Abstract
The mechanisms by which interleukin-6 (IL-6) family cytokines, which utilize the common receptor signaling subunit gp130, influence monocyte/macrophage development remain unclear. Here we have utilized macrophages devoid of either gp130-dependent STAT1/3 (gp130ΔSTAT/ΔSTAT) or extracellular signal-regulated kinases 1 and 2 (ERK1/2) mitogen-activated protein (MAP) kinase (gp130Y757F/Y757F) activation to assess the individual contribution of each pathway to macrophage formation. While the inhibition by IL-6 of macrophage colony-stimulating factor (M-CSF)-induced colony formation observed in gp130wt/wt mice was abolished in gp130ΔSTAT/ΔSTAT mice, inhibition of macrophage colony formation was enhanced in gp130Y757F/Y757F mice. In gp130ΔSTAT/ΔSTAT bone marrow-derived macrophages (BMMs), both IL-6- and M-CSF-induced ERK1/2 tyrosine phosphorylation was enhanced. By contrast, tyrosine phosphorylation of ERK1/2 in response to M-CSF was reduced in gp130Y757F/Y757F BMMs, and the pattern of ERK1/2 activation in gp130 mutant BMMs correlated with their opposing responsiveness to M-CSF-induced proliferation. When compared to the level of expression in gp130wt/wt BMMs, c-fms expression was elevated in gp130ΔSTAT/ΔSTAT BMMs but reduced in gp130Y757F/Y757F BMMs. Finally, an ERK1/2 inhibitor suppressed M-CSF-induced BMM proliferation, and this result corresponded to a reduction in c-fms expression. Collectively, these results provide a functional and causal correlation between gp130-dependent ERK MAP kinase signaling and c-fms gene activation, a finding that provides a potential mechanism underlying the inhibition of M-CSF-dependent macrophage development by IL-6 family cytokines in mice.
The formation of macrophages involves the proliferation, differentiation, and functional maturation of multipotential hematopoietic progenitor cells from the bone marrow into monocytes and, ultimately, macrophages, a process that is primarily regulated by macrophage colony-stimulating factor (M-CSF, also known as CSF-1) (36). The central role that M-CSF plays in monocyte/macrophage development has been formally demonstrated by the severe depletion of macrophages in M-CSF-deficient osteopetrotic (op/op) mice (40) and, more recently, in mice subjected to targeted inactivation of the M-CSF receptor gene, c-fms (5). During mouse development, expression of c-fms is an early and robust marker of cells belonging to the macrophage lineage, and studies on the transcriptional regulation of c-fms have identified promoter elements underlying its lineage-specific activity (14, 15).
Members of the interleukin-6 (IL-6) cytokine family, in particular, IL-6, IL-11, and leukemia inhibitory factor (LIF), are multifunctional cytokines which also regulate the production of myelomonocytic cells (4, 27). In vitro studies with the M1 murine monocytic leukemia cell line revealed that IL-6 and LIF induce the terminal differentiation of these cells into macrophages, suggesting that these factors promote macrophage development (25). However, the IL-6 family cytokines appear to act as both positive and negative regulators of the proliferation and differentiation of primary human and mouse macrophages (4, 17, 27), with recent evidence demonstrating that this may occur by regulating the expression of c-fms (3). IL-6 family cytokines share gp130 as a signaling receptor β (Rβ) subunit (19), and receptor binding of IL-6 or IL-11 leads to gp130 homodimerization (13, 29), whereas receptor binding of all other family members induces heterodimerization of gp130 with one of the highly related Rβ subunits, LIF-Rβ or oncostatin M-Rβ (10, 28). In all cases, β subunit dimerization triggers activation of two major signaling cascades, namely the SHP2/extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase (9) and the STAT1/3 (11) pathways. We have recently reported the generation of two strains of mice homozygous for knock-in mutations in gp130 which render gp130 incapable of activating either the STAT1/3 (in gp130ΔSTAT/ΔSTAT mice) (6) or the SHP2/ERK MAP kinase (in gp130Y757F/Y757F mice) (38) pathway. These mice display a wide variety of penetrant phenotypes, which in part mimic phenotypic pathologies observed in knockout mice lacking individual cytokines of the IL-6 family or their receptors, thereby genetically identifying the signaling pathway responsible for transducing a specific IL-6 family cytokine-mediated biological response. Surprisingly, the gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F mice also display novel phenotypes, which most likely are a consequence of disturbing the otherwise normal balanced activation of these two signaling pathways (6, 38).
To dissect the contribution of each of the two gp130-dependent signaling cascades to the regulation of macrophage development by IL-6 family cytokines, we have studied macrophage populations derived from gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F mice. We report here that the absence of gp130-dependent SHP2/ERK MAP kinase activation in macrophage colony-forming cells (M-CFCs) from gp130Y757F/Y757Fmice enhanced the inhibition of M-CSF-induced macrophage colony formation by IL-6. In contrast, IL-6 does not inhibit M-CSF-induced macrophage colony formation by M-CFCs from gp130ΔSTAT/ΔSTAT mice, where the absence of gp130-mediated STAT activation amplified signaling through the SHP2/ERK MAP kinase pathway. Similarly, bone marrow-derived macrophages (BMMs) from gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F mice also display opposing responsiveness to M-CSF-dependent proliferation and differentiation. Moreover, we provide evidence that the level of c-fms expression is directly related to the net output of ERK MAP kinase signaling emanating from gp130, thereby providing a molecular mechanism by which the IL-6 family cytokines modulate M-CSF-dependent macrophage development.
MATERIALS AND METHODS
Mice.
The generation of mice homozygous for the gp130ΔSTAT mutation (gp130ΔSTAT/ΔSTAT mice) (6) and the gp130Y757F mutation (gp130Y757F/Y757F mice) (38) and of the compound gp130Y757F/Y757F:IL-6−/− mice (38) has been described previously. Experiments were conducted on adult mice 8 to 16 weeks of age and included wild-type littermate controls. Mice were housed under specific-pathogen-free conditions.
Cytokines and reagents.
Recombinant murine M-CSF was a kind gift from Nic Nicola (Walter and Eliza Hall Institute, Melbourne, Australia). Recombinant human IL-6 and human IL-11 were generously provided by Glenn Begley (Amgen, Thousand Oaks, Calif.) and Lorraine Robb (Walter and Eliza Hall Institute), respectively. Commercially available antibodies were purchased as follows: the phospho-specific ERK1/2 antibody was purchased from Cell Signaling Technology (Beverly, Mass.), the ERK1 antibody was from Santa Cruz Biotechnology (Santa Cruz, Calif.), and the fluorescein isothiocyanate (FITC)-conjugated CD11b (Mac-1) antibody and streptavidin-phycoerythrin (PE) were purchased from Pharmingen (San Diego, Calif.). The Fms antibody was generously provided by Nicholas Wilson (University of Melbourne, Melbourne, Australia), and the biotinylated F4/80 antibody was a kind gift from Ken Harder (Ludwig Institute for Cancer Research, Melbourne, Australia). The MEK1 inhibitor U0216 and the p38 MAP kinase inhibitor SB203580 were obtained from Calbiochem (San Diego, Calif.).
Hematopoietic progenitor cell assays.
Clonal cultures of hematopoietic progenitor cells were assayed in semisolid agar containing 0.3% agar, Dulbecco's modified Eagle's medium (DMEM), and 20% fetal calf serum (FCS). Bone marrow cells were flushed from femurs in phosphate-buffered saline (PBS) containing 2% FCS, and cell counts revealed that the levels of cellularity (per femur) were comparable among the genotypes used in this study (data not shown). Briefly, triplicate cultures of either whole blood (1 μl) or unfractionated adult bone marrow cells (2.5 × 104 to 5 × 104) were stimulated with cytokines either individually or in multiple combinations at the indicated concentrations (see figure legends). For M-CSF stimulation of cultures containing either peripheral blood or bone marrow cells, maximal colony stimulation was achieved at a concentration of M-CSF of ≥5 ng/ml. Cultures were incubated for 7 days (bone marrow) or 14 days (blood) at 37°C in a humidified atmosphere of 10% CO2 in air, and colonies containing 50 or more cells were enumerated by using a dissection microscope. Agar cultures were then fixed with 2.5% gluteraldehyde and sequentially stained with Luxol Fast Blue and hematoxylin.
Reconstitution experiments.
One day prior to bone marrow transfer, gp130wt/wt C57BL/6 Ly5.1-recipient mice were injected intraperitoneally with 300 μg of the CD4-depleting monoclonal antibody GK1.5 (kindly provided by Y. Zhang, Walter and Eliza Hall Institute). Flow cytometric analysis of peripheral blood from GK1.5-treated mice has previously demonstrated 98% depletion of peripheral CD4+ T lymphocytes (22). Unfractionated bone marrow cells (5 × 106) from Ly5.2 gp130wt/wt, gp130ΔSTAT/ΔSTAT, and gp130Y757F/Y757F mice were then injected intravenously into lethally γ-irradiated (twice at 5.5 Gy) adult Ly5.1 recipients (4 or 5 per donor). Irradiated recipients were maintained on oral antibiotics (170 mg of enrofloxacin [Baytril] per liter; Bayer, Leverkusen, Germany). Flow cytometric analysis indicated that 89% ± 2.1% (mean ± standard deviation [SD]) of peripheral blood leukocytes obtained from chimeras expressed Ly5.2 donor cells 3 months posttransfer.
BMMs.
Bone marrow cells were flushed from femurs into DMEM supplemented with 15% FCS, 20% L-cell conditioned (LC) media (as a source of M-CSF), 2 mM glutamax, and 0.075% sodium bicarbonate (Life Technologies, Rockville, Md.) and were cultured in flasks at 37°C with 10% CO2. The following day, nonadherent cells were transferred into new flasks, and BMMs were used after 7 to 10 days of culture. Prior to experiments for cytokine stimulation, cells were washed in PBS and then starved in DMEM plus 1% FCS for 6 h before being stimulated.
Western blot analysis.
Prior to lysis, BMMs were either grown continuously in LC media or were washed and starved of cytokine before being stimulated. All stimulations were performed at 37°C with M-CSF at a concentration of 20 ng/ml and IL-6 at a concentration of 100 ng/ml at specified times (see figure legends). Cells were lysed in ice-cold lysis buffer (50 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1% glycerol, 1% Triton X-100, 50 mM NaF, 0.1% sodium dodecyl sulfate, 2 mM EDTA, 2 mM orthovanadate, and complete protease inhibitors [Roche Diagnostics Corporation, Indianapolis, Ind.]), and cell lysates were cleared of debris by centrifugation. Cell lysates were separated on sodium dodecyl sulfate-10% polyacrylamide gels and then transferred to nitrocellulose membranes. Membranes were blocked in TBST (300 mM NaCl, 10 mM Tris-HCl [pH 7.5], 0.5% Tween 20) containing 1% bovine serum albumin and incubated with primary antibodies according to the manufacturer's recommendations; subsequently they were incubated with the appropriate secondary antibodies (anti-mouse or -rabbit) coupled to horseradish peroxidase (Bio-Rad, Hercules, Calif). Proteins were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, N.J.).
Northern blot analysis.
Total RNA was extracted from BMMs and unfractionated bone marrow cells with an RNeasy kit (QIAGEN, Hilden, Germany). RNA (10 to 20 μg) was run on 1% agarose gels and transferred onto nitrocellulose, following which membranes were hybridized with a radiolabeled full-length c-fms cDNA probe (kindly provided by Nicholas Wilson, University of Melbourne). Membranes were stripped and reprobed with a radiolabeled glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe (kindly provided by Megan Baldwin, Ludwig Institute for Cancer Research) to confirm equal loading and transfer of RNA samples.
Proliferation and cell viability assays.
BMMs were washed, starved of LC media, and then harvested by scraping. Cells were then added to flat-bottom 96-well tissue culture plates at 2 × 104 cells per well in triplicate containing 100 μl of DMEM supplemented with 15% FCS, M-CSF (20 ng/ml), and/or IL-6 (100 ng/ml) as indicated. In experiments that used MAP kinase inhibitors, SB203580 was used at a final concentration of 30 μM, and U0216 was used at a final concentration of 10 μM. To measure proliferation, after 2 days of culture at 37°C, 1 μCi of [3H]thymidine (ICN Biomedicals Australasia, Seven Hills, Australia) was pulsed for the last 3 h of culture, and [3H]thymidine incorporation was measured in a TopCountNXT microplate scintillation counter (Packard, Meriden, Conn.). To measure cell viability, after 4 days of culture at 37°C, 10 μl of MTT (dimethyl thiazole diphenyl tetrazolium bromide; Sigma, St. Louis, Mo.) at a concentration of 5 mg/ml was added to the wells and incubated for a further 4 h at 37°C. A total of 200 μl of isopropanol-0.04 N HCl was then added to each well, and absorbance was measured at 550 nm. Absorbance readings were normalized against those at day 0 for 2 × 104 cells per well in triplicate.
Flow cytometry.
BMMs grown continuously in LC medium were harvested and resuspended in ice-cold PBS plus 2% FCS (staining solution). For staining with the biotinylated F4/80 and FITC-conjugated CD11b (Mac-1) antibodies, cells were initially incubated in Fc-block (1:100 dilution) on ice for 30 min, following which they were incubated with antibodies (1:200 dilution) on ice for 30 min. Cells were then washed twice in staining solution and incubated with streptavidin-PE (1:500 dilution) for a further 30 min on ice. Stained cells were then washed and immediately analyzed on a FACScan flow cytometer (Becton Dickinson, Mountain View, Calif.).
Transfections and reporter assays.
RAW cells were maintained in DMEM containing 10% FCS. Cells were transiently cotransfected with 10 μg of the pGLΔ3.5fms firefly luciferase reporter construct (14), 0.5 μg of the pRL-TK Renilla luciferase control reporter vector (Promega, Madison, Wis.), and 20 μg of either a hemagglutinin (HA)-tagged wild-type ERK1 construct (kindly provided by Ken Harder, Ludwig Institute for Cancer Research) or the granulocyte-CSF receptor (G-CSFR/gp130wt) or G-CSFR/gp130ΔSTAT chimeric receptor constructs (7) by electroporation as previously described (14). For experiments with the receptor constructs, transfected cell cultures were incubated with G-CSF (100 ng/ml) in the absence or presence of U0216 (10 μM) for 48 h prior to their lysis. Cell lysates were assayed for firefly luciferase activity and normalized against Renilla luciferase activity by the Dual-Luciferase Reporter assay system (Promega).
RESULTS
IL-6-induced suppression of macrophage colony formation is enhanced in the absence of gp130-dependent SHP2/ERK activation.
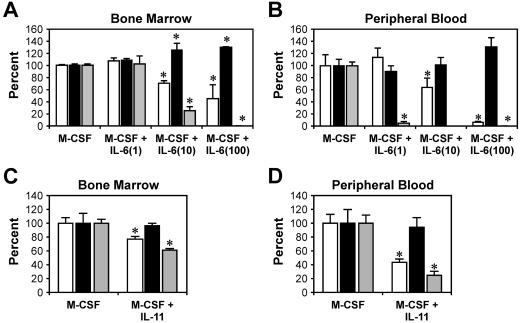
Previous studies have indicated that activation of gp130 by IL-6 family cytokines inhibited M-CSF-dependent development of murine macrophage populations in vitro (4). In order to identify the signaling pathway responsible for transducing this activity, we initially examined the capacity of IL-6 to modulate M-CSF-dependent in vitro colony formation from bone marrow and peripheral blood M-CFCs derived from mice lacking either gp130-dependent STAT1/3 (gp130ΔSTAT/ΔSTAT) or SHP2/ERK (gp130Y757F/Y757F) activation. The frequencies of M-CSF-responsive M-CFCs in gp130ΔSTAT/ΔSTAT andgp130Y757F/Y757F bone marrow (94 ± 8 and 87 ± 13 M-CFCs/5 ×104 marrow cells, respectively) were similar to that in wild-type (gp130wt/wt) bone marrow (92 ± 9 M-CFCs/5 ×104 marrow cells). In peripheral blood, there was a significant increase in the circulating concentration of M-CFCs in gp130ΔSTAT/ΔSTAT mice (341 ± 70 M-CFC/μl) compared to that in gp130wt/wt mice (217 ± 19 M-CFC/μl, P < 0.05) and gp130Y757F/Y757F mice (196 ± 37 M-CFC/μl, P < 0.05). The presence of IL-6 (100 ng/ml) inhibited M-CSF-stimulated macrophage colony formation by gp130wt/wt M-CFCs, and peripheral blood M-CFCs were more sensitive to this inhibition than bone marrow M-CFCs (90 and 55% reduction in colony numbers, respectively) (Fig. 1A and B). However, IL-6 (100 ng/ml) did not suppress the proliferation of gp130ΔSTAT/ΔSTAT bone marrow and peripheral blood M-CFCs. Rather, IL-6 recruited additional M-CFCs to form colonies from gp130ΔSTAT/ΔSTAT bone marrow and peripheral blood (Fig. 1A and B). In contrast, macrophage colony formation by gp130Y757F/Y757F M-CFCs was completely abolished by IL-6 at a concentration of 100 ng/ml (Fig. 1A and B). Lower concentrations of IL-6 also led to a greater inhibition of macrophage colony formation by gp130Y757F/Y757F M-CFCs compared to that from gp130wt/wt M-CFCs. We also examined the role of IL-11 in regulating M-CSF-dependent macrophage colony formation since IL-11, like IL-6, signals exclusively via a gp130 homodimer. Although IL-11 reduced gp130wt/wt and gp130Y757F/Y757F macrophage colony formation (by 20 and 40% in bone marrow and by 55 and 76% in peripheral blood, respectively) (Fig. 1C and D), this inhibition was not as great as that seen with IL-6. Furthermore, IL-11 (100 ng/ml) had little or no effect on M-CSF-induced macrophage colony formation by gp130ΔSTAT/ΔSTAT bone marrow and peripheral blood M-CFCs.
FIG. 1.
Effects of IL-6 family cytokines on macrophage progenitor formation in gp130ΔSTAT/ΔSTAT mice (black bars), gp130Y757F/Y757F mice (grey bars) and wild-type littermates (white bars). In vitro macrophage colony formation of bone marrow cells (A and C) and peripheral blood (B and D) in semisolid agar containing M-CSF (10 ng/ml) and various concentrations of IL-6 (1 to 100 ng/ml) (A and B) or IL-11 (100 ng/ml) (C and D) is shown. In the presence of IL-6 or IL-11 alone, no colony formation was observed in cultures containing bone marrow or peripheral blood progenitors from mice of any genotype (data not shown). Data are presented as a percentage (± SD) of maximal stimulation achieved with 10 ng of M-CSF/ml alone and are representative of the results of 1 of at least 3 separate experiments. For each genotype, n was 3. *, P < 0.05 versus data from wild-type littermates.
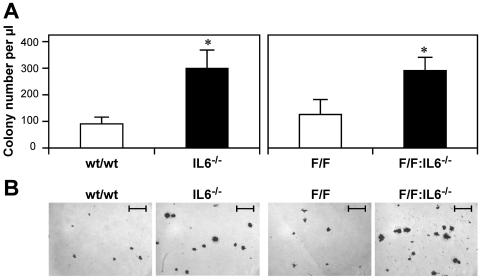
Considering that these observations indicate a major role for IL-6 in suppressing macrophage colony formation in vitro, we hypothesized that the absence of IL-6 in gp130wt/wt and gp130Y757F/Y757F mice might result in an increase in M-CFC levels in vivo. Therefore, we enumerated the concentration of M-CFCs in peripheral blood of gp130wt/wt and IL-6-deficient (IL-6−/−) mice and of gp130Y757F/Y757F and compound gp130Y757F/Y757F:IL-6−/− mice. As shown in Fig. 2A, we observed significant increases in the concentration of peripheral blood M-CFCs from IL-6−/− mice compared to the level in gp130wt/wt mice and from gp130Y757F/Y757F:IL-6−/− mice compared to the level in gp130Y757F/Y757F mice. Furthermore, the size of M-CSF-responsive colonies was greater in peripheral blood cultures from IL-6−/− and gp130Y757F/Y757F:IL-6−/− mice compared to the size of the colonies in cultures from gp130wt/wt and gp130Y757F/Y757F mice, respectively (Fig. 2B). Collectively, these observations suggest that the inhibitory actions of IL-6 and, to a lesser extent, IL-11 on macrophage colony formation by M-CFCs were reciprocally affected by the absence of either one of the two major intracellular gp130 signaling cascades.
FIG. 2.
Analysis of macrophage progenitors in the peripheral blood of wild-type (wt/wt) and gp130Y757F/Y757F (F/F) mice on an IL-6-deficient background (IL-6−/−). (A) In vitro macrophage colony formation of peripheral blood in semisolid agar containing M-CSF (10 ng/ml). Data are presented as the mean ± SD from the results of 1 of at least 3 representative experiments. For each genotype, n was 4. *, P < 0.05 for IL-6−/− and gp130Y757F/Y757F:IL-6−/− mice versus data from wild-type and gp130Y757F/Y757F mice, respectively. (B) Photomicrograph of peripheral blood macrophage colonies from wild-type, IL-6−/−, gp130Y757F/Y757F, and gp130Y757F/Y757F:IL-6−/− mice. Size bars, 100 μm.
Inhibition of macrophage colony formation by IL-6 is intrinsic to the hematopoietic progenitors and not conditioned by the hematopoietic microenvironment.
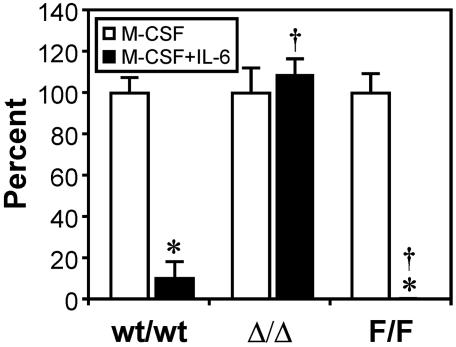
We next examined whether the inhibitory action of IL-6 on macrophage progenitor formation was an intrinsic property of macrophage progenitors or was indirectly mediated by a conditioning effect of the stromal microenvironment. Accordingly, bone marrow cells from gp130wt/wt, gp130ΔSTAT/ΔSTAT, and gp130Y757F/Y757F mice were injected into lethally irradiated gp130wt/wt recipient mice. Three months after transplantation, flow cytometric analysis of peripheral blood leukocytes in recipient mice indicated that mice had been reconstituted with between 77.9 and 97.3% donor cells (data not shown). Semisolid agar assays of peripheral blood M-CFCs from mice reconstituted with gp130wt/wt and gp130Y757F/Y757F bone marrow revealed that in the presence of IL-6, M-CSF-stimulated macrophage colony formation was inhibited by 88 and 100%, respectively (Fig. 3). In contrast, no inhibition by IL-6 on macrophage colony formation was observed for peripheral blood M-CFCs from mice reconstituted with gp130ΔSTAT/ΔSTAT bone marrow. These results are consistent with the relative sensitivities of gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F peripheral blood macrophage progenitors to IL-6 (Fig. 1B) and therefore support the notion that the inhibitory action of IL-6 is directly on the macrophage progenitors.
FIG. 3.
Effects of IL-6 on macrophage progenitors in mice reconstituted with gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F bone marrow. Wild-type recipient mice were reconstituted with bone marrow cells from gp130ΔSTAT/ΔSTAT (Δ/Δ) mice, gp130Y757F/Y757F (F/F) mice, and wild-type (wt/wt) littermates as described in Materials and Methods. In vitro macrophage colony formation in the presence of M-CSF (10 ng/ml) with or without IL-6 (100 ng/ml) was assessed on peripheral blood from recipient mice 3 months posttransplantation. For each group of recipient mice, n was 4. Data are presented as a percentage (± SD) of maximal stimulation achieved with 10 ng of M-CSF/ml alone. *, P < 0.05 versus data from stimulation with M-CSF alone within the same recipient group; †, P < 0.05 versus data from recipient mice reconstituted with wild-type bone marrow cells and stimulated with M-CSF and IL-6.
Altered response of gp130 mutant M-CFCs to M-CSF.
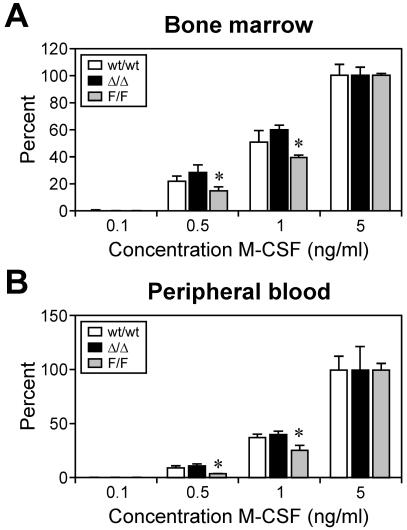
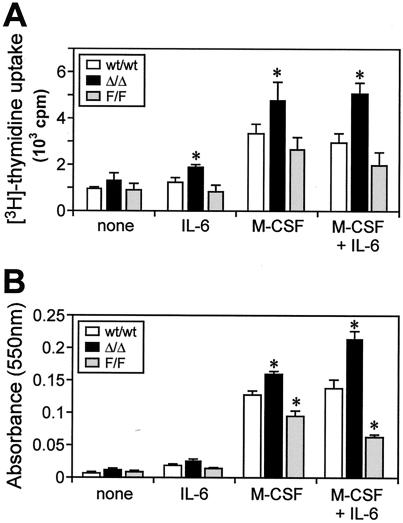
The increase in macrophage colony size in cultures of M-CFCs from mice lacking IL-6 (Fig. 2) suggested that the in vitro proliferative potential (in response to M-CSF) of M-CFCs was suppressed, at least in part, by basal levels of IL-6-driven gp130 signaling. Therefore, we next assessed the sensitivity of gp130-mutant M-CFCs to M-CSF in vitro, in which maximal colony formation was induced at 5 ng of M-CSF/ml. At lower concentrations of M-CSF (≤1 ng/ml), the size and number of macrophage colonies formed from gp130Y757F/Y757F bone marrow and peripheral blood M-CFCs were significantly reduced compared to those of colonies formed from gp130wt/wt and gp130ΔSTAT/ΔSTAT bone marrow and peripheral blood M-CFCs (Fig. 4 and data not shown). Furthermore, in the presence of M-CSF alone, the proliferation of gp130Y757F/Y757F bone marrow macrophages was reduced compared to proliferation of gp130wt/wt macrophages, as measured by [3H]thymidine incorporation (Fig. 5A). In contrast, the proliferation of gp130ΔSTAT/ΔSTAT macrophages was greater. Consistent with the responsiveness of bone marrow M-CFCs to IL-6 (Fig. 1), the presence of IL-6 in M-CSF-containing cultures further inhibited the proliferation of gp130Y757F/Y757F macrophages but augmented the proliferation of gp130ΔSTAT/ΔSTAT macrophages (Fig. 5A). Notably, cell viability, as measured by MTT assays (Fig. 5B), in M-CSF-containing cultures with or without IL-6 was comparable to the proliferation of the corresponding BMM cultures (Fig. 5A). Since viable cell number is the sum of proliferation and survival, we therefore reasoned that the opposing viability of gp130ΔSTAT/ΔSTAT (increased) and gp130Y757F/Y757F (reduced) BMMs, with respect to gp130wt/wt BMMs, was due to differences in cell proliferation and not survival. Indeed, the phosphorylation and kinase activity levels of the serine and threonine kinase Akt, which plays a critical role in the survival of macrophages (24), among gp130wt/wt and gp130 mutant BMMs stimulated with either IL-6 or M-CSF were comparable (data not shown), suggesting that Akt-mediated survival signaling in macrophages was unaffected by the gp130Y757F and gp130ΔSTAT mutations.
FIG. 4.
Dose response of macrophage progenitor formation in gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F mice. Cells from bone marrow (A) and peripheral blood (B) of gp130ΔSTAT/ΔSTAT (Δ/Δ) mice, gp130Y757F/Y757F (F/F) mice, and wild-type (wt/wt) littermates were plated in semisolid agar containing various concentrations of M-CSF (0.1 to 5 ng/ml). Data are presented as a percentage (± SD) of maximal stimulation achieved with 5 ng of M-CSF/ml alone. *, P < 0.05 versus data from wild-type littermates at the corresponding concentration of M-CSF.
FIG. 5.
Altered proliferation of gp130 mutant macrophages. BMMs from gp130ΔSTAT/ΔSTAT (Δ/Δ) mice, gp130Y757F/Y757F (F/F) mice, and wild-type (wt/wt) littermates were cultured in media containing either IL-6 (100 ng/ml), M-CSF (10 ng/ml), or both for 2 to 4 days, following which proliferation was measured by [3H]thymidine incorporation assays (A), and cell viability was measured by MTT assays (B). Data are expressed as the mean ± SD for each triplicate from the results of 1 of at least 3 representative experiments. *, P < 0.05 versus data from wild-type BMMs grown under the same conditions.
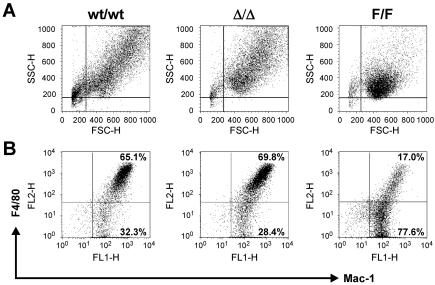
The M-CSF-induced macrophage differentiation program is characterized by a progressive decrease in proliferative capacity accompanied by morphological changes. Thus, we reasoned that the altered growth rates of gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F BMMs in M-CSF might reflect different stages of macrophage differentiation compared to growth in gp130wt/wt BMMs. We therefore performed flow cytometric characterization of the BMM populations in the presence of M-CSF to assess their morphology by cell size (forward scatter) and granularity (side scatter), both of which increase during macrophage differentiation. Macrophages from gp130Y757F/Y757F cultures were smaller and less granular than those from gp130wt/wt and gp130ΔSTAT/ΔSTAT cultures (Fig. 6A). The frequency of gp130Y757F/Y757F macrophages expressing the mature macrophage surface marker F4/80 was significantly lower than that of gp130wt/wt and gp130ΔSTAT/ΔSTAT macrophages. In contrast, levels of expression of Mac-1, a myeloid cell-specific integrin, were similar among the three genotypes (Fig. 6B). Collectively, the above observations suggest that the absence of gp130-dependent SHP2/ERK MAP kinase activation in gp130Y757F/Y757F BMMs leads to a reduction or delay in their M-CSF-induced proliferation and differentiation, whereas the absence of gp130-dependent STAT signaling in gp130ΔSTAT/ΔSTAT BMMs appears to augment their responsiveness to M-CSF.
FIG. 6.
Phenotypic analysis of gp130 mutant macrophages. (A and B) BMMs from gp130ΔSTAT/ΔSTAT (Δ/Δ) mice, gp130Y757F/Y757F (F/F) mice, and wild-type (wt/wt) littermates were cultured in 20% LC medium, and cells were subjected to dual-color flow cytometric analysis. In panel A, the x axis represents forward scatter (cellular size) and the y axis represents side scatter (granularity). Macrophage populations are presented in the upper right quadrant. In panel B, the x axis represents FITC intensity (Mac-1), and the y axis represents PE intensity (F4/80).
Reciprocal negative regulation between gp130-dependent STAT3 and ERK MAP kinase signaling pathways correlates with the M-CSF-dependent growth responsiveness of gp130 mutant BMMs.
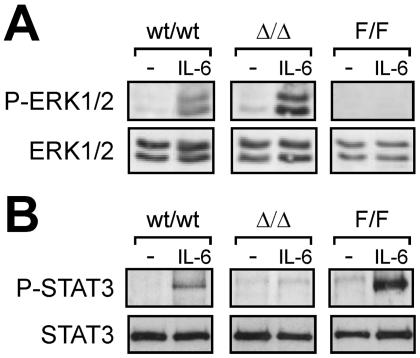
One of the unexpected findings reported here was the failure of IL-6 to inhibit gp130ΔSTAT/ΔSTAT M-CSF-dependent macrophage colony formation and BMM proliferation, which was in contrast to the exaggerated IL-6-mediated inhibition observed on gp130Y757F/Y757F macrophage colony formation and BMM proliferation (Fig. 1 and 5). These observations are reminiscent of the opposing capacities of gp130 mutant mice unable to elicit either gp130-dependent STAT3 or SHP2/ERK MAP kinase activation to mount an intestinal wound-healing response (38) or to balance the Th1 versus Th2 immune response (31). In both reports, the absence of either STAT3 binding or SHP2 binding to gp130 resulted in the enhanced IL-6-induced activation of the ERK MAP kinase and STAT3 pathways, respectively. To determine whether a similar reciprocal negative regulatory mechanism also occurred between the STAT3 and ERK MAP kinase pathways in gp130 mutant macrophages, we compared the levels of tyrosine phosphorylated ERK1/2 MAP kinase in gp130ΔSTAT/ΔSTAT BMMs and STAT3 in gp130Y757F/Y757F BMMs to that in gp130wt/wt cells. Indeed, as shown in Fig. 7, ERK1/2 tyrosine phosphorylation in gp130ΔSTAT/ΔSTAT BMMs was enhanced upon IL-6 stimulation compared to the level in gp130wt/wt BMMs. Conversely, IL-6-induced STAT3 tyrosine phosphorylation was enhanced in gp130Y757F/Y757F BMMs compared to the level in gp130wt/wt BMMs. As expected, no IL-6-induced STAT3 tyrosine phosphorylation and ERK1/2 tyrosine phosphorylation was detected in gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F BMMs, respectively. Therefore, the reciprocal M-CSF responsiveness of gp130 mutant BMMs relative to gp130wt/wt BMMs (i.e., reduced in gp130Y757F/Y757F cells and enhanced in gp130ΔSTAT/ΔSTAT cells) correlates directly with the extent of ERK MAP kinase signaling and inversely with the degree of STAT3 activation. Together with previous observations in the liver and intestine of gp130 mutant mice (38), these data suggest that the reciprocal negative regulation between these two pathways may be of general importance for the biological response of cells to gp130-dependent intracellular signals.
FIG. 7.
Reciprocal negative regulation between gp130-dependent STAT3 and ERK MAP kinase activation in gp130 mutant macrophages. BMMs from gp130ΔSTAT/ΔSTAT (Δ/Δ) mice, gp130Y757F/Y757F (F/F) mice, and wild-type (wt/wt) littermates were starved (−) and then stimulated with IL-6 for 5 min. Cell lysates were run out on 10% polyacrylamide gels, transferred to nitrocellulose membranes, and then immunoblotted with either anti-phospho-ERK1/2 (A) and anti-phospho-STAT3 (B) antibodies (upper panels) or anti-ERK1/2 (A) and anti-STAT3 (B) antibodies (lower panels), as indicated.
Altered M-CSF-dependent responsiveness of gp130 mutant BMMs correlates with differential regulation of c-fms expression.
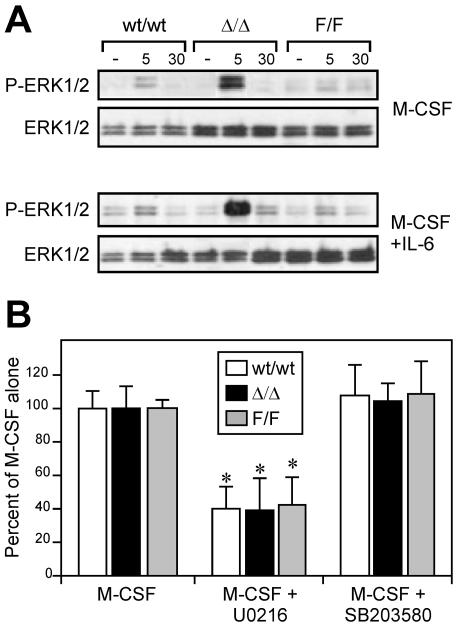
Activation of the Ras/MAP kinase pathway has been suggested to play an important role in M-CSF-induced mitogenesis (1, 23). Since the proliferation of gp130Y757F/Y757F and gp130ΔSTAT/ΔSTAT BMMs in response to M-CSF was reduced and elevated, respectively, compared to that of gp130wt/wt BMMs (Fig. 5A), we examined whether alterations in gp130-dependent ERK MAP kinase signaling affected M-CSF-induced activation of ERK MAP kinase. Compared to levels in gp130wt/wt BMMs, M-CSF-induced ERK1/2 tyrosine phosphorylation was enhanced in gp130ΔSTAT/ΔSTAT BMMs and reduced in gp130Y757F/Y757F BMMs (Fig. 8A). Consistent with a further increase in the proliferation of gp130ΔSTAT/ΔSTAT macrophages by the presence of IL-6 in M-CSF-containing cultures (Fig. 5A), we detected an increase in the level of ERK1/2 tyrosine phosphorylation in gp130ΔSTAT/ΔSTAT BMMs stimulated with M-CSF plus IL-6 compared to the level with M-CSF alone (Fig. 8A). By contrast, no alteration was detected in the level of M-CSF-induced ERK1/2 tyrosine phosphorylation in gp130wt/wt and gp130Y757F/Y757F BMMs by the presence of IL-6. This observation may reflect the limited sensitivity of the Western blot analysis, since IL-6-induced ERK1/2 tyrosine phosphorylation was weaker in gp130wt/wt and gp130Y757F/Y757F BMMs than the strong phosphorylation in gp130ΔSTAT/ΔSTAT BMMs (Fig. 7A). Nonetheless, the importance of ERK MAP kinase activation for M-CSF-driven proliferative signaling was confirmed in experiments assessing the proliferation of gp130wt/wt and gp130 mutant BMMs in the presence of the ERK1/2 MAP kinase-specific inhibitor U0216, in which proliferation of BMMs from all three genotypes was inhibited by approximately 40% (Fig. 8B). In contrast, the addition of the p38 MAP kinase inhibitor SB203580 had no effect on M-CSF-stimulated macrophage proliferation.
FIG. 8.
Altered M-CSF-dependent ERK MAP kinase activation in gp130 mutant macrophages. (A) BMMs from gp130ΔSTAT/ΔSTAT (Δ/Δ) mice, gp130Y757F/Y757F (F/F) mice, and wild-type (wt/wt) littermates were starved (−) and then stimulated with M-CSF or M-CSF plus IL-6 for the times (minutes) indicated. Cell lysates were immunoblotted with either an anti-phospho-ERK1/2 antibody or an anti-ERK1/2 antibody, as indicated. (B) BMMs were grown in M-CSF (10 ng/ml) in the presence of the ERK1/2 inhibitor U0216 or the p38 MAP kinase inhibitor SB203580 for 2 days, following which proliferation was measured by a [3H]thymidine incorporation assay. Proliferation is presented as a percentage ± SD of maximal stimulation achieved with M-CSF alone. *, P < 0.05 versus data from BMMs within the same genotype grown in M-CSF alone.
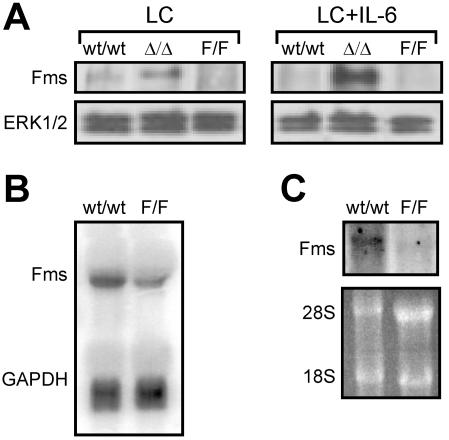
We next assessed the expression levels of c-fms in gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F BMMs in order to explore whether the altered M-CSF-induced ERK1/2 MAP kinase activation in gp130 mutant macrophages was due to corresponding alterations in c-fms expression (Fig. 9). Western blot analysis revealed a higher abundance of Fms protein in gp130ΔSTAT/ΔSTAT BMMs than in gp130wt/wt BMMs grown in medium containing M-CSF (Fig. 9A, LC). Furthermore, the presence of IL-6 in culture medium led to a marked increase in the amount of Fms protein in gp130ΔSTAT/ΔSTAT BMMs but not in gp130wt/wt BMMs (Fig. 9A, compare panels LC and LC plus IL-6). Fms expression in gp130Y757F/Y757F BMMs grown in medium containing M-CSF with or without IL-6 was below the level of detection by Western blot analysis (Fig. 9A). However, Northern blot analysis indicated that c-fms was expressed, albeit at a lower level, in gp130Y757F/Y757F BMMs compared to gp130wt/wt BMMs (Fig. 9B). Similarly, the amount of c-fms RNA transcript in the bone marrow compartment of gp130Y757F/Y757F mice was also severalfold lower than in bone marrow from gp130wt/wt mice (Fig. 9C), confirming that the reduced expression of c-fms in cultured gp130Y757F/Y757F BMM populations was not the result of their ex vivo expansion. Thus, the data presented in Fig. 8 and 9 correlate with alterations in the M-CSF-dependent proliferative response of gp130 mutant macrophages with the level of ERK MAP kinase activation, which in turn corresponds with the differential expression of c-fms. In addition, our observations further support the notion that ERK1/2 are the primary MAP kinases promoting M-CSF-dependent mitogenesis.
FIG. 9.
Altered expression of c-fms in gp130 mutant macrophages. (A) BMMs were grown in 20% LC media in the absence or presence of IL-6 (100 ng/ml) as indicated. Cell lysates were run out on 10% polyacrylamide gels, transferred to nitrocellulose membranes, and then immunoblotted with an anti-Fms antibody (top panels) or anti-ERK1/2 antibody (bottom panels). (B) Ten micrograms of total RNA from BMMs grown continuously in 20% LC media was hybridized with a radiolabeled full-length c-fms cDNA fragment. The membrane was stripped and reprobed for expression of the GAPDH gene. (C) Twenty micrograms of total RNA from unfractionated bone marrow was hybridized with the c-fms cDNA probe. Ethidium bromide staining revealed similar amounts of RNA analyzed.
The c-fms gene is a direct target of the gp130-dependent SHP2/ERK MAP kinase pathway.
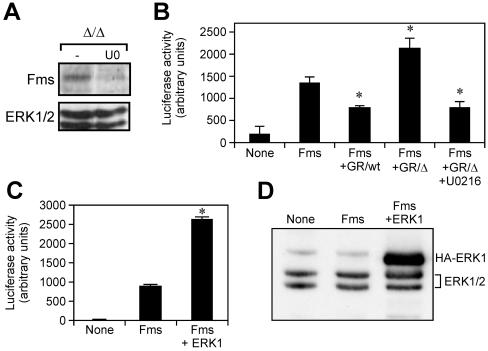
The reciprocal negative regulation among the two pathways emanating from gp130 provides a unifying explanation for the opposing biological effects of IL-6 on gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F BMMs. Our observation in Fig. 9A that the presence of IL-6 in gp130ΔSTAT/ΔSTAT BMM cultures increased Fms expression suggested that the altered capacity of IL-6 to modulate the M-CSF responsiveness of gp130 mutant macrophages could correlate directly with the extent of gp130-dependent ERK MAP kinase activation, since gp130-dependent STAT activation is abolished in gp130ΔSTAT/ΔSTAT mice. In support of this notion, addition of the ERK MAP kinase inhibitor U0216 to cultured gp130ΔSTAT/ΔSTAT BMMs led to a severalfold reduction in the level of Fms protein (Fig. 10A), which suggests that the relative overexpression of Fms in gp130ΔSTAT/ΔSTAT BMMs compared to gp130wt/wt BMMs was due to enhanced activation of the ERK MAP kinase pathway.
FIG. 10.
Regulation of the c-fms promoter by gp130-dependent ERK1/2 MAP kinase activation. (A) BMMs were grown in 20% LC media in the absence or presence of the ERK1/2 inhibitor U0216 for 1 h, and cell lysates were subjected to Western blot analysis as described in the legend of Fig. 9A. (B and C) Transcriptional activation of the c-fms-luciferase reporter construct upon transient cotransfection with the G-CSFR/gp130wt (GR/wt) or G-CSFR/gp130ΔSTAT (GR/Δ) chimeric receptor constructs (B) and HA-tagged wild-type ERK1 construct (C) in RAW cells. In panel B, transfected cells were incubated with G-CSF (100 ng/ml) in the absence or presence of the ERK1/2 MAP kinase inhibitor U0216 for 48 h and assayed for luciferase activity. The data presented were corrected for transfection efficiency by reference to the activity of Renilla luciferase driven by the thymidine kinase promoter in a cotransfected plasmid. *, P < 0.05 versus basal c-fms-luciferase activity in cells transfected with the c-fms-luciferase reporter construct alone (Fms). (D) Cell lysates from transfected RAW cells were run out on 10% polyacrylamide gels, transferred to nitrocellulose membranes, and then immunoblotted with an anti-ERK1/2 antibody; overexpressed, HA-tagged ERK1 (HA-ERK1; upper band) runs above the endogenous ERK1/2 (middle and lower bands).
In order to test directly whether gp130-dependent ERK1/2 activation enhanced c-fms promoter activity, we assayed the transcriptional activation of a c-fms promoter-luciferase reporter construct (14) following its transient transfection in RAW macrophage cells, which also expressed either a chimeric G-CSFR/gp130wt or G-CSFR/gp130ΔSTAT receptor construct (7). In unstimulated RAW cells, the 3.5-kb proximal c-fms promoter showed a high level of basal activity (Fig. 10B), which was consistent with previous findings (14). However, c-fms-luciferase activity was reduced by 40% upon ligand-induced activation of the G-CSFR/gp130wt receptor. In contrast, c-fms reporter activity was elevated by 55% upon activation of the G-CSFR/gp130ΔSTAT receptor. Moreover, the presence of the ERK1/2 MAP kinase inhibitor U0216 resulted in a threefold reduction in promoter activity upon G-CSFR/gp130ΔSTAT receptor activation. A role for ERK MAP kinase in the transcriptional activation of the c-fms promoter was further demonstrated upon transient cotransfection of cells with constructs for the c-fms promoter-luciferase reporter and wild-type ERK1, overexpression of which has been shown to enhance gene expression (32). As shown in Fig. 10C and 10D, overexpression of wild-type ERK1 increased c-fms reporter activity by 2.5-fold relative to that in cells transfected with the c-fms promoter-luciferase reporter alone. Therefore, we propose that the increased M-CSF responsiveness of BMMs following activation of the gp130ΔSTAT receptor is attributable to enhanced ERK MAP kinase activation which is the direct consequence of the incapacity of the gp130ΔSTAT receptor to activate the STAT pathway. Moreover, our data presented here identify a unique mechanism whereby the differential regulation of the c-fms gene is dependent upon the level of gp130-dependent ERK1/2 MAP kinase activation.
DISCUSSION
We report here the use of mice deficient in gp130-dependent activation of either STAT1/3 (gp130ΔSTAT/ΔSTAT) or SHP2/ERK MAP kinase (gp130Y757F/Y757F) to demonstrate that the capacity of IL-6 family cytokines to inhibit M-CSF-dependent proliferation and differentiation of hematopoietic cells is inversely related to their ability to activate gp130-dependent ERK MAP kinase signaling. Hence, IL-6 and, to a lesser extent, IL-11 exhibited greater inhibition of macrophage colony formation (in response to M-CSF) from peripheral blood and bone marrow M-CFCs from gp130Y757F/Y757F mice than from gp130wt/wt mice. By contrast, in gp130ΔSTAT/ΔSTAT mice, where gp130-dependent ERK MAP kinase activation was enhanced, IL-6 and IL-11 failed to inhibit macrophage colony formation by M-CFCs but, rather, augmented colony formation. Similarly, the presence of IL-6 in liquid cultures enhanced the proliferation of gp130ΔSTAT/ΔSTAT BMMs in response to M-CSF, whereas M-CSF-stimulated proliferation of gp130Y757F/Y757F BMMs was reduced. In all cases, the inhibition by IL-11 was less pronounced than inhibition by IL-6, an observation which is consistent with that made by Clutterbuck et al. (4), and may reflect reduced surface expression of IL-11 receptor α (IL-11Rα) compared to expression of IL-6Rα on macrophage progenitor cells. We also note that peripheral blood M-CFCs from gp130wt/wt and gp130Y757F/Y757F mice were more sensitive than bone marrow M-CFCs to IL-6-induced inhibition of macrophage colony formation. The inhibitory effect of IL-6 has been previously shown to be inversely proportional to the proliferative capacity of the macrophage progenitor, in which case peripheral blood progenitors produce smaller colonies (i.e., have lower proliferative potential) than bone marrow progenitors and are more sensitive to IL-6 inhibition (4). Although the molecular explanation for this differential IL-6 sensitivity is unknown, it most likely reflects a reprogramming of the transcriptional machinery within a progenitor cell during differentiation or maturation, which is associated with a reduced proliferative capacity, to a growth inhibitory response by IL-6.
In gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F BMMs, we found that functional elimination of one signaling pathway resulted in enhanced activation of the remaining pathway in response to IL-6. This observation is consistent with the emerging theme that the gp130-dependent SHP2/ERK MAP kinase and STAT1/3 pathways negatively regulate each other (31, 38). We have previously shown that the absence of gp130-dependent STAT1/3 signaling causes up-regulation of IL-6-induced SHP2 and ERK1/2 tyrosine phosphorylation due to impaired STAT-mediated induction of SOCS1 and SOCS3, two key negative regulators of gp130 signaling (6). Conversely, the gp130Y757F substitution mutation removes the phosphotyrosine residue that simultaneously provides a binding site for SHP2 and SOCS3, and therefore disrupts the capacity of SOCS3 to negatively regulate gp130-dependent signaling (35) and of SHP2 to dephosphorylate critical tyrosine residues in the gp130-JAK signaling complex (18). Therefore, the collective loss of these negative feedback mechanisms may augment JAK activity and subsequent STAT3 activation in gp130Y757F/Y757F mice. Importantly, this reciprocal negative regulation between the STAT and ERK MAP kinase pathways highlights how the overall balance of positive and negative signaling cascades can mediate specific biological responses to IL-6 family cytokines. Indeed, recent examples include the findings that mucosal wound healing (38) and the Th1 versus Th2 immune response (31) are influenced by the balance of gp130-dependent SHP2/ERK MAP kinase and STAT3 activation.
The other major finding uncovered by our study presented here is the regulation of c-fms gene expression by IL-6 family cytokines via the gp130-dependent ERK MAP kinase pathway. The c-fms promoter contains several conserved Ets/AP-1 growth factor response elements (15), and the synergistic actions of the Ras pathway and Ets family transcription factors to induce expression of M-CSF-responsive genes have been demonstrated (12, 41). Furthermore, activation of the Ras/ERK MAP kinase pathway is necessary for both Ets-2 gene expression and protein activation (8), which in turn positively regulates Ras-dependent proliferation in response to M-CSF (20, 21). The Ras pathway has been shown to be a signaling component downstream of Fms that is necessary for M-CSF-driven mitogenesis (2, 21), which is also supported by our observation here that the ERK1/2 MAP kinase-specific inhibitor U0216 reduced M-CSF-dependent proliferation of BMMs. However, here we provide compelling evidence for a situation where the Ras-MAP kinase pathway (activated via gp130) can also be functionally upstream of Fms, thereby providing a link by which IL-6 family cytokines suppress the responsiveness of macrophages to M-CSF. Thus, we speculate that a reduction in gp130-dependent ERK1/2 MAP kinase signaling, as is the case for gp130Y757F/Y757F macrophages, might suppress the activity of Ets family members, and therefore the level of c-fms expression, which in turn leads to a reduced biological response to M-CSF-induced signaling. Detailed analysis of the ability of gp130-dependent ERK MAP kinase signals to induce the expression of downstream effectors of M-CSF-stimulated proliferative signaling, such as Ets-2 and AP-1, and induce c-fms promoter activity in the absence of specific (i.e., Ets/AP-1) binding sites, will ultimately test the validity of this model.
Several recent studies have highlighted the importance of STAT3 signaling during maturation and activation of macrophages. A study by Takeda et al. (37) revealed that IL-6 not only failed to inhibit the M-CSF-driven proliferation of STAT3-deficient BMMs but, rather, enhanced their proliferation. Notably, this observation correlates with our data presented here for gp130ΔSTAT/ΔSTAT macrophages and is therefore consistent with our interpretation that exaggerated gp130-dependent ERK MAP kinase activation, as a consequence of an absence in STAT3 signaling, enhances these macrophage activities. In contrast, it has also been reported that constitutive activation of STAT3 in macrophages inhibited their proliferation (30). While these reports provide compelling evidence for a STAT3-specific role in mediating antiproliferative effects on macrophages, they provide little evidence as to how altering the activation status of STAT3 elicits these actions. In light of our observations here, we propose that altering the level of STAT3 activation, for example by gene-specific inhibition or expression of a constitutively active STAT3, may affect target gene regulation indirectly by unbalancing the net output of complex signaling networks. Specifically, we identify here that, at least in the context of gp130 activation, the signaling output from the ERK MAP kinase cascade is inversely related to the extent of STAT3 activation.
The inhibition of M-CSF-stimulated macrophage formation by IL-6 family cytokines contrasts with the synergistic actions of this cytokine family with other hematopoietic growth factors, such as stem cell factor and IL-3, on promoting the proliferation and differentiation of multipotential (immature) hematopoietic progenitors (16, 39). Furthermore, IL-6 promotes the formation of macrophages in cultures containing granulocyte-M-CSF (17, 27) and has also been shown to enhance the expansion of primitive macrophage-committed (pre-CFU-M) progenitors (26). These observations suggest differences in the transcriptional programming of immature and committed (i.e., CFU-M) hematopoietic progenitors to respond to gp130 signaling, with the latter primed for inhibition by IL-6 family cytokines. Indeed, such a notion is supported by recent observations by Clutterbuck et al. (4) on the preferential inhibitory action of IL-6 on macrophage progenitors displaying a reduced proliferative capacity, which itself is a hallmark of committed cells undergoing differentiation and maturation. Although a molecular mechanism underlying these opposing effects remains to be elucidated, in both situations it is likely that the effect of IL-6 is via the regulation of c-fms expression (3). In this regard, it is therefore not surprising that the human and mouse c-fms promoter regions contain binding sites for transcription factors that mediate opposing effects on the transcriptional regulation of c-fms (33). With the recent generation of mice expressing a c-fms-enhanced green fluorescent protein reporter gene within the monocyte/macrophage lineage throughout development (34), the tools are now in place for further in vivo analyses of the transcriptional machinery controlling both negative and positive regulation of c-fms during macrophage development.
Acknowledgments
We are grateful to Graham Lieschke and Ken Harder for valuable discussions and critical reading of the manuscript, and we thank Nicholas Wilson and Tony Burgess for critical reading of the manuscript. We also thank Elsbeth Richardson and Maureen Nerrie for technical assistance.
This work was supported in part by a research grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Bortner, D. M., M. Ulivi, M. F. Roussel, and M. C. Ostrowski. 1991. The carboxy-terminal catalytic domain of the GTPase-activating protein inhibits nuclear signal transduction and morphological transformation mediated by the CSF-1 receptor. Genes Dev. 5:1777-1785. [DOI] [PubMed] [Google Scholar]
- 2.Buscher, D., R. A. Hipskind, S. Krautwald, T. Reimann, and M. Baccarini. 1995. Ras-dependent and -independent pathways target the mitogen-activated protein kinase network in macrophages. Mol. Cell. Biol. 15:466-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chomarat, P., J. Banchereau, J. Davoust, and A. K. Palucka. 2000. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat. Immunol. 1:510-514. [DOI] [PubMed] [Google Scholar]
- 4.Clutterbuck, R., R. Powles, J. Millar, and D. Catovsky. 2000. Interleukin-6 and other gp130-dependent cytokines selectively inhibit proliferation of macrophage-lineage hemopoietic progenitor cells. Exp. Hematol. 28:1120-1128. [DOI] [PubMed] [Google Scholar]
- 5.Dai, X. M., G. R. Ryan, A. J. Hapel, M. G. Dominguez, R. G. Russell, S. Kapp, V. Sylvestre, and E. R. Stanley. 2002. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99:111-120. [DOI] [PubMed] [Google Scholar]
- 6.Ernst, M., M. Inglese, P. Waring, I. K. Campbell, S. Bao, F. J. Clay, W. S. Alexander, I. P. Wicks, D. M. Tarlinton, U. Novak, J. K. Heath, and A. R. Dunn. 2001. Defective gp130-mediated signal transducer and activator of transcription (STAT) signaling results in degenerative joint disease, gastrointestinal ulceration, and failure of uterine implantation. J. Exp. Med. 194:189-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst, M., U. Novak, S. E. Nicholson, J. E. Layton, and A. R. Dunn. 1999. The carboxyl-terminal domains of gp130-related cytokine receptors are necessary for suppressing embryonic stem cell differentiation: involvement of STAT3. J. Biol. Chem. 274:9729-9737. [DOI] [PubMed] [Google Scholar]
- 8.Fowles, L. F., M. L. Martin, L. Nelsen, K. J. Stacey, D. Redd, Y. M. Clark, Y. Nagamine, M. McMahon, D. A. Hume, and M. C. Ostrowski. 1998. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol. Cell. Biol. 18:5148-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukada, T., M. Hibi, Y. Yamanaka, M. Takahashi-Tezuka, Y. Fujitani, T. Yamaguchi, K. Nakajima, and T. Hirano. 1996. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5:449-460. [DOI] [PubMed] [Google Scholar]
- 10.Gearing, D. P., C. J. Thut, T. VandeBos, S. D. Gimpel, P. B. Delaney, J. King, V. Price, D. Cosman, and M. P. Beckmann. 1991. Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J. 10:2839-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhartz, C., B. Heesel, J. Sasse, U. Hemmann, C. Landgraf, J. Schneider-Mergener, F. Horn, P. C. Heinrich, and L. Graeve. 1996. Differential activation of acute phase response factor/STAT3 and STAT1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. I. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J. Biol. Chem. 271:12991-12998. [DOI] [PubMed] [Google Scholar]
- 12.Guidez, F., A. C. Li, A. Horvai, J. S. Welch, and C. K. Glass. 1998. Differential utilization of Ras signaling pathways by macrophage colony-stimulating factor (CSF) and granulocyte-macrophage CSF receptors during macrophage differentiation. Mol. Cell. Biol. 18:3851-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton, D. J., A. A. Hilton, A. Raicevic, S. Rakar, M. Harrison-Smith, N. M. Gough, C. G. Begley, D. Metcalf, N. A. Nicola, and T. A. Willson. 1994. Cloning of a murine IL-11 receptor alpha-chain: requirement for gp130 for high affinity binding and signal transduction. EMBO J. 13:4765-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himes, S. R., H. Tagoh, N. Goonetilleke, T. Sasmono, D. Oceandy, R. Clark, C. Bonifer, and D. A. Hume. 2001. A highly conserved c-fms gene intronic element controls macrophage-specific and regulated expression. J. Leukoc. Biol. 70:812-820. [PubMed] [Google Scholar]
- 15.Hume, D. A., X. Yue, I. L. Ross, P. Favot, A. Lichanska, and M. C. Ostrowski. 1997. Regulation of CSF-1 receptor expression. Mol. Reprod. Dev. 46:46-52. [DOI] [PubMed] [Google Scholar]
- 16.Ikebuchi, K., G. G. Wong, S. C. Clark, J. N. Ihle, Y. Hirai, and M. Ogawa. 1987. Interleukin 6 enhancement of interleukin 3-dependent proliferation of multipotential hemopoietic progenitors. Proc. Natl. Acad. Sci. USA 84:9035-9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen, J. H., J. C. Kluin-Nelemans, J. Van Damme, G. J. Wientjens, R. Willemze, and W. E. Fibbe. 1992. Interleukin 6 is a permissive factor for monocytic colony formation by human hematopoietic progenitor cells. J. Exp. Med. 175:1151-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H., T. S. Hawley, R. G. Hawley, and H. Baumann. 1998. Protein tyrosine phosphatase 2 (SHP-2) moderates signaling by gp130 but is not required for the induction of acute-phase plasma protein genes in hepatic cells. Mol. Cell. Biol. 18:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishimoto, T., S. Akira, M. Narazaki, and T. Taga. 1995. Interleukin-6 family of cytokines and gp130. Blood 86:1243-1254. [PubMed] [Google Scholar]
- 20.Klappacher, G. W., V. V. Lunyak, D. B. Sykes, D. Sawka-Verhelle, J. Sage, G. Brard, S. D. Ngo, D. Gangadharan, T. Jacks, M. P. Kamps, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 2002. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 109:169-180. [DOI] [PubMed] [Google Scholar]
- 21.Langer, S. J., D. M. Bortner, M. F. Roussel, C. J. Sherr, and M. C. Ostrowski. 1992. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol. Cell. Biol. 12:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawlor, K. E., I. K. Campbell, K. O'Donnell, L. Wu, and I. P. Wicks. 2001. Molecular and cellular mediators of interleukin-1-dependent acute inflammatory arthritis. Arthritis Rheum. 44:442-450. [DOI] [PubMed] [Google Scholar]
- 23.Lioubin, M. N., G. M. Myles, K. Carlberg, D. Bowtell, and L. R. Rohrschneider. 1994. Shc, Grb2, Sos1, and a 150-kilodalton tyrosine-phosphorylated protein form complexes with Fms in hematopoietic cells. Mol. Cell. Biol. 14:5682-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, H., H. Perlman, L. J. Pagliari, and R. M. Pope. 2001. Constitutively activated Akt-1 is vital for the survival of human monocyte-differentiated macrophages: role of Mcl-1, independent of nuclear factor (NF)-κB, Bad, or caspase activation. J. Exp. Med. 194:113-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lord, K. A., A. Abdollahi, S. M. Thomas, M. DeMarco, J. S. Brugge, B. Hoffman-Liebermann, and D. A. Liebermann. 1991. Leukemia inhibitory factor and interleukin-6 trigger the same immediate early response, including tyrosine phosphorylation, upon induction of myeloid leukemia differentiation. Mol. Cell. Biol. 11:4371-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metcalf, D. 1997. Murine hematopoietic stem cells committed to macrophage/dendritic cell formation: stimulation by Flk2-ligand with enhancement by regulators using the gp130 receptor chain. Proc. Natl. Acad. Sci. USA 94:11552-11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitani, H., N. Katayama, H. Araki, K. Ohishi, K. Kobayashi, H. Suzuki, K. Nishii, M. Masuya, K. Yasukawa, N. Minami, and H. Shiku. 2000. Activity of interleukin 6 in the differentiation of monocytes to macrophages and dendritic cells. Br. J. Haematol. 109:288-295. [DOI] [PubMed] [Google Scholar]
- 28.Mosley, B., C. De Imus, D. Friend, N. Boiani, B. Thoma, L. S. Park, and D. Cosman. 1996. Dual oncostatin M (OSM) receptors: cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J. Biol. Chem. 271:32635-32643. [DOI] [PubMed] [Google Scholar]
- 29.Murakami, M., M. Hibi, N. Nakagawa, T. Nakagawa, K. Yasukawa, K. Yamanishi, T. Taga, and T. Kishimoto. 1993. IL-6-induced homodimerization of gp130 and associated activation of a tyrosine kinase. Science 260:1808-1810. [DOI] [PubMed] [Google Scholar]
- 30.O'Farrell, A. M., Y. Liu, K. W. Moore, and A. L. Mui. 1998. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J. 17:1006-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtani, T., K. Ishihara, T. Atsumi, K. Nishida, Y. Kaneko, T. Miyata, S. Itoh, M. Narimatsu, H. Maeda, T. Fukada, M. Itoh, H. Okano, M. Hibi, and T. Hirano. 2000. Dissection of signaling cascades through gp130 in vivo: reciprocal roles for STAT3- and SHP2-mediated signals in immune responses. Immunity 12:95-105. [DOI] [PubMed] [Google Scholar]
- 32.Park, J. H., and L. Levitt. 1993. Overexpression of mitogen-activated protein kinase (ERK1) enhances T-cell cytokine gene expression: role of AP1, NF-AT, and NF-KB. Blood 82:2470-2477. [PubMed] [Google Scholar]
- 33.Reddy, M. A., B. S. Yang, X. Yue, C. J. Barnett, I. L. Ross, M. J. Sweet, D. A. Hume, and M. C. Ostrowski. 1994. Opposing actions of c-ets/PU. 1 and c-myb protooncogene products in regulating the macrophage-specific promoters of the human and mouse colony-stimulating factor-1 receptor (c-fms) genes. J. Exp. Med. 180:2309-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasmono, R. T., D. Oceandy, J. W. Pollard, W. Tong, P. Pavli, B. J. Wainwright, M. C. Ostrowski, S. R. Himes, and D. A. Hume. 2003. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101:1155-1163. [DOI] [PubMed] [Google Scholar]
- 35.Schmitz, J., M. Weissenbach, S. Haan, P. C. Heinrich, and F. Schaper. 2000. SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. J. Biol. Chem. 275:12848-12856. [DOI] [PubMed] [Google Scholar]
- 36.Stanley, E. R., K. L. Berg, D. B. Einstein, P. S. Lee, F. J. Pixley, Y. Wang, and Y. G. Yeung. 1997. Biology and action of colony-stimulating factor-1. Mol. Reprod. Dev. 46:4-10. [DOI] [PubMed] [Google Scholar]
- 37.Takeda, K., B. E. Clausen, T. Kaisho, T. Tsujimura, N. Terada, I. Forster, and S. Akira. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10:39-49. [DOI] [PubMed] [Google Scholar]
- 38.Tebbutt, N. C., A. S. Giraud, M. Inglese, B. Jenkins, P. Waring, F. J. Clay, S. Malki, B. M. Alderman, D. Grail, F. Hollande, J. K. Heath, and M. Ernst. 2002. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat. Med. 8:1089-1097. [DOI] [PubMed] [Google Scholar]
- 39.Tsuji, K., S. D. Lyman, T. Sudo, S. C. Clark, and M. Ogawa. 1992. Enhancement of murine hematopoiesis by synergistic interactions between steel factor (ligand for c-kit), interleukin-11, and other early acting factors in culture. Blood 79:2855-2860. [PubMed] [Google Scholar]
- 40.Wiktor-Jedrzejczak, W., A. Bartocci, A. W. Ferrante, Jr., A. Ahmed-Ansari, K. W. Sell, J. W. Pollard, and E. R. Stanley. 1990. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc. Natl. Acad. Sci. USA 87:4828-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, B. S., C. A. Hauser, G. Henkel, M. S. Colman, C. Van Beveren, K. J. Stacey, D. A. Hume, R. A. Maki, and M. C. Ostrowski. 1996. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol. Cell. Biol. 16:538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]