Abstract
The rRNA gene cluster consists of multiple transcription units. Half of these are active, while the other half are transcriptionally inactive. Previously, in vivo studies have demonstrated that silencing of ribosomal DNA (rDNA) is mediated by the chromatin remodeling NoRC (nucleolar remodeling complex). To explore the mechanisms underlying NoRC-directed silencing of rDNA transcription, we investigated the effect of recombinant NoRC on RNA polymerase I transcription on reconstituted chromatin templates. We show that NoRC interacts with the transcription terminator factor (TTF-I), and this interaction is required both for the binding of TTF-I to its promoter-proximal target site and for the recruitment of NoRC to the promoter. After association with the rDNA promoter, NoRC alters the position of the promoter-bound nucleosome, thereby repressing RNA polymerase I transcription. This NoRC-directed rDNA repression requires the N terminus of histone H4. Repression is effective before preinitiation complex formation and as such is unable to exert an effect upon activated rDNA genes. Furthermore, the early steps of rDNA repression do not depend on DNA and histone modifications. These results reveal an important role for TTF-I in recruiting NoRC to rDNA and an active role for NoRC in the establishment of rDNA silencing.
RNA polymerase I transcription in mammalian cells accounts for 35 to 65% of total cellular transcription, depending on the metabolic activity of a given cell. To achieve high levels of ribosomal DNA (rDNA) transcription, mammalian cells harbor about 400 gene copies that are arranged in tandem repeats. Even in metabolically active cells, however, only half of the genes are actively transcribed, whereas the remaining genes are maintained in a silenced state. This ratio of active to inactive genes is stably propagated throughout the cell cycle and is independent of the transcriptional activity of the cell (4). Herein, the term “rDNA silencing” defines the mechanisms that establish and maintain the transcriptionally inactive rDNA genes in the cell.
During replication, the chromatin states are erased and the newly replicated daughter strands are repackaged into nucleosomes (14). We previously demonstrated that ATP-dependent chromatin remodeling at the rDNA promoter is required in order to activate RNA polymerase I (Pol I) transcription on chromatin templates. This remodeling reaction is triggered by the transcription termination factor TTF-I (12, 13), a multifunctional protein that mediates transcription termination and DNA replication fork arrest (7, 8). TTF-I binds to a target site (T0) upstream of the rDNA promoter, which has the characteristics of a promoter element and was previously shown to be essential for transcriptional activation in vivo (2, 9, 15). TTF-I-dependent nucleosome positioning establishes a specific promoter architecture that is compatible with, if not a prerequisite for, preinitiation complex formation (13). Upon searching for the remodeling activity that alters the chromatin structure of the rDNA promoter, we identified and purified NoRC (nucleolar remodeling complex), a member of the ISWI family of ATP-dependent chromatin remodeling complexes (20). NoRC consists of Tip5 (TTF-I-interacting protein 5) and Snf2H, the mammalian homolog of the ATPase ISWI. NoRC has been shown to be the key player for establishing the silent state of rRNA genes. Overexpression of Tip5 results in histone deacetylation, de novo DNA methylation, and transcriptional repression of a transfected rDNA reporter plasmid (17, 22). These studies not only revealed an essential role for NoRC in heterochromatin formation and transcriptional silencing but also provided evidence for a link between chromatin remodeling, histone modifications, and de novo methylation.
To elucidate the process of rDNA silencing, we established an in vitro model system that allowed the molecular analysis of NoRC-mediated silencing of rDNA transcription. We reconstituted NoRC from recombinant Tip5 and Snf2H and assayed its function in ATPase assays, chromatin remodeling, promoter targeting, and rDNA transcription on chromatin templates. We found that NoRC interacts with the N-terminal part of TTF-I, and this interaction enables TTF-I to bind to its cognate sequence upstream of the gene promoter. Furthermore, the interaction between TTF-I and NoRC brings NoRC to the rDNA promoter. NoRC binds near the promoter and subsequently represses rDNA transcription. This repression requires the presence of NoRC prior to the recruitment of the transcription initiation factors and depends upon the presence of the histone H4 tail. The results suggest that TTF-I may recruit different remodeling activities to rDNA which either activate or repress Pol I transcription. Whereas the activating chromatin remodeling activity remains to be characterized, we propose that NoRC plays a direct role in the silencing of rDNA.
MATERIALS AND METHODS
Expression and purification of recombinant proteins.
The Snf2H expression clone pFastBac1:Snf2H(Flag) (1) was kindly provided by R.E. Kingston. Expression and purification of Snf2H was performed as described previously (1), with minor modifications. Briefly, binding to M2 agarose (Sigma) was performed for 12 h at 4°C in buffer EX-500 (500 mM KCl, 10 mM Tris-HCl [pH 7.6], 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 0.05% Nonidet P-40, and protease inhibitors). The beads were washed twice with EX-1000 and EX-500, and proteins were eluted for 3 h at 4°C with EX-300 that contained 200 μg of Flag peptide per ml.
Myc epitope-tagged Tip5 (20) was subcloned into pFastBac1 (Life Technologies, Inc.), expressed in Sf9 cells, and purified by immunoaffinity chromatography with α-myc antibodies (9E10). Proteins were eluted for 3 h at room temperature with EX-300 that contained 200 μg of myc peptide per ml.
NoRC was prepared by coexpression of Tip5 and Flag-tagged Snf2H in Sf9 cells. After separation of free Snf2H from NoRC on a BioRex70 column (Bio-Rad), the complex was precipitated with anti-Flag antibodies. After washing with buffer EX-500, NoRC was eluted for 3 h at 4°C with EX-300 that contained 200 μg of Flag peptide per ml.
Full-length TTF-I cDNA was inserted into the pFastBac-HTb vector (Life Technologies, Inc.), and the protein was expressed in Sf9 cells. Histidine-tagged TTF-I and TTFΔN185 were purified on a heparin column (Bio-Rad), followed by purification with Ni-NTA agarose according to the manufacturer’s instructions (Qiagen). ACF (ATP-utilizing chromatin assembly factor) was expressed and purified as described previously (10). Expression, purification, and refolding of histones were all performed as described previously (3).
ATPase, nucleosome mobility, and electrophoretic mobility shift assays.
ATPase assays and nucleosome mobility experiments were performed as described previously (5, 11).
Chromatin assembly, supercoiling analysis, and MNase digestion.
Nucleosomes were assembled from DNA and recombinant histones by salt gradient dialysis (11). Chromatin was purified by ultracentrifugation (SW-41 [Beckman] at 45,000 rpm, 14 h) in a 15 to 30% sucrose gradient. For DNA supercoiling analysis, chromatin (1 μg) was incubated for 1 h with 10 U of topoisomerase I (Promega) and deproteinized with 50 μg of proteinase K for 1 h at 50°C. DNA was separated by electrophoresis at 100 V for 24 h in 1.2% Tris-glycine agarose gels containing 3.3 μM chloroquine and was visualized by ethidium bromide staining. Chromatin fractions exhibiting a high density of DNA supercoils were used for chromatin and transcription assays. For micrococcal nuclease (MNase) digestion, 300 ng of chromatinized DNA was digested for 20 to 180 s and deproteinized with proteinase K, and purified DNA was separated on 1.3% agarose gels.
MNase footprinting.
Purified nucleosomal arrays (300 ng) were digested with 1.5 U of MNase (Sigma) for 40 s. Reactions were conducted in 10 mM Tris-HCl (pH 7.6), 80 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.5 mM ATP, and 200 ng of bovine serum albumin per μl and were then stopped by the addition of 0.2 volumes of stop buffer (4% sodium dodecyl sulfate [SDS], 100 mM EDTA, 1 μg of glycogen, 10 μg of proteinase K). DNA was purified and analyzed by a single round of PCR (denaturation, 5 min at 95°C; annealing, 2 min at 56°C; extension, 1 min at 72°C) using radioactively labeled oligonucleotides that hybridized to the rDNA promoter. Primer extension fragments were resolved on 8% sequencing gels and quantified with a PhosphorImager and Aida software.
In vitro transcription assay.
Transcription was performed on an rDNA minigene (pMrWT-T) that represented a fusion of mouse promoter and terminator sequences. pMrWT-T contains rDNA promoter sequences from positions −170 to +155 and including the upstream terminator T0 at position −170 and a 3.5-kb 3′-terminal rDNA fragment (from positions +57 to +3643) containing 10 terminator elements (T1 through T10). The promoter and the terminator elements are separated by a transcribed region of 686 bp. Twenty-five-microliter transcription assays contained 12 mM Tris-HCl (pH 7.9); 0.1 mM EDTA; 0.5 mM dithiothreitol; 2 mM MgCl2; 80 mM KCl; 10 mM creatine phosphate; 12% glycerol; 0.66 mM each of ATP, UTP, GTP, 0.01 mM CTP; 1.5 μCi of [α-32P]CTP; 20 ng of either naked DNA or chromatin; and 5 μl of nuclear extract proteins that had been partially purified by chromatography on DEAE-Sepharose CL-6B (DEAE-280 [18]). Transcription assay reactions were incubated for 1 h at 30°C with purified TTF-I, NoRC, Snf2H, and ACF. Reactions were stopped by the addition of 25 μl of stop buffer (20 μg of glycogen, 2% SDS, 10 μg of proteinase K, 100 mM EDTA) and incubation for 1 h at 40°C. Transcripts were purified by ethanol precipitation and analyzed on 4.5% polyacrylamide gels.
RESULTS
NoRC is a nucleosome-dependent ATPase that mobilizes nucleosomes.
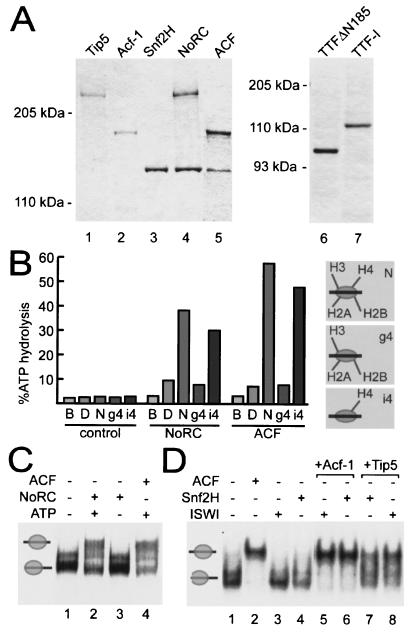
To assess the function of NoRC in chromatin remodeling and rDNA transcription, we coexpressed Snf2H (1) and Tip5 in Sf9 cells and purified the complex. The properties of immunopurified NoRC were compared with those of recombinant Snf2H and ACF (10) (Fig. 1A). To evaluate the function of the recombinant NoRC complex, we conducted ATPase assays in the presence of either DNA or nucleosomes (Fig. 1B). NoRC and ACF displayed a low constitutive level of ATPase activity in the absence of substrate (Fig. 1B, column labeled B). ATPase activity was stimulated poorly in the presence of free DNA (Fig. 1B, column labeled D). However, in the presence of nucleosomal DNA (Fig. 1B, column labeled N), the ATPases of NoRC and ACF were strongly activated. Previous studies have established that nucleosome remodeling by ISWI-containing complexes requires the N terminus of histone H4 (3, 20). Consistent with these studies, the histone H4 tail is required for activation of the ATPase activity of both NoRC and ACF (Fig. 1B, column labeled i4). In contrast, nucleosomes lacking the N terminus of histone H4 did not stimulate the ATPase activity of NoRC and ACF (Fig. 1B, column labeled g4).
FIG. 1.
NoRC reconstitution and functional analysis. (A) Purification of recombinant Tip5, Acf-1, Snf2H, NoRC, ACF, TTFΔN185, and TTF-I. The proteins were expressed in Sf9 cells, purified, resolved by SDS- polyacrylamide gel electrophoresis and stained with Coomassie blue. (B) ATPase assay with recombinant NoRC and ACF (0.5 pmol) in the absence of substrate (B, buffer) or in the presence of 300 ng of DNA (labeled D) and 300 ng of the indicated recombinant octamers reconstituted into nucleosomal arrays (N, nucleosomes reconstituted from full-length histones; g4, histone H4 lacks the N-terminal tail, the other histones are full-length proteins; i4, histone H4 is a full-length protein, the other histones lack the N-terminal tail). The bars show the percentage of hydrolyzed ATP. (C) Nucleosome sliding assay. Purified nucleosomes (0.1 pmol), positioned at the end of the DNA fragment (lane 1), were incubated for 90 min with 0.1 pmol of recombinant NoRC in the presence (lane 2) or absence (lane 3) of ATP or with ACF (0.1 pmol, lane 4). Nucleosome positions were analyzed by native gel electrophoresis. The positions of end-positioned and center-positioned nucleosomes are indicated on the left. (D) Nucleosome sliding assay using nucleosomes positioned at the end of the DNA fragment (lane 1). Nucleosomes were incubated with ISWI or Snf2H alone (lanes 3 and 4), in the presence of Acf-1 (lanes 5 and 6), or in the presence of Tip5 (lanes 7 and 8) and the indicated ATPase. A control reaction with ACF (lane 2) indicates the position of the central nucleosome.
In order to characterize the nucleosome remodeling activity of NoRC, we used the “nucleosome sliding” assay that monitors ATP-dependent changes of nucleosome positions (11). On mononucleosomes positioned at the ends of the DNA fragments, cellular NoRC moves nucleosomes to a central position (20). Recombinant NoRC and ACF exhibit similar properties in this assay, i.e., induced ATP-dependent movement of nucleosomes positioned at the end of the rDNA fragment to a more central position (Fig. 1C). Snf2H, like ISWI (18), did not mobilize nucleosomes positioned at the end of the rDNA fragment (Fig. 1D, lanes 3 and 4). Strikingly, the nucleosome was moved to the center of the DNA fragment (Fig. 1D, lanes 5 to 8) if the large subunits of ACF and NoRC, i.e., Tip5 or Acf1, were added to Snf2H or ISWI, respectively. This finding demonstrates that the large subunits of both ACF (6) and NoRC determine the directionality of nucleosome movement.
Interaction of NoRC with TTF-I enables DNA binding of TTF-I and chromatin remodeling at the rDNA promoter.
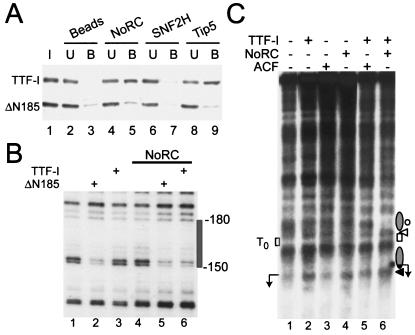
Tip5 was initially identified in a yeast two-hybrid screen as a protein that interacts with TTF-I. The N-terminal 185 amino acids of TTF-I were found to be required for interaction with Tip5. TTFΔN185, a deletion mutant that lacks the N terminus, was incapable of interacting with Tip5 (unpublished data). This finding indicates that sequences within amino acids 1 to 185 mediate the association of TTF-I with Tip5. To test whether this interaction was direct, we incubated recombinant full-length TTF-I and TTFΔN185 with immobilized NoRC, Tip5, or Snf2H, respectively, and bound TTF-I was visualized by Western blotting. Consistent with the N terminus of TTF-I mediating the interaction with NoRC, full-length TTF-I, but not TTFΔN185, was retained by both Tip5 and NoRC (Fig. 2A).
FIG. 2.
NoRC-TTF-I interaction triggers chromatin remodeling at the rDNA promoter. (A) Pull-down assay. NoRC, Snf2H, and Tip5 were immobilized on beads and incubated with a mixture of full-length TTF-I and N-terminally truncated TTF-I (ΔN185). TTF-I in the input (I), unbound (U), and bound to the beads (B) was analyzed by Western blotting. (B) Chromatin was incubated with TTFΔN185 (ΔN185, lanes 2 and 5) and TTF-I (lanes 3 and 6) in the absence or presence of NoRC. TTF binding to T0 was detected by partial MNase digestion of the DNA and primer extension footprinting. The gray box indicates the TTF binding site. (C) Mapping of MNase cleavage sites by indirect end labeling of the rDNA promoter reconstituted into chromatin. Chromatin, in the presence of ATP, was incubated with TTF-I, ACF, or NoRC (lanes 2 to 4) and TTF-I in the presence of ACF or NoRC (lanes 5 and 6). Each reaction was digested with 1 U of MNase for 30 s, and purified DNA was digested with NdeI and analyzed by Southern blotting. The positions of rearranged nucleosomes (gray ellipse), the TTF-I binding site, and the transcription start site (arrow) are indicated. Protected (circles) or enhanced (white and black triangles) MNase cleavage sites are indicated on the right. Position +22, the 3′ boundary of the nucleosome positioned at the rDNA promoter, is marked by the black triangle.
The assembly of DNA into nucleosomes usually reduces the accessibility for specific DNA-binding factors (21). TTFΔN185, however, is able to recognize its DNA binding site on the surface of a nucleosome (13). In contrast, full-length TTF-I does not bind to naked DNA (19). The available data suggest that the N-terminal region of TTF-I is a negative regulatory domain that inhibits DNA binding (16, 19). To examine whether the interaction with NoRC would uncover the DNA binding activity of TTF-I, we monitored binding of TTF-I to chromatin. For the experiment shown in Fig. 2B, we incubated reconstituted nucleosomal arrays with TTF-I in the presence or absence of NoRC, and TTF-I binding was assayed by MNase footprinting. Consistent with previous results, TTFΔN185 recognized the cognate site T0 even when it was packaged into chromatin. Upon TTF-I binding, a diagnostic band at position −150 was protected (Fig. 2B, lane 2). Neither NoRC alone nor full-length TTF-I altered the pattern of MNase-sensitive sites (lanes 3 and 4). Significantly, in the presence of NoRC, TTF-I was able to bind to its target sequence T0 and thus protect the binding site from MNase digestion. This indicates that the interaction with NoRC facilitates binding of full-length TTF-I to the upstream terminator T0 (lane 6). This experiment shows that DNA binding of TTF-I is regulated by protein-protein interactions and suggests a mechanism by which TTF-I may recruit NoRC to the rDNA promoter.
To establish whether recruitment of NoRC by TTF-I leads to rearrangement of the chromatin structure at the rDNA promoter, we performed partial MNase digestion and indirect end-labeling experiments with reconstituted nucleosomal arrays. Incubation of rDNA-chromatin with TTF-I, ACF, or NoRC in the presence of ATP did not significantly alter the MNase digestion pattern at the rDNA promoter (Fig. 2C, lanes 1 to 4). However, in the presence of NoRC, full-length TTF-I bound to its target site, T0. Binding of TTF-I generated an MNase-sensitive site flanking the TTF-I binding site (lane 6). In addition, adjacent regions were protected from MNase digestion, indicating that in the presence of both NoRC and TTF-I, nucleosomes were specifically positioned upstream and downstream of T0. In contrast to NoRC, ACF did not facilitate TTF-I binding and the chromatin structure remained unchanged (lane 5). This finding underscores the specificity of the interaction between TTF-I and NoRC and demonstrates that this interaction has important functional consequences: it facilitates TTF-I binding to rDNA, recruits NoRC to the rDNA promoter, and alters the position of nucleosomes at the rDNA promoter.
TTF-I-dependent targeting of NoRC increases the rate of chromatin remodeling at the rDNA promoter.
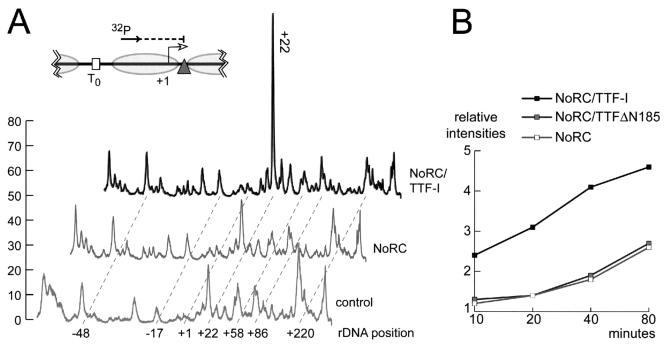
To more directly demonstrate alterations in nucleosome positioning, we mapped the boundaries of the promoter-bound nucleosomes by MNase digestion and primer extension by using a radioactively labeled oligonucleotide that was positioned within the realm of the nucleosome (Fig. 3A). The intensity of individual MNase cleavage sites correlates with the number of nucleosomes positioned next to the MNase-sensitive sites. In the absence of NoRC and TTF-I, multiple nucleosome positions without a preferred positioning site were observed (Fig. 3A). The pattern of MNase cleavage sites was not altered in the presence of TTF-I or TTFΔN185 (data not shown). After incubation with saturating concentrations of NoRC and TTF-I, the pattern of MNase cleavage sites was significantly altered (Fig. 3A). The most pronounced feature was the enhancement of the MNase-sensitive site at nucleotide position +22, which corresponds to the 3′ boundary of the nucleosome positioned at the rDNA promoter. Nucleosome remodeling reactions by TTFΔN185 were indistinguishable from remodeling reactions that were triggered by full-length TTF-I (data not shown).
FIG. 3.
Chromatin remodeling and TTF-I-dependent recruitment of NoRC to the rDNA promoter. (A) Mapping nucleosome boundaries at the rDNA promoter. Chromatin was incubated in the presence of NoRC and NoRC plus TTF-I for 90 min. Nucleosome positions were mapped by partial MNase digestion and primer extension of the purified DNA. Primer extension products were resolved on 8% sequencing gels and quantified with a PhosphorImager. The graph shows the positions and relative intensities of the MNase cleavage sites. The 3′ boundary of the remodeled nucleosome is indicated (+22). The relative position of the oligonucleotide used for primer extension is shown, and the MNase-sensitive sites on the rDNA promoter relative to the transcription start site are indicated. (B) Quantitative analysis of NoRC-dependent nucleosome remodeling. The N terminus of TTF-I directs NoRC interaction and increases the nucleosome remodeling kinetics. Nucleosome remodeling was performed as described in panel A, but in contrast to that experiment, limiting amounts of the NoRC complex were used. Analysis of NoRC-dependent kinetics of nucleosome remodeling in the presence of TTF-I and the N-terminally truncated TTFΔN185 followed. The appearance of the MNase cleavage site at position +22 was quantified relative to that of the input material (chromatin in the absence of remodeler). Relative intensities of the MNase cleavage site at position +22, correlating with overall nucleosome remodeling, was plotted in a graph versus the reaction time.
In order to monitor the kinetics of NoRC recruitment and nucleosome remodeling, we performed the assays with limiting amounts of NoRC. NoRC concentrations were chosen that remodel about 50% of the nucleosomes in the presence of TTF-I within 80 min. The assays were stopped after 10, 20, 40, and 80 min, and nucleosome positions were analyzed by MNase footprinting and primer extension. Changes of the MNase-sensitive sites at position +22 were quantified with a PhosphorImager and correlated with chromatin that had been incubated in the absence of NoRC. The diagram in Fig. 3B shows the quantitation of nucleosomes positioned at position +22 during incubation with NoRC, NoRC and TTF-I, or NoRC and TTFΔN185. NoRC alone did not significantly increase the relative intensity of the MNase-sensitive site at position +22. Similarly, the kinetics of NoRC plus TTFΔN185 was almost indistinguishable from that of NoRC alone, demonstrating that TTFΔN185 did not enhance NoRC-mediated remodeling at the rDNA promoter. However, in the presence of full-length TTF-I, nucleosomes were remodeled two- to threefold more efficiently than was true with NoRC or NoRC plus TTFΔN185. This result highlights the role of the N-terminal region of TTF-I in targeting NoRC to rDNA and demonstrates that the interaction of TTF-I and NoRC promotes nucleosome remodeling.
NoRC specifically represses rDNA transcription in chromatin.
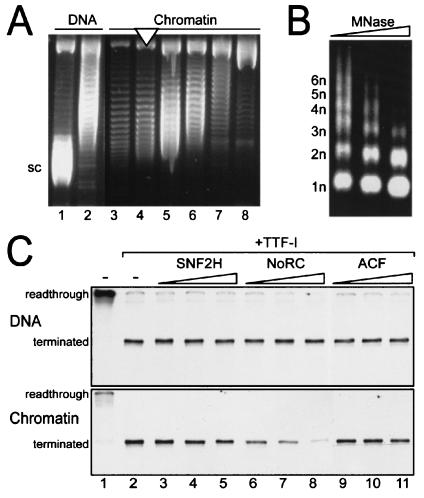
Recent in vivo experiments have demonstrated that NoRC is associated with inactive ribosomal gene copies. Moreover, overexpression of Tip5 causes silencing of rDNA transcription (17, 22). In order to study the function of NoRC on rDNA transcription in vitro, we reconstituted an rDNA minigene into chromatin by using either purified or recombinant histone octamers. The nucleosomal templates were purified in a sucrose gradient, and the nucleosome density was monitored by DNA supercoiling and MNase digestion (Fig. 4A and B). Naked DNA or nucleosomal templates were incubated with a partially purified nuclear extract from mouse cells (DEAE-280 fraction [18]) that contained all the factors required for transcription. This fraction, however, lacks TTF-I; hence, long readthrough transcripts were synthesized from naked DNA templates (Fig. 4C, DNA panel, lane 1). Upon the addition of TTF-I, transcription was terminated 686 nucleotides downstream of the initiation site (lane 2). A different result was obtained for chromatin templates. Consistent with previous data, transcription carried out with preassembled chromatin templates was repressed and required the presence of TTF-I to activate transcription (Fig. 4C, chromatin panel, lanes 1 and 2).
FIG. 4.
NoRC silences rDNA transcription for chromatin templates. (A) Supercoiling assay. Nucleosomes were assembled on pMrWT-T by salt dialysis and purified in a sucrose gradient. Individual fractions were incubated with topoisomerase I, and the topoisomer distribution of the purified DNA was visualized on agarose gels containing chloroquine. Supercoiled DNA (sc, lane 1), partially relaxed DNA (lane 2), and fractions with decreasing nucleosome density (lanes 3 to 8) are shown. The nucleosomal fraction used for the experiments is indicated by a white triangle. (B) MNase digestion. The indicated chromatin fraction was digested with increasing amounts of MNase. Purified DNA was visualized by agarose gel electrophoresis and ethidium bromide staining. The regular fragment ladder indicative of the nucleosomal array is indicated (1n through 6n). (C) Transcription assay. A minigene (pMrWT-T) containing the rDNA promoter and the termination region was used for in vitro transcription. DNA and chromatin were incubated with the transcription extract in the absence or presence of TTF-I (lanes 1 and 2). Readthrough transcription in the absence and terminated transcription in the presence of TTF is indicated on the left. Increasing amounts of Snf2H (lanes 2 to 4; 25, 50, and 100 fmol, respectively), NoRC (lanes 6 to 8; 25, 50, and 100 fmol, respectively) and ACF (9 to 11; 25, 50, and 100 fmol, respectively), were incubated with TTF-I, resulting in terminated transcripts. Transcription was performed for naked DNA and for chromatin templates as indicated.
To examine the function of NoRC in rDNA transcription, we compared the effects of NoRC, Snf2H, and ACF in in vitro transcription assays. Transcription assays were performed on naked DNA and on chromatin templates with purified chromatin remodeling complexes that exhibited identical activities with respect to ATP hydrolysis (data not shown). For naked DNA, exogenous Snf2H, NoRC, and ACF had no effect upon rDNA transcription (Fig. 4C, DNA panel). For chromatin templates, however, a concentration-dependent repression of transcription by NoRC, but not by Snf2H or ACF, was observed (Fig. 4C, chromatin panel). This result demonstrates that NoRC specifically silences rDNA transcription on chromatin templates in vitro.
N terminus of histone H4 is required for NoRC-dependent transcriptional repression.
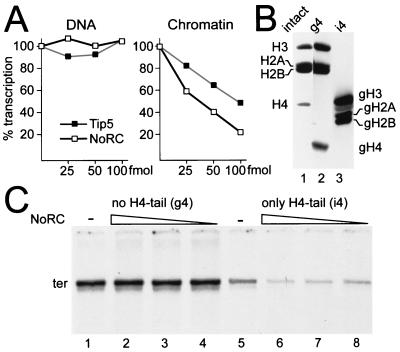
The results described above suggest a specific role for NoRC in repressing rDNA transcription. The results also raise the question of whether nucleosome remodeling is involved in this process. To uncouple the effects of Tip5 on rDNA transcription from nucleosome remodeling, we compared the effect of increasing the amounts of NoRC or Tip5 on transcription at naked rDNA and chromatin templates (Fig. 5A). Again, neither NoRC nor Tip5 affected transcription for free DNA. However, for chromatin templates, Tip5 repressed rDNA transcription, albeit to a lower extent. We also observed repression with recombinant NoRC reconstituted with an ATPase-deficient Snf2H mutant (data not shown). This finding suggests either that (i) Tip5 on its own may bind to nucleosomal rDNA and inhibit Pol I transcription or that (ii) Snf2H present in the transcription extract associates with Tip5, yielding a functional NoRC complex.
FIG. 5.
rDNA repression correlates with nucleosome remodeling. (A) Transcription experiments performed with free DNA and chromatin that used increasing amounts of NoRC (25 to 100 fmol) and Tip5 (25 to 100 fmol). Transcription reactions were quantified and plotted versus the protein concentration. (B) Histone octamers were reconstituted from appropriate combinations of full-length and tailless recombinant histones as indicated. A Coomassie blue-stained, SDS-15% polyacrylamide gel of purified octamers is shown. Recombinant octamers with all histone tails (intact, lane 1), lacking the tail of histone H4 (globular, g4; lane 2) and containing only the histone H4 tail (intact, i4; lane 3) are shown. (D) Transcription assay. Nucleosomal arrays were analyzed for transcription in the presence of TTF-I and decreasing amounts of NoRC (100, 50, and 25 fmol; lanes 2 to 4 and 6 to 8). Transcription was assayed with the nucleosomal arrays either lacking only the histone H4 N terminus (lanes 1 to 4) or containing only the histone H4 N terminus and lacking the N termini of H3, H2A, and H2B (lanes 5 to 8).
To distinguish between the two possibilities, we performed transcription assays for nucleosomal templates that lacked the N terminus of histone H4 or for all histone N termini except that of histone H4 (Fig. 5B). As nucleosome remodeling by ISWI- and Snf2H-containing complexes is known to require the N terminus of histone H4 (3), we expected this experimental approach to reveal whether binding of Tip5 alone or the chromatin remodeling causes transcriptional repression. Significantly, NoRC did not affect Pol I transcription for nucleosomes lacking the histone H4 tail (Fig. 5C, lanes 1 to 4). In contrast, transcription for templates lacking all histone tails except the N terminus of histone H4 was repressed (lanes 5 to 8). These results demonstrate that the N terminus of histone H4 is required for rDNA repression and suggest that the ATPase activity of NoRC may be required for transcriptional repression.
NoRC affects transcription initiation and has no effect on postinitiation events.
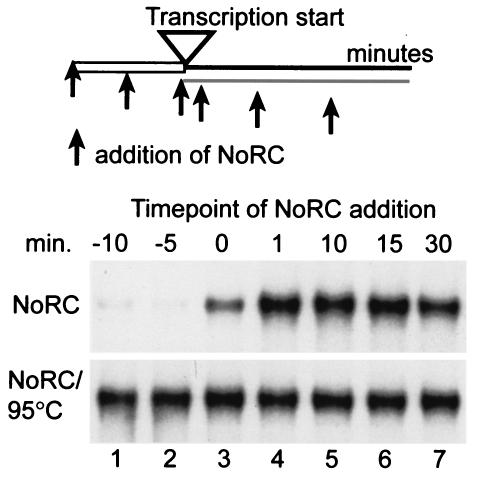
If NoRC exerted a specific effect on rDNA transcription initiation, NoRC should repress RNA polymerase I transcription prior to initiation complex formation and should not affect transcription elongation. To address this issue, we added NoRC at different time points to either the chromatin template or the transcription reaction (Fig. 6, lanes 1 to 7). Preincubation of the template with NoRC before or at the same time as the addition of transcription factors drastically repressed rDNA transcription (lanes 1 to 3). Addition of NoRC after incubation of the template with the transcription factors had no effect on rDNA transcription (lanes 4 to 7). Control reactions that used heat-inactivated NoRC did not affect transcription (lanes 1 to 7, NoRC/95°C). This experiment shows that NoRC acts prior to transcription initiation and establishes a repressive promoter structure without affecting transcription elongation.
FIG. 6.
Transcription silencing requires NoRC prior to preinitiation complex formation. NoRC (50 fmol, NoRC panel) or heat-denatured NoRC (NoRC/95°C panel) were preincubated with the chromatin template (−10, −5 min; lanes 1 and 2) or added to the transcription reaction at different time points (0, 1, 10, 15, and 30 min; lanes 3 to 7), as indicated. The transcription reactions were incubated for 60 min in the presence of TTF-I, and the transcripts were analyzed on 4.5% polyacrylamide gels.
DISCUSSION
Previous in vitro transcription studies of chromatin templates revealed that binding of TTF-I to the terminator T0 mediates ATP-dependent nucleosome remodeling, a finding which correlates with the efficient transcription initiation on otherwise repressed nucleosomal rRNA gene templates (12, 13). This finding indicated that TTF-I activates transcription by recruiting remodeling factors that place nucleosomes at the promoter into a position that allows transcription factor binding and initiation complex formation. A search for proteins that interact with TTF-I revealed the presence of Tip5, the large subunit of the Snf2H-containing remodeling complex NoRC (20). In contrast to our expectations, overexpression of Tip5 did not activate Pol I transcription but instead induced a closed heterochromatic chromatin state at the rDNA promoter that inhibited Pol I transcription in vivo. These results established a link between chromatin remodeling, histone modification, DNA methylation, and transcriptional silencing (17, 22); the results also indicated a key role for NoRC in the establishment or propagation of rRNA gene silencing.
In the course of this study, we investigated the functional relationship between NoRC and TTF-I and studied the role of NoRC with rDNA transcription in vitro. We have shown that recombinant NoRC causes transcriptional repression with chromatin templates in vitro, a finding which indicates that the nucleosome remodeling complex plays a central role in rRNA gene silencing. This result appears to be contrary to previous data, which showed that TTF-I-mediated chromatin remodeling is required for Pol I transcription for nucleosomal templates (12, 13). To reconcile these apparently contradictory results, we postulate a dual function of TTF-I for the regulation of Pol I transcription. For active genes, TTF-I may recruit an unknown activator that counteracts repressive chromatin structures and facilitates the formation of transcription initiation complexes. Alternatively, for genes to be silenced, TTF-I recruits NoRC to rDNA, which in turn leads to the formation of a transcriptionally repressed promoter. Together, the in vivo and the in vitro studies reveal a key function of TTF-I for rDNA transcription, in terms of both gene activation and gene silencing. We hypothesize that opposing chromatin modifiers exist that are recruited by TTF-I and determine the activity status of a given rDNA repeat.
The interdependence of TTF-I and NoRC for rDNA transcription is also shown by the ability of NoRC to facilitate the binding of TTF-I to DNA. Previous studies have established that the DNA binding activity of full-length TTF-I is masked. However, DNA binding activity is recovered by partial proteolytic digestion or deletion of the N-terminal region of TTF-I (16, 19). These findings suggested that the N terminus of TTF-I blocks the DNA binding domain, presumably via intra- or intermolecular protein-protein interactions. We found that the interaction of TTF-I with Tip5 uncovers the DNA binding domain and enables TTF-I to bind to its target sequence, T0, in chromatin. Binding of TTF-I to its promoter-proximal target site, in turn, is a prerequisite for the recruitment of NoRC to the rDNA promoter and transcriptional repression on chromatin templates.
NoRC-mediated rDNA repression depends on Tip5 and the N terminus of histone H4. Chromatin-specific transcriptional repression may result either from the passive association of NoRC with the rDNA promoter and/or from the active remodeling of the promoter-proximal nucleosome. Nucleosome remodeling and NoRC binding would establish an inaccessible chromatin structure, thereby inhibiting the binding of initiation factors. Several lines of experimental evidence suggest that NoRC-mediated transcriptional silencing in vitro is due to active chromatin remodeling. First, transcriptional repression was observed only for chromatin templates and not for naked DNA. Second, targeting of NoRC to the rDNA promoter leads to active nucleosome rearrangement. Third, NoRC-dependent rDNA repression occurs prior to initiation factor binding and cannot revert to an activated state in chromatin. Fourth, repression requires the tail of histone H4. NoRC did not repress transcription for nucleosomal templates that lack the tail of histone H4 but did inhibit Pol I transcription for histone octamers containing only the tail of histone H4. The requirement of the histone H4 tail for ISWI/Snf2H- and NoRC-mediated nucleosome movement has been demonstrated previously (3). On the other hand, NoRC reconstituted with the Snf2H ATPase mutant did repress rDNA transcription, similar to the behavior of Tip5, implying that nucleosome remodeling is not required. Upon TTF-I binding, however, rDNA chromatin is rearranged by chromatin remodeling activities in the transcription extract (12) and does not allow direct distinguishing between the active and passive repression mechanisms. As repression depends on the N terminus of histone H4, the passive mechanism would require the specific interaction of Tip5 with this histone tail. However, electrophoretic mobility shift assays that were performed with intact nucleosomes and nucleosomes lacking the H4 tail did not reveal preferential binding of Tip5 or NoRC to the N terminus of histone H4 (data not shown), thus arguing against a specific interaction. This result, taken together with data demonstrating that overexpression of an ATPase-deficient Snf2H mutant abolished TIP5-mediated transcriptional repression in vivo (R. Santoro and I. Grummt, unpublished data), underscores the possible role of Snf2H remodeling in NoRC-mediated silencing.
NoRC-mediated transcriptional repression was accompanied by specific positioning of a nucleosome on the rDNA promoter. Presumably, this specific nucleosome position is incompatible with transcription factor binding and initiation complex formation. In support of this presumption, NoRC repressed transcription when added prior to transcription initiation, i.e., before transcription factor bound to the promoter. When added to preformed transcription initiation complexes, NoRC affected neither transcription elongation nor the termination reaction. This means that, once activated, NoRC does not reverse the open chromatin structure and does not inhibit Pol I transcription.
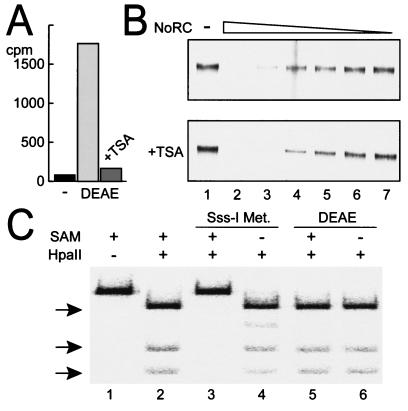
In vivo data demonstrate that NoRC-dependent rDNA repression is associated with histone deacetylation and de novo DNA methylation. Tip5 interacts with the DNA methyltransferases Dnmt1 and Dnmt3a and with the histone deacetylases (HDAC) HDAC1 and HDAC4. Furthermore, overexpression of Tip5 leads to hypoacetylation of histones and methylation of rDNA (17, 22). These findings, together with the observation that in vivo silent genes are methylated and exhibit heterochromatic features, suggest that NoRC establishes or propagates the silent state of rDNA by triggering heterochromatin formation. The in vitro experiments allow the elucidation of the order of events in the process of rDNA silencing. We showed herein that NoRC repressed rDNA transcription without modifying histones or methylating DNA. To reconcile these results, we investigated whether NoRC mediates acetylation and deacetylation of histones or DNA methylation in the in vitro transcription system. However, none of these experiments revealed an effect of NoRC on histone or DNA modification (Fig. 7).
FIG. 7.
Histone deacetylation and DNA methylation do not influence NoRC-mediated silencing of the rDNA gene. (A) The use of incubation-acetylated histones labeled with radioactive H3-acetyl-SCoA with the partially purified transcription extract (DEAE) leads to histone deacetylation. The addition of 500 nM trichostatin A (TSA) to the extract blocks histone deacetylase activities in the extract. (B) Transcription assay on chromatin templates in the presence of TTF-I, with decreasing amounts of NoRC (lanes 2 to 7), conducted in the absence (upper panel) or presence of 500 nM TSA (+TSA panel). (C) DNA methylation assay, probing the labeled rDNA promoter fragment (lane 1) with the methylation-sensitive enzyme HpaII. Methylation was performed under transcription conditions for 60 min with bacterial Sss-I methylase (New England Biolabs; lanes 3 and 4) and the DEAE extract (lanes 5 and 6) in the presence or absence of the substrate S-adenosylmethionine (SAM) as indicated. DNA was purified, digested with HpaII (lanes 2 to 6), and separated on a 5% polyacrylamide gel. Unmethylated digestion products are indicated by arrows.
NoRC seems to serve at least two functions. First, it serves as a remodeling complex that is recruited to the rDNA promoter and alters its chromatin structure. This step is most important for the establishment of the silent state of ribosomal genes because it prevents transcription factor binding and, consequently, transcription initiation. Second, NoRC may function as a scaffold that coordinates the activities of macromolecular complexes that modify histones, methylate DNA, and establish a closed heterochromatic chromatin state. This step is most likely required in order to maintain and propagate the repressed state during cell cycle progression.
Acknowledgments
We thank R. E. Kingston and H. Y. Fan for the Snf2H clone, and we thank Josephine Sutcliffe for careful reading of the manuscript.
R.S. and A.N. were supported by grants from Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Aalfs, J. D., G. J. Narlikar, and R. E. Kingston. 2001. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 276:34270-34278. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, E., and M. R. Paule. 1988. Promoter occlusion during ribosomal RNA transcription. Cell 54:985-992. [DOI] [PubMed] [Google Scholar]
- 3.Clapier, C. R., G. Längst, D. F. Corona, P. B. Becker, and K. P. Nightingale. 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753-761. [DOI] [PubMed] [Google Scholar]
- 5.Corona, D. F., G. Längst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 6.Eberharter, A., S. Ferrari, G. Längst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 16:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerber, J. K., E. Gogel, C. Berger, M. Wallisch, F. Muller, I. Grummt, and F. Grummt. 1997. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90:559-567. [DOI] [PubMed] [Google Scholar]
- 8.Grummt, I., H. Rosenbauer, I. Niedermeyer, U. Maier, and A. Ohrlein. 1986. A repeated 18-bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell 45:837-846. [DOI] [PubMed] [Google Scholar]
- 9.Henderson, S., and B. Sollner-Webb. 1986. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell 47:891-900. [DOI] [PubMed] [Google Scholar]
- 10.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Längst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 12.Längst, G., P. B. Becker, and I. Grummt. 1998. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 17:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Längst, G., T. A. Blank, P. B. Becker, and I. Grummt. 1997. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 16:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucchini, R., and J. M. Sogo. 1995. Replication of transcriptionally active chromatin. Nature 374:276-280. [DOI] [PubMed] [Google Scholar]
- 15.McStay, B., and R. H. Reeder. 1990. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes Dev. 4:1240-1251. [DOI] [PubMed] [Google Scholar]
- 16.Sander, E. E., S. W. Mason, C. Munz, and I. Grummt. 1996. The amino-terminal domain of the transcription termination factor TTF-I causes protein oligomerization and inhibition of DNA binding. Nucleic Acids Res. 24:3677-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santoro, R., J. Li, and I. Grummt. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 32:393-396. [DOI] [PubMed] [Google Scholar]
- 18.Schnapp, G., B. R. Graveley, and I. Grummt. 1996. TFIIS binds to mouse RNA polymerase I and stimulates transcript elongation and hydrolytic cleavage of nascent rRNA. Mol. Gen. Genet. 252:412-419. [DOI] [PubMed] [Google Scholar]
- 19.Smid, A., M. Finsterer, and I. Grummt. 1992. Limited proteolysis unmasks specific DNA-binding of the murine RNA polymerase I-specific transcription termination factor TTFI. J. Mol. Biol. 227:635-647. [DOI] [PubMed] [Google Scholar]
- 20.Strohner, R., A. Németh, P. Jansa, U. Hofmann-Rohrer, R. Santoro, G. Längst, and I. Grummt. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20:4892-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widom, J. 1998. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27:285-327. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, Y., R. Santoro, and I. Grummt. 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21:4632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]