Abstract
SCL/TAL1 is a hematopoietic-specific transcription factor of the basic helix-loop-helix (bHLH) family that is essential for erythropoiesis. Here we identify the erythroid cell-specific glycophorin A gene (GPA) as a target of SCL in primary hematopoietic cells and show that SCL occupies the GPA locus in vivo. GPA promoter activation is dependent on the assembly of a multifactorial complex containing SCL as well as ubiquitous (E47, Sp1, and Ldb1) and tissue-specific (LMO2 and GATA-1) transcription factors. In addition, our observations suggest functional specialization within this complex, as SCL provides its HLH protein interaction motif, GATA-1 exerts a DNA-tethering function through its binding to a critical GATA element in the GPA promoter, and E47 requires its N-terminal moiety (most likely entailing a transactivation function). Finally, endogenous GPA expression is disrupted in hematopoietic cells through the dominant-inhibitory effect of a truncated form of E47 (E47-bHLH) on E-protein activity or of FOG (Friend of GATA) on GATA activity or when LMO2 or Ldb-1 protein levels are decreased. Together, these observations reveal the functional complementarities of transcription factors within the SCL complex and the essential role of SCL as a nucleation factor within a higher-order complex required to activate gene GPA expression.
How specific patterns of gene expression are generated represents a fundamental question in understanding cell type specification. There is increasing evidence that this process is controlled by networks of interacting transcription factors and that subtle variations in protein partners may have profound consequences for gene expression programs (56). Among the crucial regulators of cell type specification are transcription factors that contain basic helix-loop-helix (bHLH) domains, such as the hematopoietic master regulator SCL/TAL-1 (6). SCL is expressed in hematopoietic stem cells, as well as multipotent, erythroid, and megakaryocytic progenitors (7, 16, 26, 47). Loss- and gain-of-function studies with different vertebrate models have shown that SCL is essential for the establishment of the hematopoietic system and that it can specify the hematopoietic cell fate when ectopically expressed (21, 34, 45, 52, 53, 55). Due to the absence of hematopoietic cells in SCL−/− mice, deciphering its role in the differentiation of particular blood cell lineages has been elusive. Recent conditional knockout experiments (which bypass the embryonic lethality observed in SCL−/− mice) have demonstrated that SCL is essential for erythroid and megakaryocytic differentiation (23, 36). In addition to its critical role during normal hematopoiesis, the SCL gene is the most frequent target of chromosomal rearrangements in patients with T-cell acute lymphoblastic leukemia (T-ALL). This leukemic phenotype is recapitulated in transgenic mice coexpressing SCL and collaborating oncogenes such as the LIM domain proteins LMO1/2 (6). Therefore, SCL is an essential regulator at several levels in the hematopoietic hierarchy and its inappropriate regulation leads to severe pathological consequences.
Like other tissue-specific bHLH factors, SCL forms E-box (CANNTG) binding heterodimers with ubiquitous bHLH partners known as E-proteins, which include products of the E2A gene (E12 and E47), HEB, and E2-2 (27). In erythroid cells, SCL is found in a multifactorial complex (SCL complex) with E47, LMO2, Ldb1, and GATA-1 (68). Although potential binding sites for the SCL complex are found in erythroid genes such as GATA-1 and EKLF (1, 66), functional dissection of the mechanism of action of SCL on erythroid targets remains to be documented. For example, the importance of the N-terminal transactivation domain and the basic domain of SCL remains controversial, as they are both dispensable for the genetic rescue of specification of the hematopoietic cell fate (44) and for c-kit transcription activation (31) and yet DNA binding-defective mutants of SCL fail to rescue the maturation of definitive hematopoietic lineages in SCL−/− ES cells (44) and to induce erythroid differentiation in established cell lines (3). In more primitive hematopoietic progenitors, GATA-2 can function within the SCL complex (as has been observed in the context of the c-kit promoter) (31). This study also identified Sp1 as a novel component of the SCL complex (consistent with the importance of Sp1 in hematopoietic gene regulation) (57). In leukemic cells, SCL also associates with E2A gene products LMO2 and Ldb1 (22); in this cellular context, however, SCL and LMO1/2 may inhibit the normal functions of E-proteins, which are crucial regulators of lymphoid cell differentiation. Thus, the functions of SCL may differ depending on the cellular context and target genes. To clarify our understanding of the functions played by SCL in different hematopoietic lineages, it is crucial to define the mechanisms by which SCL and its partners regulate the expression of candidate target genes in these cellular compartments.
We previously demonstrated that ectopic expression of SCL in TF-1 cells, a bipotent cell line that can be induced to differentiate along the erythroid or monocyte/macrophage lineages, increases cell surface expression of the erythroid marker glycophorin A (GPA) and renders the induction of erythroid differentiation more efficient (26). GPA is one of the most abundant erythrocyte membrane proteins, and its highly glycosylated sialic acid-rich extracellular domain is predominantly responsible for the negative charge of the red cell membrane. Despite the recognition that SCL collaborates with its partners to activate transcription and determine the hematopoietic fate (31, 33, 68), there is little evidence for the formation of a functional high-molecular-weight SCL complex with regulatory sequences of physiological target genes. In the present report, we show that the SCL complex determines GPA gene expression and that the main function of SCL is to assemble this complex on target gene regulatory elements to activate transcription.
MATERIALS AND METHODS
DNA constructs.
Expression vectors for SCL and its partners, as well as SCL point and deletion mutants, were formerly described (31, 44). The murine stem cell virus (MSCV)-neo and MSCV-YFP vectors, expressing SCL or an antisense (AS) RNA of SCL (AS-SCL), were constructed as previously detailed (30). Vectors encoding AS-LMO2 and AS-Ldb1 were generated by subcloning the LMO2 and Ldb1 cDNAs in reverse orientation into the MSCV-neo multiple-cloning site. Expression vectors encoding FOG (Friend of GATA) (62), GATA-1V205→G (13), and E47-bHLH (46) were generously provided by Stuart H. Orkin (Harvard Medical School, Boston, Mass.), John D. Crispino (Ben May Institute for Cancer Research, Chicago, Ill.), and Jacques Drouin (Institut de Recherches cliniques de Montréal, Montreal, Quebec, Canada), respectively. For retroviral infections, the FOG and E47-bHLH cDNAs were subcloned into the MSCV-neo vector. GPA promoter fragments −456, −116, −84, and −79 were PCR amplified (using forward primers [−456 to −440, −116 to −100, −84 to −68, and −75 to 60, respectively] and a reverse primer [+56 to +40]) from human genomic DNA. Amplified fragments were digested with BglII/KpnI and ligated upstream of the luciferase gene in the pXPIII plasmid (31). Nucleotide positions are numbered relative to the transcription initiation site as described by Rahuel and colleagues (48, 49). GPA promoter point mutations were generated by three-step PCR and resulted in the nucleotide substitutions indicated (see Fig. 3). Vectors encoding GST-GATA-1, GST-LMO2, and GST-SCL were generated by cloning PCR-amplified cDNAs into the pGex2T plasmid (Amersham Pharmacia Biotech, Piscataway, N.J.), while the origin of the GST-Sp1 vector was described previously (31). All vectors were verified by sequencing.
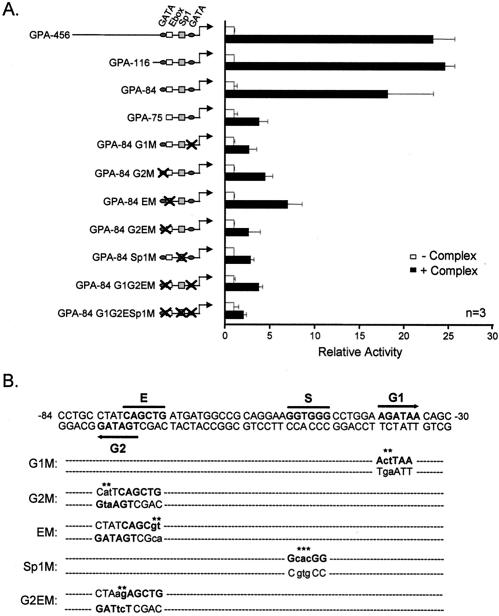
FIG. 3.
cis elements required for GPA promoter activation by SCL and its partners. (A) Deletion or point mutations in two GATA sites (G1 at position −36 and G2 at −74), an Sp1 motif (S at position −48), and an E-box element (E at position −70) impair activation by the SCL complex. NIH 3T3 cells were transfected with mutant GPA reporter constructs in the absence (open bars) or the presence (solid bars) of the SCL complex. Results are shown as luciferase activity levels relative to that of each reporter vector transfected alone; luciferase activities were normalized by cotransfection of CMV-β-Gal and are representative of n independent experiments. The basal promoter activity of the mutant GPA reporters was identical to that of GPA-456 (1,000 normalized relative light units on average, which is comparable to the results seen with the empty pXPIII vector). (B) The GPA promoter sequence from position −84 to position −30 is indicated. The G1, G2, S, and E sites are indicated, and the mutations that were introduced into these motifs are designated by asterisks. Note that the G2 and E elements partially overlap on opposite strands.
Cells and retroviral infections.
TF-1 cells were grown as described previously (30). NIH 3T3 and BOSC23 cells were maintained in Iscove's modified Dulbecco's medium (IMDM) (GIBCO Invitrogen Corporation, Burlington, Ontario, Canada) containing 10% fetal calf serum (FCS) (GIBCO Invitrogen Corporation). For TF-1 cell infections (Fig. 1B and C), high-titer viruses encoding SCL or AS-SCL were produced by transient transfection into Bing cells (as formerly detailed) (30), which were irradiated and cogrown with TF-1 cells for 24 h in the presence of 8 μg of Polybrene/ml. Otherwise, the viruses encoding SCL, ΔbSCL, SCL-ΔNt, AS-Ldb1, AS-LMO2, FOG, and E47-bHLH and their MSCV control were produced by transfection of the 293 GPG retroviral packaging cell line (42). The viruses were concentrated by ultracentrifugation, and TF-1 cells were then grown for 24 h in the presence of concentrated viruses and 8 μg of Polybrene/ml. Following the infections, the cells were recovered and polyclonal populations were analyzed 1 week after selection in G418 at 1 mg/ml. For infections of primary hematopoietic cells, fetal livers from E14.5 embryos were dissected, disaggregated into single-cell suspensions, and washed in IMDM containing 10% FCS. The cells were then incubated overnight with control (MSCV-YFP) and AS-SCL-expressing (AS-SCL-YFP) retroviruses in the presence of 4 μg of Polybrene/ml. Following infection, cells were washed and cultured in IMDM containing 10% FCS, 1U of erythropoietin/ml, 100 ng of Steel factor/ml, and 50 ng of interleukin-3/ml. After 24 h, infected cells were analyzed by fluorescence-activated cell sorter (FACS) and reverse transcription-PCR (RT-PCR) as described below.
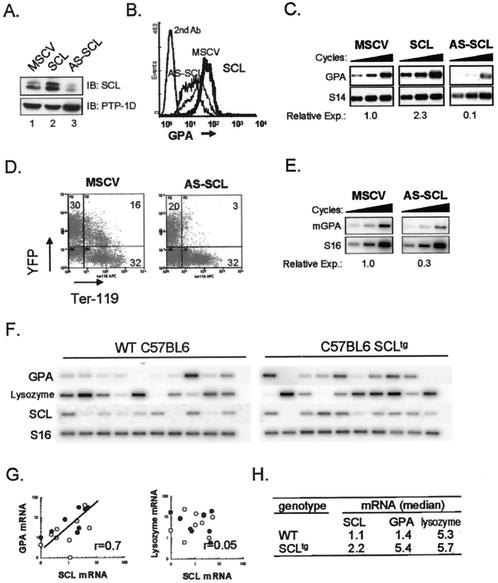
FIG. 1.
SCL levels determine GPA expression. (A) Immunoblotting (IB) with an anti-SCL antibody (upper panel) shows the expression of SCL in TF-1 cells transduced with control retroviruses (MSCV) or viruses encoding SCL or AS-SCL. The blots were stripped and reprobed with an antibody directed against the PTP-1D phosphatase (lower panel) as a loading control. (B) GPA and SCL levels correlate in erythroid cells. The indicated TF-1 transfectants were analyzed by FACS for GPA expression (thick line, SCL transfectant; dotted line, MSCV empty vector; black line, AS-SCL transfectant). Cells labeled with the secondary antibody alone (2nd Ab) were included as negative controls for staining. For simplicity, the 2nd Ab is shown for MSCV control cells only and is representative of the secondary antibody labeling obtained with the SCL and AS-SCL transfectants. (C) GPA mRNA expression in TF-1 cell transfectants was determined by RT-PCR analysis following 18, 20, and 22 cycles of amplification. S14 was used as a normalization control. Amplified fragments were revealed through hybridization with internal oligonucleotide probes. Following normalization with S14, the relative expression (Relative Exp.) of GPA for each transfectant was quantified compared to that of the MSCV control, which was set at 1.0. (D) Expression of an AS-SCL in fetal liver cells decreases Ter119 labeling. Fetal liver cells were infected with the control YFP and YFP-AS-SCL retroviruses and analyzed by FACS for the expression of YFP and Ter119. (E) Reduced GPA mRNA levels in AS-SCL-expressing fetal liver cells. Aliquots of YFP+ cells (infected as described for panel D) were subjected to RT-PCR to measure endogenous GPA expression. S16 was used as a control for the amount of cDNA. The relative expression of GPA mRNA was quantified (as described for panel C) following normalization with S16 signals. (F) Increased GPA expression in pluripotent colonies from SCL transgenic mice. RT-PCR analysis was performed on pluripotent colonies (CFU-GEMM) derived from WT and SCLtg mice (2). (G) Correlation between GpA and SCL mRNA levels. Data shown in panel F were quantified using ImageQuant software and are expressed as ratios over S16 controls (○, distribution from WT colonies; •, SCLtg mouse colonies). The coefficient of correlation between SCL and GPA mRNA levels (r) is shown. There was no correlation with lysozyme mRNA levels (r = 0.05). (H) Increased GPA and SCL levels in colony cells from SCLtg mice. Data represent the medians of the distributions shown in panel F for mRNA expression in individual colonies from bone marrow cells of SCLtg and age-matched control mice.
FACS analysis.
TF-1 cell samples were stained with rat anti-human GPA (YTH 89.1 [immunoglobulin M {IgM}]; Serotec, Oxford, United Kingdom) in staining buffer (phosphate-buffered saline [PBS] with 2% FCS) followed by treatment with a secondary fluorescein isothiocyanate-conjugated goat anti-rat antibody (CALTAG, Burlingame, Calif.). Fetal liver cells were labeled with a phycoerythrin-conjugated murine Ter119 antibody (Pharmingen, Mississauga, Ontario, Canada) in staining buffer. A FACSCalibur apparatus (Becton-Dickinson, San Jose, Calif.) was used to assess fluorescein isothiocyanate, phycoerythrin, and yellow fluorescent protein (YFP) fluorescence on stained cells. Dead cells were excluded by adding 1 μg of propidium iodide/ml prior to detection.
RT-PCR analysis.
Total RNA from TF-1 or fetal liver cells was prepared as detailed previously (25) and was reverse transcribed using a Superscript first-strand cDNA synthesis system (GIBCO Invitrogen Corporation). For PCR amplifications, 2 μl of a cDNA sample was added to mixtures containing 1 μM of forward and reverse primers, 20 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 5% dimethyl sulfoxide, 0.2 mM deoxynucleoside triphosphate, and 1.25 U of TaqDNA polymerase. Samples were amplified for 18, 20, and 22 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s), and PCR products were migrated on a 1.5% agarose gel, transferred on nylon membranes, and hybridized with internal probes. The blots were exposed to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.). Human GPA, murine GPA, SCL, and lysozyme mRNA levels were quantified using ImageQuant software (Molecular Dynamics) and are expressed as ratios over hS14 or mS16 signals. Data for oligonucleotides used for amplifications are presented in Table 1.
TABLE 1.
Oligonucleotides used for RT-PCR and ChIP analysis
| Oligonucleotide | Sequence
|
||
|---|---|---|---|
| Forward | Reverse | Internal | |
| Human GPA | ATTGTCAGCAATTGTGAGCATA | TGATCACTTGTCTCTGGATTTT | ATATGCAGCCACTCCTAGAGC |
| Human S14 | GGCAGACCGAGATGATACCTCA | CAGGTCCAGGGGTCTTGGTCC | GACCTGGGTATCACCGCCCT |
| Murine GpA | ATATGAATTCCTGGGAAGGATGCTTTGC | ATATGGATCCTCCACTGCAAGGAAAGGG | GGAAGTTGCTTTCTTGAATA |
| Murine S16 | AGGAGCGATTTGCTGGTGTGA | GCTACCAGGGCCTTTGAGATG | AAATTTATGCCATCCGACAGTC |
| Lysozyme | CCAAGGTCTATGAACGTTGTGA | TGCCATCATTACACCAGTATCG | GTCAGCCTGGCCGACTGTAAGTCTC |
| SCL | ATTGATGTACTTCATGGCAAGG | TCCCCATATGAGATGGAGATTT | TGGAGATTTCTGATGGTCCTCACACCAAA |
| GPA-ChIP | ATTAGGTACCTCCATGTATCTTTATT | TTAAAGATCTCCTGAGATCATGAGCT | CCCCTGCCTATCAGCTGATGATGGCC |
| HPRT-ChIP | TGAGGCAAAAATAGAGGCTCA | TCCCAAGACCTTGCACTACC | TGTACAAAACTACAGAGCAG |
Colony assays.
Clonal cultures were performed using freshly extracted bone marrow cells from wild-type or SIL-SCL transgenic (SCLtg) mice [line A (5)3SCL] (2) in 1% methylcellulose-10% FCS-200 μg of transferrin/ml-2% bovine serum albumin-5 ng of interleukin-3/ml-1 U of erythropoietin/ml-100 ng of Steel factor/ml-5 × 10−5 M α-monothioglycerol. Colony formation was monitored at appropriate times, and day 7 mixed hematopoietic colonies were picked, washed in PBS, and subjected to RNA extraction and RT-PCR.
Transfections and nuclear extracts.
Transactivation assays were performed essentially as previously described (31). NIH 3T3 cells were transfected using calcium phosphate 24 h after plating 30,000 cells/well in 12-well culture plates. GPA reporter constructs were kept at 1.5 μg/well, while 100 ng of cytomegalovirus (CMV)-β-galactosidase (CMV-β-Gal)/well was included in each transfection mixture as an internal control for normalization. Expression vector doses are indicated in the figure legends. The total DNA was kept constant at 4.5 μg/well with pGem4 (Promega, Madison, Wis.). Luciferase and β-Gal activities were measured 36 h posttransfection. All luciferase values were normalized using β-Gal values. Results are shown as the means ± standard deviations (SD) of one experiment performed in triplicate and are representative of three or more independent experiments (depicted in the figures).
TF-1 cell nuclear extracts were prepared as previously described (30). For BOSC23 cell extracts, the cells (4.2 × 106) were first plated and then transfected 24 h later with the expression vectors for LMO2 and Ldb1 (11.25 μg) as well as for GATA-1, E47, and SCL or SCL (2.25 μg) mutants. At 36 h after transfection, the cells were harvested, washed twice in cold PBS, and subjected to nuclear extraction as indicated above.
Gel shift, pulldown, and chromatin immunoprecipitation (ChIP) assays.
Binding reactions were performed at room temperature for 15 min in the presence of 0.5 μg of poly(dI-dC) in 20 mM HEPES (pH 7.5)-50 mM KCl-1 mM dithiothreitol-1 mM EDTA-5% glycerol-10 μg of bovine serum albumin-15,000 cpm of double-stranded probe-1 to 20 μg of TF-1 or BOSC23 cell nuclear extract in a total volume of 20 μl. The sequences of the GPA-84 probe and mutant promoter fragments used for competition experiments are indicated (see Fig. 3). For antibody supershift assays, 1 μg of the following affinity-purified antisera was used: goat anti-GATA-1 (M20), mouse anti-E2A (YAE), rabbit anti-E47 (N-649), goat anti-Ldb1/CLIM-2 (N-18), and rabbit anti-Sp1 (PEP-2) antibodies (all from Santa Cruz Biotechnology Inc., Santa Cruz, Calif.). The BTL73 mouse anti-SCL antibody was kindly provided by D. Mathieu (Institut de Génétique Moléculaire, Montpellier, France). As a control, equal amounts of species-matched serum Ig (Sigma, St. Louis, Mo.) were added to the binding reactions. Protein complexes were resolved by electrophoresis at 150 V on 4% polyacrylamide gels in 0.5× Tris-borate-EDTA at 4°C.
For pulldown assays, glutathione S-transferase (GST), GST-SCL, GST-LMO2, GST-GATA-1, and GST-Sp1 were purified from bacteria and coupled to glutathione Sepharose beads (Amersham Pharmacia Biotech). A TNT-coupled reticulocyte lysate system (Promega) was used to translate SCL, GATA-1, LMO2, Ldb1, and luciferase in vitro in the presence of [35S]methionine. Labeled proteins (15 μl) were incubated with 2 μg of immobilized GST fusion proteins in 400 μl of binding buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1% Nonidet P-40 [NP-40], 5 mM dithiothreitol, 10% glycerol, 200 μg of ethidium bromide/ml) for 2 h at 4°C with agitation and then centrifuged for 1 min at 3,000 × g. Samples were washed three times with binding buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and visualized and quantified using ImageQuant software (Molecular Dynamics).
ChIP assays were performed essentially as described previously (31, 59). Briefly, exponentially growing TF-1 cells were fixed by incubation with formaldehyde (1% final) for 10 min at room temperature. After the formaldehyde was quenched with glycine (0.125 M final concentration), cells were sequentially washed and sonicated to make chromatin extracts (ranging in size from 500 to 1,000 bp) as formerly detailed (31). Protein concentrations were determined by Bradford staining, and 500 μg of chromatin extract was incubated overnight at 4°C with specific antisera against SCL and its partners or control Ig (which are described above). An aliquot of chromatin extract was kept for isolation of input DNA. Samples were then precipitated by the addition of Pansorbin cells (Calbiochem, San Diego, Calif.) for 30 min at 4°C. Precipitated chromatin samples were sequentially washed and eluted as previously detailed (31) and incubated overnight at 65°C to reverse cross-linking. After RNA and proteins were degraded (using 30 μg of RNase A for 30 min at 37°C and 120 μg of proteinase K for 2 to 3 h at 37°C), DNA was phenol-chloroform extracted and precipitated with ethanol in the presence of 10 μg of tRNA as a carrier. PCRs were then performed on precipitated samples as described previously (31). PCR products were migrated on a 1.5% agarose gel, transferred to nylon membranes, and hybridized with internal oligonucleotide probes. Oligonucleotide data are indicated in Table 1.
RESULTS
GPA expression is dependent on the levels of SCL.
Hoang et al. previously demonstrated that overexpression of SCL in TF-1 cells increases the expression of GPA (26). To clarify the mechanism through which SCL regulates gene expression within the erythroid lineage, we performed experiments to address whether GPA might be a downstream target of SCL. We first utilized retrovirus-mediated gene transfer to increase or decrease SCL levels in TF-1 cells. Retroviruses containing an empty MSCV-neo genome or encoding either SCL or AS-SCL were generated and used to infect exponentially growing TF-1 cells. Compared to the results seen with mock-infected TF-1 cells (Fig. 1A, lane 1), SCL protein levels were increased by 2.3-fold in SCL infected cells (lane 2) whereas they were diminished 5-fold in cells expressing AS-SCL (lane 3). To address the question of whether GPA expression depends on SCL levels, we monitored GPA protein and mRNA expression by flow cytometry and RT-PCR analysis, respectively. Cells overexpressing SCL exhibited an increase in cell surface expression of GPA, as shown by labeling with an anti-GPA antibody (Fig. 1B). Increased GPA expression occurred at the transcriptional level, since GPA mRNA levels were twofold higher following ectopic SCL expression (Fig. 1C). Conversely, decreased SCL protein levels in AS-SCL-infected cells were associated with a severe 10-fold reduction in GPA protein and mRNA expression compared to that seen with mock-infected cells (Fig. 1B and C).
To determine whether this correlation was also observed in primary hematopoietic cells, we next infected primary E14.5 fetal liver cells with control (MSCV-YFP) and AS-SCL (AS-SCL-YFP)-expressing retroviruses, which also encode the fluorogenic protein YFP, allowing for analysis of infected cells in the YFP+ fraction. It has recently been demonstrated that the Ter119 antibody, which specifically labels cells of the erythroid lineage, recognizes an epitope on mouse GPA (5). Flow cytometric analysis of viable fetal liver cells following gene transfer revealed a dramatic reduction in Ter119 reactivity in the YFP+ fraction of AS-SCL-infected cells compared to that seen with control samples, as the proportion of Ter119+ cells dropped from 16 to 3% (Fig. 1D). In contrast, the proportion of Ter119-reactive cells in the uninfected population (YFP−) was 32% for both AS-SCL and control MSCV cells. In AS-SCL-infected cells, this reduction in Ter119 labeling is concomitant with a threefold decrease in GPA mRNA expression as determined by RT-PCR analysis (Fig. 1E). Together, these results demonstrate a close correlation in SCL and GPA levels in an erythroid progenitor cell line (TF-1) and in primary fetal liver erythroid cells. In both cell types, the level of GPA per cell decreases when SCL levels are lowered, as flow cytometry analysis reveals fluorescence signals at the single-cell level.
To assess whether GPA expression in primary hematopoietic cells increases following SCL gain of function, we next analyzed SCL and GPA expression in multipotent colonies (CFU-GEMM) derived from bone marrows of wild-type or SIL-SCLtg mice, which express SCL ubiquitously (2). Total RNA was extracted from individually harvested multipotent colonies, and gene expression was assessed by RT-PCR analysis. Gene expression within single multipotent colonies was normalized on the basis of the control S16 mRNA level (Fig. 1F) and quantified as the ratio over S16 (Fig. 1G). This analysis revealed a linear relationship between the expression levels of SCL and GPA in individual colonies from both wild-type and SCLtg mouse bone marrows, with a correlation coefficient of 0.7. Furthermore, in colonies from SCLtg mice, which exhibit on average a twofold-increased level of SCL, GPA expression was increased fourfold whereas levels of the myeloid marker lysozyme remained constant (Fig. 1H). Together, data shown in Fig. 1 indicate a close correlation between SCL and GPA levels in the TF1 cell line and in primary hematopoietic cells.
The GPA promoter is activated by a complex containing SCL, E47, LMO2, Ldb1, and GATA-1.
Increased GPA expression in SCLtg mouse colonies might be due to a direct effect of the SCL transgene on GPA expression or to an indirect increase in the erythroid content of each colony. To address the question of whether GPA is a direct target of SCL, we performed transactivation assays using heterologous NIH 3T3 cells and a reporter vector in which the GPA promoter region (from position −456 to position +56) was inserted in front of the luciferase gene (GPA-456). Using this assay, Lécuyer et al. previously demonstrated that the regulation of c-kit promoter sequences by SCL requires its integration within a multifactorial complex (SCL complex) containing E47, LMO2, Ldb1, and GATA-1/-2 (31). When transfected on its own, the GPA-456 reporter shows a low background level of luciferase expression comparable to the promoter-less reporter pXPIII (data not shown). GATA-1 could activate GPA-456 on its own by three- to fourfold (Fig. 2A) (consistent with a previous report on the glycophorin B gene [GPB] promoter, which is highly homologous to the GPA promoter) (49). In contrast, expression of SCL and E47 had no effect on promoter activity. When we coexpressed SCL with E47, LMO2, Ldb1, and GATA-1, strikingly, the GPA promoter was synergistically activated by 20- to 25-fold over its basal level (Fig. 2A and B). Omission of either one of the expression vectors from the transfection mixtures severely reduced promoter activation, demonstrating that each partner was required for synergistic transactivation (Fig. 2B). These results suggest that SCL and its partners regulate GPA expression through direct activation of the proximal GPA promoter.
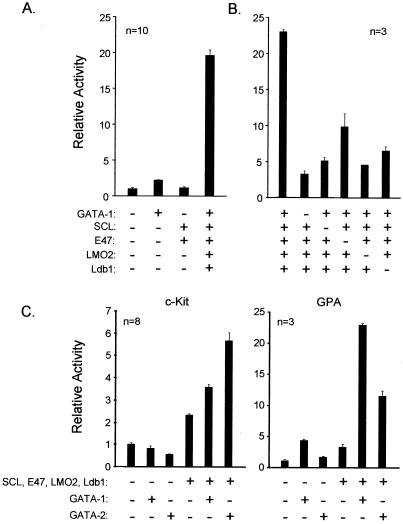
FIG. 2.
A complex containing SCL, E47, LMO2, Ldb1, and GATA-1 (SCL complex) activates the GPA promoter. (A) The GPA promoter is synergistically activated by SCL, E47, LMO2, Ldb1, and GATA-1. NIH 3T3 cells were transfected with the GPA-456 reporter (1,500 ng) and the indicated expression vectors: SCL, E47, and GATA-1 (150 ng) and LMO2 and Ldb1 (750 ng). (B) Each factor of the SCL complex is required for full GPA promoter activation. (C) Complexes containing GATA-1 or GATA-2 show various levels of transactivation efficiency depending on the target promoter. NIH 3T3 cells were transfected with the c-kit (left panel) or GPA (right panel) reporter constructs and the indicated complexes containing either GATA-1 or GATA-2. For panels A to C, + and − indicate inclusion and omission of specific expression vectors. For all transfections, the total amount of transfected DNA was kept constant (using pGem4) at 4.5 μg. Results are shown as luciferase activity levels relative to those of the reporter vector transfected alone and represent the averages ± SD of triplicate determinations and are representative of n independent experiments. Luciferase reporter activities were normalized to that of an internal control (CMV-β-Gal; 100 ng).
Both GATA-1 and GATA-2 can associate in complexes with SCL, although it is not known whether distinct complexes show differences in target gene specificity. GATA-1 is a master regulator of erythroid development (69), while GATA-2 plays a crucial role in maintaining a normal pool of hematopoietic progenitor cells (60). Therefore, we tested whether complexes containing either GATA-1 or GATA-2 showed differences in specificity in activating progenitor or erythroid cell targets of the SCL complex (i.e., c-kit or GPA, respectively). In the context of the c-kit promoter, we found that activation was more efficient with GATA-2-containing complexes compared to activation by those with GATA-1 (Fig. 2C, left panel) (31). With the GPA promoter, however (and the same amount of GATA expression vector [150 ng] as was used in the c-kit promoter analysis), we found that GATA-1-containing complexes were much more efficient than GATA-2 complexes (Fig. 2C, right panel). This functional specificity concurs with the known biological functions of these two GATA factors.
The SCL complex activates the GPA promoter through an E-box motif, two GATA binding sites, and a Sp1 binding site.
We next sought to identify the cis elements that were required to recruit the SCL complex to the GPA promoter. We first generated a series of GPA promoter 5′ deletion mutants to identify the minimal promoter sequence that remained maximally activated by the SCL complex. As shown in Fig. 3A, a promoter segment lacking the sequence up to position −84 (GPA-84) was still maximally activated and further deletion up to position −75 (GPA-75) resulted in a dramatic decrease in activation. Segments of the proximal GPA and GPB promoters were previously shown to contain sequences necessary for erythroid-specific expression (references 48 and 49 and data not shown). These studies had highlighted the presence of functionally important GATA motifs at positions −36 (G1) and −74 (G2) and an Sp1 binding site at −48 (Sp1) (Fig. 3B). An E-box sequence at position −70 (E) was also previously characterized, although its involvement in erythroid cell-specific expression of the glycophorin promoters remained unclear (10). The E and G2 elements are overlapping between positions −79 and −70 of the promoter, the G2 site being arranged in an opposite orientation relative to the GPA gene (Fig. 3B). In the GPA-75 construct, which shows a severe decrease in activation, both of these motifs were affected by the deletion, demonstrating that they are important for responsiveness to the SCL complex (Fig. 3A). To address whether the G1, G2, Sp1, and E elements were required for activation by the SCL complex, point mutations were introduced into these motifs as indicated in Fig. 3B. Promoter activation was greatly reduced when mutations were introduced into each of these elements (G1 M, G2 M, E M, and Sp1 M) and was completely abolished when all of the sites were simultaneously mutated (Fig. 3A). The requirement for an Sp1 binding site for promoter activation by the SCL complex is not unexpected, since Lécuyer et al. have previously demonstrated that Sp1 helps to recruit the SCL complex to the proximal c-kit promoter in hematopoietic progenitor cells (31). These results demonstrate that several cis elements collaborate to confer transcription activation by the SCL complex to the GPA promoter.
SCL and its partners form a low-mobility complex on the GPA promoter.
We next addressed the question of whether the SCL complex assembles on the GPA proximal promoter sequence. For this we performed electrophoretic mobility shift assays (EMSA) using the GPA-84 probe, which spans position −84 to position −32 of the GPA promoter (Fig. 3B). We initially performed a titration experiment (in which various concentrations of TF-1 cell nuclear extracts were incubated with the GPA-84 probe) and found that a very-low-mobility complex was formed on this probe at higher concentrations of protein extract (Fig. 4A). This slowly migrating complex was not observed on probes containing the G1 and G2E motifs alone (data not shown). Addition of specific antibodies against SCL, E2A, Ldb1, GATA-1, and Sp1 supershifted or disrupted the migration of the low-mobility complex (Fig. 4B, lanes 2 to 6 and 10), whereas control Ig had no effect (lanes 7 to 8 and 11). This demonstrates that the SCL complex can indeed directly associate with the GPA promoter in vitro.
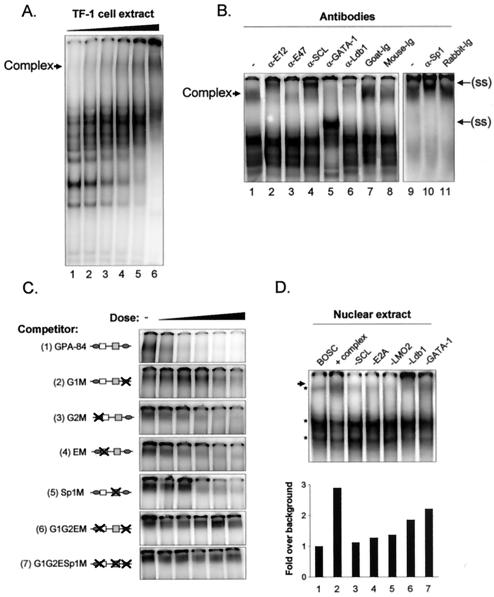
FIG. 4.
The SCL complex associates with the GPA promoter in vitro. (A) A low-mobility complex from TF-1 cell nuclear extracts forms on the GPA promoter. EMSA were performed using the GPA-84 probe (−84 to −30) and increasing amounts of TF-1 cell nuclear extracts (1 to 20 μg). (B) SCL and its partners form a low-mobility complex on the GPA-84 promoter sequence. Supershift assays were performed using the antibodies against partners of the SCL complex (lanes 1 to 6 and 10) or control species-matched antiserum (lanes 7 to 8 and 11). The binding reaction was performed with 10 μg (lanes 1 to 8) or 20 μg (lanes 9 to 11) of TF-1 nuclear extracts. The arrow on the left points to the SCL complex, and the arrows on the right point to the supershifted (ss) complexes. −, no antibody. (C) Binding of the SCL complex requires the G1, G2, S, and E motifs. The indicated competitor fragments were titrated into binding reactions at 3-, 10-, 30-, 100-, and 300-fold molar excess compared to the labeled GPA-84 probe. −, control. (D) Each partner is required to form a complex on the GPA promoter. EMSA were performed with GPA-84 (lanes 1 to 7) and nuclear extracts of untransfected BOSC cells (lane 1) or cells transfected with SCL and its partners (lane 2 to 7). Where indicated (−), particular expression vectors were omitted from the transfection mixtures (lanes 3 to 7). The asterisks indicate complexes formed in control untransfected BOSC23 cells, and the arrow points to the SCL complex. The binding intensities of the SCL complexes were quantified using ImageQuant software.
To assess the contribution of the G1, G2, Sp1, and E-box elements for binding of the SCL complex, we next compared the ability of wild-type and mutant GPA promoter fragments to compete with the binding of the SCL complex to the GPA-84 probe. Competitor fragments were titrated into the binding reactions in amounts ranging from 3- to 300-fold molar excess relative to the 32P-labeled GPA-84 probe. The wild-type competitor efficiently displaced the binding of the complex (Fig. 4C, top panel). In contrast, competitor fragments mutated in the proximal GATA site (G1 M) was 30-fold less efficient (second panel from the top) and mutants with mutations in the G2, E, and Sp1 motifs (third through fifth panels) were 10-fold less efficient than the wild-type competitor. As expected, combined mutations in these cis elements completely abolished the ability of the promoter fragments to compete with the binding of the SCL complex to the GPA-84 probe (sixth and seventh panels from the top). In addition to being required for GPA promoter activation, therefore, the G1, G2, E, and Sp1 motifs are also required for direct binding of the SCL complex to the GPA promoter.
EMSA with TF-1 cell nuclear extracts allowed us to demonstrate that the SCL complex associates with the GPA promoter. To assess whether SCL and its partners are necessary and sufficient to form a complex on the GPA promoter, we next sought to reconstitute the complex by ectopic expression in heterologous cells. When EMSA were performed with the GPA-84 probe and nuclear extracts of BOSC23 cells transfected with SCL and its partners, we observed the appearance of a low-mobility complex that was distinct from the background seen with untransfected BOSC23 extracts (Fig. 4D, lanes 1 and 2). To determine whether each partner was required for complex formation, we prepared extracts of BOSC23 cells in which the expression vector for each factor was sequentially omitted from the transfection mixtures. We found that subtracting either partner compromised the formation of the complex on the GPA-84 probe (lanes 3 to 7). Together, these results indicate that consistent with their crucial contribution in promoter transactivation, the presence of all SCL partners is required for the formation of a complex on the GPA promoter.
Partners of the SCL complex occupy the GPA promoter in hematopoietic cells.
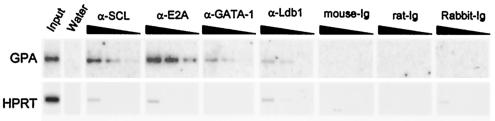
To address the question of whether SCL and its partners indeed associate with the GPA promoter in vivo in hematopoietic cells, we next performed ChIP assays with TF-1 cell chromatin extracts (31). Exponentially growing TF-1 cells were treated with formaldehyde, and fragmented chromatin was then subjected to immunoprecipitation with antibodies directed against SCL and its partners. Following immunoprecipitation, cross-linking was reversed and associated DNA fragments were purified, serially diluted, and subjected to PCR with primers specific for the GPA promoter region (as well as for the promoter segment of the ubiquitously expressed hypoxanthine phosphoribosyltransferase [HPRT] gene as a control). To confirm the specificity of amplification, fragments were hybridized with 32P-labeled internal oligonucleotide probes. We found that antibodies against SCL, E2A, and GATA-1 were able to precipitate the proximal GPA promoter 10- to 15-fold more efficiently than the HPRT promoter region, whereas control Ig did not bring down these sequences (Fig. 5). In contrast, immunoprecipitation with the anti-Ldb1 antibody showed a modest twofold enrichment of the GPA promoter compared to the levels seen with HPRT. Since Ldb1 is a ubiquitously expressed, it is possible that this factor regulates the expression of both tissue-specific and more widely expressed genes. Nevertheless, these results demonstrate that partners of the SCL complex directly and specifically associate with the GPA promoter in hematopoietic cells.
FIG. 5.
Partners of the SCL complex associate with the GPA promoter in vivo. TF-1 cell chromatin extracts were subjected to immunoprecipitation with anti-SCL, α-E2A, -GATA-1, and -Ldb1 antibodies and control Ig. Precipitated chromatin was heated overnight at 65°C to reverse cross-linking, and DNA molecules were purified and subjected to PCR analysis to test for the presence of the GPA and HPRT promoter sequences. Input chromatin represents 1.25% of the amount used in each immunoprecipitation; 3 fivefold serial dilutions of the immunoprecipitated samples were used for amplification. Following electrophoresis and transfer, PCR fragments were hybridized with an internal oligonucleotide probe.
Multiple interactions between partners of the complex.
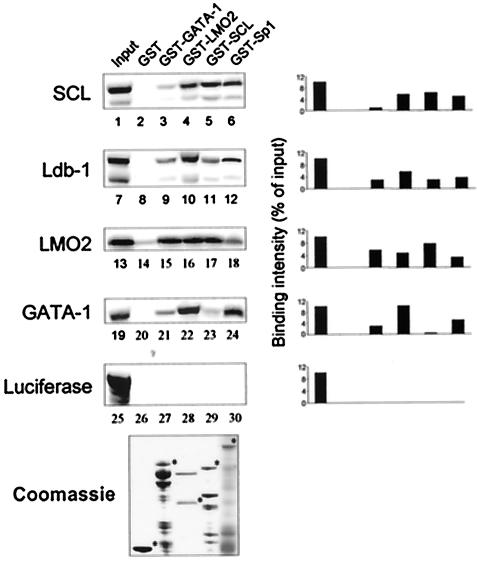
Multiprotein complexes formed on DNA are stabilized by protein-protein interactions. To define the network of physical interactions that occur between SCL and its partners, we next performed in vitro pulldown assays. Bacterially produced fusion proteins GST-GATA-1, GST-LMO2, GST-SCL, and GST-Sp1 were immobilized on Sepharose beads and incubated with [35S]methionine-labeled SCL, Ldb1, LMO2, and GATA-1. The binding reactions were performed in the presence of a high concentration of ethidium bromide (200 μg/ml) to ensure that contaminant DNA molecules did not indirectly bridge protein interactions. Columns containing GST alone and reactions with 35S-labeled luciferase (Fig. 6, lanes 25 to 30) were included as negative controls. In addition to confirming previously documented interactions, novel homo- and heterotypic interactions between partners of the complex were observed. SCL was found to interact efficiently with LMO2 and Sp1 and with itself (lanes 4 to 6 and 17) with binding intensities of 4 to 6% of the input, whereas it did not interact with GATA-1 (lanes 3 and 23). Indeed, the interaction of SCL with LMO2 and Sp1 has been previously documented and SCL has also been shown to interact with itself in the yeast two-hybrid system (31, 67, 71). We also observed that GATA-1 interacted strongly with LMO2 and Sp1 (lanes 15, 22, and 24; 7 to 14% of input), while it could also associate with itself more weakly (lane 21; 3% of input), as previously described (14, 31, 35, 43). In addition to its strong interaction with GST-LMO2 (lane 10; 5% of input), we found that Ldb1 could also bind to columns containing GST-GATA-1, GST-SCL, and GST-Sp1 (lanes 9, 11, and 12; 3% of input), albeit with lower efficiency. Finally, we identified a novel high-efficiency interaction of LMO2 with itself (lane 16; 8% of input). Together, these results demonstrate that there are multiple direct physical interactions between partners of the SCL complex that might serve to stabilize their association into a higher order complex on the GPA promoter.
FIG. 6.
Interactions between partners of the SCL complex. Pulldown experiments were performed with immobilized GST, GST-GATA-1, GST-LMO2, GST-SCL, and GST-Sp1 as well as with 35S-labeled SCL, Ldb1, LMO2, GATA-1, and luciferase. Protein signals were quantified using ImageQuant software, and binding efficiency levels (percentages of input) were calculated in comparison to those of input samples (10%) after subtraction of background GST signals. The lower panel shows bacterially expressed GST fusion proteins, which were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue.
SCL domain requirements for GPA gene regulation.
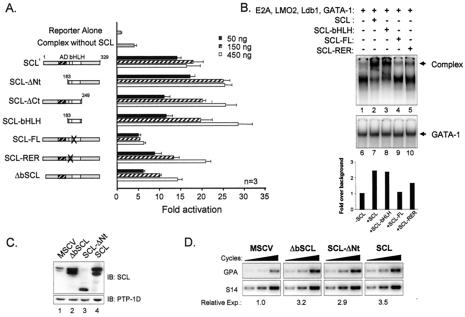
Like most transcription factors, the SCL protein has a modular structure with a basic DNA binding region and a HLH protein interaction motif as well as an N-terminal transactivation domain that is absent in shorter SCL isoforms. To ascertain which domains of SCL are required for its function, different SCL mutants were tested for their ability to activate the GPA promoter in collaboration with the other partners of the complex. As previously observed with the c-kit promoter, the bHLH domain of SCL was sufficient for GPA promoter activation and a putative N-terminal transactivation domain was deleted without affecting SCL function in this assay (Fig. 7A). In addition, the integrity of the HLH domain of SCL was crucial for its function since point mutations in helix 1 (SCL-FL), which are known to disrupt interactions with E2A and Sp1 (31, 44), abolished SCL activity. Since the GPA promoter required an E-box for its full activation by the SCL complex, we tested whether SCL mutants with point mutations (SCL-RER) or a deletion (ΔbSCL) from the basic domain, which renders them unable to bind to DNA (31, 44), would still be functionally active. These DNA binding-defective mutants were less efficient than wild-type SCL at lower doses of expression vector, although they were active at higher doses. These results suggest that DNA binding by SCL is important for maximal GPA promoter activation, although it is not essential, which is similar to findings regarding the intermediate effect on promoter activation that were observed upon mutation of the GPA E-box motif (Fig. 3A).
FIG. 7.
SCL domain requirements for GPA promoter activation and binding. (A) SCL domains required for GPA promoter activation. NIH 3T3 cells were cotransfected with the GPA-84 reporter and complexes containing the indicated doses (50 to 450 ng) of expression vectors encoding SCL point or deletion mutants (44). In all samples, the total amount of transfected DNA was kept constant (using pGem4) at 4.5 μg. The numbers correspond to amino acid residues of SCL. The helix 1 mutations were F-L→ A—A; the basic region mutations were RER→AAA. Open boxes, bHLH; AD, putative activation domain (hatched box); n, number of representative experiments. (B) The bHLH domain of SCL is necessary and sufficient to nucleate a complex on the GPA promoter. EMSA were performed with the GPA-84 probe and nuclear extracts of BOSC cells (10 μg) transfected with complexes containing wild-type or mutant forms of SCL. The arrowhead points to SCL-containing complexes. The binding intensities of the complexes were quantified using ImageQuant software. (C) Immunoblotting (IB) analysis of extracts of TF-1 cells transduced with control (MSCV) retroviruses or viruses encoding SCL, ΔbSCL, or SCL-ΔNt. Blots were sequentially hybridized with the antibody indicated to the right of each panel. (D) Induction of endogenous GPA expression by SCL mutants. GPA mRNA expression in TF-1 cell transfectants (following normalization with S16 signals) was determined by RT-PCR as described for Fig. 1C.
We next determined whether the mutants that were tested in our functional assay were able to form complexes on the GPA promoter. For this, EMSA were performed with the GPA-84 probe and nuclear extracts of BOSC23 cells expressing different SCL mutants and the other partners of the complex (Fig. 7B). As predicted from our promoter activation assays, we found that complexes containing full-length SCL or only the bHLH domain of SCL were formed efficiently on the GPA promoter (Fig. 7B, lanes 2 and 3). In contrast, the binding of the complex formed with the SCL-FL mutant (lane 4) was comparable to the binding observed when SCL was absent from the extracts (lane 1). Finally, when extracts containing the SCL-RER mutant were tested we found that complexes were formed with intermediate efficiency (Fig. 7B, lane 5), being stronger than that seen with SCL-FL yet weaker than that seen with either wild-type SCL or the bHLH domain of SCL. In control EMSA performed with a probe containing a consensus GATA motif, no variation in GATA DNA binding activity between these samples (lanes 6 to 10) was observed. Together, these findings demonstrate that the integrity of the HLH domain of SCL is crucial for efficient promoter activation and complex formation on GPA promoter sequences.
To assess whether the transactivation and DNA binding domains of SCL were required for the induction of endogenous gene GPA expression, we next infected TF-1 cells with viruses encoding SCL, ΔbSCL, and SCL-ΔNt and monitored GPA mRNA expression by RT-PCR analysis. Immunoblotting was first performed to confirm that retrovirus-mediated gene delivery resulted in efficient overexpression of SCL, ΔbSCL, and SCL-ΔNt (Fig. 7C, lanes 2 to 4) compared to that seen with mock-infected cells (lane 1). Interestingly, we found that both ΔbSCL and SCL-ΔNt could induce endogenous gene GPA expression at levels that were similar to those of wild-type SCL (from 3- to 3.5-fold) (Fig. 7D). The high levels of expression attained with retroviral infection (Fig. 7C) may explain why the ΔbSCL is as efficient as wild-type SCL, since transactivation assays revealed a difference between the two proteins at low doses but not at high doses (Fig. 7A). Finally, in consistency with the results of transient assays, the integrity of the putative transactivation domain of SCL is dispensable for the induction of endogenous gene GPA expression. We therefore conclude that SCL functions mainly as a nucleation factor for a multifactorial complex endowed with a capacity to drive erythroid gene expression.
LMO2, Ldb1, GATA factors, and E47 are required for SCL complex assembly on DNA and for gene GPA expression in hematopoietic cells.
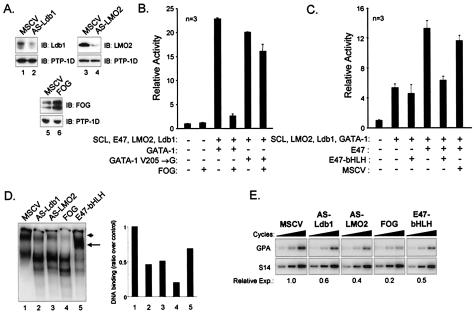
Transient reporter assays with GPA regulatory sequences indicate that LMO2, Ldb1, E47, and GATA-1 comprise a transcriptionally active SCL complex. To determine whether the same partners are required for GPA expression in chromatin, we next sought to interfere with members of the complex through diverse strategies. For LMO2 and Ldb1, we generated stable TF-1 cells lines exhibiting reduced LMO2 or Ldb1 protein expression through retrovirus-mediated delivery of AS RNA molecules (AS-LMO2 or AS-Ldb1) (Fig. 8A, lanes 1 to 4). For GATA factors, our results indicate that both GATA-1 and GATA-2 can contribute to the activity of the SCL complex although GATA-1 is more active on the GPA promoter. TF-1 cells express GATA-1 and GATA-2, and both factors interact with FOG (62), a modulator of GATA activity (20, 62). In the context of the GPA promoter, our transactivation assay revealed that FOG coexpression drastically reduced GPA promoter activation by SCL and its partners (Fig. 8B). This inhibitory effect of FOG was due to its direct interaction with GATA-1, since the GATA-1V205→G mutant, which is deficient with respect to FOG interaction (13), efficiently replaced normal GATA-1 in the SCL complex to activate the GPA promoter while conferring resistance to inhibition by FOG (Fig. 8D). We therefore generated a TF-1 cell line stably expressing FOG to repress both GATA-1 and GATA-2 activities (Fig. 8A, lanes 5 and 6). Finally, E47 is part of a family of widely expressed proteins that have overlapping functions comparable to GATA factors. We therefore assessed the activity of a truncated E47 protein, comprising the bHLH domain of E47 (E47-bHLH), within the SCL complex. In sharp contrast to the results seen with the SCL-bHLH construct, this truncated protein fails to collaborate with other members of the SCL complex in transient assays (Fig. 8C), indicating that the N-terminal domain of E47 is essential for transcription activation by the SCL complex. Furthermore, this truncated protein was found to be dominantly negative over wild-type E47 (Fig. 8C). We therefore stably expressed the truncated E47-bHLH mutant in TF-1 cells.
FIG. 8.
Loss of function of Ldb1 and LMO2 or overexpression of FOG or the bHLH domain of E47 leads to a disruption of the SCL complex and to decreased GPA activation. (A) Immunoblotting (IB) of extracts from control TF-1 cells (MSCV) or cells expressing AS-Ldb1, AS-LMO2, and FOG. Hybridizations were performed with the antibody indicated to the right of each panel, while detection of PTP-1D served as a loading control. Note the important reduction of Ldb1 and LMO2 expression levels in AS-Ldb1 and AS-LMO2 cells and the increased FOG expression level in FOG-infected cells. (B) FOG inhibits GPA promoter activation by the SCL complex through specific interaction with GATA-1. Transactivation assays were performed using the GPA-84 reporter and mixtures supplemented with the indicated expression vectors (150 ng of GATA-1 or GATA-1V205→G; 900 ng of FOG) as described for Fig. 2. V205→G, point mutation in the N-terminal zinc finger of GATA-1 that disrupts interaction with FOG (13). (C) The bHLH domain of E47 is nonfunctional and acts as a dominant-negative inhibitor of the SCL complex. Transactivation assays were done using the GPA-84 reporter and mixtures containing the indicated expression vectors (150 ng of E47, E47-bHLH, or empty MSCV-neo vector). E47-bHLH, the bHLH domain of E47 from amino acids 518 to 610. For panels B and C, + and − indicate inclusion and omission of specific expression vectors. For each sample, 100 ng of CMV-β-Gal was added as an internal control; the total amount of DNA was kept constant (using pGem4) at 4.5 μg. Results represent the averages ± SD of triplicate determinations and are representative of n independent experiments. (D) Extracts (10 μg) from TF-1 cells infected with control (MSCV) or AS-Ldb1-, AS-LMO2-, FOG-, or E47-bHLH-expressing retroviruses were subjected to EMSA with the GPA-84 probe. The arrowhead indicates the usual low-mobility SCL complex, whereas the arrow points to a faster-migrating complex observed in TF-1 cells expressing the bHLH domain of E47. The binding intensities of the low-mobility complexes were quantified using ImageQuant software. (E) Reduced expression of the endogenous gene GPA in TF-1 cells expressing AS-Ldb1, AS-LMO2, FOG, and E47-bHLH. After normalization with S16 signals, GPA mRNA expression in TF-1 cell infectants was assessed by RT-PCR as described for Fig. 1C. Relative Exp., relative expression.
Since the assembly of a low-mobility complex containing SCL and its partners was required for GPA activation (Fig. 4), we first determined the consequences of AS-Ldb1, AS-LMO2, FOG, and E47-bHLH expression for endogenous TF-1 cell complexes by performing gel shift assays with the GPA-84 probe. While a low-mobility complex was revealed in nuclear extracts from parental TF-1 cells (Fig. 8D, lanes 1), decreased LMO2 and Ldb1 protein (AS-Ldb1 and AS-LMO2) levels led to a reduction in DNA binding by the SCL complex (Fig. 8D, lanes 1 to 3). Similarly, FOG overexpression severely disrupted complex formation (lanes 4) and ectopic expression of E47-bHLH shifted the migration of the DNA bound SCL complex towards higher mobility (lanes 5), indicating that this truncated mutant is capable of DNA binding and that it displaces the endogenous wild-type protein from the SCL complex (thus acting as a dominant-negative inhibitor of wild-type E47).
Finally, these interventions significantly decreased GPA mRNA expression as assessed by RT-PCR analysis (Fig. 8E), resulting in a twofold reduction when Ldb1 and LMO2 protein levels are decreased or when E47 binding is displaced by E47-bHLH. Furthermore, there was a fivefold decrease in GPA mRNA levels when GATA factors were sequestered by FOG. Strikingly, these reductions in GPA mRNA levels closely correlate with the decrease in DNA binding observed by EMSA (compare Fig. 8D and E). Taken together, these results strengthen the view that LMO2, Ldb1, GATA factors, and E proteins (more specifically, E47) are important components of SCL-containing complexes and that they are indeed required for GPA gene expression in hematopoietic cells.
DISCUSSION
The present study provides genetic and functional evidence that the erythroid gene GPA is a direct target of a multifactorial complex containing SCL, E47, LMO2, Ldb1, GATA-1, and Sp1. Our observations also reveal functional specialization within the complex, as SCL is required as a nucleation factor to assemble the complex on target regulatory elements, GATA-1 provides a DNA binding function, and E47 provides a potential transactivation function.
SCL in erythropoiesis.
The catastrophic consequences of SCL gene ablation in mice, which results in early embryonic lethality due to a complete absence of hematopoietic cells (45, 52, 53, 55), has complicated the assessment of the roles that SCL might play in the maturation of particular blood cell lineages. Several lines of evidence point to SCL as an important regulator of erythropoiesis. First, during development SCL is expressed in both primitive and definitive erythroid cells of the yolk sac blood islands and fetal liver (15, 29, 47). Analysis of hematopoietic precursors has shown that SCL is highly expressed in committed erythroid progenitors (BFU-E and CFU-E/proerythroblasts), whereas it becomes down regulated in terminally differentiated red cells (7, 26, 29). Therefore, the pattern of SCL expression suggests that it might be involved in the initial stages of commitment or consolidation of the erythroid cell fate with respect to pluripotent progenitors. Second, enforced SCL expression in hematopoietic cell lines and primary bone marrow cells favors erythroid differentiation (3, 17, 26, 63). Third, the genetic rescue of SCL-deficient mice and recent conditional gene targeting experiments have demonstrated that SCL is required for proper erythroid differentiation in vivo (23, 36, 54). Indeed, Sanchez et al. (54) showed that a transgene driving SCL expression in stem cells was able to rescue early hematopoietic progenitors in SCL−/− embryos; however, these mice still exhibited a defect in erythroid differentiation that resulted in embryonic lethality, demonstrating that sustained SCL expression is required for erythropoiesis. In recent conditional knockout studies in which floxed SCL alleles were lacking in mice expressing an interferon-inducible Cre recombinase, two groups have demonstrated that SCL inactivation leads to a complete block of erythroid and megakaryocytic cell maturation, seemingly without affecting hematopoietic stem cells (23, 36). Interestingly, Mikkola et al. (36) observed that a population of Ter119+ CD71+ cells (representing normal erythroid precursors) disappears following SCL inactivation, leading to the accumulation of an abnormal Ter119lo/− CD71+ population. Since Ter119 recognizes an epitope on murine GPA (5), the observation made by Mikkola et al. provides additional genetic evidence for the importance of SCL in driving GPA expression during murine erythropoiesis (shown herein). It seems, therefore, that once SCL has specified the hematopoietic cell fate from uncommitted mesodermal precursors, its sustained expression is not required for stem cell function but becomes required anew for the generation of red blood cells and megakaryocytes. Together, these results clearly demonstrate the essential role played by SCL in activating the transcription of erythrocyte-specific genes and driving the erythroid lineage.
Glycophorin genes and GPA promoter regulation.
The human glycophorins A, B, and E are part of a family of erythrocyte-specific membrane glycoproteins, which contribute to the expression of blood group antigens and determine the invasion and growth of parasites such as the malaria pathogen Plasmodium falciparum (11, 12). GPA is thought to form complexes with other erythroid membrane components (such as band 3, ankyrin, and protein 4.2), and their association appears to regulate the mechanical properties of the red cell membrane (8, 24). GPA-deficient human red blood cells show decreased sulfate anion transport due to the association between GPA and band 3, the human erythrocyte anion transporter (9). The GPA, GPB, and GPE genes are clustered on chromosome 4q28-q31 and seem to have evolved from successive duplications of the gene GPA (40). The cis-regulatory elements found here to be important for GPA promoter activation by the SCL complex are perfectly conserved in the GPB and GPE promoters (48), suggesting that these genes might also be direct targets of the SCL complex.
Previous studies of glycophorin promoter regulation, mainly focusing on the GPB promoter, demonstrated that these sites are protected from DNase I digestion in the presence of erythroid cell extracts and are required for promoter function (10, 48, 49). While the authors found that GATA-1 was the main factor binding to the G1 and G2 sites in EMSA and that the Sp1 motif was important for promoter activity, they did not identify other partners of the SCL complex as potential regulators of the glycophorin genes (49). Using a probe encompassing a longer segment of the GPA promoter (GPA-84), we demonstrate the existence of a large protein complex containing SCL and its partners. We show that within the GPA proximal promoter, the most crucial determinant for binding of the SCL complex is the G1 motif (followed by the Sp1, G2, and E elements) and that all these sites are required for optimal binding. We further complement these findings with the demonstration (through ChIP) that partners of the SCL complex indeed occupy the GPA promoter in vivo in hematopoietic cells. These findings further underscore the importance of Sp1 within the SCL complex, since Lécuyer et al. previously demonstrated that activation of the c-kit promoter by the SCL complex is critically dependent on the presence of a consensus GC-box and that Sp1 physically interacts with multiple partners of the complex (31).
Involvement of LMO2 and Ldb1 in erythroid gene regulation.
The results presented in this report reveal (using transient transactivation assays, ChIP, and AS-mediated loss of function in TF-1 cells) the importance of LMO2 and Ldb1 as essential partners within the SCL complex and establish their requirement for the appropriate regulation of an erythrocyte-specific gene. These findings contrast with those of a previous study suggesting that LMO2 and Ldb1 are negative regulators of erythropoiesis, as their enforced expression was shown to hinder terminal erythroid differentiation of G1ER cells (65), a GATA-1-deficient cell line blocked at the proerythroblast stage of differentiation. While these studies may appear contradictory at face value, previous analyses of Chip, the Drosophila orthologue of Ldb1, may help to reconcile these findings. Chip is a widely expressed regulator of several crucial processes, including embryonic segmentation (38), neuronal development (50), and dorsoventral patterning of the Drosophila wing (18). During wing morphogenesis, Chip associates within complexes containing the LIM-homeodomain protein Apterous (as well as the Drosophila LMO protein) and maintaining the appropriate stoichiometry of these complexes is crucial for proper wing development (18, 37, 51, 64). In this context, both overexpression and loss-of-function mutations of the Chip gene lead to the same phenotypic abnormalities in wing morphogenesis (18). Therefore, if the stoichiometry characteristics of LMO2- and Ldb1-containing complexes are similarly tightly regulated during erythropoiesis, enforced expression of these factors (as performed by Visvader et al.) (65) might interfere with endogenous complexes through sequestration mechanisms and lead to the same outcome as loss-of-function approaches, which were utilized in the present study. In support of this hypothesis, a recent report by Xu and colleagues (70), who identified the protein 4.2 gene as a erythroid target of SCL and its partners, showed that enforced expression of wild-type or a dominant-negative version of Ldb1 perturbed activation of the protein 4.2 gene (consistent with the view that the stoichiometry of these complexes is indeed important for erythroid gene regulation).
In Drosophila, Chip was initially identified as an important regulator of enhancer-promoter communication (38), a property that seemingly relies on its ability to self-dimerize and to interact with several families of regulators, including LIM domain- and homeodomain-containing proteins (58). Furthermore, it has recently been shown that proper patterning of the Drosophila nervous system depends on the ability of Chip to interact physically with Pannier, a Drosophila orthologue of GATA-1, and with bHLH factors of the Achaete/Scute complex (50). Our results extend these findings to show that mammalian Ldb1 also directly interacts with GATA family members and bHLH factors. Interestingly, Ldb1 gene ablation in mice results in severe patterning defects and, among other phenotypes, compromises the development of yolk sac blood islands (39). This hematopoietic phenotype is most likely caused by defects in gene regulation by the SCL complex at the onset of hematopoiesis. We also provide evidence that LMO proteins can self-associate (in similarity to the homodimerization of the LIM proteins CRP and MLP) (4, 19). Therefore, this network of interactions most likely modulates the assembly, targeting, and activity of the SCL complex at different levels in the hematopoietic hierarchy.
Functional specialization within the SCL complex.
SCL is an important regulator at several positions in the hematopoietic hierarchy. Whether its molecular mode of action differs in different hematopoietic lineages or populations remains ill defined. In the present study, we found differences in the mechanisms by which SCL regulates the gene GPA versus our previous analysis of the gene c-kit, which constitute erythroid and stem or progenitor cell targets of SCL, respectively. First, we found that maximal GPA promoter activation (at lower concentrations of expression vector) and assembly of the SCL complex on GPA promoter sequences requires an E-box motif and SCL DNA binding activity, although at higher concentrations of SCL (as observed following enforced expression in TF-1 cells) SCL DNA binding mutants are active. In contrast, Lécuyer et al. previously showed that activation of the c-kit promoter by the SCL complex was E-box independent and did not require SCL DNA binding (31). This mechanistic difference is consistent with the observation that SCL DNA binding-defective mutants rescue hematopoietic cell commitment in SCL−/− embryonic stem cells although they are unable to restore the proper maturation of definitive erythroid cells (44). Therefore, the requirement for SCL DNA binding is one characteristic that might delineate SCL function at the onset of hematopoiesis and during erythropoiesis. It is possible that a higher level of affinity of DNA binding by the SCL complex is required for the proper activation of the erythroid program, which would be provided in part by SCL itself and by other partners of the complex, such as GATA and Sp/XKLF family members. It will be possible to assess whether the necessity of SCL DNA binding is a broad difference that distinguishes erythroid and stem cell targets of SCL through the identification and molecular characterization of additional SCL target genes.
Unlike the results seen with SCL, the bHLH domain of E47 is unable to replace the function of the full-length protein during GPA promoter activation. Moreover, this truncated protein is dominant negative over wild-type E47 both in transient assays and in chromatin, as it likely competes for DNA binding with endogenous E proteins. This finding contrasts with that of a previous study of the POMC promoter, in which E47-bHLH was shown to form a functional tripartite complex with NeuroD and Pitx-1 (46). Besides the C-terminally located bHLH domain, the E47 protein harbors two distinctive activation domains in its N terminus (designated AD1 and AD2) which are absent from E47-bHLH. These domains are highly conserved in other ubiquitously expressed bHLH factors such as E12, E2-2, and HEB, and E. Lécuyer and T. Hoang have observed that HEB can functionally replace E47 within the SCL complex (unpublished data). Therefore, it is likely that the transactivation domains of E47 are required for the proper function of the SCL complex (although our results do not exclude the possibility that an unknown function of the N-terminal moiety of E47 might be involved). Interestingly, it has recently been shown that the AD1 domain serves as a recruitment motif for the SAGA histone acetyltransferase complex (32). Since infection of TF-1 cells with E47-bHLH causes an important shift in the mobility of the SCL complex, it is possible that this mutant hinders the recruitment of additional regulatory factors (such as elements of the SAGA complex) to SCL target genes. Further investigation will be required to evaluate this possibility.
Transcription regulation by the SCL complex in different hematopoietic compartments.
During differentiation, transcription factor complexes may undergo dynamic changes in composition, a view described as a cocktail party scenario by Sieweke and Graf (56). For example, our observations identified a requirement for Sp1 as a member of the SCL complex in c-Kit-expressing cells and in erythroid cells (reference 31 and the present study) and there is no evidence for the involvement of Sp1 in T cells (41). By evolving in such a manner, the activity and target gene specificity of such multifactorial complexes might be modulated by environmental cues and favor differentiation towards particular cell fates. This type of mechanism would seem energetically cost effective for an organism, as it would bypass the requirement for major dismantling events as a prerequisite to the commencement and shutdown of different programs of gene expression. In this respect, our finding that FOG can inhibit promoter activation by the SCL complex demonstrates how cofactors can modulate the activity of higher-order transcription factor complexes through interactions with specific components. Since the GATA-FOG interaction is essential for erythroid and megakaryocytic cell differentiation (13, 61), our findings suggest that during differentiation into these lineages a proportion of the GATA factor pool is recruited into FOG-containing complexes, thus enabling GATA factors to exert functions that are independent of SCL complexes. Furthermore, our observation that SCL complexes containing GATA-1 or GATA-2 demonstrate preferential activation efficiency for erythroid or stem cell targets, respectively, suggests that SCL-containing complexes may evolve dynamically during hematopoiesis to favor the maintenance of pluripotency or to consolidate differentiation towards specific lineages.
At the onset of hematopoiesis, when GATA-2 is the predominant GATA family member, SCL complexes would be required for the activation of stem cell targets such as c-kit (31), which would favor the maintenance of an undifferentiated phenotype. At this stage, SCL complexes may also start to weakly activate the expression of erythroid targets such as GPA, GATA-1, and EKLF, which would help to prime stem and progenitor cells for their eventual commitment towards the erythroid-megakaryocytic pathways. The activity of such a complex would account for the multilineage gene expression priming that is thought to precede the commitment of hematopoietic stem cells into different lineages (28). Since the level of GATA-1 expression increases in progenitors of the erythroid lineage and that of GATA-2 is down regulated, the replacement of GATA-2 by GATA-1 within the SCL complex might delineate a point at which the activation of erythrocyte-specific genes is engaged more robustly. In the T lineage, however, GATA-3 is preferentially expressed and may substitute for GATA-2 within the SCL complex to drive the expression of T-cell-specific genes (41). Therefore, subtle variations in composition seem to modulate the specificity of action of SCL-containing complexes and may also account for the differential requirements for SCL DNA binding activity in different hematopoietic compartments. This type of mechanism is most likely a recurrent theme in cell fate determination in many other tissues.
Acknowledgments
Rachid Lahlil and Eric Lécuyer contributed equally to this work.
We thank Peter D. Aplan for SIL-SCL transgenic mice as well as Stuart H. Orkin, Catherine Porcher, John D. Crispino, and Jacques Drouin for providing expression vectors for SCL mutants, the GATA-1 V205→R mutant, FOG, and E47-bHLH, respectively.
This work was supported in part by a grant from the Canadian Institute for Health Research (CIHR) to T.H.; by a studentship from the CIHR (E.L.), and by a fellowship from the Leukemia Research Fund of Canada (S.H.).
REFERENCES
- 1.Anderson, K. P., S. C. Crable, and J. B. Lingrel. 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95:1652-1655. [PubMed] [Google Scholar]
- 2.Aplan, P. D., C. A. Jones, D. S. Chervinsky, X. Zhao, M. Ellsworth, C. Wu, E. A. McGuire, and K. W. Gross. 1997. An scl gene product lacking the transactivation domain induces bony abnormalities and cooperates with LMO1 to generate T-cell malignancies in transgenic mice. EMBO J. 16:2408-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aplan, P. D., K. Nakahara, S. H. Orkin, and I. R. Kirsch. 1992. The SCL gene product: a positive regulator of erythroid differentiation. EMBO J. 11:4073-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber, S., and P. Caroni. 1996. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 10:289-300. [DOI] [PubMed] [Google Scholar]
- 5.Auffray, I., S. Marfatia, K. de Jong, G. Lee, C. H. Huang, C. Paszty, M. J. Tanner, N. Mohandas, and J. A. Chasis. 2001. Glycophorin A dimerization and band 3 interaction during erythroid membrane biogenesis: in vivo studies in human glycophorin A transgenic mice. Blood 97:2872-2878. [DOI] [PubMed] [Google Scholar]
- 6.Begley, C. G., and A. R. Green. 1999. The SCL gene: from case report to critical hematopoietic regulator. Blood 93:2760-2770. [PubMed] [Google Scholar]
- 7.Brady, G., F. Billia, J. Knox, T. Hoang, I. R. Kirsch, E. B. Voura, R. G. Hawley, R. Cumming, M. Buchwald, and K. Siminovitch. 1995. Analysis of gene expression in a complex differentiation hierarchy by global amplification of cDNA from single cells. Curr. Biol. 5:909-922. (Erratum, 10:1201). [DOI] [PubMed] [Google Scholar]
- 8.Bruce, L. J., S. Ghosh, M. J. King, D. M. Layton, W. J. Mawby, G. W. Stewart, P. A. Oldenborg, J. Delaunay, and M. J. Tanner. 2002. Absence of CD47 in protein 4.2-deficient hereditary spherocytosis in man: an interaction between the Rh complex and the band 3 complex. Blood 100:1878-1885. [DOI] [PubMed] [Google Scholar]
- 9.Bruce, L. J., J. D. Groves, Y. Okubo, B. Thilaganathan, and M. J. Tanner. 1994. Altered band 3 structure and function in glycophorin A- and B-deficient (MkMk) red blood cells. Blood 84:916-922. [PubMed] [Google Scholar]
- 10.Camara-Clayette, V., C. Rahuel, O. Bertrand, and J. P. Cartron. 1999. The E-box of the human glycophorin B promoter is involved in the erythroid-specific expression of the GPB gene. Biochem. Biophys. Res. Commun. 265:170-176. [DOI] [PubMed] [Google Scholar]
- 11.Chasis, J. A., and N. Mohandas. 1992. Red blood cell glycophorins. Blood 80:1869-1879. [PubMed] [Google Scholar]
- 12.Chishti, A. H., J. Palek, D. Fisher, G. J. Maalouf, and S. C. Liu. 1996. Reduced invasion and growth of Plasmodium falciparum into elliptocytic red blood cells with a combined deficiency of protein 4.1, glycophorin C, and p55. Blood 87:3462-3469. [PubMed] [Google Scholar]
- 13.Crispino, J. D., M. B. Lodish, J. P. MacKay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 14.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elefanty, A. G., C. G. Begley, L. Hartley, B. Papaevangeliou, and L. Robb. 1999. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood 94:3754-3763. [PubMed] [Google Scholar]
- 16.Elefanty, A. G., C. G. Begley, D. Metcalf, L. Barnett, F. Kontgen, and L. Robb. 1998. Characterization of hematopoietic progenitor cells that express the transcription factor SCL, using a lacZ “knock-in” strategy. Proc. Natl. Acad. Sci. USA 95:11897-11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elwood, N. J., H. Zogos, D. S. Pereira, J. E. Dick, and C. G. Begley. 1998. Enhanced megakaryocyte and erythroid development from normal human CD34+ cells: consequence of enforced expression of SCL. Blood 91:3756-3765. [PubMed] [Google Scholar]
- 18.Fernandez-Funez, P., C. H. Lu, D. E. Rincon-Limas, A. Garcia-Bellido, and J. Botas. 1998. The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 17:6846-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feuerstein, R., X. Wang, D. Song, N. E. Cooke, and S. A. Liebhaber. 1994. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc. Natl. Acad. Sci. USA 91:10655-10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox, A. H., C. Liew, M. Holmes, K. Kowalski, J. Mackay, and M. Crossley. 1999. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18:2812-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gering, M., A. R. Rodaway, B. Gottgens, R. K. Patient, and A. R. Green. 1998. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 17:4029-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grutz, G. G., K. Bucher, I. Lavenir, T. Larson, R. Larson, and T. H. Rabbitts. 1998. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 17:4594-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, M. A., D. J. Curtis, D. Metcalf, A. G. Elefanty, K. Sourris, L. Robb, J. R. Gothert, S. M. Jane, and C. G. Begley. 2003. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc. Natl. Acad. Sci. USA 100:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassoun, H., T. Hanada, M. Lutchman, K. E. Sahr, J. Palek, M. Hanspal, and A. H. Chishti. 1998. Complete deficiency of glycophorin A in red blood cells from mice with targeted inactivation of the band 3 (AE1) gene. Blood 91:2146-2151. [PubMed] [Google Scholar]
- 25.Herblot, S., A.-M. Steff, P. Hugo, P. D. Aplan, and T. Hoang. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-Ta chain expression. Nat. Immunol. 1:138-144. [DOI] [PubMed] [Google Scholar]
- 26.Hoang, T., E. Paradis, G. Brady, F. Billia, K. Nakahara, N. N. Iscove, and I. R. Kirsch. 1996. Opposing effects of the basic helix-loop-helix transcription factor SCL on erythroid and monocytic differentiation. Blood 87:102-111. [PubMed] [Google Scholar]
- 27.Hsu, H. L., L. Huang, J. T. Tsan, W. Funk, W. E. Wright, J. S. Hu, R. E. Kingston, and R. Baer. 1994. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol. Cell. Biol. 14:1256-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774-785. [DOI] [PubMed] [Google Scholar]
- 29.Kallianpur, A. R., J. E. Jordan, and S. J. Brandt. 1994. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood 83:1200-1208. [PubMed] [Google Scholar]
- 30.Krosl, G., G. He, M. Lefrancois, F. Charron, P. H. Romeo, P. Jolicoeur, I. R. Kirsch, M. Nemer, and T. Hoang. 1998. Transcription factor SCL is required for c-kit expression and c-Kit function in hemopoietic cells. J. Exp. Med. 188:439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lécuyer, E., S. Herblot, M. Saint-Denis, R. Martin, C. G. Begley, C. Porcher, S. H. Orkin, and T. Hoang. 2002. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood 100:2430-2440. [DOI] [PubMed] [Google Scholar]
- 32.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4:63-73. [DOI] [PubMed] [Google Scholar]
- 33.Mead, P. E., A. E. Deconinck, T. L. Huber, S. H. Orkin, and L. I. Zon. 2001. Primitive erythropoiesis in the Xenopus embryo: the synergistic role of LMO-2, SCL and GATA-binding proteins. Development 128:2301-2308. [DOI] [PubMed] [Google Scholar]
- 34.Mead, P. E., C. M. Kelley, P. S. Hahn, O. Piedad, and L. I. Zon. 1998. SCL specifies hematopoietic mesoderm in Xenopus embryos. Development 125:2611-2620. [DOI] [PubMed] [Google Scholar]
- 35.Merika, M., and S. H. Orkin. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkola, H. K., J. Klintman, H. Yang, H. Hock, T. M. Schlaeger, Y. Fujiwara, and S. H. Orkin. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547-551. [DOI] [PubMed] [Google Scholar]
- 37.Milan, M., and S. M. Cohen. 1999. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol. Cell 4:267-273. [DOI] [PubMed] [Google Scholar]
- 38.Morcillo, P., C. Rosen, M. K. Baylies, and D. Dorsett. 1997. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 11:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay, M., A. Teufel, T. Yamashita, A. D. Agulnick, L. Chen, K. M. Downs, A. Schindler, A. Grinberg, S. P. Huang, D. Dorward, and H. Westphal. 2003. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130:495-505. [DOI] [PubMed] [Google Scholar]
- 40.Onda, M., S. Kudo, A. Rearden, M. G. Mattei, and M. Fukuda. 1993. Identification of a precursor genomic segment that provided a sequence unique to glycophorin B and E genes. Proc. Natl. Acad. Sci. USA 90:7220-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono, Y., N. Fukuhara, and O. Yoshie. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol. 18:6939-6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osada, H., G. Grutz, H. Axelson, A. Forster, and T. H. Rabbitts. 1995. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc. Natl. Acad. Sci. USA 92:9585-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porcher, C., E. C. Liao, Y. Fujiwara, L. I. Zon, and S. H. Orkin. 1999. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126:4603-4615. [DOI] [PubMed] [Google Scholar]
- 45.Porcher, C., W. Swat, K. Rockwell, Y. Fujiwara, F. W. Alt, and S. H. Orkin. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47-57. [DOI] [PubMed] [Google Scholar]
- 46.Poulin, G., M. Lebel, M. Chamberland, F. W. Paradis, and J. Drouin. 2000. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 20:4826-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulford, K., N. Lecointe, K. Leroy-Viard, M. Jones, D. Mathieu-Mahul, and D. Y. Mason. 1995. Expression of TAL-1 proteins in human tissues. Blood 85:675-684. [PubMed] [Google Scholar]
- 48.Rahuel, C., J. F. Elouet, and J. P. Cartron. 1994. Post-transcriptional regulation of the cell surface expression of glycophorins A, B, and E. J. Biol. Chem. 269:32752-32758. [PubMed] [Google Scholar]
- 49.Rahuel, C., M. A. Vinit, V. Lemarchandel, J. P. Cartron, and P. H. Romeo. 1992. Erythroid-specific activity of the glycophorin B promoter requires GATA-1 mediated displacement of a repressor. EMBO J. 11:4095-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramain, P., R. Khechumian, K. Khechumian, N. Arbogast, C. Ackermann, and P. Heitzler. 2000. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol. Cell 6:781-790. [DOI] [PubMed] [Google Scholar]
- 51.Rincon-Limas, D. E., C. H. Lu, I. Canal, and J. Botas. 2000. The level of DLDB/CHIP controls the activity of the LIM homeodomain protein apterous: evidence for a functional tetramer complex in vivo. EMBO J. 19:2602-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robb, L., N. J. Elwood, A. G. Elefanty, F. Kontgen, R. Li, L. D. Barnett, and C. G. Begley. 1996. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15:4123-4129. [PMC free article] [PubMed] [Google Scholar]
- 53.Robb, L., I. Lyons, R. Li, L. Hartley, F. Kontgen, R. P. Harvey, D. Metcalf, and C. G. Begley. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92:7075-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez, M. J., E. O. Bockamp, J. Miller, L. Gambardella, and A. R. Green. 2001. Selective rescue of early haematopoietic progenitors in Scl−/− mice by expressing Scl under the control of a stem cell enhancer. Development 128:4815-4827. [DOI] [PubMed] [Google Scholar]
- 55.Shivdasani, R. A., E. L. Mayer, and S. H. Orkin. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373:432-434. [DOI] [PubMed] [Google Scholar]
- 56.Sieweke, M. H., and T. Graf. 1998. A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev. 8:545-551. [DOI] [PubMed] [Google Scholar]
- 57.Tenen, D. G., R. Hromas, J. D. Licht, and D. E. Zhang. 1997. Transcription factors, normal myeloid development, and leukemia. Blood 90:489-519. [PubMed] [Google Scholar]
- 58.Torigoi, E., I. M. Bennani-Baiti, C. Rosen, K. Gonzalez, P. Morcillo, M. Ptashne, and D. Dorsett. 2000. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc. Natl. Acad. Sci. USA 97:2686-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tremblay, M., S. Herblot, E. Lécuyer, and T. Hoang. 2003. Regulation of pTα gene expression by a dosage of E2A, HEB, and SCL. J. Biol. Chem. 278:12680-12687. [DOI] [PubMed] [Google Scholar]
- 60.Tsai, F. Y., and S. H. Orkin. 1997. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89:3636-3643. [PubMed] [Google Scholar]
- 61.Tsang, A. P., Y. Fujiwara, D. B. Hom, and S. H. Orkin. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 63.Valtieri, M., A. Tocci, M. Gabbianelli, L. Luchetti, B. Masella, L. Vitelli, R. Botta, U. Testa, G. L. Condorelli, and C. Peschle. 1998. Enforced TAL-1 expression stimulates primitive, erythroid and megakaryocytic progenitors but blocks the granulopoietic differentiation program. Cancer Res. 58:562-569. [PubMed] [Google Scholar]
- 64.van Meyel, D. J., D. D. O'Keefe, L. W. Jurata, S. Thor, G. N. Gill, and J. B. Thomas. 1999. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol. Cell 4:259-265. [DOI] [PubMed] [Google Scholar]
- 65.Visvader, J. E., X. Mao, Y. Fujiwara, K. Hahm, and S. H. Orkin. 1997. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl. Acad. Sci. USA 94:13707-13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vyas, P., M. A. McDevitt, A. B. Cantor, S. G. Katz, Y. Fujiwara, and S. H. Orkin. 1999. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126:2799-2811. [DOI] [PubMed] [Google Scholar]
- 67.Wadman, I., J. Li, R. O. Bash, A. Forster, H. Osada, T. H. Rabbitts, and R. Baer. 1994. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 13:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wadman, I. A., H. Osada, G. G. Grutz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss, M. J., G. Keller, and S. H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 8:1184-1197. [DOI] [PubMed] [Google Scholar]
- 70.Xu, Z., S. Huang, L.-S. Chang, A. D. Agulnick, and S. J. Brandt. 2003. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23:7585-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, X. F., and P. D. Aplan. 1999. The hematopoietic transcription factor SCL binds the p44 subunit of TFIIH. J. Biol. Chem. 274:1388-1393. [DOI] [PubMed] [Google Scholar]