Abstract
FoxO transcription factors and sirtuin family deacetylases regulate diverse biological processes, including stress responses and longevity. Here we show that the Caenorhabditis elegans sirtuin SIR-2.4—homolog of mammalian SIRT6 and SIRT7 proteins—promotes DAF-16–dependent transcription and stress-induced DAF-16 nuclear localization. SIR-2.4 is required for resistance to multiple stressors: heat shock, oxidative insult, and proteotoxicity. By contrast, SIR-2.4 is largely dispensable for DAF-16 nuclear localization and function in response to reduced insulin/IGF-1-like signaling. Although acetylation is known to regulate localization and activity of mammalian FoxO proteins, this modification has not been previously described on DAF-16. We find that DAF-16 is hyperacetylated in sir-2.4 mutants. Conversely, DAF-16 is acetylated by the acetyltransferase CBP-1, and DAF-16 is hypoacetylated and constitutively nuclear in response to cbp-1 inhibition. Surprisingly, a SIR-2.4 catalytic mutant efficiently rescues the DAF-16 localization defect in sir-2.4 null animals. Acetylation of DAF-16 by CBP-1 in vitro is inhibited by either wild-type or mutant SIR-2.4, suggesting that SIR-2.4 regulates DAF-16 acetylation indirectly, by preventing CBP-1-mediated acetylation under stress conditions. Taken together, our results identify SIR-2.4 as a critical regulator of DAF-16 specifically in the context of stress responses. Furthermore, they reveal a novel role for acetylation, modulated by the antagonistic activities of CBP-1 and SIR-2.4, in modulating DAF-16 localization and function.
Author Summary
Sensing and responding appropriately to environmental insults is a challenge facing all organisms. In the roundworm C. elegans, the FoxO protein DAF-16 moves to the nucleus in response to stress, where it regulates gene expression and plays a key role in ensuring organismal survival. In this manuscript, we characterize SIR-2.4 as a novel factor that promotes DAF-16 function during stress. SIR-2.4 is a member of a family of proteins called sirtuins, some of which promote increased lifespan in model organisms. Worms lacking SIR-2.4 show impaired DAF-16 nuclear recruitment, DAF-16–dependent gene expression, and survival in response to a variety of stressors. SIR-2.4 regulates DAF-16 by indirectly affecting levels of a modification called acetylation on DAF-16. Overall, our work has revealed SIR-2.4 to be a key new factor in stress resistance and DAF-16 regulation in C. elegans. Future studies will address whether mammalian SIR-2.4 homologs SIRT6 and SIRT7 act similarly towards mammalian FoxO proteins.
Introduction
Elucidation of mechanisms regulating stress resistance and longevity has been aided tremendously by the use of invertebrate models. FoxO transcription factors regulate multiple biological processes in many organisms [1]. In C. elegans and Drosophila, increased FoxO activity promotes longevity, fat storage, and stress resistance [2]. Mammals possess four FoxO homologs, with partially redundant and distinct functions: FoxO1, FoxO3A, FoxO4, and FoxO6 [1]. These proteins regulate apoptosis, cell cycle arrest, oxidative defense, DNA repair, metabolism, differentiation, stem cell function, and tumor suppression in a cell type- and context-specific manner [1].
FoxO activity is tightly controlled, and subcellular localization is a principal mechanism of FoxO regulation [3]. In this context, insulin/IGF-1-like signaling (IIS) is the major influence on FoxO function. IIS leads to FoxO phosphorylation and cytoplasmic segregation in a complex with 14-3-3 chaperone proteins. Conversely, stress stimuli promote nuclear translocation of FoxO proteins by multiple mechanisms, including activation of stress kinases that modify FoxO proteins on residues distinct from those phosphorylated in IIS [2]. In response to oxidative insult, mammalian FoxO proteins are acetylated [4], [5], [6], [7], mono-ubiquitylated [8], and phosphorylated [9], [10], [11], [12]. Overall, these post-translation modifications function as a “FoxO code”, providing a means by which FoxO activity is finely regulated in response to various stimuli to promote altered metabolism, stress responses, or cell death [3].
The sirtuins are an evolutionarily conserved protein family impacting many biological processes, including longevity, stress responses, metabolism, and cancer [13]. Sirtuins modify target proteins by means of their NAD+-dependent lysine deacetylase and ADP-ribosyltransferase activities. Lysine acetylation has emerged as a post-translational modification with a key role in modulating protein function, akin to phosphorylation [14]. Mammals possess seven sirtuins, SIRT1-SIRT7. The C. elegans genome encodes four sirtuins, SIR-2.1 through SIR-2.4, corresponding to mammalian SIRT1 (SIR-2.1), SIRT4 (SIR-2.2 and SIR-2.3), and SIRT6/7 (SIR-2.4) [15]. SIR-2.1 has been implicated in numerous physiologic processes, including stress responses [16], [17], [18], [19], [20], [21], [22], [23]. In contrast, functions of other worm sirtuins are largely uncharacterized [24].
In different organisms, sirtuins modulate FoxO activity via diverse means. In C. elegans, there are reports that SIR-2.1 extends longevity in a DAF-16-dependent manner [21], [25], [26], [27]. However, this is currently a disputed finding [28], [29]. In mammals, SIRT1 directly deacetylates FoxO proteins in response to oxidative stress. The effect of FoxO deacetylation is somewhat controversial [4], [6], [7], [30], [31], but it is likely that the overall outcome is to promote DNA repair and cell cycle arrest while inhibiting apoptosis [3]. SIRT2 also deacetylates FoxO proteins to inhibit adipocytic differentiation [32], [33] and regulate levels of intracellular reactive oxygen species (ROS) [34]. SIRT1 and SIRT2-mediated deacetylation of FoxO1 promotes nuclear accumulation of this protein [5], [32]. Acetylation of FoxO1 has also been reported to attenuate DNA binding, to promote AKT-mediated FoxO1 phosphorylation, and to direct FoxO1 to nuclear PML bodies [7], [35]. The mitochondrial sirtuin SIRT3 has also been proposed to modulate FoxO function [36], [37].
Here, we characterize functions of the C. elegans sirtuin SIR-2.4, a protein about which little is currently known. We find that SIR-2.4 plays a crucial role in promoting DAF-16 transcriptional activity and stress-induced nuclear localization, and is required for normal stress resistance in the worm. However, SIR-2.4 is largely dispensable for DAF-16 nuclear localization and function in the context of reduced IIS. We show directly for the first time that DAF-16 itself is acetylated, by the acetyltransferase CBP-1. SIR-2.4 attenuates DAF-16 acetylation in a non-catalytic activity-dependent manner, and catalytic function of SIR-2.4 is dispensable for regulation of DAF-16 localization in response to stress. Acetylation of DAF-16 by CBP-1 is inhibited in the presence of SIR-2.4. Our results indicate acetylation plays a key role in regulating DAF-16 localization and function, and that levels of this modification are modulated by the antagonistic functions of CBP-1 and SIR-2.4.
Results
SIR-2.4 is required for efficient stress-induced DAF-16 nuclear localization
Mammalian SIRT6 and SIRT7 proteins both promote genotoxic stress resistance [38], [39]. We therefore tested a potential role for SIR-2.4 in stress resistance and DAF-16 regulation. We generated an RNAi construct encoding nucleotides 1–467 of the SIR-2.4 open reading frame in the RNAi vector L4440. sir-2.4 knockdown (KD) resulted in no obvious defects under basal conditions. However, sir-2.4 RNAi severely impaired stress-induced DAF-16 nuclear localization (Figure 1A). sir-2.4 RNAi inhibited DAF-16 nuclear translocation in response to either heat shock or oxidative insult by ∼50% shortly after stress induction (Figure 1B). At later timepoints, DAF-16 did translocate to the nucleus in sir-2.4 KD worms (see below and data not shown).
Figure 1. SIR-2.4, but not SIR-2.1, is required for stress-induced DAF-16 nuclear localization.
TJ356 animals carrying an integrated daf-16::gfp array were fed either vector control or sir-2.4 RNAi bacteria for at least one generation before being subjected to heat-shock or oxidative stress. (A) Images of TJ356 animals grown on control or sir-2.4 RNAi bacteria after 15 min heat-shock. (B) Quantification of DAF-16::GFP nuclear accumulation in response to heat-shock (35°C for 15 min.) or oxidative stress (1.5 mM H2O2 for 1 hr). Worms were scored for the presence or absence of GFP accumulation within the intestinal nuclei (n = 120 or greater for all treatments). An animal was scored as having nuclear GFP if one or more intestinal nuclei contained DAF-16-GFP. (C–D) Time course analysis of DAF-16::GFP nuclear accumulation in response to stress. TJ356 or EQ200 [sir-2.4(n5137); daf-16::gfp] animals grown on either control or sir-2.1 RNAi bacteria were subjected to (C) heat-shock (35°C) or (D) oxidative stress (1.5 mM H2O2). Worms were scored for GFP accumulation within the head hypodermic nuclei at day 1 of adulthood (n = 30∼50) every 5–30 min.
sir-2.4 deletion is reported to confer lethality/sterility (National Bioresource Project, Japan). However, during the course of analyzing the effects of sir-2.4 RNAi, we obtained a viable strain with a deletion removing all but the initial 9 amino acids of the SIR-2.4 open reading frame (kind gift of H.R. Horvitz; see Materials and Methods section for a complete description of this strain). As with sir-2.4 RNAi, sir-2.4 KO animals showed no apparent defects under unstressed conditions. However, like sir-2.4 KD worms, sir-2.4 knockouts (KOs) showed significantly delayed stress-induced DAF-16 nuclear translocation in response to oxidative stress (Figure 1C; p<0.001 by Poisson regression analysis) and heat shock (Figure 1D; p<0.001). We conclude that SIR-2.4 is dispensable for viability and fertility, but plays a crucial role in directing DAF-16 to the nucleus in response to stress, particularly at early time points following stress induction.
It has been reported that SIR-2.1 and 14-3-3 proteins act in concert to activate DAF-16 [21], [26]. To examine whether SIR-2.1 plays a role in directing DAF-16 to the nucleus in response to stress, we assessed the effect of sir-2.1 KD on DAF-16 nuclear localization. sir-2.1 KD alone had little impact on DAF-16 nuclear recruitment in response to either oxidative insult (Figure 1C; p<0.72) or heat stress (Figure 1D; p<0.44), indicating that SIR-2.1 does not play a major role in stress-induced DAF-16 nuclear localization, consistent with published data [17]. Moreover, KD of sir-2.1 in the context of sir-2.4 mutation did not produce any statistically significant additional delay in DAF-16 nuclear recruitment versus sir-2.4 KO alone (Figure 1C–1D; p<0.89 and p<0.13 for oxidative and heat stress, respectively). We conclude that SIR-2.4, but not SIR-2.1, plays a major role in promoting rapid nuclear recruitment of DAF-16 in response to oxidative stress or heat shock. These results imply that SIR-2.1 and SIR-2.4 act in distinct pathways to influence DAF-16 functions.
While stress-induced subcellular translocation of DAF-16 was affected in all cell types by sir-2.4 inhibition, we did note that sir-2.4 KD and KO had a bigger impact in head hypodermis cells than in intestinal cells (data not shown). SIR-2.4 was very weakly expressed in most cell types, but showed much stronger expression in a subset of head and tail neurons as well as in a subset of somatic gonad cells (Figure S1). We have not yet formally assessed the tissue requirements for SIR-2.4 function in the regulation of DAF-16 localization, though our functional data suggest a cell autonomous mechanism (see below).
SIR-2.4 promotes DAF-16–dependent transcription under basal and stress conditions
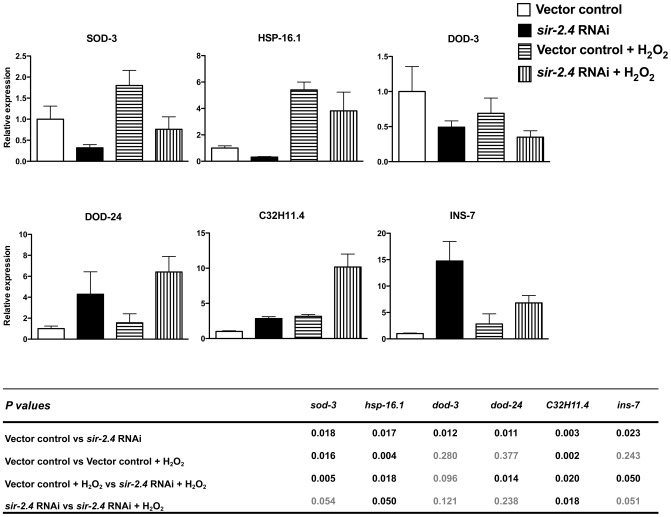
DAF-16 carries out its functions by transcriptional regulation of a large number of target genes [40], [41], [42]. The role of SIR-2.4 in DAF-16-dependent gene expression was tested in the context of six well-known DAF-16 targets. We confirmed the published role of DAF-16 in regulating expression of all six of these genes (Figure S2A). Under both basal and stress conditions, sir-2.4 RNAi led to decreased mRNA levels of three genes positively regulated by DAF-16 (SOD-3, HSP-16.1, and DOD-3), (Figure 2, top row). Expression of these DAF-16 targets was similarly attenuated in the sir-2.4 KO strain (Figure S2B). Conversely, expression of three genes negatively regulated by DAF-16 was greatly increased by sir-2.4 RNAi (DOD-24, C32H11.4, and INS-7) (Figure 2, bottom row). We conclude that SIR-2.4 promotes DAF-16 transcriptional function under both basal and oxidative stress conditions.
Figure 2. SIR-2.4 is required for optimal DAF-16–dependent gene expression.
Wild-type N2 animals fed on either vector control or sir-2.4 RNAi bacteria from the time of hatching were exposed to 10 mM H2O2 for 80 min. Relative mRNA levels of SOD-3, HSP-16.1, DOD-3, DOD-24, C32H11.4, and INS-7 were measured by quantitative RT-PCR and the means of three different sample sets are shown. Relative mRNA levels were normalized against ACT-1 (beta-actin). Error bars: ± STD. Statistical significance as determined by two-tailed t-test is shown in the table below; significant differences are represented in black font.
SIR-2.4 is required for normal stress resistance
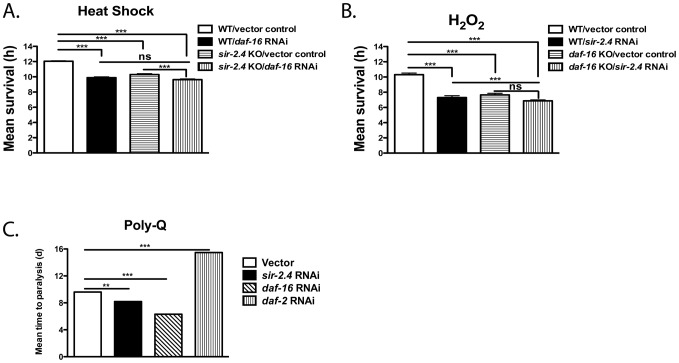
DAF-16 is a key regulator of stress responses in C. elegans [43]. We therefore tested the impact of SIR-2.4 on stress resistance. sir-2.4 KO and sir-2.4 KD worms were hypersensitive to heat shock (Figure 3A, Table S1) and oxidative insult (Figure 3B, Table S1). Simultaneous inhibition of both sir-2.4 and daf-16 increased stress sensitivity to a similar extent as observed in daf-16 single mutants, and the degree of hypersensitivity conferred by either single KO/KD alone was similar (Figure 3A–3B, Figure S2C–S2D), suggesting that SIR-2.4 and DAF-16 modulate stress resistance via a common pathway. Conversely, overexpression of SIR-2.4 did not produce increased stress resistance (Figure S3A–S3B; Table S1); hence SIR-2.4 levels are not limiting for DAF-16 regulation and stress resistance.
Figure 3. SIR-2.4 promotes stress resistance.
(A) Wild-type N2 or sir-2.4(n5137) worms grown on vector control or daf-16 RNAi bacteria were subjected to heat-shock at 35°C and scored for viability every 1-2 hours. (B) Wild-type N2 or daf-16(mu86) worms grown on vector control or sir-2.4 RNAi bacteria were treated with 1.5 mM H2O2 and scored for viability every 1–2 hours. Data are mean survival±SEM, in hours for (A–B). ***, p<0.001; ns, p>0.05. See Table S1 for statistical analysis. (C) AM140 worms expressing a poly-Q tract (Q35::YFP) were seeded on the RNAi bacteria indicated and scored for poly-Q induced paralysis every other day. Data are mean time to paralysis in days±SEM. ***, p<0.001; **, p<0.01; ns, p>0.05.
Expression of fluorescently tagged polyglutamine (polyQ) repeat-containing proteins in C. elegans body wall muscle causes paralysis that is antagonized by DAF-16 [44], [45], a model for proteotoxicity occurring in human neurodegenerative diseases such as Huntington's disease. The role of SIR-2.4 in proteotoxicity resistance was assessed. Worms expressing 35 glutamine residues conjugated to YFP in body wall muscle (unc-54p::Q35::YFP) were grown on daf-2 RNAi, daf-16 RNAi, sir-2.4 RNAi, or control bacteria. daf-2 RNAi delayed, and both daf-16 and sir-2.4 RNAi accelerated, the onset of Q35::YFP-induced paralysis (Figure 3C). Thus, like DAF-16, SIR-2.4 is required for resistance to multiple stressors, including proteotoxic injury.
SIR-2.4 is largely dispensable for DAF-16 function in response to reduced IIS
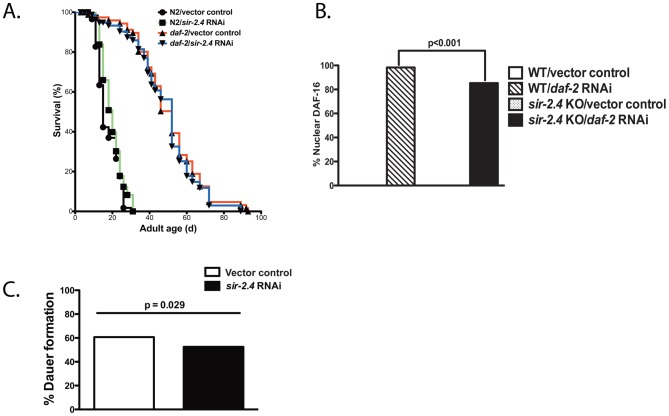
Reduced IIS promotes DAF-16 nuclear localization and functions independently of exogenous stress. We performed several assays to determine whether SIR-2.4 regulates DAF-16 in the context of IIS. Many manipulations that reduce IIS increase lifespan in C. elegans. However, neither sir-2.4 RNAi nor SIR-2.4 overexpression affected the lifespan of wild-type worms (Figure 4A and Figure S3C, Table S2), nor did sir-2.4 RNAi suppress increased longevity of daf-2 (e1370) insulin/IGF-I-like receptor mutants (Figure 4A, Table S2). sir-2.4 deletion or sir-2.4 RNAi minimally impacted DAF-16 nuclear translocation induced by reduced IIS (Figure 4B and Figure S2E), in contrast to its potent impact on stress-induced DAF-16 relocalization. Moreover, sir-2.4 RNAi only slightly impaired dauer formation, a process antagonized by IIS (Figure 4C). We conclude that that the effects of SIR-2.4 on DAF-16 are largely independent of IIS, and are most functionally significant in the context of stress.
Figure 4. Minimal impact of SIR-2.4 on IIS-induced longevity, DAF-16 nuclear localization induced by reduced IIS, and dauer formation.
(A) Lifespan analysis of wild-type (N2) animals or daf-2(e1370) mutants grown on vector control bacteria (black or red) or sir-2.4 RNAi bacteria (green or blue) at 20°C. (B) DAF-16 nuclear localization was assessed in TJ356 (expressing daf-16::gfp in WT background) or EQ200 (expressing daf-16::gfp in sir-2.4(n5137) background) animals. Animals were fed with either vector control or daf-2 RNAi from the time of hatching. Worms were scored for the presence or absence of GFP accumulation within the head hypodermic nuclei as day 1 adult (n = 116 or greater) under unstressed condition. P-values were calculated by Pearson's chi-square test. (C) daf-2(e1370) mutants (P0) were fed with control or sir-2.4 RNAi bacteria at 20°C. F1 eggs were then moved to 22°C for 72 hours prior to being scored for dauer formation (n = 336 or greater). P-values were calculated by Pearson's chi-square test.
SIR-2.4 regulates DAF-16 acetylation and localization independently of its catalytic function
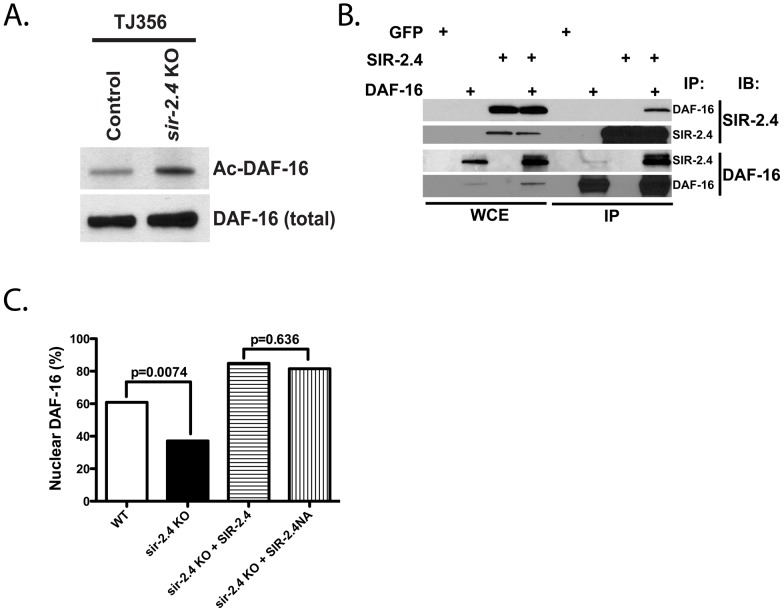
In mammals, SIRT1 and other sirtuins deacetylate FoxO proteins to promote stress responses and other processes [4], [6], [30], [31], [32], [33], [36]. Although DAF-16 interacts with the acetyltransferases CBP and p300 [46], acetylation of DAF-16 itself has never been demonstrated. Given our data linking SIR-2.4 to stress resistance and DAF-16 localization and function, we tested whether DAF-16 was acetylated, and whether SIR-2.4 might play a role in regulating levels of this modification. Indeed, we were able to detect DAF-16 acetylation in C. elegans, and levels of this modification were elevated in sir-2.4 KO (Figure 5A) or KD worms (Figure S4A). Moreover, reciprocal co-immunoprecipitation studies revealed that SIR-2.4 binds to DAF-16 in mammalian cells (Figure 5B), suggesting that SIR-2.4 might interact with DAF-16 to deacetylate this protein. To test this hypothesis, we performed in vitro deacetylation assay with purified SIR-2.4 proteins and pre-acetylated DAF-16 as substrates. However, in multiple experiments we were unable to detect significant deacetylation of DAF-16 by SIR-2.4 in vitro, despite efficiently deacetylating histones with mammalian SIRT1 and SIRT6 in parallel under identical conditions (data not shown).
Figure 5. SIR-2.4 interacts with DAF-16 and promotes DAF-16 deacetylation and function independently of catalytic activity.
(A) sir-2.4 deletion promotes DAF-16 hyperacetylation. DAF-16 acetylation was assessed in control or sir-2.4 KO worms by acetyl-lysine immunoprecipitation followed by GFP immunoblot. (B) SIR-2.4 and DAF-16 interact. Plasmids encoding FLAG-tagged SIR-2.4 and HA-tagged DAF-16 were transfected into 293T cells as indicated (GFP, negative control). Immunoprecipitation and immunoblotting were performed as shown. (C) Rescue of DAF-16 nuclear localization with a catalysis-defective sir-2.4 mutant. Stable transgenic strains of sir-2.4(n5137) were generated expressing either wild-type SIR-2.4 or the sir-2.4 N124A mutant. Worms were scored for GFP accumulation within the head hypodermic nuclei as day 1 adult (n = 50 or greater) after 20 min of heat-shock at 35°C. P-values were calculated by Pearson's chi-square test.
Given this result, the requirement for SIR-2.4 catalytic function in DAF-16 localization was directly tested in worms. As shown above, sir-2.4 KO worms showed impaired DAF-16 nuclear localization in response to stress. This defect was rescued by transgenic expression of wild-type SIR-2.4 (Figure 5C). Curiously, expression of the SIR-2.4 N124A mutant, bearing a mutation at highly conserved residues predicted based on homology to greatly diminish catalytic function [47], restored DAF-16 relocalization as efficiently as wild-type SIR-2.4. Unfortunately, co-overexpression of DAF-16 together with chromosomally integrated SIR-2.4 caused worms to be very sick and produce few progeny, precluding a direct assessment of DAF-16 acetylation in rescued animals (data not shown). We conclude that SIR-2.4 regulates DAF-16 acetylation and localization independently of its catalytic activity, potentially by preventing the action of an acetyltransferase on DAF-16 and/or recruiting another deacetylase.
Potential cooperativity between SIR-2.1 and SIR-2.4 with respect to regulation of DAF-16 acetylation was assessed (Figure S4B). As before, sir-2.4 KO caused a large increase in DAF-16 acetylation. Interestingly sir-2.1 RNAi caused a modest but detectable increase in DAF-16 acetylation. In sir-2.1 KD;sir-2.4 KO animals, levels of DAF-16 acetylation resembled those in the sir-2.4 KO. Thus, SIR-2.1 does impact DAF-16 acetylation, perhaps by directly deacetylating DAF-16, as has been shown for its homolog mammalian SIRT1. However, in contrast to SIR-2.4, the impact of SIR-2.1 on DAF-16 acetylation is quantitatively insufficient to promote significant DAF-16 nuclear localization (our results and [17]). Alternatively, it is possible that SIR-2.1 and SIR-2.4 modulate acetylation on different DAF-16 lysine residue that regulate divergent aspects of DAF-16 function.
SIR-2.4 blocks CBP1-dependent DAF-16 acetylation
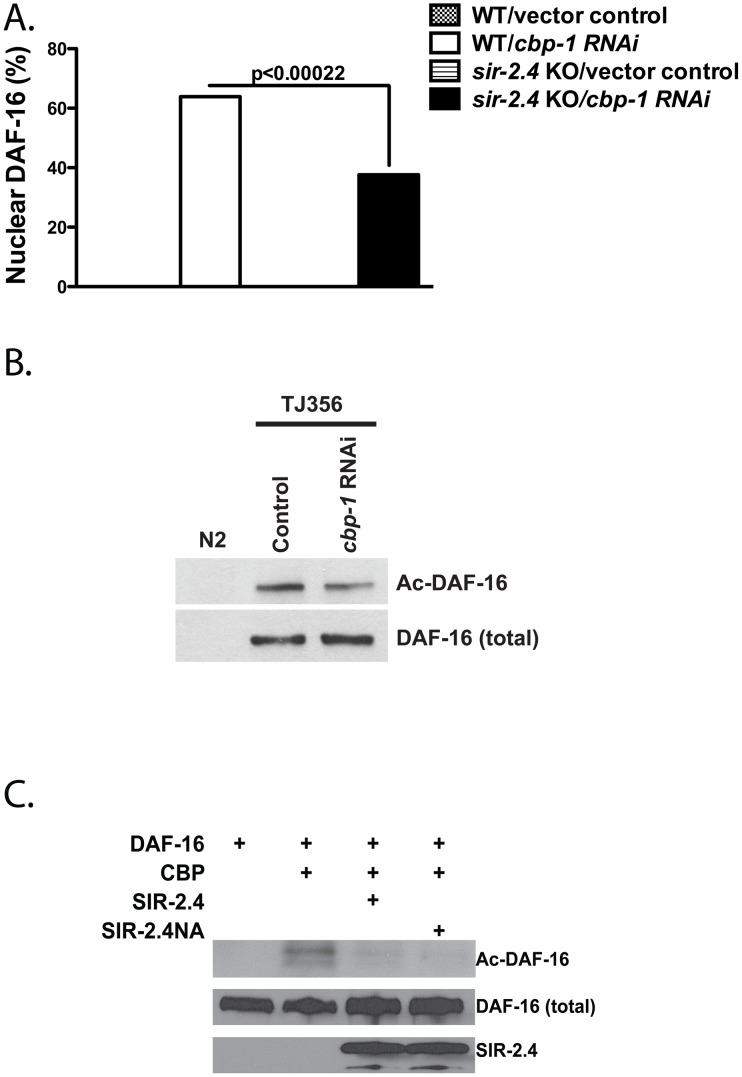
Acetylation regulates of FoxO proteins in a context-specific manner [3]. To identify the enzyme that acetylates DAF-16 in vivo, five C. elegans acetyltransferases – CBP-1 (p300/CBP), PCAF-1 (p300/CBP-associated factor), MYS-1, MYS-2 and MYS-3 (MYST histone acetyltransferases) – were examined for potential impacts on nuclear translocation and acetylation of DAF-16. RNAi KD of cbp-1 induced dramatic, virtually complete DAF-16 nuclear translocation under basal, unstressed conditions (Figure 6A). In contrast, inhibition of pcaf-1 or simultaneous KD of all three MYST acetyltransferases had no effect on DAF-16 localization (data not shown). This effect of cbp-1 RNAi on DAF-16 nuclear localization was largely epistatic to the impact of sir-2.4 KO (Figure 6A). These results are consistent with a recent report showing that mammalian CBP promotes cytoplasmic localization of FoxO3A [48]. Consistent with a role for CBP-1 in acetylating DAF-16 in vivo, we found that DAF-16 is hypo-acetylated in cbp-1 KD animals (Figure 6B). To identify CBP-1 target sites on DAF-16, we performed mass spectrometry analysis on recombinant DAF-16 treated with CBP-1 in vitro (Figure 6C). We identified four CBP-1 dependent acetylation sites on DAF-16: K248, K253, K375 and K379 (Table S3). Together, these findings suggest that DAF-16 is acetylated by CBP-1 in vivo to promote its nuclear exclusion under basal conditions.
Figure 6. SIR-2.4 inhibits CBP1-mediated DAF-16 acetylation.
(A) DAF-16 nuclear localization was assessed in TJ356 (expressing daf-16::gfp in WT background) or EQ200 (expressing daf-16::gfp in sir-2.4(n5137) background) animals. Animals (n = 90 or greater) were scored for DAF-16::GFP nuclear translocation as described in Figure 4B. (B) cbp-1 KD decreases DAF-16 acetylation. DAF-16 acetylation was assessed in TJ356 worms grown on either control or cbp-1 RNAi by acetyl-lysine immunoprecipitation followed by GFP immunoblot. (C) SIR-2.4 blocks CBP1-dependent DAF-16 acetylation in vitro. Purified DAF-16 was incubated with CBP in the presence of WT SIR-2.4 or the SIR-2.4 NA mutant at 37°C. DAF-16 acetylation levels were assessed as described in (B).
It has been reported that cbp-1 RNAi confers stress sensitivity and reduced lifespan [49]. We confirmed that cbp-1 RNAi caused hypersensitivity to heat shock (Figure S5A) and oxidative insult (Figure S5B). Moreover, as previously reported, cbp-1 RNAi caused worms to be very short-lived (Figure S5C). We conclude that the constitutive nuclear localization of DAF-16 occurring in the context of cbp-1 inhibition is inadequate to promote stress resistance or increased longevity, perhaps due to loss of cbp-1 acetylation of other key targets necessary for health and normal lifespan, potentially in many different cell types. Consistent with this hypothesis, CBP-1 is widely express in most or all somatic cells of the worms [50], [51], [52].
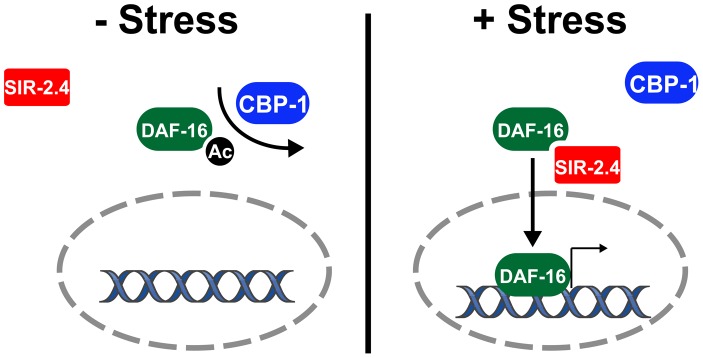
The DEK protein binds histones to prevent p300- and PCAF-mediated histone acetylation [53]. To test whether SIR-2.4 might exert its effect on DAF-16 through a similar mechanism, we examined the effect of the presence of SIR-2.4 on CBP-mediated DAF-16 acetylation in vitro. Purified DAF-16 and CBP were incubated with or without SIR-2.4. The presence of SIR-2.4 blocked CBP-mediated acetylation of DAF-16 (Figure 6C). A similar result was also obtained when DAF-16 and CBP were incubated with the SIR-2.4 N124A catalytic mutant (Figure 6C). We conclude that SIR-2.4 inhibits CBP-mediated DAF-16 acetylation likely through protein-protein interaction, independent of deacetylase function (Figure 7).
Figure 7. Model: SIR-2.4 promotes DAF-16 deacetylation and function during stress.
See text for details.
Discussion
We have shown that the sirtuin SIR-2.4 plays a crucial role in promoting DAF-16 activity and stress resistance. In response to multiple forms of stress, SIR-2.4 promotes DAF-16 nuclear recruitment and function, and ultimately organismal survival. SIR-2.4 is also required for optimal DAF-16 deacetylation and transcriptional activity under basal, unstressed conditions. In contrast, SIR-2.4 does not greatly impact DAF-16 nuclear localization, dauer formation, or longevity due to reduced IIS. We find that SIR-2.4 associates with DAF-16 and negatively regulates DAF-16 acetylation by interfering with the ability of CBP-1 to acetylate DAF-16 under stress conditions. Although mammalian FoxO proteins are acetylated in a functionally significant manner [3], to our knowledge this is the first demonstration that this modification is conserved in C. elegans. These data suggest that deacetylation likely plays two distinct roles in modulating DAF-16 function, in promoting optimal DAF-16 transcriptional activity under both basal and stress conditions, and in promoting DAF-16 nuclear recruitment following stress. We propose a model in which SIR-2.4 does not interact with DAF-16 under unstressed conditions. This allows CBP-1 to acetylate DAF-16 and sequester DAF-16 in the cytosol. In response to stress, SIR-2.4 binds to DAF-16 to block CBP-1-depedent acetylation and promote DAF-16 nuclear translocation and gene expression (Figure 7).
There are several important issues raised by our results. First, our data suggest that deacetylation promotes DAF-16 nuclear localization and function in response to stress, but is largely dispensable in the context of reduced IIS, suggesting that regulation of this modification may be involved in adjusting DAF-16 activity in the face of different organismal requirements. Our mass spectrometry analysis has identified four acetylation sites on DAF-16 (Table S3). Notably, acetylation at one of these (K248), is also present at homologous lysine residues in FoxO1 (K262) and FoxO3A (K259) [4], [54], [55], [56]. Future studies mutating these acetylation sites individually and in combinations will be required to identify specific residues involved in regulating DAF-16 nuclear recruitment and transcriptional activity.
Our results provide further evidence that multiple sirtuins regulate FoxO proteins by diverse mechanisms to achieve different functional outcomes. In mammals, several sirtuins directly deacetylate FoxO proteins [4], [6], [30], [31], [32], [33], [36]. In worms, SIR-2.1 promotes DAF-16 function to increase stress resistance via 14-3-3 proteins [26]. We find SIR-2.4 suppresses DAF-16 acetylation and is critical in the organismal response to a variety of stressors. Whereas SIR-2.4 is required for optimal DAF-16 nuclear recruitment in response to stress, SIR-2.1 is dispensable for this process (Figure 1C-1D) [17]. It has been reported that SIRT1 promotes survival of cerebellar granular neurons in response to low extracellular potassium levels in a catalytic activity-independent fashion [57]. Thus, sirtuins may regulate stress responses through mechanisms other than direct covalent substrate modification. Consistent with this idea, we found that SIR-2.4 regulates DAF-16 function independent of its catalytic function, by preventing CBP-1-mediated DAF-16 acetylation. We note that SIRT6 was recently shown to be capable of binding NAD+ and undergoing a conformational change in the absence of substrate; based on this finding, it has been speculated that SIRT6 might act as an NAD+ metabolite sensor, independently of catalytic activity [58]. This hypothesis is consistent with our data regarding the SIRT6 homolog SIR-2.4. Chromatin modifying complexes frequently involve multiple enzymes with apparently redundant biochemical activities, including multiple histone deacetylases; analogously SIR-2.4 likely associate with other deacetylases to regulate DAF-16. SIR-2.1 associates with DAF-16, although SIR-2.1 was not shown to deacetylate DAF-16 [21], [26]. DAF-16 deacetylases could in principle be SIR-2.1 or other worm sirtuins, or non-sirtuin deacetylases. However, the fact that SIR-2.1 does not play a major role in stress-induced DAF-16 nuclear recruitment strongly suggests that SIR-2.1 is not likely the major DAF-16 deacetylase.
FoXO proteins has been shown to interact with acetyltransferases CBP and p300 in mammalian cells [46], although it was previously unknown whether these proteins directly acetylate DAF-16 in worms. In this study, we showed that inactivation of cbp-1 by RNAi results in constitutive nuclear localization of DAF-16, and that CBP-1 can directly acetylate DAF-16 on multiple residues in vitro (Figure 6C, Table S3). However, cbp-1 KD only modestly reduces the level of DAF-16 acetylation in vivo (Figure 6B). Since the pan-acetyl-lysine antibody we used in these experiments is not able to discriminate among DAF-16 species acetylated at different sites, or singly versus multiply acetylated DAF-16 species, it is possible that CBP-1 is not the only acetyltransferase that acetylates DAF-16 in vivo. Thus the effect of cbp-1 KD on DAF-16 acetylation may be partially obscured by CBP-1-independent acetylation of DAF-16. Nevertheless, CBP-1 has a major impact on DAF-16 localization (Figure 6A).
Finally, it is currently unclear whether SIR-2.4 regulates DAF-16 in a cell-autonomous matter, or whether SIR-2.4 is required in only a subset of cells to carry out this function. The tissue expression pattern of endogenous SIR-2.4 is ambiguous. A SIR-2.4::GFP translational fusion protein shows weak expression in most cell types, but much stronger expression in a subset of head and tail neurons as well as in a subset of somatic gonad cells (Figure S1). Thus, SIR-2.4 could conceivably regulate DAF-16 in neurons to influence the systemic hormonal milieu – e.g., by reducing INS-7 expression – thereby impacting DAF-16 localization and function throughout the animal. However, our observation that DAF-16 is hyperacetylated in extracts from whole worms, together with our biochemical data, supports the hypothesis that SIR-2.4 regulates DAF-16 acetylation in most tissues of the animal cell autonomously. Overall, SIR-2.4 is a novel regulator of DAF-16 localization and function in response to stress in C. elegans.
Materials and Methods
Strains
CF1041: daf-2(e1370)III, TJ356: zIs356 [daf-16::gfp + rol-6], AM140: rmIs132[unc-54p::Q35::yfp], MT18068: sir-2.4(n5137)I, EQ 137: iqEx47 [sir-2.4p::sir-2.4::gfp + rol-6], EQ158: iqEx50 [sir-2.4p::sir-2.4 + myo-3p::rfp], EQ 200: sir-2.4(n5137)I; zIs356 [daf-16::gfp + rol-6], EQ205: sir-2.4(n5137)I; zIs356 [daf-16::gfp + rol-6]; iqEx59 [sir-2.4p::sir-2.4NA + myo-3p::rfp], EQ211: sir-2.4(n5137)I; zIs356 [daf-16::gfp + rol-6]; iqEx60 [sir-2.4p::sir-2.4 + myo-3p::rfp].
All strains used were maintained and handled as described previously [59]. TJ356, AM140 and CF1041 were obtained from the Caenorhabditis Genetic Center. For the generation of transgenic animals, plasmid DNA mixes were injected into the gonad of young adult hermaphrodite animals, using the standard method described previously [60]. F1 progeny were selected on the basis of the roller phenotype. Individual F2 progenies were isolated to establish independent lines. For the generation of the EQ137 (SIR-2.4::GFP overexpressor) strain, plasmid DNA containing a mixture of 100 ng/ml of sir-2.4p::sir-2.4::gfp and 50 ng/ml of pRF4 (rol-6) constructs was injected into N2 animals. For the generation of EQ158 (native SIR-2.4 overexpressor), plasmid DNA containing a mixture of 30 ng/ml of native SIR-2.4 driven by its own promoter and 50 ng/ml of coinjection marker myo-3p::rfp was injected into N2 animals. For the generation of EQ211, plasmid DNA containing a mixture of 30 ng/ml of sir-2.4p::sir-2.4 and 80 ng/ml of myo-3p::rfp constructs was injected into TJ356 animals. For the generation of EQ205, plasmid DNA containing a mixture of 30 ng/ml of sir-2.4p::sir-2.4NA and 80 ng/ml of myo-3p::rfp constructs was injected into TJ356 animals.
RNA–interference (RNAi) experiments
HT115 bacteria transformed with RNAi vectors (L4440) expressing dsRNA of the genes indicated were grown at 37°C in LB with 10 mg/mL tetracycline and 50 mg/mL carbenicillin, then seeded onto NG-carbenicillin plates and supplemented with 100 µL 0.1 M IPTG. The sir-2.4 RNAi construct was generated by cloning nucleotides 1–467 of the SIR-2.4 cDNA into the L4440 vector. The identity of all RNAi clones was verified by sequencing the inserts using M13-forward primer. Eggs were added to plates and transferred to new plates every 3–6 days.
sir-2.4 deletion mutant analysis
The deletion in a sir-2.4 mutant strain (kind gift of the Horvitz laboratory) was mapped by PCR, and found to encompass 1,929 bp of chromosome I (5990775–5992703), which encodes nucleotides 28–879 of the SIR-2.4 spliced mRNA (data available upon request).
DAF-16 nuclear localization assay
For quantification of DAF-16::GFP localization, synchronized eggs from TJ356 animals (i.e. transgenic animals expressing DAF-16::GFP) or other strains as indicated were seeded onto either vector control or appropriate RNAi plates. For stress response experiments, day 1 adults were washed with M9 three times and transferred to new plates or and subjected to heat shock (35°C) or oxidative stress (1.5 mM H2O2 in M9). GFP localization was then analyzed using an Olympus BX61 fluorescent microscope at 40× or 100× magnification. For time-course analysis, worms were scored for the presence or absence of GFP accumulation within the nuclei of head hypodermis cells (n = 30∼50) in a blinded fashion every 5–30 min. An animal was scored as having nuclear GFP if more than one head hypodermic nuclei contained DAF-16::GFP. For single time point experiments, worms were blindly scored for the presence or absence of GFP accumulation within the nuclei of indicated cells (n = 120 or greater). P values were calculated by Poisson regression (time-course assays) or chi-square test (single time point assays).
Quantitative RT–PCR analysis
Total RNA was isolated from approximately 5,000 day 1 adult worms, and cDNA was generated from 4 µg of RNA using Superscript III RT (Invitrogen). Real-time qRT-PCR experiments were performed using the Power SYBR Green PCR Master mix (Applied Biosystems) and the Chromo 4 system (MJ Research). Relative mRNA level of the genes of interest were calculated and normalized against an internal control (ACT-1; worm β-actin). Primer sequences were (all 5′-3′): SOD-3 (GTTTCAGCGCGACTTCGGTTCCCT, CGTGCTCCCAAACGTCAATTCCAA); DOD-3 (AAAAAGCCATGTTCCCGAAT, GCTGCGAAAAGCAAGAAAAT); DOD-24 (TGTCCAACACAACCTGCATT, TGTGTCCCGAGTAACAACCA); C32H11.4 (TTACTTCCCATCGCCAAAGT, CAATTCCGGCGATGTATGAT); HSP-16.1 (GATCAAAAGTTTGCCATAAATCTC, TTCAGTCTTTAATTCTTGTTCTCC); INS-7 (TCGTTGTGGAAGAAGAATACATTC, TTAAGGACAGCACTGTTTTCG); and ACT-1 (CTACGAACTTCCTGACGGACAAG, CCGGCGGACTCCATACC).
Stress assays
For thermotolerance assays, eggs from N2 worms were transferred to plates seeded with vector control, daf-16 RNAi, or sir-2.4 RNAi bacteria and grown to day 1 of adulthood. Worms were then transferred to plates without any food and heat-shocked at 35°C. Viability was determined at the indicated timepoints; death was determined by the lack of movement after prodding. For oxidative stress assays, eggs from N2 worms were transferred to plates seeded with vector control, daf-16 RNAi, or sir-2.4 RNAi bacteria and grown to day 3 of adulthood. Worms were then transferred to 24-well plates and soaked in 1.5 mM H2O2 in M9 media. Viability was determined at the indicated timepoints as above. For stress assays, a Kaplan–Meier survival analysis with a log-rank test was performed, and a P-value of 0.05 was considered statistically significant.
Paralysis assay
Synchronized eggs from AM140 animals (i.e. transgenic animals expressing Q35::YFP) were seeded on either vector control or the indicated RNAi bacteria. Animals were scored for polyQ-induced paralysis every other day during adulthood. Paralyzed worms were identified as those failing to make forward or backward movement in response to stimulation by plate-tapping and tail-prodding; these worms still exhibited pharyngeal pumping. For the paralysis assay, a Kaplan–Meier survival analysis with a log-rank test was performed.
Lifespan analysis
Lifespan analysis were conducted at 20°C as described previously [61], [62]. Strains were grown at 20°C for at least two generations without starvation prior to lifespan analysis. At least 60 worms were used for each experiment. In all experiments, the pre-fertile period of adulthood was used as t = 0 for lifespan analysis. Stata 8 software was used for statistical analysis to determine the means and percentiles. In all cases, P-values were calculated using the log-rank (Mantel-Cox) method.
Assessment of DAF-16 acetylation in worms
∼15,000 synchronized day 1 adult worms grown at 20°C were harvested by washing three times with cold M9 buffer and once with HB-high salt buffer (10 mM HEPES, pH 7.9; 10 mM KCl; 1.5 mM MgCl2; 0.1 mM EDTA; 0.5 mM EGTA; 44 mM Sucrose; 100 mM NaCl; 0.5% Triton X-100). Worm pellets were then resuspended in 3× volume of HB-high salt buffer supplemented with Protease Inhibitor Cocktail Complete (Roche), 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM nicotinamide and 1 µM trichostatin A. Pellets were immediately frozen and stored in liquid nitrogen. Frozen suspensions were thawed, homogenized with a Dounce homogenizer (30 strokes with pestle B), and centrifuged at 14,000×g at 4°C for 20 minutes. Supernatants were collected and total protein concentrations were quantified by Bradford assay. For immunoprecipitation, 30 µl of anti-acetyl-lysine agarose beads (Immunechem; ICP0388) were added to 1 mg of protein extract with 100 µg/ml of ethidium bromide and incubated with gentle shaking at 4°C overnight. The beads were then washed 3 times with HB-high salt buffer supplemented with 50 mg/ml ABESF, 1 mM sodium orthovanadate, 1 mM nicotinamide and 1 µM trichostatin A. before being subjected to western blotting analysis. The samples were subjected to SDS-PAGE and transferred to a PVDF membrane (Millipore). The membrane was washed three times with TBS containing 0.1% Tween 20 (TBST). After blocking with TBST containing 5% nonfat milk for 60 min, the membrane was incubated with the primary antibody indicated (e.g. anti-GFP, Abcam, #AB6556) at 4°C for 12 h and washed three times with TBST. The membrane was then probed with HRP-conjugated secondary antibody for 1 h at room temperature and washed with TBST three times. Finally, the immunoblots were developed using a chemiluminescent substrate (Millipore) and visualized by autoradiography.
Assessment of SIR-2.4/DAF-16 interaction
10 cm dishes of HEK293T cells were transfected using TransIT-293 (Mirus) with plasmids encoding HA-tagged DAF-16 (1 µg), FLAG-tagged SIR-2.4 (9 µg), both DAF-16 and SIR-2.4 plasmids together, or a plasmid encoding GFP (5 µg), to assess transfection efficiency. Cells were harvested 48 hours post transfection, and were lysed by rotation at 4°C for 20 minutes in lysis buffer (LB; 150 mM NaCl, 1% Triton X-100, 0.5% NP40, 50 mM Tris pH 7.4, 10% glycerol, with Protease Inhibitor Cocktail Complete-EDTA free (Roche) [63], followed by brief sonication. 1 mg of whole cell extract from each cell line was pre-cleared by slow rotation with 50 µl of protein G conjugated agarose beads. For immunoprecipitation, either 25 µl of M2-agarose beads (Sigma) (anti-FLAG IP) or 25 µl of anti-HA-agarose beads (Roche) (anti-HA IP) was added to pre-cleared WCE, and IPs were incubated overnight by slow rotation at 4°C. After incubation, beads were pelleted and washed three times in LB. FLAG-tagged proteins were eluted in 80 µl of FLAG elution buffer (150 ng/µl 3×FLAG peptide (Sigma), 10 mM Tris-HCl (pH 8.0) and 150 mM NaCl) at 4°C for 4 hours. 40 µl of eluate was loaded on an SDS-PAGE gel. For HA IPs, 80 µl of Laemmli buffer was added to the anti-HA-agarose beads, and beads were boiled at 100°C for 5 minutes; 40 µl was loaded on a gel. DAF16 and SIR2.4 interaction was assessed by immunoblot using anti-DAF-16 (Santa Cruz) or anti-HA antibodies.
In vitro DAF-16 acetylation assay
3×FLAG tagged DAF-16, CBP or SIR-2.4 were each transfected and expressed in 293T cells. These proteins were then purified with Anti-FLAG M2 Affinity Gel (Sigma, A2220). 0.5 µg of purified DAF-16 was incubated with 0.2 µg of purified CBP with or without 0.5 µg of purified SIR-2.4 in the presence of HAT buffer (50 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 1 mM dithiothreitol, 10%glycerol) supplemented with 200 µM of acetyl-CoA. The reactions were incubated at 37°C for 3 hours and analyzed by western blot. Acetylated DAF-16 was queried with anti-acetyl-lysine antibody (Immunechem; ICP0380) and total level of DAF-16 were assessed with anti-FLAG antibody (Sigma, #F3165).
Supporting Information
Expression pattern of sir-2.4 in C. elegans. Transgenic lines expressing a SIR-2.4 translational GFP fusion (sir-2.4p::sir-2.4::gfp) were utilized to analyze the expression pattern of sir-2.4 in C. elegans. Images of (a–b) L3 or (c–e) late L4/young adult stage transgenic animals (EQ137) expressing GFP protein under control of the sir-2.4 promoter. sir-2.4 is highly expressed in a subset of head and tail neurons beginning at early larval stage, indicated by white arrows. High expression of sir-2.4 is also found in spermathecal-uterine valve (sp-ut valve) cells beginning at L4 larval stage, indicated by white arrows. The yellow arrow indicates the nuclear accumulation of SIR-2.4::GFP fusions in these cells. It is worth noting that very weak expression of sir-2.4 is found ubiquitously in most tissues, although it is difficult to capture in these images.
(PDF)
Effects of sir-2.4 RNAi or deletion on gene expression, stress resistance, and IIS-mediated DAF-16 translocation. (A–B) Expression of the genes indicated was measured in worms of the indicated genotypes as in Figure 2. (C) Thermotolerance was assessed in worms of the indicated genotypes as in Figure 3A. (D) Oxidative stress resistance was assessed in worms of the indicated genotypes as in Figure 3B. (E) TJ356 animals were fed with either vector control, daf-2 RNAi, a 1∶1 mix of daf-2 and sir-2.4 RNAi, or a 1∶1 mix of daf-2 and yk615e RNAi bacteria from the time of hatching. yk615e is a gene randomly selected as a negative control in double RNAi experiments. Animals (n = 125 or greater) were scored for DAF-16::GFP nuclear translocation as described in Figure 4B.
(PDF)
Minimal impact of SIR-2.4 overexpression on stress resistance or lifespan. Transgenic animals overexpressing (A) a SIR-2.4::GFP fusion protein or (B) native untagged SIR-2.4 protein and N2 controls were exposed to 35°C heat stress. Viability was then scored at the timepoints indicated. (C) Survival curves of wild-type (N2) animals or transgenic animals overexpressing native SIR-2.4 (EQ158) at 20°C. All statistical details are presented in Tables S1 and S2.
(PDF)
sir-2.4 RNAi promotes DAF-16 hyperacetylation. (A) DAF-16 acetylation was assessed in control or sir-2.4 RNAi worms by acetyl-lysine immunoprecipitation followed by GFP immunoblot as described in Figure 5A. (B) DAF-16 acetylation was assessed in sir-2.4 KO, sir-2.1 RNAi, or double loss-of function animals as indicated.
(PDF)
cbp-1 loss of function confers stress sensitivity and shortened lifespan. Mean survival of cbp-1 RNAi worms in response to heat shock (A) or peroxide stress (B). (C) Lifespan curves of wildtype or sir-2.4 KO animals in the presence of cbp-1 or control RNAi bacteria.
(PDF)
Effects of sir-2.4 expression on stress resistance. Animals indicated were exposed to heat or oxidative stress. Mean survival ± SEM, in hours, observed in the stress analysis is shown. 75th percentile is the time at which the fraction of animals alive reaches 0.25. ‘n’ indicates the number of animals scored in the each experiment. P-Values calculated by pair-wise comparisons to vector control of the same experiment. We used Stata 8 software for statistical analysis and to determine means and percentiles. The logrank (Mantel-Cox) test was used to test the hypothesis that the survival functions among groups were equal. a P-Values calculated by pair-wise comparisons to N2 grown on vector control of the same experiment. b Compared to N2 grown on the same RNAi bacteria. c Compared to the same mutants grown on vector control. ‘*’ indicates the sets of experiments plotted and shown in Figures.
(PDF)
Effects of SIR-2.4 on lifespan. Adult mean lifespan ± SEM, in days, observed in lifespan analyses. Lifespan experiments were carried out at 20°C. 75th percentile is the age at which the fraction of animals alive reaches 0.25. ‘n’ shows the number of observed deaths relative to total number of animals started at day 1. The difference between these numbers represents the number of animals censored during the experiment, and includes animals that exploded, bagged (i.e. exhibited internal progeny hatching), or crawled off the plates. a p-Values calculated by pair-wise comparisons to N2 control of the same experiment. b p-Values calculated by pair-wise comparisons to mutants fed with control bacteria of the same experiment. We used Stata 8 software for statistical analysis and to determine means and percentiles. The logrank (Mantel-Cox) test was used to test the hypothesis that the survival functions among groups were equal.
(PDF)
Peptides representing unique acetylated sequences from the FOXO transcription factor are shown with additional information. A“#” sign indicates the site of acetylation in the sequence. The site position refers to which lysine residue on FOXO the identified acetylation site represents. The mass to charge ratio (m/z) and charge state of the peptide as they were observed in the mass spectrometer are given. The mass error specifies the difference between the observed m/z ratio and the theoretical m/z ratio of a peptide, reported in part per million (PPM). The XCorr is the cross correlation score between the theoretical and observed MS2 spectra for the matched peptide. The unique ΔCorr is the difference between the XCorr of the top ranking peptide match (the reported peptide) and the XCorr of the next closest ranked peptide of a unique primary sequence (not simply an alternate placement of the acetylation site), normalized by the XCorr of the top ranking peptide match. All reported peptides passed a cutoff of a false discovery rate (FDR) <0.1%, based on the target-decoy strategy [61]. See Text S1 for materials and methods used in these studies.
(DOCX)
Supplemental materials and methods.
(DOCX)
Acknowledgments
The authors thank H. R. Horvitz for providing the MT18068 strain; the Ruvkun lab for providing the daf-16 RNAi plasmid; the Caenorhabditis Genetic Center for the TJ356, AM140, and CF1041 strains; and the National Biosource Project of Japan (Dr. Mitani) for the sir-2.4 (tm2817) strain. Scott Pletcher is gratefully acknowledged for helpful discussions and comments on the manuscript. Hsiu-yin Chiang is gratefully acknowledged for statistical analysis. The authors also thank Carol Mousigan for technical assistance.
Funding Statement
This work was supported by R01 grants from NIA (AG028516; A-LH) and NIGMS (GM101171; DBL), a research grant from the American Federation for Aging Research (DBL), a Rackham Predoctoral Fellowship (W-CC), an NIA training grant (AG000114; DXT), and a pilot award from the Nathan Shock Center at the University of Michigan (P30 AG013283; DBL). DBL is a New Scholar in Aging of the Ellison Medical Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Arden KC (2008) FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene 27: 2345–2350. [DOI] [PubMed] [Google Scholar]
- 2. Yen K, Narasimhan SD, Tissenbaum HA (2011) DAF-16/Forkhead box O transcription factor: many paths to a single Fork(head) in the road. Antioxid Redox Signal 14: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calnan DR, Brunet A (2008) The FoxO code. Oncogene 27: 2276–2288. [DOI] [PubMed] [Google Scholar]
- 4. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, et al. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015. [DOI] [PubMed] [Google Scholar]
- 5. Frescas D, Valenti L, Accili D (2005) Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem 280: 20589–20595. [DOI] [PubMed] [Google Scholar]
- 6. van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, et al. (2004) FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J Biol Chem 279: 28873–28879. [DOI] [PubMed] [Google Scholar]
- 7. Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, et al. (2005) FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab 2: 153–163. [DOI] [PubMed] [Google Scholar]
- 8. van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, et al. (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073. [DOI] [PubMed] [Google Scholar]
- 9. Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, et al. (2006) A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125: 987–1001. [DOI] [PubMed] [Google Scholar]
- 10. Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, et al. (2004) FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J 23: 4802–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, et al. (2005) JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A 102: 4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sunayama J, Tsuruta F, Masuyama N, Gotoh Y (2005) JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J Cell Biol 170: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finkel T, Deng CX, Mostoslavsky R (2009) Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spange S, Wagner T, Heinzel T, Kramer OH (2009) Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol 41: 185–198. [DOI] [PubMed] [Google Scholar]
- 15. Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798. [DOI] [PubMed] [Google Scholar]
- 16. Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, et al. (2011) The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet 7: e1002235 doi:10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heidler T, Hartwig K, Daniel H, Wenzel U (2010) Caenorhabditis elegans lifespan extension caused by treatment with an orally active ROS-generator is dependent on DAF-16 and SIR-2.1. Biogerontology 11: 183–195. [DOI] [PubMed] [Google Scholar]
- 18. Pasco MY, Catoire H, Parker JA, Brais B, Rouleau GA, et al. (2010) Cross-talk between canonical Wnt signaling and the sirtuin-FoxO longevity pathway to protect against muscular pathology induced by mutant PABPN1 expression in C. elegans. Neurobiol Dis 38: 425–433. [DOI] [PubMed] [Google Scholar]
- 19. Catoire H, Pasco MY, Abu-Baker A, Holbert S, Tourette C, et al. (2008) Sirtuin inhibition protects from the polyalanine muscular dystrophy protein PABPN1. Hum Mol Genet 17: 2108–2117. [DOI] [PubMed] [Google Scholar]
- 20. Greiss S, Hall J, Ahmed S, Gartner A (2008) C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev 22: 2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, et al. (2006) C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev 127: 741–747. [DOI] [PubMed] [Google Scholar]
- 22. Viswanathan M, Kim SK, Berdichevsky A, Guarente L (2005) A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell 9: 605–615. [DOI] [PubMed] [Google Scholar]
- 23. Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, et al. (2005) Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet 37: 349–350. [DOI] [PubMed] [Google Scholar]
- 24. Mair W, Panowski SH, Shaw RJ, Dillin A (2009) Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE 4: e4535 doi:10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tissenbaum HA, Guarente L (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230. [DOI] [PubMed] [Google Scholar]
- 26. Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L (2006) C. elegans SIR-2.1 Interacts with 14-3-3 Proteins to Activate DAF-16 and Extend Life Span. Cell 125: 1165–1177. [DOI] [PubMed] [Google Scholar]
- 27. Viswanathan M, Guarente L (2011) Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature 477: E1–2. [DOI] [PubMed] [Google Scholar]
- 28. Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, et al. (2011) Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477: 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lombard DB, Pletcher SD, Canto C, Auwerx J (2011) Ageing: longevity hits a roadblock. Nature 477: 410–411. [DOI] [PubMed] [Google Scholar]
- 30. Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, et al. (2004) Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A 101: 10042–10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, et al. (2004) Mammalian SIRT1 represses forkhead transcription factors. Cell 116: 551–563. [DOI] [PubMed] [Google Scholar]
- 32. Jing E, Gesta S, Kahn CR (2007) SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 6: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang F, Tong Q (2009) SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell 20: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang F, Nguyen M, Qin FX, Tong Q (2007) SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 6: 505–14. [DOI] [PubMed] [Google Scholar]
- 35. Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, et al. (2005) Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A 102: 11278–11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, et al. (2008) SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int J Biol Sci 4: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, et al. (2009) Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 119: 2758–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E (2008) Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J Physiol Pharmacol 59 Suppl 9: 201–212. [PubMed] [Google Scholar]
- 39. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, et al. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329. [DOI] [PubMed] [Google Scholar]
- 40. Lee SS, Kennedy S, Tolonen AC, Ruvkun G (2003) DAF-16 target genes that control C. elegans life-span and metabolism. Science 300: 644–647. [DOI] [PubMed] [Google Scholar]
- 41. McElwee J, Bubb K, Thomas JH (2003) Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121. [DOI] [PubMed] [Google Scholar]
- 42. Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- 43. Mukhopadhyay A, Oh SW, Tissenbaum HA (2006) Worming pathways to and from DAF-16/FOXO. Exp Gerontol 41: 928–934. [DOI] [PubMed] [Google Scholar]
- 44. Hsu AL, Murphy CT, Kenyon C (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145. [DOI] [PubMed] [Google Scholar]
- 45. Morley JF, Brignull HR, Weyers JJ, Morimoto RI (2002) The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A 99: 10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nasrin N, Ogg S, Cahill CM, Biggs W, Nui S, et al. (2000) DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc Natl Acad Sci U S A 97: 10412–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finnin MS, Donigian JR, Pavletich NP (2001) Structure of the histone deacetylase SIRT2. Nat Struct Biol 8: 621–625. [DOI] [PubMed] [Google Scholar]
- 48. Senf SM, Sandesara PB, Reed SA, Judge AR (2011) p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am J Physiol Cell Physiol 300: C1490–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang M, Poplawski M, Yen K, Cheng H, Bloss E, et al. (2009) Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol 7: e1000245 doi:10.1371/journal.pbio.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eastburn DJ, Han M (2005) A gain-of-function allele of cbp-1, the Caenorhabditis elegans ortholog of the mammalian CBP/p300 gene, causes an increase in histone acetyltransferase activity and antagonism of activated Ras. Mol Cell Biol 25: 9427–9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, et al. (2007) Highx-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237 doi:10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, et al. (2003) Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol 68: 159–169. [DOI] [PubMed] [Google Scholar]
- 53. Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, et al. (2006) Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett 580: 3217–3222. [DOI] [PubMed] [Google Scholar]
- 54. Qiang L, Banks AS, Accili D (2010) Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem 285: 27396–27401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brent MM, Anand R, Marmorstein R (2008) Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 16: 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang F, Chan CH, Chen K, Guan X, Lin HK, et al. (2012) Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 31: 1546–1557. [DOI] [PubMed] [Google Scholar]
- 57. Pfister JA, Ma C, Morrison BE, D'Mello SR (2008) Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS ONE 3: e4090 doi:10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, et al. (2011) Structure and biochemical functions of SIRT6. J Biol Chem 286: 14575–14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Apfeld J, Kenyon C (1999) Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809. [DOI] [PubMed] [Google Scholar]
- 62. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- 63. Ahn BH, Kim HS, Song S, Lee IH, Liu J, et al. (2008) A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A 105: 14447–14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression pattern of sir-2.4 in C. elegans. Transgenic lines expressing a SIR-2.4 translational GFP fusion (sir-2.4p::sir-2.4::gfp) were utilized to analyze the expression pattern of sir-2.4 in C. elegans. Images of (a–b) L3 or (c–e) late L4/young adult stage transgenic animals (EQ137) expressing GFP protein under control of the sir-2.4 promoter. sir-2.4 is highly expressed in a subset of head and tail neurons beginning at early larval stage, indicated by white arrows. High expression of sir-2.4 is also found in spermathecal-uterine valve (sp-ut valve) cells beginning at L4 larval stage, indicated by white arrows. The yellow arrow indicates the nuclear accumulation of SIR-2.4::GFP fusions in these cells. It is worth noting that very weak expression of sir-2.4 is found ubiquitously in most tissues, although it is difficult to capture in these images.
(PDF)
Effects of sir-2.4 RNAi or deletion on gene expression, stress resistance, and IIS-mediated DAF-16 translocation. (A–B) Expression of the genes indicated was measured in worms of the indicated genotypes as in Figure 2. (C) Thermotolerance was assessed in worms of the indicated genotypes as in Figure 3A. (D) Oxidative stress resistance was assessed in worms of the indicated genotypes as in Figure 3B. (E) TJ356 animals were fed with either vector control, daf-2 RNAi, a 1∶1 mix of daf-2 and sir-2.4 RNAi, or a 1∶1 mix of daf-2 and yk615e RNAi bacteria from the time of hatching. yk615e is a gene randomly selected as a negative control in double RNAi experiments. Animals (n = 125 or greater) were scored for DAF-16::GFP nuclear translocation as described in Figure 4B.
(PDF)
Minimal impact of SIR-2.4 overexpression on stress resistance or lifespan. Transgenic animals overexpressing (A) a SIR-2.4::GFP fusion protein or (B) native untagged SIR-2.4 protein and N2 controls were exposed to 35°C heat stress. Viability was then scored at the timepoints indicated. (C) Survival curves of wild-type (N2) animals or transgenic animals overexpressing native SIR-2.4 (EQ158) at 20°C. All statistical details are presented in Tables S1 and S2.
(PDF)
sir-2.4 RNAi promotes DAF-16 hyperacetylation. (A) DAF-16 acetylation was assessed in control or sir-2.4 RNAi worms by acetyl-lysine immunoprecipitation followed by GFP immunoblot as described in Figure 5A. (B) DAF-16 acetylation was assessed in sir-2.4 KO, sir-2.1 RNAi, or double loss-of function animals as indicated.
(PDF)
cbp-1 loss of function confers stress sensitivity and shortened lifespan. Mean survival of cbp-1 RNAi worms in response to heat shock (A) or peroxide stress (B). (C) Lifespan curves of wildtype or sir-2.4 KO animals in the presence of cbp-1 or control RNAi bacteria.
(PDF)
Effects of sir-2.4 expression on stress resistance. Animals indicated were exposed to heat or oxidative stress. Mean survival ± SEM, in hours, observed in the stress analysis is shown. 75th percentile is the time at which the fraction of animals alive reaches 0.25. ‘n’ indicates the number of animals scored in the each experiment. P-Values calculated by pair-wise comparisons to vector control of the same experiment. We used Stata 8 software for statistical analysis and to determine means and percentiles. The logrank (Mantel-Cox) test was used to test the hypothesis that the survival functions among groups were equal. a P-Values calculated by pair-wise comparisons to N2 grown on vector control of the same experiment. b Compared to N2 grown on the same RNAi bacteria. c Compared to the same mutants grown on vector control. ‘*’ indicates the sets of experiments plotted and shown in Figures.
(PDF)
Effects of SIR-2.4 on lifespan. Adult mean lifespan ± SEM, in days, observed in lifespan analyses. Lifespan experiments were carried out at 20°C. 75th percentile is the age at which the fraction of animals alive reaches 0.25. ‘n’ shows the number of observed deaths relative to total number of animals started at day 1. The difference between these numbers represents the number of animals censored during the experiment, and includes animals that exploded, bagged (i.e. exhibited internal progeny hatching), or crawled off the plates. a p-Values calculated by pair-wise comparisons to N2 control of the same experiment. b p-Values calculated by pair-wise comparisons to mutants fed with control bacteria of the same experiment. We used Stata 8 software for statistical analysis and to determine means and percentiles. The logrank (Mantel-Cox) test was used to test the hypothesis that the survival functions among groups were equal.
(PDF)
Peptides representing unique acetylated sequences from the FOXO transcription factor are shown with additional information. A“#” sign indicates the site of acetylation in the sequence. The site position refers to which lysine residue on FOXO the identified acetylation site represents. The mass to charge ratio (m/z) and charge state of the peptide as they were observed in the mass spectrometer are given. The mass error specifies the difference between the observed m/z ratio and the theoretical m/z ratio of a peptide, reported in part per million (PPM). The XCorr is the cross correlation score between the theoretical and observed MS2 spectra for the matched peptide. The unique ΔCorr is the difference between the XCorr of the top ranking peptide match (the reported peptide) and the XCorr of the next closest ranked peptide of a unique primary sequence (not simply an alternate placement of the acetylation site), normalized by the XCorr of the top ranking peptide match. All reported peptides passed a cutoff of a false discovery rate (FDR) <0.1%, based on the target-decoy strategy [61]. See Text S1 for materials and methods used in these studies.
(DOCX)
Supplemental materials and methods.
(DOCX)