Abstract
U12-dependent introns are spliced by the so-called minor spliceosome, requiring the U11, U12, and U4atac/U6atac snRNPs in addition to the U5 snRNP. We have recently identified U6-p110 (SART3) as a novel human recycling factor that is related to the yeast splicing factor Prp24. U6-p110 transiently associates with the U6 and U4/U6 snRNPs during the spliceosome cycle, regenerating functional U4/U6 snRNPs from singular U4 and U6 snRNPs. Here we investigated the involvement of U6-p110 in recycling of the U4atac/U6atac snRNP. In contrast to the major U6 and U4/U6 snRNPs, p110 is primarily associated with the U6atac snRNP but is almost undetectable in the U4atac/U6atac snRNP. Since p110 does not occur in U5 snRNA-containing complexes, it appears to be transiently associated with U6atac during the cycle of the minor spliceosome. The p110 binding site was mapped to U6 nucleotides 38 to 57 and U6atac nucleotides 10 to 30, which are highly conserved between these two functionally related snRNAs. With a U12-dependent in vitro splicing system, we demonstrate that p110 is required for recycling of the U4atac/U6atac snRNP.
Small nuclear ribonucleoproteins (snRNPs) are essential cofactors in nuclear mRNA splicing that participate in the assembly, catalysis, and disassembly of the spliceosome (for reviews, see references 7, 13, and 36). Splicing of the canonical introns of the GT-AG type requires five small nuclear RNAs (snRNAs), U1, U2, U4, U5, and U6, together with more than 100 protein factors (10, 12, 41). In addition, a minor-type spliceosome was discovered that contains a different set of snRNAs: U11, U12, U4atac, and U6atac; U5 snRNA is present in both the major and minor spliceosomes (32). Surprisingly, the minor snRNAs share only a little sequence similarity with their counterparts of the major spliceosome, but structurally and functionally, U11, U12, U4atac, and U6atac snRNAs correspond directly to the major-type U1, U2, U4, and U6 snRNAs, respectively (reviewed in references 6, 28, 33, and 38). Altogether, there are more than 400 U12-dependent introns in the human genome (17), and there is evidence that U12-type introns provide an additional means of regulating gene expression at the mRNA splicing level (23).
The dynamic nature of the spliceosome becomes particularly apparent when following the U4 and U6 snRNPs throughout the spliceosome cycle (5). Most of the U4 and U6 snRNPs are stably base paired with each other; before entering the assembling spliceosome, the U5 snRNP joins the U4/U6 snRNP, resulting in the U4/U6.U5 tri-snRNP. In the active spliceosome, the U4-U6 base pairing is disrupted and isomerizes to a mutually exclusive U6-U2 interaction. Finally, after splicing catalysis, the spliceosome disassembles and the U4 and U6 snRNPs are released in their singular forms. Therefore, recycling is necessary to restore the original U4-U6 base pairing. Significantly, all of these dynamic RNA-RNA interactions are conserved between the major and minor snRNAs (9, 11, 14, 26, 29). Although the same basic RNA network operates in both types of spliceosomes, we still know very little of how their protein constituents compare. Most of the protein components appear to be identical between the major- and minor-type spliceosomal snRNPs (20, 21, 26, 37), with the possible exception of a 70,000-molecular-weight homolog found in the U11/U12 snRNP (37).
We have recently identified a human U6-specific RNA binding protein, U6-p110 (called p110 in the following), which is specifically required during the snRNP recycling phase, mediating the reassembly of the postspliceosomal singular U4 and U6 snRNPs (3). p110 is functionally related to the splicing factor PRP24 from Saccharomyces cerevisiae (24, 27), which is known to function in U4-U6 RNA annealing and U4/U6 snRNP recycling (8, 25, 34). p110 associates only transiently with the U6 and U4/U6 snRNPs but is absent from the U4/U6.U5 tri-snRNP and spliceosomes (3).
Here we report that p110 also functions in recycling of the U4atac/U6atac snRNP. We have identified a difference in the recycling phase between the major and minor spliceosomes, in that p110 stably interacts only with the U6atac snRNP but is almost undetectable in the U4atac/U6atac snRNP. Because U6 and U6atac are structurally equivalent yet greatly diverge in their primary sequences, we searched also for the common elements in the U6 and U6atac snRNAs that are recognized by the p110 protein. Comparing the sequence requirements for p110 binding revealed a short sequence of 20 nucleotides that is highly homologous between the U6 and U6atac snRNAs. Finally, we demonstrate directly that in vitro recycling of the U4atac/U6atac snRNP requires p110 protein.
MATERIALS AND METHODS
DNA oligonucleotides.
The following DNA oligonucleotides were used in this study: U1-F, 5′-GGG GAG ATA CCA TGA TCA CG-3′; U1-R, 5′-GTC GAG TTT CCC ACA TTT GG-3′; U2-F, 5′-CTC GGC CTT TTG GCT AAG AT-3′; U2-R, 5′-TGC AAT ACC AGG TCG ATG C-3′; U4-F, 5′-AGC TTT GCG CAG TGG CAG T-3′; U4-R, 5′-CCG TAG AGA CTG TCA AAA ATT GC-3′; U5-F, 5′-TGG TTT CTC TTC AGA TCG CAT A-3′; U5-R, 5′-CCA AGG CAA GGC TCA AAA-3′; U6-F, 5′-CGC TTC GGC AGC ACA TAT AC-3′; U6-R, 5′-AAA ATA TGG AAC GCT TCA CGA-3′; U4atac-F, 5′-AAC CAT CCT TTT CTT GGG GTT G-3′; U4atac-R, 5′-TAT TTT TCC AAA AAT TGC ACC AA-3′; U6atac-F, 5′-TGT ATG AAA GGA GAG AAG GTT A-3′; U6atac-R, 5′-AAA AAC GAT GGT TAG ATG C-3′; U2 49-27, 5′-ATA AGA ACA GAT ACT ACA CTTG A-3′; SP6-U4atac 1-16, 5′-ATT TAG GTG ACA CTA TAG AAC CAT CCT TTT CTT G-3′; NsiI-U4atac 131-113, 5′-ATG CAT ATT TTT CCA AAA ATT GCA-3′; T7-U6atac 1-18, 5′-TAA TAC GAC TCA CTA TAG GTG TTG TAT GAA AGG AGA-3′; T7-U6atac 38-59, 5′-TAA TAC GAC TCA CTA TAG ACA AGG ATG GAA-3′; DraI-U6atac 125-105, 5′-TTT AAA AAC GAT GGT TAG ATG CCA-3′.
RNA analysis.
RNA was separated by electrophoresis in denaturing polyacrylamide-urea gels (8% acrylamide, 0.42% bisacrylamide, 50% urea, 1× Tris-borate-EDTA buffer). For 3′-terminal labeling, RNA was incubated with 10 μCi of [32P]pCp and 50 U of T4 RNA ligase (Roche) in the provided buffer, containing in addition 10% dimethyl sulfoxide (5 h, 18°C). Northern blot analysis of RNA was done as described previously (2). The PCR products of the U1, U2, U4, U5, U6, U4atac, and U6atac snRNAs were amplified by reverse transcription-PCR or from plasmid templates with the corresponding forward primer-reverse primer pairs. Digoxigenin-labeled probes directed against the snRNAs mentioned above were obtained by multiple-cycle extension of the R primers, including the corresponding PCR products as templates and PCR digoxigenin-labeling mixture (Roche).
Immunoprecipitation.
For immunoprecipitation experiments, a 50-μl packed volume of protein A Sepharose CL-4B (Amersham Pharmacia Biotech) in 50 μl of N100 (50 mM Tris-HCl [pH 8.0] 100 mM NaCl, 0.05% NP-40, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride) was incubated with 175 μl of immune or preimmune rabbit serum with rotation overnight at 4°C and washed five times with 1 ml of N100. The beads were then incubated for 4 h together with 500 μl of N100 and 500 μl of HeLa nuclear extract (4C Biotec) and then washed five times with 1 ml of N200 buffer (containing 200 mM NaCl). Coprecipitated RNAs were eluted in 300 μl of PK buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 0.5% sodium dodecyl sulfate) for 10 min at 80°C, phenolized, precipitated, and analyzed by Northern blotting.
Constructs, in vitro transcription, and in vitro RNA binding of p110.
Human U4 and U6 snRNAs were transcribed from SP6-U4 (35) and SP6-U6 (4), respectively. The template for in vitro transcription of U4atac snRNA was obtained by reverse transcription-PCR with total RNA from HeLa nuclear extract and primers SP6-U4atac 1-16 and NsiI-U4atac 131-113. The template for in vitro transcription of U6atac snRNA was prepared in the same way with primers T7-U6atac 1-18 and DraI-U6atac 125-105. The U4atac PCR product was cloned into the pCR2.1TOPO vector (Invitrogen), the U6atac PCR product was cloned into the pQE-30UA vector (Qiagen), and both were confirmed by sequence analysis. After linearization with NsiI, U4atac snRNA was transcribed in vitro by SP6 RNA polymerase. U6atac snRNA was transcribed by T7 RNA polymerase from a DraI-cut U6atac plasmid template. The template for U6atac 38-125 was generated by PCR with the T7-U6atac 38-59 and U6atac-R primers. Templates for the U6 and U6atac snRNA derivatives containing short internal regions were transcribed in vitro from double-stranded oligonucleotide templates. All RNAs were 3′ end labeled with [32P]pCp. Expression in SF21 insect cells and purification of recombinant p110 protein and the RNA binding assays were described previously (3).
Native gel assays of p110-RNA binding.
U4atac, U6atac, U4, and U6 snRNAs were transcribed in vitro in the presence of [α-32P]UTP. To study the affinity of the RNA-protein interaction, 49 fmol of in vitro-transcribed RNA or preannealed RNA duplex was incubated with the indicated molar excess of N-terminally tagged glutathione S-transferase-p110 (J. Medenbach, unpublished data) in a 12-μl reaction mixture containing 20 mM HEPES-KOH (pH 7.5), 140 mM KCl, 2.0 mM MgCl2, 1 mM DTT, 83.3 ng of yeast tRNA per μl, and 3.3 U of RNasin (Promega) per μl for 30 min at 30°C. The p110-RNA complexes were analyzed by native gel electrophoresis (6% acrylamide-0.075% bisacrylamide-0.5× Tris-borate-EDTA buffer run at 1.4 V/cm and 4°C). Quantitation of the protein-free and p110-bound RNA was performed with a Gel-Pro Analyzer 3.0 (Media Cybernetics).
To prepare U4atac/U6atac duplex RNA, 4.7 pmol of 32P-labeled U6atac snRNA was mixed with a 10-fold molar excess of unlabeled U4atac snRNA in a 24-μl reaction mixture containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT. After a 1-min incubation at 80°C, the reaction mixture was allowed to cool down slowly to 30°C. U4/U6 duplex RNA was prepared under the same conditions as described for the atac snRNAs, except that a 1,000-fold molar excess of oligonucleotide U6-R was added.
p110 immunodepletion from HeLa nuclear extract.
For p110 immunodepletion, a 100-μl volume of packed protein A Sepharose beads was incubated overnight at 4°C with polyclonal anti-p110 antiserum. Beads were then washed five times with 1 ml each of N100 buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.05% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT), followed by addition of 400 μl of nuclear extract (4C Biotec) and incubation at 4°C for 2 h. As a control, mock-depleted extract was prepared in parallel with nonimmune serum. The efficiency of p110 depletion was analyzed by Western blotting as described previously (3).
U4atac/U6atac snRNP recycling in vitro.
In vitro splicing reaction mixtures with unlabeled SCN4AENH1 pre-mRNA (10 ng/25-μl reaction mixture) in mock-depleted nuclear extract or in nuclear extract depleted of p110 were prepared as previously described (15, 32, 39). Before the reaction, U2-dependent splicing was blocked by incubation with 1.2 mM U2 49-27 oligonucleotide for 20 min at 30°C. For complementation assays, recombinant p110 protein was added to depleted extract (25, 50, or 100 ng/25-μl reaction mixture). After splicing for 1.5 h at 30°C, the entire reaction mixture (50 μl) was fractionated by CsCl gradient centrifugation as previously described (18), with a CsCl-buffer D solution with a density of 1.55 g/ml and containing 15 mM MgCl2. Five fractions of 0.2 ml each were obtained (fraction 5 including the pellet), and RNA was purified and analyzed on an 8% denaturing polyacrylamide gel, followed by Northern hybridization with digoxigenin-labeled U4atac and U6atac probes.
RESULTS
p110 is primarily associated with the singular U6atac snRNP but not the U4atac/U6atac snRNP.
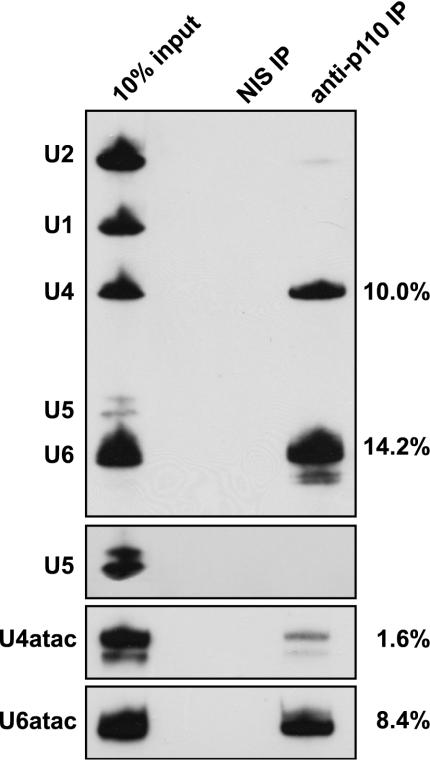
By immunoprecipitation with anti-p110 antibodies from both HeLa nuclear and S100 extracts, we had previously found U4 and U6 as p110-associated snRNAs (3). Since U4atac and U6atac snRNAs occur at a much lower abundance, we tested whether they are associated with p110 by using anti-p110 immunoprecipitates from HeLa nuclear extract and detecting coprecipitated snRNAs by denaturing gel electrophoresis and Northern hybridization (Fig. 1, anti-p110 IP lane). Nonimmune antiserum was used in control immunoprecipitations (NIS IP lane). Consistent with our previous analysis (3), anti-p110 antibodies, but not the nonimmune antiserum, efficiently precipitated both U4 and U6 snRNAs. In contrast, no signals for U1, U2, or U5 snRNA were detectable. U6atac snRNA was coimmunoprecipitated by anti-p110 antibodies, with an efficiency comparable to that of the U6 snRNA; however, only background levels of U4atac could be detected. We have carried out similar coimmunoprecipitation assays with HeLa S100 extract and by using primer extension instead of Northern detection, which gave essentially the same results (data not shown). We conclude that—in contrast to U4 and U6 snRNAs—p110 is primarily associated with the U6atac snRNP but is almost undetectable in the U4atac/U6atac snRNP.
FIG. 1.
Human p110 protein is associated primarily with the singular U6atac snRNP but not the U4atac/U6atac di-snRNP. Immunoprecipitation (IP) from HeLa nuclear extract was done with anti-p110 antibodies (anti-p110 IP lane) or nonimmune antiserum (NIS IP lane). RNA was prepared from the immunoprecipitates and 10% of the input (10% input lane), followed by Northern hybridization analysis with a mixed probe detecting U1, U2, U4, U5, and U6 snRNAs or probes specific for U4atac and U6atac. Since U5 snRNA was more difficult to detect than the other snRNAs, an additional, longer exposure is shown for this snRNA in the upper middle gel. In addition, percent immunoprecipitation efficiencies are listed on the right.
snRNA sequence requirements for p110 binding.
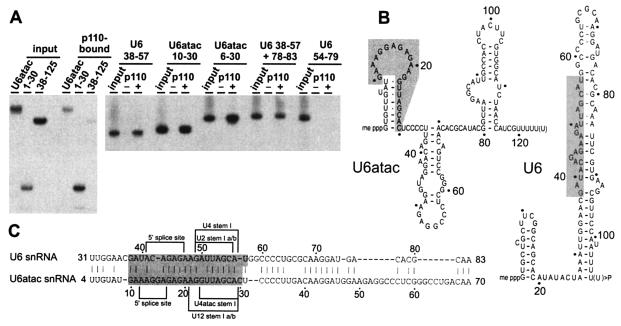
After identifying the U6atac snRNA as a second p110 target, we determined the RNA sequence requirements for p110 binding of U6atac snRNA (Fig. 2). In our previous study (3), we had mapped the U6 snRNA region required for p110 binding to an internal region composed of nucleotides 38 to 57 and 78 to 83. Importantly, U6 and U6atac have only 45% overall sequence identity, and therefore, comparing the sequence requirements for p110 RNA binding on these two functionally related snRNAs should reveal more about the RNA binding specificity of p110.
FIG. 2.
(A) U6 and U6atac snRNA sequence requirements for p110 binding. (Left part) 32P-labeled full-length U6atac snRNA and derivatives U6atac 1-30 and 38-125 (see panel B for a schematic representation) were incubated with recombinant His-tagged p110 protein, precipitated with Ni-nitrilotriacetic acid agarose, and subjected to direct RNA analysis on a denaturing gel. For each RNA, 20% of the input and the precipitate (p110 bound) are shown. (Right part) The following 32P-labeled RNAs were incubated with or without His-tagged p110 protein (− and + lanes): U6 38-57, U6atac 10-30, U6atac 6-30, U6 38-57 + 78-83, and U6 54-79. Twenty percent of the input is shown in each case. (B) Schematic secondary-structure models of U6 and U6atac snRNAs (19, 22, 31). Shading indicates the snRNA region required for p110 binding. (C) Sequence alignment of U6 nucleotides 31 to 83 and U6atac nucleotides 4 to 70 (by pairwise BLAST; www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html). The p110 binding region is shaded. In addition, sequences important for 5′ splice site interaction (5′ splice site), U4 base pairing (U4 or U4atac stem I), and U2 base pairing (U2 or U12 stem Ia/b) are indicated.
Full-length U6atac snRNA, 5′-terminal U6atac nucleotides 1 to 30 (corresponding to the 5′ stem-loop), and 3′-terminal nucleotides 38 to 125 were transcribed by T7 RNA polymerase, labeled with [32P]pCp, and incubated with recombinant His-tagged p110 protein (Fig. 2A; panel B is a schematic representation of the RNAs). Bound RNAs were selected on Ni-nitrilotriacetic acid agarose; both full-length U6atac and the short 5′-terminal fragment (U6atac 1-30) efficiently bound p110, whereas the long 3′-terminal fragment (U6atac 38-125) bound only at background levels.
Comparing U6 and U6atac by sequence alignment (Fig. 2C) revealed that U6atac nucleotides 10 to 30 represent the longest stretch of sequence homology between U6 and U6atac (76% identity). Therefore, we also tested two smaller U6atac derivatives for p110 binding, U6atac 6-30 and U6atac 10-30, in direct comparison with the corresponding U6 region, U6 38-57 (Fig. 2A). With the His tag precipitation assay, we found that each of these short RNAs efficiently bound p110. As previously reported (3), the slightly longer U6 38-57 + 78-83 RNA was positive in p110 binding and the intramolecular stem-loop of U6 (U6 54-79) was negative. In sum, these assays mapped the p110 binding site to U6atac nucleotides 10 to 30 and corresponding U6 nucleotides 38 to 57.
p110 binds to U4/U6 and U4atac/U6atac duplex RNAs.
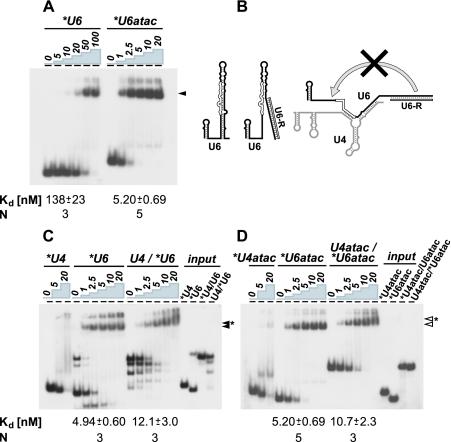
We had initially detected p110 protein in both the endogenous U6 and U4/U6 snRNPs (3). In the case of the atac snRNPs, in contrast, we found p110 to be present almost exclusively in the singular U6atac snRNP (see above and Fig. 1). To investigate this interesting difference further, we assayed p110-RNA binding by native gel electrophoresis, titrating the p110 concentration for each RNA binding substrate (molar excess indicated above the lanes) and determining the apparent dissociation constant, Kd (Fig. 3).
FIG. 3.
In vitro binding of p110 protein to singular and duplex forms of U6 and U6atac snRNAs. The apparent Kd values are shown below the corresponding panels. Standard deviations were derived from three to five independent experiments (N, number of experiments). The signals above the marked complexes are mostly due to material that had not entered the gel. (A) Binding to singular U6 and U6atac snRNAs. 32P-labeled U6 and U6atac snRNAs were incubated without or with recombinant p110 protein (molar excesses are indicated above the lanes). RNA-protein complex formation was analyzed by native gel electrophoresis and visualized by autoradiography. The mobilities of the p110 complexes with U6 and U6atac snRNAs are marked by an arrowhead. (B) Structural alterations of singular U6 and U4/U6 duplex snRNAs by binding of the oligonucleotide U6-R. The secondary-structure model of the singular U6 snRNA (19) is shown with the p110 binding region indicated by open lines. Binding of oligonucleotide U6-R is expected to open up the extended 3′ stem of the singular form of U6. For the U4/U6 duplex, annealing of oligonucleotide U6-R should prevent the intramolecular interaction of U6 (19) and thereby stabilize the protein-free U4/U6 duplex. (C) Binding to singular U6 and U4/U6 duplex snRNAs. 32P-labeled U4 snRNA, 32P-labeled U6 snRNA, and the U4/U6 snRNA duplex (with the U6 snRNA 32P labeled) were incubated without or with recombinant p110 protein (molar excesses are indicated above the lanes). RNA-protein complex formation was analyzed as described for panel A. The input RNAs are also shown (input lanes: 32P-labeled U4 and U6 snRNAs, U4/32P-U6, and for comparison, 32P-U4/U6 snRNA duplex). The mobilities of the p110-U6 complex (filled arrowhead) and the p110 complex with the U4/U6 duplex snRNA (filled arrowhead with asterisk) are marked. The multiple lower bands were observed only after oligonucleotide annealing and most likely reflect different conformations of U6 and U4/U6 snRNAs. (D) Binding to singular U6atac and U4atac/U6atac duplex snRNAs. 32P-labeled U4atac snRNA, 32P-labeled U6atac snRNA, and the U4/U6 snRNA duplex (with the U6atac snRNA 32P labeled) were incubated without or with recombinant p110 protein (molar excesses are indicated above the lanes). RNA-protein complex formation was analyzed as described for panel A. The input RNAs are also shown (input lanes: 32P-labeled U4atac and U6atac snRNAs, U4atac/32P-U6atac, and for comparison, 32P-U4atac/U6atac snRNA duplex). The mobilities of the p110-U6atac complex (open arrowhead) and the p110 complex with the U4atac/U6atac duplex snRNA (open arrowhead with asterisk) are marked.
Initially we compared p110 binding by U6 and U6atac snRNAs (Fig. 3A). 32P-labeled, in vitro-transcribed U6 and U6atac RNAs were incubated in the absence of or with increasing amounts of recombinant p110 protein, followed by native gel electrophoresis. For the p110 complexes of U6 and U6atac snRNAs, quantitative evaluation gave Kd values of 138 ± 23 and 5.20 ± 0.69 nM, respectively. We conclude that p110 has an approximately 25-fold higher affinity for U6atac than for U6 snRNA.
Next we directly compared the singular U6 and U6atac snRNAs with the corresponding snRNA duplexes (U4/U6 and U4atac/U6atac) in terms of their affinity for p110 protein; in addition, p110 binding to U4 and U4atac snRNAs was assayed (Fig. 3C and D). 32P-labeled, in vitro-transcribed RNAs were incubated in the absence of or with increasing amounts of recombinant p110 protein and then subjected to native gel electrophoresis. The U4/U6 and U4atac/U6atac snRNA duplexes were 32P labeled in their U6 (U6atac) component and were formed in vitro (see Materials and Methods). Note that in Fig. 3C, both singular U6 and the U4/U6 duplex had a DNA oligonucleotide bound at the 3′-terminal region of U6, which was necessary for efficient formation of the U4/U6 snRNA duplex (schematic in Fig. 3B). Binding of oligonucleotide U6-R apparently prevents destabilization of the U4/U6 duplex by intramolecular base pairing (see also reference 19). Only in this way were we able to directly compare the affinities of p110 for the singular and duplex snRNAs. The p110 complexes of singular forms of U6 and U6atac snRNAs and the corresponding duplex forms could be resolved in this gel system (compare Fig. 3C and D).
In sum, the following apparent Kd values were derived: U6 (with U6-3′ oligonucleotide bound), 4.94 ± 0.60 nM; U4/U6 (with U6-3′ oligonucleotide bound), 12.1 ± 3.0 nM; U6atac, 5.20 ± 0.69 nM; U4atac/U6atac, 10.7 ± 2.3 nM.
In contrast to these specific U6- or U6atac-containing complexes, only at the highest protein concentration did the U4 and U4atac snRNAs form complexes of diffuse mobility, which probably reflect nonspecific p110 binding.
In conclusion, the affinity of p110 for either singular U6 or U6atac snRNA is significantly higher than that for the corresponding duplex form. For the normal and atac versions of U6, the Kd values differ approximately twofold. More importantly, we have observed a strong effect of binding of the 3′-terminal oligonucleotide to U6 on p110 affinity. The Kd was lowered more than 25-fold (from 138 to 4.94 nM), most likely reflecting the extensive structural alteration introduced by oligonucleotide binding (schematic in Fig. 3B).
p110 is required for U4atac/U6atac snRNP recycling.
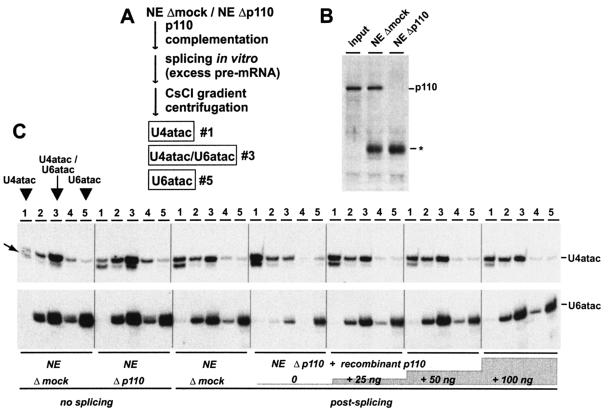
To determine whether p110 plays a role in recycling of the minor-type spliceosome, we developed an assay similar to the one we have used to assess the recycling status of the U4 and U6 snRNAs (3) (Fig. 4A contains a schematic outline). In vitro splicing reaction mixtures were prepared with SCN4AENH1 pre-mRNA. This pre-mRNA substrate contains a single ATAC-type intron, derived from the human voltage-gated skeletal muscle sodium channel α subunit (SCN4A) pre-mRNA, and has a purine-rich enhancer inserted at the end of the second exon (39).
FIG. 4.
p110 is required for in vitro recycling of the U4atac/U6atac snRNP. (A) Schematic representation of in vitro recycling assay (see text and reference 3). (B) p110 immunodepletion of nuclear extract (NE). Aliquots of untreated nuclear extract (input), and nuclear extract after mock depletion (NE Δmock) and p110 depletion (NE Δp110) were analyzed by Western blotting with anti-p110 antibodies. The asterisk indicates the heavy-chain immunoglobulin G band. (C) In vitro recycling of the U4atac/U6atac snRNP. Mock-depleted nuclear extract (NE Δmock) and nuclear extract depleted of p110 (NE Δp110) without prior incubation were fractionated by CsCl gradient centrifugation. Similarly, mock-depleted nuclear extract (NE Δmock) and nuclear extract depleted of p110 (NE Δp110) were analyzed after a 1.5-h splicing reaction with SCN4AENH1 pre-mRNA under recycling conditions. In addition, incubations were done in extract depleted of p110 and then complemented with 25, 50, or 100 ng of recombinant p110 protein per 25-μl reaction mixture (as indicated). For each reaction mixture, the distribution of U4atac and U6atac throughout the entire CsCl gradient was determined. RNA was prepared from fractions 1 to 5 (top to bottom; as indicated at the top) and analyzed by denaturing gel electrophoresis and Northern blotting with separate probes for U4atac (top) and U6atac (bottom). The positions of the U4atac/U6atac snRNP (fraction 3) and the singular forms of U4atac (fraction 1) and U6atac (fraction 5) within the CsCl gradient are shown for the first panel on the left. The electrophoretic mobilities of the U4atac and U6atac snRNAs are marked on the right. The minor form of U4atac snRNA (31) is indicated by the arrow on the left.
As previously done for a major-type intron (3; data not shown), we first established conditions for the SCN4AENH1 pre-mRNA under which we expected splicing to be limited by recycling. Splicing activity was assayed at three different pre-mRNA concentrations, ranging between 0.2 and 10 ng/25-μl reaction mixture, and in mock-depleted extract, extract depleted of p110, and extract depleted of p110 and then complemented with p110 protein (50 ng/25-μl reaction mixture). Immunodepletion with anti-p110 antibodies routinely resulted in selective removal of at least 95% of the p110, as determined by Western blot analysis (Fig. 4B). Thereby, splicing factors including the atac snRNPs should become limiting for the minor spliceosome. The overall in vitro splicing activity of 32P-labeled SCN4AENH1 pre-mRNA was relatively low, as previously reported (39; data not shown). Interestingly, the addition of recombinant p110 protein to p110-depleted extract did not significantly change the splicing activity at the low pre-mRNA concentration but resulted in a twofold stimulation of splicing at the high pre-mRNA concentration (0.2 versus 10 ng/25-μl reaction mixture; data not shown), suggesting that the p110-dependent recycling step indeed becomes limiting for splicing of the ATAC-type intron.
By using these in vitro splicing conditions and the SCN4AENH1 pre-mRNA, we investigated the recycling status of the U4atac and U6atac snRNAs (Fig. 4C). We assayed at a pre-mRNA concentration of 10 ng/25-μl reaction mixture, which is approximately seven times higher than the pre-mRNA level normally used in in vitro splicing of this U12-dependent intron (39). Following the in vitro splicing reactions, the relative levels of U4atac/U6atac and the free U4atac and U6atac snRNPs were quantitatively measured by CsCl density gradient centrifugation. Under these highly stringent conditions, core complexes of snRNPs are characteristically stable and fractionate according to their density, which is determined by their RNA/protein ratio (16). We confirmed this also for the minor U4atac/U6atac snRNP (Fig. 4C, NEΔmock/no splicing). Nuclear extract was fractionated by CsCl density gradient centrifugation, and RNA was prepared from the resulting five fractions (numbered 1 to 5, from top to bottom) and analyzed by Northern blot hybridization with probes specific for the U4atac and U6atac snRNAs. Clearly, the U4atac and U6atac snRNAs cofractionated with a peak in fraction 3, representing the U4atac/U6atac core snRNP and containing approximately 70% of the total U4atac material and half of the total U6atac material. U6atac shows an additional peak at the bottom of the gradient, representing approximately one-third of the total U6atac material and most likely consisting of material derived from free U6atac snRNP. Only a very small part (5%) of the U4atac material fractionated in fraction 1, where no U6atac snRNA could be detected; we conclude that this material at the top of the gradient represents the singular U4atac snRNP, similar to what we had observed for the singular U4 snRNP (3). In sum, we were able to quantitatively monitor by CsCl density gradient centrifugation the relative levels of base-paired U4atac/U6atac, free U4atac, and free U6atac snRNPs. For each assay described in the following, the splicing reaction mixture was first fractionated on a CsCl gradient and then the U4atac and U6atac distribution was determined by Northern blot hybridization and quantitatively evaluated. We considered fractions 1, 3, and 5 to be indicative of the abundance of free U4atac, base-paired U4atac/U6atac, and free U6atac snRNPs, respectively.
Interestingly, there is a minor U4atac form, which had been noted previously by Tarn and Steitz (31) and which is slightly shortened at the 5′ end (Fig. 4C, minor band marked by arrow). Most of this 5′-shortened U4atac occurs in fraction 1, where no U6atac snRNA could be detected, and very little is present in the di-snRNP form. We interpreted this to mean that the 5′-shortened U4atac snRNA is blocked in di-snRNP formation, most likely as a result of the truncation of U4atac/U6atac stem II.
Next we compared the distributions of U4atac and U6atac in mock-depleted (NEΔmock) and p110-immunodepleted (NEΔp110) nuclear extract without splicing incubation (Fig. 4C, NEΔmock and Δp110/no splicing). Quantitation of the relative distributions of U4atac and U6atac in the singular and base-paired forms showed no dramatic difference: Under both conditions, the U4atac/U6atac di-snRNP represents the major form of these two snRNAs and there is very little singular U4atac snRNP.
To assay for a role of p110 in atac snRNP recycling, we then determined the snRNP distribution after splicing for 1.5 h with an excess of ATAC pre-mRNA (10 ng/25-μl reaction mixture), that is, under recycling conditions in which we expect multiple rounds of the spliceosome cycle to occur. The splicing incubation was done in mock-depleted nuclear extract (NEΔmock), in nuclear extract depleted of p110 (NEΔp110), and in nuclear extract depleted of p110 and then complemented with 25, 50, or 100 ng of recombinant p110 protein (Fig. 4C, postsplicing). Compared with that in the mock-depleted extract, the postspliceosomal snRNP distribution was dramatically altered in extract depleted of p110 (panels NEΔmock/postsplicing and NEΔp110/postsplicing). Most (60%) of the U4atac was converted to the singular U4atac snRNP, and most (72%) of the U6atac was shifted to the bottom of the gradient, representing the singular U6atac form; only approximately 20% of the U4atac and U6atac remained in the di-snRNP form. The most likely explanation for these changes is that the U4atac/U6atac snRNPs are consumed during assembly of the minor spliceosome and in SCN4AENH1 pre-mRNA splicing. As a result of SCN4AENH1 pre-mRNA splicing and the recycling block, postspliceosomal free U4atac and U6atac snRNPs accumulate.
Since the steady-state level of U6atac snRNA after splicing appears to be lower in p110-depleted than in mock-depleted extract, we have tested whether the stability of U6atac snRNA might be affected by p110 depletion. Control experiments showed, however, that the total amount of U6atac snRNA was not reduced significantly in comparison to other snRNAs (data not shown); therefore, part of the U6atac snRNA appears to be lost in the form of precipitates during the splicing reaction in extract depleted of p110.
When we complemented with recombinant p110 protein, these shifts in the distribution of both U4atac and U6atac could be reversed in a concentration-dependent manner (Fig. 4C, compare postsplicing gels). Already, the addition of 50 ng of p110 protein fully restored the distribution observed in mock-depleted extract. This demonstrates that p110 functions as a U4atac/U6atac snRNP recycling factor and is responsible for the regeneration of base-paired U4atac/U6atac snRNPs from postspliceosomal free U4atac and U6atac snRNPs.
DISCUSSION
In this study, we have characterized the role of a human snRNP recycling factor, p110, in the recognition of U6atac snRNA and in the postspliceosomal regeneration of the U4atac/U6atac snRNP. We had recently identified p110 as a U6-specific RNA binding protein on the basis of [32P]pCp labeling of coimmunoprecipitated snRNAs (3). At that time, we had failed to detect U6atac, most likely because of its low abundance, which is 2 orders of magnitude below that of the corresponding major snRNA (40).
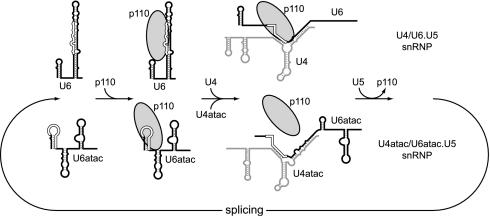
Interestingly, we detected p110 primarily in the singular U6atac snRNP but only at background levels in the U4atac/U6atac di-snRNP (Fig. 1); in contrast, p110 can associate with both the U6 and U4/U6 snRNPs (3) (Fig. 5 contains a schematic representation). This indicates a difference in protein composition between the major- and minor-type snRNPs, which is unexpected, considering that the assembly of the U4/U6 snRNP and its minor-type counterpart follows a highly conserved hierarchical pathway (21). Since in our previous study (3) we had not found U5 snRNA in p110 immunoprecipitates, we concluded that p110 associates only transiently with the U6atac snRNP. Why does p110 appear not to remain associated with the U4atac/U6atac snRNP, in contrast to the U4/U6 snRNP? As we show here, p110 is able to recognize the U4atac/U6atac snRNA duplex (Fig. 3C), and therefore differences in RNA binding to singular versus duplex forms cannot explain the apparent absence of p110 from the U4atac/U6atac di-snRNP. Perhaps the large excess of the abundant U6 snRNA over U6atac snRNA efficiently competes for p110 binding. Alternatively, subtle differences in the RNP structure between the major and minor di-snRNPs may result in destabilization of p110 binding in the minor di-snRNP.
FIG. 5.
Model of U4/U6 and U4atac/U6atac snRNP recycling. Binding and release of p110 are schematically represented during the postspliceosomal transitions from the singular to the di-snRNP structures and further to the tri-snRNP complexes. The p110 binding regions are indicated by open lines for both the singular and duplex snRNA structures.
Here we have also mapped and compared the RNA binding site for p110 in the U6 and U6atac snRNAs (Fig. 2C contains a sequence comparison). Significantly, this 20-nucleotide region coincides with the sequence most highly conserved between U6 and U6atac; moreover, the same region is engaged in the U6(atac)-5′ splice site interaction, as well as in base pairing with U4(atac) (in the di-snRNP) and with U2 or U12 (in the U2/U6 and U12/U6atac structures, respectively, of the active spliceosome). In sum, this short region serves multiple functions during sequential stages of snRNP and spliceosome assembly and recycling.
In contrast to the high sequence conservation of the p110 binding site in the U6 and U6atac snRNAs, we note that, surprisingly, they differ greatly in their secondary structures (Fig. 2B, current secondary-structure models). Whereas p110 requires most of the first stem-loop of U6atac for binding (nucleotides 10 to 30), the corresponding U6 region (nucleotides 38 to 57) lies within the extended irregular 3′ stem-loop structure. Further studies should therefore reveal the common structural characteristics of the p110 binding site.
We have observed a strong effect of p110 depletion on U6atac recycling. After splicing of a U12-dependent intron, both U4atac and U6atac snRNAs accumulated in their singular forms (Fig. 4C). This establishes a novel role for the recycling factor p110. Since the majority of p110 has recently been localized to the Cajal bodies (30), we would expect—in analogy to the U4/U6 snRNP—that U4atac/U6atac snRNP recycling also takes places in this nuclear subcompartment. Despite this clear effect on recycling, we found only a relatively minor effect on in vitro splicing of SCN4AENH1 pre-mRNA (data not shown). Since p110 has been depleted by more than 95%, we believe that this is likely due to the experimental difficulties with the U12-dependent pre-mRNA that is spliced inefficiently in vitro. However, we have also noted that for both the major U4/U6 snRNPs and the minor-type equivalents, the distribution of singular and di-snRNPs was not altered after RNA interference-induced knockdown of p110, suggesting that in vivo alternative mechanisms of U4-U6 annealing may substitute for a defect in the p110 system. Nevertheless, p110 knockdown (to approximately 10% of the normal levels) resulted in growth inhibition and subsequently cell death (data not shown). Therefore, p110 may have other functions in addition to U4/U6 recycling. It will be important in future studies to reproduce snRNP recycling with more purified components, comparing the U4/U6 snRNP and the minor-type counterpart. Such experiments should also help to dissect the contributions of the p110 protein and the LSm protein complex (1) in di-snRNP assembly.
Acknowledgments
We thank George Miloshev (Institute of Molecular Biology, Sofia, Bulgaria) for help with quantitation, Jingyi Hui for providing the U1 snRNA probe, and Jan Medenbach for critical reading of the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Lührmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, M., and A. Bindereif. 1999. Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: function of the Sm site in core RNP formation and nuclear localization. Nucleic Acids Res. 27:3986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, M., S. Schreiner, A. Damianov, R. Reddy, and A. Bindereif. 2002. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 21:2724-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindereif, A., T. Wolff, and M. R. Green. 1990. Discrete domains of human U6 snRNA required for the assembly of U4/U6 snRNP and splicing complexes. EMBO J. 9:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 6.Burge, C. B., R. A. Padgett, and P. A. Sharp. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2:773-785. [DOI] [PubMed] [Google Scholar]
- 7.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 8.Ghetti, A., M. Company, and J. Abelson. 1995. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA 1:132-145. [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, S. L., and R. A. Padgett. 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271:1716-1718. [DOI] [PubMed] [Google Scholar]
- 10.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Incorvaia, R., and R. A. Padgett. 1998. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA 4:709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kambach, C., S. Walke, and K. Nagai. 1999. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9:222-230. [DOI] [PubMed] [Google Scholar]
- 14.Kolossova, I., and R. A. Padgett. 1997. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA 3:227-233. [PMC free article] [PubMed] [Google Scholar]
- 15.Krainer, A. R., T. Maniatis, B. Ruskin, and M. R. Green. 1984. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36:993-1005. [DOI] [PubMed] [Google Scholar]
- 16.Lelay-Taha, M. N., I. Reveillaud, J. Sri-Widada, C. Brunel, and P. Jeanteur. 1986. RNA-protein organization of U1, U5 and U4-U6 small nuclear ribonucleoproteins in HeLa cells. J. Mol. Biol. 189:519-532. [DOI] [PubMed] [Google Scholar]
- 17.Levine, A., and R. Durbin. 2001. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 29:4006-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lücke, S., T. Klöckner, Z. Palfi, M. Boshart, and A. Bindereif. 1997. Trans mRNA splicing in trypanosomes: cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 16:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mougin, A., A. Gottschalk, P. Fabrizio, R. Lührmann, and C. Branlant. 2002. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cells and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J. Mol. Biol. 317:631-649. [DOI] [PubMed] [Google Scholar]
- 20.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Lührmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nottrott, S., H. Urlaub, and R. Lührmann. 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 21:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padgett, R. A., and G. C. Shukla. 2002. A revised model for U4atac/U6atac snRNA base pairing. RNA 8:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel, A. A., M. McCarthy, and J. A. Steitz. 2002. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 21:3804-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rader, S. D., and C. Guthrie. 2002. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8:1378-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghunathan, P. L., and C. Guthrie. 1998. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279:857-860. [DOI] [PubMed] [Google Scholar]
- 26.Schneider, C., C. L. Will, O. V. Makarova, E. M. Makarov, and R. Lührmann. 2002. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol. 22:3219-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon, K. W., and C. Guthrie. 1991. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA binding motifs. Genes Dev. 5:773-785. [DOI] [PubMed] [Google Scholar]
- 28.Sharp, P. A., and C. B. Burge. 1997. Classification of introns: U2-type or U12-type. Cell 91:875-879. [DOI] [PubMed] [Google Scholar]
- 29.Shukla, G. C., A. J. Cole, R. C. Dietrich, and R. A. Padgett. 2002. Domains of human U4atac snRNA required for U12-dependent splicing in vivo. Nucleic Acids Res. 30:4650-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanek, D., S. D. Rader, M. Klingauf, and K. M. Neugebauer. 2002. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 160:505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarn, W. Y., and J. A. Steitz. 1996. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273:1824-1832. [DOI] [PubMed] [Google Scholar]
- 32.Tarn, W. Y., and J. A. Steitz. 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84:801-811. [DOI] [PubMed] [Google Scholar]
- 33.Tarn, W. Y., and J. A. Steitz. 1997. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 22:132-137. [DOI] [PubMed] [Google Scholar]
- 34.Vidaver, R. M., Fortner, D. M., Loos-Austin, L. S., and D. A. Brow. 1999. Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics 153:1205-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wersig, C., and A. Bindereif. 1990. Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res. 18:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Will, C. L., and R. Lührmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 37.Will, C. L., C. Schneider, R. Reed, and R. Lührmann. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284:2003-2005. [DOI] [PubMed] [Google Scholar]
- 38.Wu, Q., and A. R. Krainer. 1999. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol. 19:3225-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, Q., and A. R. Krainer. 1998. Purine-rich enhancers function in the AT-AC pre-mRNA splicing pathway and do so independently of intact U1 snRNP. RNA 4:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, Y.-T., E. C. Scharl, C. M. Smith, and J. A. Steitz. 1999. The growing world of small nuclear ribonucleoproteins, p. 487-524. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 41.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]