAetc(in which Genbank > Rfam) > known miRNA > repeat > exon > intron. Program and Parameters: Software developed by BGI-tag2annotation
Known miRNA expression profile
We aligned small RNA tags to the miRNA precursor/mature miRNA of corresponding species in miRBase14.0. To show detailed information of alignment, including structure of known miRNA precursor, length and count of tags from the sample, etc, click the miRNA id in the left table to see detailed information of that miRNA. Note: Only part of the known miRNA alignment are shown in this report.
Novel miRNA prediction
The characteristic hairpin structure of miRNA precursor was used to predict novel miRNA. We used the predictive software Mireap to predict novel miRNA by exploring the secondary structure, and the minimum free energy of the unannotated small RNA tags which could be mapped to genome. Mireap can be accessed from the following link: http://sourceforge.net/projects/mireap/
Program and parameters
Software developed by BGI- Mireap Minimal miRNA sequence length (18); Maximal miRNA sequence length (25); Minimal miRNA reference sequence length (20); Maximal miRNA reference sequence length (23); Maximal copy number of miRNAs on reference (20); Maximal free energy allowed for a miRNA precursor (-18 kcal/mol); Maximal space between miRNA and miRNA* (300); Minimal base pairs of miRNA and miRNA* (16); Maximal bulge of miRNA and miRNA* (4); Maximal asymmetry of miRNA/miRNA* duplex (4); Flank sequence length of miRNA precursor (20); ram and Parameters: blastall -p blastn -FF -e 0.01.
Differential expression of known miRNA
Comparison of the known miRNA expression between two samples to determine the differentially expressed miRNA. The procedures are shown as below: (1) Normalize the expression of miRNA in two samples (control and treatment) to get the expression of transcript per million(TPM). Normalization forum:Normalized expression = Actual miRNA count/Total count of clean reads*1000000; (2) Calculate fold-change and P-value from the normalized expression according the Bayesian method developed by Audic and Claverie (1997) [28]. Then generate the log2 ratio plot and scatter plot.
Fold-change forum: Fold change = log 2 (treatment/control)
P-value forum:
The N1 and x represent total counts of clean reads and normalized expression level of a given miRNA in sRNA library of + S sample, respectively. The N2 and y represent total counts of clean reads and normalized expression levels of a given miRNA in sRNA library of -S sample, respectively.
Quantitative RT-PCR validation of miRNAs
Sulfur-deprivation procedure is the same as previously described. Both total RNA from 72 h cells cultivated in TAP and TAP-S were isolated respectively for quantitative RT-PCR using the TRIZOL reagent (Invitrogen, Life Technologies, Carlsbad, CA). Low molecular weight RNA and high molecular weight RNA were separated with 4 M LiCl. sRNAs were polyadenylated at 37°C for 60 min in a 50 μl reaction mixture with 1.5 μg of total RNA, 1 mM ATP, 2.5 mM MgCl2, and 4 U poly(A) polymerase (Takara, Japan). Poly (A)-tailed sRNA was recovered by phenol/chloroform extraction and ethanol precipitation. The sRNAs were dissolved, treated with RNase-free DnaseI (Takara, Japan) and reversely transcribed using poly (T) adapter. Real-time PCR was performed using SYBRR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan) and all the primers used were as listed in Additional file 2: Table S1. For each reaction, 1 μL of diluted cDNA (equivalent to 100 pg of total RNA) was mixed with 10 μL of 2 × SYBR green reaction mix (SYBRR Green qRT-PCR Master Mix; Takara, Japan), and 5 pmol of the forward and the reverse primers were added to make a final volume of 20 μL. The conditions for the PCR amplification were as follows: polymerase activation was conducted at 95°C for 30 s; followed by 40 cycles at 95°C for 5 s, 60°C for 31 s. The specificity of the primer amplicons was tested by analysis of a melting curve. The U4 snoRNA was used as a reference gene in the real-time PCR detection of miRNAs. The data was analyzed using the 2--ΔΔCt program, all with an R2 above 0.998. For this quantitative RT-PCR analysis, 3 technical replicates and 2 biological replicates were used.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LS contributed to execute experiments and write the manuscript, ZH contributed to design the research plans and write the manuscript. Both authors read and approved the final manuscript.
Supplementary Material
Figure S1 Mapping of small RNAs in the + S (a) and -S (b) libraries to genome by SOAP. Y axis represents the number of small RNA tags that locate on each chromosome. The numbers of sRNAs on the sense strand of chromosome are positive (shown in blue), and those on the antisence strand of chromosome are negative (shown in red). X axis shows the chromosomes.
Table S1 The primers used in this experiment.
Contributor Information
Longfei Shu, Email: Longfei.Shu@eawag.ch.
Zhangli Hu, Email: huzl@szu.edu.cn.
Acknowledgements
We thank the BGI (Beijing Genomics Institute) for their Solexa sequencing. Also thank Prof. Craig Thompson and Dr. W. Lindsey White for language corrections. This work was financially supported by the National Natural Science Foundation of China (Grant No.31070323, 41176106), and Shenzhen Grant Plan for Science and Technology.
References
- Grossman A, Takahashi H. Macronutrient utilization by photosynthetic eukaryotes and the fabric of interactions. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:163–210. doi: 10.1146/annurev.arplant.52.1.163. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell. 2004;3:1331–1348. doi: 10.1128/EC.3.5.1331-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AV, Thomas-Hall SR, Malnoe A, Timmins M, Mussgnug JH, Rupprecht J, Kruse O, Hankamer B, Schenk PM. The transcriptome of photo-biological hydrogen production induced by sulphur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2008;7:1965–1979. doi: 10.1128/EC.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR. RNA-Seq Analysis of Sulfur-Deprived Chlamydomonas Cells Reveals Aspects of Acclimation Critical for Cell Survival. Plant Cell. 2010;22:2058–2084. doi: 10.1105/tpc.109.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling C, Fiehn O. Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol. 2005;139:1995–2005. doi: 10.1104/pp.105.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A, Zhang LP, Forestier M, Ghirardi ML, Seibert M. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2000;122(1):127–135. doi: 10.1104/pp.122.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi ML, Zhang JP, Lee JW, Flynn T, Seibert M, Greenbaum E, Melis A. Microalgae: a green source of renewable H2. Trends Biotechnol. 2000;18(12):506–511. doi: 10.1016/S0167-7799(00)01511-0. [DOI] [PubMed] [Google Scholar]
- Ghirardi ML, Posewitz MC, Maness PC, Dubini A, Yu J, Seibert M. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu Rev Plant Biol. 2007;58:71–91. doi: 10.1146/annurev.arplant.58.032806.103848. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang X-J, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1130. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- Li RQ, Li YR, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen S, Yang P, Ma Z, Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009;10:R6. doi: 10.1186/gb-2009-10-1-r6. doi:10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Van de Peer Y, Rouze P. The small RNA world of plants. New Phytol. 2006;171:451–468. doi: 10.1111/j.1469-8137.2006.01806.x. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Shu L, Hu Zhangli. Small silencing RNAs in Chlamydomonas reinhardtii. Minerva Biotecnologica. 2010;22(1):29–37. [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MJ, Bartel B, Bartel DP. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Jung J-H, Seo PJ, Park C-M. MicroRNA biogenesis and function in higher plants. Plant Biotechnol Rep. 2009;3:111–126. doi: 10.1007/s11816-009-0085-8. [DOI] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi YJ. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58(1):157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann EC, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe DC. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58(1):165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- James EG, Attila M, David CB, Janneke B. RNA silencing of hydrogenase(-like) genes and investigation of their physiological roles in the green alga Chlamydomonas reinhardtii. Biochem J. 2010;431(3):345–351. doi: 10.1042/BJ20100932. [DOI] [PubMed] [Google Scholar]
- Chang CW, Moseley JL, Wykoff D, Grossman AR. The LPB1 gene is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation. Plant Physiol. 2005;138:319–329. doi: 10.1104/pp.105.059550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Pollock SV, Pootakham W, Grossman AR. The central role of a SNRK2 kinase in sulfur deprivation responses. Plant Physiol. 2008;147:216–227. doi: 10.1104/pp.108.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos Z, Várkonyi Z, Hagio M, Kovács L, Masamoto K, Itoh S, Wada H. Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone QB in the photosystem II reaction center. Biochemistry. 2002;41:3796–3802. doi: 10.1021/bi011884h. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Mapping of small RNAs in the + S (a) and -S (b) libraries to genome by SOAP. Y axis represents the number of small RNA tags that locate on each chromosome. The numbers of sRNAs on the sense strand of chromosome are positive (shown in blue), and those on the antisence strand of chromosome are negative (shown in red). X axis shows the chromosomes.
Table S1 The primers used in this experiment.