Abstract
The genetic basis of sexual isolation that contributes to speciation is one of the unsolved questions in evolutionary biology. Drosophila ananassae and Drosophila pallidosa are closely related, and postmating isolation has not developed between them. However, females of both species discriminate their mating partners, and this discrimination contributes to strong sexual isolation between them. By using surgical treatments, we demonstrate that male courtship songs play a dominant role in female mate discrimination. The absence of the song of D. pallidosa dramatically increased interspecies mating with D. ananassae females but reduced intraspecies mating with D. pallidosa females. Furthermore, genetic analysis and chromosomal introgression by repeated backcrosses to D. pallidosa males identified possible loci that control female discrimination in each species. These loci were mapped on distinct positions near the Delta locus on the middle of the left arm of the second chromosome. Because the mate discrimination we studied is well developed and is the only known mechanism that prevents gene flow between them, these loci may have played crucial roles in the evolution of reproductive isolation, and therefore, in the speciation process between these two species.
What kind of genetic mechanisms control speciation? Understanding the genetic basis of reproductive isolation is one of the best ways to answer this question. However, we have not yet completely understood how many and what types of genes are involved in reproductive isolation, and how they are responsible for speciation, except in limited cases (1–4). In monkeyflowers (Mimulus), some loci for eight floral traits mediating reproductive isolation between sympatric two species have been mapped (5). Genetic analyses of hybrid male sterility and hybrid inviability in Drosophila also show the existence of many loci, and these studies suggest that loci affecting postmating isolation have spread widely in various regions across the genome (6–9). However, genetic studies of premating isolation, especially for mating discrimination, have been limited to the chromosomal level (10–15), despite the fact that this may be a primary cause of speciation in many animal taxa (16, 17). We do not know whether changes in a small number of genes having relatively large effects can directly affect premating isolation, or whether accumulation of such changes with minor effects gradually produces the species-specific phenotypes for isolation. Although some loci controlling sex pheromone differences (18, 19) or sexual behaviors (20) have been identified, which may contribute to sexual isolation, it has not been shown whether these loci really cause sexual isolation between closely related species.
Drosophila ananassae and Drosophila pallidosa are sexually isolated species (21–24). D. ananassae is cosmopolitan; its distribution has been expanded by human activities, and it is found in sympatry with D. pallidosa in the Tongan and Fijian islands in Melanesia (25, 26). Because there was not enough work to estimate the history of their associations in the past, it is unclear whether they evolved in sympatry or allopatry. Nevertheless, the important thing is that they do not exhibit apparent postmating isolation such as hybrid inviability or sterility in either the F1 or F2 generation (21–24). Hybrids and their progeny can be produced in the laboratory for genetic analysis.
The two species are genetically distinct in nature (24, 26). Ecological isolation, such as differences of niche, also seems to be insufficient to explain the absence of observed hybridization in the wild because these two species were captured together in the same traps in Tonga and Fiji (25). Thus, it seems that the most well developed reproductive isolation mechanism that can prevent gene flow between the two species is sexual (ethological) isolation (24). On the basis of these observations and the morphological similarity between them, Bock and Wheeler (26) suggested that the phylogenetic separation of D. ananassae and D. pallidosa must have been a recent event in the speciation of the melanogaster group. Thus, we suppose that these flies are excellent materials to study the genetic basis of sexual isolation and speciation.
D. ananassae and D. pallidosa are not distinguished easily by their morphological characters (26). Males from both species actively court heterospecific females as well as conspecific females (24, 27). Thus, the strong sexual isolation between them may well depend on female discrimination on the basis of male courtship signals such as courtship song by wing vibration, cuticular hydrocarbons (sex pheromones), male courtship behavior itself, and so on (20, 28). Significant differences between these two species are seen in some parameters of male courtship songs (29) and major cuticular hydrocarbons (27, 30), which may affect female discrimination.
In this report, we present several lines of evidence that female discrimination is based on male courtship song, which causes sexual isolation between the two species. Next, we identify a region controlling female discrimination by chromosomal introgression methods. This study mapped possible loci that control female discrimination in each species leading to sexual isolation.
Materials and Methods
Fly Stocks.
All flies were kept in 3 × 10.5-cm vials containing Drosophila standard yeast/cornmeal/glucose medium under a 14/10-day light/dark cycle (light, 7:00–21:00) at 25 ± 1°C with a relative humidity of 55 ± 5%. Four wild strains of each species were used: HW (Hawaii), TNG (Tonga), NAN84 (Fiji), and C347 (Thailand) as D. ananassae; and NAN4 (Fiji), NAN57 (Fiji), TBU155 (Tonga), and VAV92 (Tonga) as D. pallidosa. Multimarker strains of D. ananassae, Pr (Prickly, short and twisted bristles) and Tr (Trident, branched crossvein in the wing) and Bl (Bristle, shortened bristles of dorsal thorax) and Lo (Lobe, bar-like eyes), were used for the second and the third chromosome marker, respectively. Dl e pi Om(2D)63/NG2 (Delta, end of L2 and costal vein fused in wing, ebony pink Optic morphology (2D) 63/second chromosome balancer) was used for the genetic mapping on the second chromosome. Dl is located in the middle of the left arm of the second chromosome and Om(2D)63 is in the right arm (cytological map, 43B). Detailed explanations on these strains and mutants are available in ref. 25.
Mating Experiments.
Mating success was measured in a “No choice” experiment in the morning (9:00–12:00), during which time flies showed highest mating activity (29). Flies were sexed under light anesthesia with diethyl ether within 12 h of their emergence. About 40 males and females were maintained separately for 4 to 6 days in food vials until use in experiments. Ten males and ten virgin females were placed in a food vial and left for 2 h. After 2 h, the males were discarded; the females were dissected within 24 h. Copulation was checked by the presence of sperm in spermathecas or seminal receptacles. All crosses were repeated 10 times. A total of 100 females were examined for their mate discrimination.
Manipulation of Flies.
Male wings are the organ used to produce the courtship song, and female aristae extended from the third antennal segment are the receiving organ for the sounds (20, 28, 31, 32). To examine the effect of male courtship songs in mate discrimination by females, we examined the mating success of wild-type females and males unable to generate sound (prepared by removing male wings), and between wild-type males and females unable to hear the sounds (prepared by removing female aristae). Two days before the experiments, all male flies were anesthetized lightly. Both wings of half the males were removed from the basement of the wing with a pair of microsurgical scissors (wingless males) and the other half of the males were used as a control (winged males). Ten wingless and 10 winged males were kept in food vials until used in the mating experiments. Both female aristae were removed completely from the basement on the third antennal segment with the microsurgical scissors. The flies with an injured antennal segment anywhere from the first to the third segment (caused by messy cutting) were excluded from the experiments.
Repeated Backcrossing.
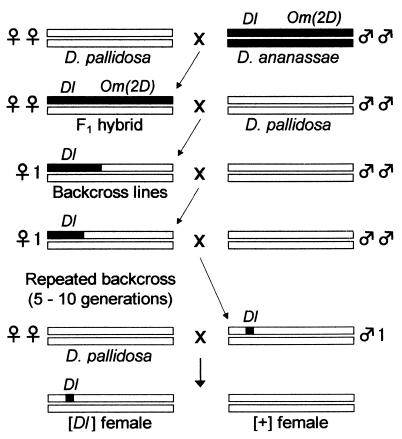
To identify the region controlling female discrimination, repeated backcrossing was performed to introgress a small region of the D. ananassae second chromosome into D. pallidosa genetic background (Fig. 1). The backcross was carried out by using Dl as the visible marker (see Results and Table 1D). Dl Om(2D) males were first crossed with D. pallidosa females to produce F1 female hybrids. [Dl Om(2D)] F1 hybrid females then were crossed with D. pallidosa males, and only [Dl] F2 females were selected. At this generation, 20 sublines were established by setting up 20 independent crosses of one [Dl] F2 female with D. pallidosa males. These 20 backcrossed lines were maintained independently by repeated backcrossing of one virgin female heterozygous for the Dl locus with D. pallidosa males. After 5 to 10 generations of backcrossing, one [Dl] backcrossed male that was heterozygous for the Dl region was crossed with virgin D. pallidosa females, and their female progeny, [Dl] (having D. ananassae Dl region) and [+] (otherwise possessing only D. pallidosa genome) were used for mating experiments.
Figure 1.
Mating scheme for repeated backcrossing. White bars represent the D. pallidosa second chromosome and black bars represent the D. ananassae second chromosome. The other chromosomes are not shown.
Table 1.
Mating success (%) in wild-type females of D. ananassae, D. pallidosa, mutant females of D. ananassae, F1 hybrid females, and recombinant females (N = 100, 10 replicates in each cross)
| Female | Male*
|
||
|---|---|---|---|
| D. ananassae | D. pallidosa | ||
| D. ananassae | |||
| HW | 82 | 6 | |
| TNG | 86 | 0 | |
| NAN84 | 88 | 0 | |
| C347 | 72 | 0 | |
| Pr; Tr | 86 | 1 | |
| Dl e pi Om(2D)63/NG2 | 60 | 0 | |
| D. pallidosa | |||
| NAN4 | 4 | 81 | |
| NAN57 | 3 | 85 | |
| VAV92 | 2 | 46 | |
| TBU155 | 2 | 96 | |
| F1 hybrid from the cross† | |||
| HW | × NAN4 | 78 | 7 |
| NAN4 | × HW | 82 | 4 |
| Pr; Tr | × NAN4 | 84 | 37 |
| NAN4 | × Pr; Tr | 90 | 32 |
| NAN4 | × Dl Om(2D) | 72 | 10 |
| Dl Om(2D) | × NAN4 | 60 | 0 |
| Recombinant (female genotype) | |||
| + | + | 13 | 50 |
| + | Om(2D) | 20 | 50 |
| Dl | + | 70 | 0 |
| Dl | Om(2D) | 63 | 18 |
Males from the same strains were used in intraspecies crosses. In interspecies and F1 hybrid crosses, HW and NAN4 males were used as D. ananassae and D. pallidosa males, respectively.
Female strain presented first.
Results
Female Discrimination and Sexual Isolation.
There exists strong sexual isolation between D. ananassae
and D. pallidosa (Table 1 D. ananassae and
D. pallidosa). In both species, wild-type females
from all of the strains used in this study showed quite low mating
success with heterospecific wild-type males, compared with high mating
success with conspecific males. To examine whether courtship song
generated by male wings could contribute to this sexual isolation, we
examined the mating success of wild-type females with wingless males
and the success of aristaless females with normal males. Female
discrimination based on male courtship songs causes mating failure
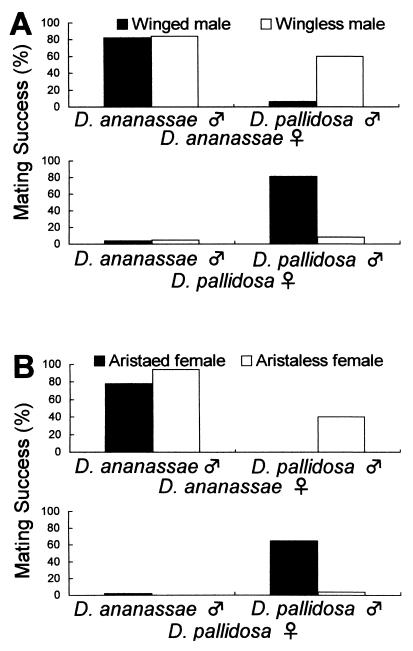
(Fig. 2). In this experiment, females
from each species showed characteristic differences in mating success
with wingless males (Fig. 2A). D.
ananassae females copulated with both winged and wingless D.
ananassae males (82% and 84%). Furthermore, D.
ananassae females showed significantly higher mating success with
wingless D. pallidosa males (which did not produce a
D. pallidosa-specific male courtship song, 60%) than with
winged D. pallidosa males (6%, χ =
63.52, P < 0.001). On the other hand, D.
pallidosa females hardly copulated with wingless males even if the
males were conspecific (8%). These results suggest that the song of
D. pallidosa males prevents D. ananassae females
from copulating with them, and facilitates D. pallidosa
females copulating with them.
=
63.52, P < 0.001). On the other hand, D.
pallidosa females hardly copulated with wingless males even if the
males were conspecific (8%). These results suggest that the song of
D. pallidosa males prevents D. ananassae females
from copulating with them, and facilitates D. pallidosa
females copulating with them.
Figure 2.
Female discrimination between D. ananassae and D. pallidosa. (A) Mating success (%) of D. ananassae HW females and that of D. pallidosa NAN4 females with winged or wingless males. Sample size in all of the combinations was 100 females. (B) Mating success (%) of aristaed and aristaless D. ananassae HW females (Upper) and that of D. pallidosa NAN4 females with males of both strains. All males used for crosses were winged normal males.
The mating of aristaless females with winged males (Fig.
2B) showed almost the same discrimination patterns
consistent with the results from wingless males (Fig.
2A). D. ananassae females having no
aristae showed significant mating success with D.
pallidosa males (34%) but females having aristae did not
copulate with D. pallidosa males at all (0%,
χ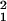 = 38.59, P < 0.001). Normal
D. pallidosa females copulated when they could receive a
conspecific courtship song (65%). Aristaless females could recognize
all male courtship signals except for sound. Visual signals such as a
pattern of wing vibration (24) and chemosensory signals such as male
sex pheromones and mechanosensory signals such as a tapping by foreleg,
etc., should be perceived, because these females have intact eyes,
organs for olfaction (third antennal segment, maxillary palp in
the mouth, distal forelegs, etc.), and bristles for mechanosensation
(20, 33). Similar results obtained by removing the organs necessary to
produce (wings) or receive (aristae) song strongly suggest that females
discriminate among their mating partners on the basis of the courtship
song, and not on the basis of a visual or a chemical signal.
= 38.59, P < 0.001). Normal
D. pallidosa females copulated when they could receive a
conspecific courtship song (65%). Aristaless females could recognize
all male courtship signals except for sound. Visual signals such as a
pattern of wing vibration (24) and chemosensory signals such as male
sex pheromones and mechanosensory signals such as a tapping by foreleg,
etc., should be perceived, because these females have intact eyes,
organs for olfaction (third antennal segment, maxillary palp in
the mouth, distal forelegs, etc.), and bristles for mechanosensation
(20, 33). Similar results obtained by removing the organs necessary to
produce (wings) or receive (aristae) song strongly suggest that females
discriminate among their mating partners on the basis of the courtship
song, and not on the basis of a visual or a chemical signal.
Genetic Control of Female Discrimination.
Does a single gene or do complex genic interactions control female discrimination? We examined the genetic basis of this discrimination. All of the F1 hybrid females between D. pallidosa and wild-type D. ananassae or mutant D. ananassae showed quite similar mating patterns with D. ananassae (Table 1 F1 hybrid from the cross). This dominant phenotype of D. ananassae made it possible to test for single-locus control of female discrimination. Chromosomal analyses showed a highly significant effect of the second chromosome on mating success (Fig. 3), corroborated by three-way ANOVA after arcsine transformation (F1,72 = 146.8, P < 0.001 for D. ananassae males and F1,72 = 19.3, P < 0.001 for D. pallidosa males). A significant effect was seen also on chromosome 3 (F1,72 = 10.2, P < 0.01 for D. ananassae males and F1,72 = 4.4, P < 0.05 for D. pallidosa males) but no interaction between chromosomes was significant. Because a similar significant effect for the second chromosome was obtained from another chromosomal analysis by using other visible markers (Bl; Lo, F1,72 = 221.7, P < 0.001 for D. ananassae males; F1,72 = 105.0, P < 0.001 for D. pallidosa males), we concluded that the locus (loci) on the second chromosome mainly controls the observed female discrimination. Furthermore, genetic mapping placed the second chromosomal factor(s) between the Dl and Om(2D) loci, closer to Dl, because both [Dl +] and [Dl Om(2D)] recombinant females dominantly displayed the D. ananassae-specific phenotype (Table 1 Recombinant).
Figure 3.
Chromosomal analysis of female discrimination (mating success). The methods and sample size are the same as shown in Fig. 2. Filled bars in female genotype represent the D. pallidosa chromosome and blanked bars represent the D. ananassae chromosome. Letters in the chromosome represent the morphological markers for the cross.
Because of the low number of dominant markers in D. ananassae, we could not narrow down the location of this locus by classical recombination mapping. So, to map the precise locus for female discrimination, next we tried to introgress the D. ananassae chromosome segments around Dl into D. pallidosa genetic background by repeated backcrosses (Fig. 1). Twenty independent backcross lines followed by 5 to 10 generations were set up (see Materials and Methods). After these backcrosses, the flies examined for mating success were believed to have almost the complete D. pallidosa genetic background, except for the introgressed Dl region on the second chromosome. If the introgressed (Dl phenotype) and nonintrogressed (wild-type) females show different discrimination patterns, it is because of the differences in the introgressed region. Surprisingly, [Dl] females in all 20 backcross lines copulated significantly more with D. ananassae males than did [+] females (Table 2). Although some [+] females showed rather high mating success with D. ananassae males, the D. ananassae chromosome segment dramatically induced D. ananassae female discrimination in all of the lines that showed high mating success with D. ananassae males. As for the mating success with D. pallidosa males, the 20 lines performed in one of two different ways: either the [Dl] females copulated significantly less with D. pallidosa males than did [+] females (class 1; lines 1, 4, 9, and 14 in Table 2) or the [Dl] females copulated easily with D. pallidosa males as well as with [+] females (class 2; the other 16 lines in Table 2). These results suggested that loci controlling female discrimination in either direction are located close to Dl on the second chromosome.
Table 2.
Mating success (%) in repeated backcross females (N = 50, 5 replicates in each cross)
| Line no. | Female genotype | Male
|
|||
|---|---|---|---|---|---|
| D. ananassae | χ2 | D. pallidosa | χ2 | ||
| 1 | D1 | 90 | 39.89** | 65 | 4.24* |
| + | 25 | 85 | |||
| 2 | D1 | 65 | 20.92** | 70 | 0.85 |
| + | 18 | 80 | |||
| 3 | D1 | 78 | 15.60** | 64 | 0.04 |
| + | 36 | 68 | |||
| 4 | D1 | 80 | 25.16** | 50 | 4.81* |
| + | 28 | 73 | |||
| 5 | D1 | 80 | 42.13** | 54 | 0.16 |
| + | 13 | 60 | |||
| 6 | D1 | 90 | 60.84** | 75 | 0.56 |
| + | 10 | 83 | |||
| 7 | D1 | 90 | 60.84** | 80 | 0.85 |
| + | 10 | 70 | |||
| 8 | D1 | 90 | 60.84** | 80 | 0.85 |
| + | 10 | 70 | |||
| 9 | D1 | 60 | 30.93** | 35 | 8.00** |
| + | 5 | 65 | |||
| 10 | D1 | 85 | 28.49** | 65 | 0.08 |
| + | 30 | 70 | |||
| 11 | D1 | 80 | 28.39** | 80 | 0.22 |
| + | 25 | 75 | |||
| 12 | D1 | 84 | 31.68** | 76 | 2.89 |
| + | 26 | 58 | |||
| 13 | D1 | 82 | 22.69** | 75 | 0.15 |
| + | 33 | 70 | |||
| 14 | D1 | 85 | 40.26** | 40 | 25.32** |
| + | 20 | 90 | |||
| 15 | D1 | 40 | 3.85* | 65 | 0.05 |
| + | 20 | 65 | |||
| 16 | D1 | 95 | 67.35** | 75 | 0.26 |
| + | 10 | 70 | |||
| 17 | D1 | 70 | 23.27** | 80 | 0.85 |
| + | 20 | 70 | |||
| 18 | D1 | 90 | 41.34** | 68 | 0 |
| + | 23 | 70 | |||
| 19 | D1 | 88 | 51.86** | 85 | 4.21* |
| + | 15 | 66 | |||
| 20 | D1 | 63 | 16.59** | 65 | 1.38 |
| + | 20 | 78 | |||
*, P < 0.05, **, P < 0.001.
Discussion
The Roles of Female Discrimination in Sexual Isolation.
The present study strongly suggests that females discriminate on the basis of male courtship songs and that this discrimination is the main cause of sexual isolation between D. ananassae and D. pallidosa. Some song parameters are known to be important between these two species (29). In a related species, Drosophila biauraria, it is the difference in interpulse interval that elicits the female rejection behaviors (34).
Although D. ananassae females seemed to show no difference in mating success with respect to the presence of conspecific male wings (Fig. 2), the roles of conspecific male songs were masked by our mating-experimental design (10 males × 10 females, 2 h), which accentuated species differences in female discrimination. A different mating test (pair-mating in 30 min) revealed that winged males mated more successfully than wingless males (M.D. and Y.O., unpublished data).
The Locus for Female Discrimination.
Fig. 4 summarizes the results of the mating experiments and estimated map locations of the loci for female discrimination in the two species. D. ananassae and D. pallidosa females mate with conspecific males (Fig. 4 A and B). F1 hybrid females showed the same mating pattern as D. ananassae females (Fig. 4 A and C), because the D. ananassae female discrimination is dominant. That all of the [Dl] backcross females showed high mating success with D. ananassae males indicates that the locus controlling D. ananassae female discrimination and Dl did not segregate after 10 backcrosses, even though there are no chromosome inversions near Dl to prevent recombination (data not shown). Thus, this locus maps very near to the Dl locus on the second chromosome (Fig. 4 D and E). On the assumption of normal recombination, the introgressed region containing the discrimination locus and Dl can be estimated to be quite small. On the assumption of low recombination in hybrid background, however, the introgressed region may be larger than the size we supposed. If so, we may not conclude that the locus is located closely to Dl. Furthermore, several genes may be included in this introgressed region and control female discrimination. Although there are no data to suggest such a low recombination rate in our study, this possibility should be considered and further analyzed. Finding of additional molecular markers in these species could help to improve estimates of the size of the genetic region we introgressed. In both cases, our data indicate that the region linked with Dl contains the dominant gene (or genes) for female discrimination.
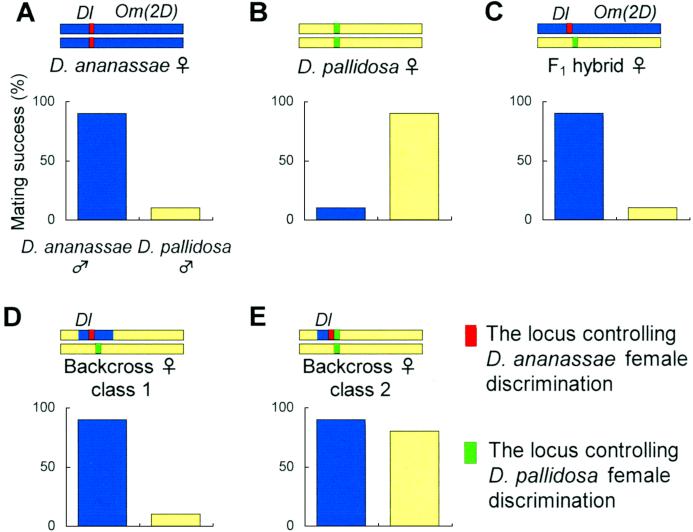
Figure 4.
Schematic model of the map position of the two loci controlling female discrimination in D. ananassae (red bar) and D. pallidosa (green bar). Upper bars on the graphs represent the genetic construction of the second chromosome, blue for the chromosome of D. ananassae and yellow for that of D. pallidosa. Other chromosomes are not shown. Graphs under the chromosomes indicate the mating success patterns in females having the second chromosome shown above. (A) Dl Om(2D) D. ananassae females. (B) Wild-type D. pallidosa females. (C) F1 hybrid females. (D) Repeated-backcross females (class 1). (E) Repeated backcross females (class 2).
Because the Dl mutation did not alter D. ananassae female discrimination (Table 1), we conclude that the high mating success with D. ananassae males in repeated-backcross females was not caused by the Dl mutation itself but rather by the introgressed segment. A possibility remains that the neurogenic gene, Dl, controls the Dl phenotype (35) itself and D. ananassae discrimination. Each species' Dl may have different roles in female discrimination. Although we cannot understand fully what kind of gene controls female discrimination until the gene is cloned, it is surprising to find that the female discrimination really is mapped in a single locus or a single cluster of some loci.
Female discrimination of D. pallidosa also was mapped close to Dl but is nonallelic to the discrimination of D. ananassae, because two types of mating patterns with D. pallidosa males were seen (Table 2). Class 1 backcross females probably have a somewhat longer introgressed D. ananassae chromosome (Fig. 4D), because of fewer backcrosses (five times in lines 9 and 14). Thus, the D. pallidosa locus was heterozygous and the D. pallidosa female type of discrimination was not seen. On the other hand, the occurrence of both female phenotypes in class 2 females may have been caused by introgression of a shorter chromosomal segment of D. ananassae (Fig. 4E). The intermediate phenotypes of D. pallidosa female discrimination (all lines except for 1, 4, 9, and 14 in Table 2) may result from a modification of the dominant D. ananassae factor by the homozygous D. pallidosa factor or vice versa, or from a longer introgression by low recombination, which means that these introgressed regions contain enough genes to show these phenotypes.
Significant effects on the third chromosome (Fig. 3) also indicate the existence of a possible locus (or loci) controlling mating success between the two species on this chromosome. However, the effects of these loci are much weaker than those on the second chromosome.
Sexual Isolation and Speciation in Different Drosophila Species Complexes.
This study maps a possible loci for female discrimination causing sexual isolation in an animal taxon. The introgressed Dl chromosomal region dramatically weakened the sexual isolation between D. ananassae and D. pallidosa. This result means that a small number of genes must have contributed to the reproductive isolation between them. Considering the facts that female discrimination is the main effect of this sexual isolation and that the only detected genetic barrier between the two species is sexual isolation, our findings suggest that the loci we mapped might have played a crucial role in the speciation process of these two species. If so, speciation could occur because of changes in a small number of genes. Interestingly, Ting et al. (36) reported a contrasting result with the present study by using two populations (Zimbabwean and cosmopolitan races) in Drosophila melanogaster. These two races show mating asymmetry and polymorphic sexual behaviors, suggesting that these two races are at an incipient stage of speciation (15, 37). Despite the limited molecular divergence between Zimbabwean and cosmopolitan races, four loci are responsible for male mating success and three others control female mating preference on one chromosome alone. In many cases of speciation, changes in a large number of loci might be required for population divergence, like the Zimbabwean and cosmopolitan races in D. melanogaster. A complex of loci seem to contribute to mating behavior in these populations—the great challenge is to clarify and quantify which signals, such as visual, acoustic, or olfactory, contribute to mating and which genes control such signals. The present study shows clearly that the locus close to Dl contributes specifically to female discrimination of courtship song and is effective for sexual isolation. In some speciation processes, like those in D. ananassae and D. pallidosa, a small number of loci with large effects might evolve rapidly to divide two populations. In the near term, molecular analysis of reproductive barriers in species where few loci seem to control these barriers, such as D. ananassae and D. pallidosa, might lead to rapid progress in our understanding of genetic mechanisms of speciation.
Acknowledgments
We thank Y. N. Tobari and Y. Tomimura for providing strains and helpful comments, and C.-I. Wu for communicating unpublished results and discussions. We also thank R. S. J. Weisburd, J.-M. Jallon, and D. R. J. Macer for their valuable advice and suggestions. This work was supported partly by the Ministry of Education, Science, Sports, and Culture of Japan, Grant-in-Aid for Scientific Research (C) (2) 09839005 (to Y.O.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Coyne J A. Nature (London) 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y-H, Ota T, Vacquier V D. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 3.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 4.Ting C-T, Tsaur S-C, Wu M-L, Wu C-I. Science. 1998;282:1501–1503. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw H D, Jr, Wilbert S M, Otto K G, Schemske D W. Nature (London) 1995;376:762–765. [Google Scholar]
- 6.Cabot E L, Davis A W, Johnson N A, Wu C-I. Genetics. 1994;137:175–189. doi: 10.1093/genetics/137.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis A W, Noonburg E, Wu C-I. Genetics. 1994;137:191–199. doi: 10.1093/genetics/137.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palopoli M, Wu C-I. Genetics. 1994;138:329–341. doi: 10.1093/genetics/138.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis A W, Wu C-I. Genetics. 1996;143:1287–1298. doi: 10.1093/genetics/143.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan C C. Genetics. 1946;31:558–573. doi: 10.1093/genetics/31.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zouros E. Genetics. 1981;97:703–718. doi: 10.1093/genetics/97.3-4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne J A. Proc Natl Acad Sci USA. 1989;86:5464–5468. doi: 10.1073/pnas.86.14.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyne J A, Crittenden A P, Mah K. Science. 1994;265:1461–1464. doi: 10.1126/science.8073292. [DOI] [PubMed] [Google Scholar]
- 14.Carracedo M C, Casares R P. Heredity. 1995;75:541–546. doi: 10.1038/hdy.1995.170. [DOI] [PubMed] [Google Scholar]
- 15.Wu C-I, Hollocher H, Begun D, Aquadro C A, Xu Y, Wu M-L. Proc Natl Acad Sci USA. 1995;92:2519–2523. doi: 10.1073/pnas.92.7.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne J A, Orr H A. Evolution (Lawrence, Kans) 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 17.Coyne J A. Evolution (Lawrence, Kans) 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- 18.Ferveur J-F. Genetics. 1991;128:293–301. doi: 10.1093/genetics/128.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferveur J-F, Jallon J-M. Genetics. 1993;133:561–567. doi: 10.1093/genetics/133.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall J C. Science. 1994;264:1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- 21.Stone W S, Wheeler M R, Wilson F D, Gerstenberg V R, Yang A H. Univ Texas Publ. 1966;6615:1–36. [Google Scholar]
- 22.Futch D G. Univ Texas Publ. 1966;6615:79–120. [Google Scholar]
- 23.Spieth H T. Univ Texas Publ. 1966;6615:133–145. [Google Scholar]
- 24.Futch D G. Evolution (Lawrence, Kans) 1973;27:456–467. doi: 10.1111/j.1558-5646.1973.tb00692.x. [DOI] [PubMed] [Google Scholar]
- 25.Tobari Y N. In: Drosophila ananassae: Genetical and Biological Aspects. Tobari Y, editor. Tokyo, Japan: Jap. Sci. Soc. Press; 1993. pp. 209–259. [Google Scholar]
- 26.Bock I R, Wheeler M R. Univ Texas Publ. 1972;7313:1–102. [Google Scholar]
- 27.Doi M, Nemoto T, Nakanishi H, Kuwahara Y, Oguma Y. J Chem Ecol. 1997;23:2067–2078. [Google Scholar]
- 28.Cobb M, Ferveur J-F. Behav Proc. 1996;36:35–54. doi: 10.1016/0376-6357(95)00052-6. [DOI] [PubMed] [Google Scholar]
- 29.Oguma Y. In: Drosophila ananassae: Genetical and Biological Aspects. Tobari Y, editor. Tokyo, Japan: Jap. Sci. Soc. Press; 1993. pp. 199–207. [Google Scholar]
- 30.Nemoto T, Doi M, Oshio K, Matsubayashi H, Oguma Y, Suzuki T, Kuwahara Y. J Chem Ecol. 1994;20:3019–3027. doi: 10.1007/BF02033708. [DOI] [PubMed] [Google Scholar]
- 31.Ewing A W. Biol Rev. 1983;58:278–292. [Google Scholar]
- 32.Eberl D F, Hardy R W, Kernan M J. J Neurosci. 2000;20:5981–5988. doi: 10.1523/JNEUROSCI.20-16-05981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson J R. Trends Genet. 1996;12:175–180. doi: 10.1016/0168-9525(96)10015-9. [DOI] [PubMed] [Google Scholar]
- 34.Tomaru M, Matsubayashi H, Oguma Y. Anim Behav. 1995;50:905–914. [Google Scholar]
- 35.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 36.Ting C-T, Takahashi A, Wu C-I. Proc Natl Acad Sci USA. 2001;98:6709–6713. doi: 10.1073/pnas.121418898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollocher H, Ting C-T, Wu M-L, Wu C-I. Genetics. 1997;147:1191–1201. doi: 10.1093/genetics/147.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]