Abstract
The methylation of intracisternal A-type particle (IAP) sequences is maintained during mouse embryogenesis. Methylation suppresses IAP expression and the potential for mutagenesis by retrotransposition, but it is not clear how methylation of these elements is maintained during the embryonic stages when the bulk of the genome is being demethylated. It has been suggested that the high levels of DNA methyltransferase-1 (Dnmt1) present during cleavage could be important for keeping IAPs methylated. To test this hypothesis, we combined mutant alleles of Dnmt1 with an agouti allele (Aiapy), which provided a coat color readout for the methylation status of the IAP insertion in the agouti locus. We found that reduction in Dnmt1 levels directly impacted methylation at this locus, leading to stable transcriptional activation of the agouti gene in the adult. Specifically, the short maternal Dnmt1 protein was important in maintaining methylation at the Aiapy locus in cleavage embryos, whereas the longer Dnmt1 isoform found in somatic cells was important in maintaining IAP methylation during the postimplantation stage. These results underscore the importance of maintaining proper maintenance of methylation patterns during gestation and suggest that interference with this process may stably affect gene expression patterns in the adult and may have profound phenotypic consequences.
Epigenetic marks such as genomic methylation have been shown to regulate gene expression and development (16, 21). Interference with proper establishment of methylation can result in a runted phenotype, tumorigenesis (7), and death (13, 22, 30). DNA methylation patterns established in the germ line during gametogenesis are largely erased in early embryogenesis and are reset after implantation. Patterns of methylation are then stably maintained through somatic cell divisions (21).
The various methyltransferases that regulate genomic methylation follow distinct expression patterns that are well coordinated in gametogenesis and embryogenesis (21). The establishment of new genomic methylation patterns in the differentiating gametes involves the de novo methyltransferases Dnmt3a and Dnmt3b and the Dnmt3-like (Dnmt3L) protein in the oocyte (1, 12). Dnmt3a and Dnmt3L are also expressed during male gametogenesis and are necessary for the completion of spermatogenesis (1, 12). After fertilization, the highly methylated paternal genome undergoes active demethylation in the male pronucleus (26, 31), whereas the maternal genome is only passively demethylated during the subsequent cleavage divisions (14, 17, 35). At the blastocyst stage, the bulk of the genome is hypomethylated except for alleles of imprinted genes and some repetitive elements which remain methylated (19, 34). High levels of a short form of Dnmt1 (Dnmt1o) are produced during oogenesis and are present in the cleavage embryo (27) but disappear after implantation. While this maternal store of enzyme may maintain the methylation of repetitive elements such as intracisternal A-type particles (IAPs), it is not clear why the bulk of the genome becomes demethylated. Dnmt1 is also expressed at high levels in spermatogonia and in leptotene spermatocytes but is not detectable in the nucleus of pachytene spermatocytes (27). This is consistent with a role for Dnmt1 in setting or maintaining paternal methylation patterns during the early stages of spermatogenesis. The somatic form of Dnmt1 mRNA (Dnmt1s) is transcribed postzygotically and, in concert with Dnmt3a and -b, has been shown to be responsible for the wave of global de novo methylation after implantation (22, 25, 30). All evidence suggests that the primary function of the longer somatic Dnmt1 isoform is to maintain global methylation after implantation of the embryo as the genome becomes remethylated by Dnmt3a and Dnmt3b. However, some de novo methylation activity has been observed in Dnmt3a−/−/3b−/− double knockout embryonic stem cells, suggesting that other methyltransferases may play a role in remethylation (24). In addition, it has been shown that methylation is important for maintaining the stability of the genome (3, 6, 7).
The genome of mice contains multiple copies of retrotransposable IAP elements, and it is well established that transposition of these elements can cause insertional inactivation as well as the ectopic activation of genes (10, 40). Also, it has been demonstrated that methylation of IAPs is an important silencing mechanism that suppresses activation and transposition of the elements (15, 45). The insertion of an IAP element into the agouti gene (designated as the Avy or Aiapy allele) has been shown to lead to ubiquitous ectopic activation of the gene. Hypomethylation of a cryptic promoter within the long terminal repeat (LTR) of the IAP drives constitutive ectopic expression of the agouti gene, leading to yellow coat color, obesity, and a high incidence of tumors (5, 28). In contrast, methylation of the IAP LTR silences the cryptic promoter and allows normal tissue-specific and regulated agouti expression, resulting in a “pseudoagouti” phenotype. The degree of yellow contribution to the fur provides an easy readout of the level of IAP methylation in the animal. The methylation state of an agouti IAP can be maternally transmitted to the next generation (a process termed transgenerational inheritance of epigenetic states) (29). For example, yellow Avy mothers are more likely to have yellow pups than pseudoagouti mothers. Insertion of an IAP element into another gene which regulates embryonic axis formation (axin) leads to maternal as well as paternal transmission of the methylation state of this IAP and an associated kinked-tail phenotype (33). Thus, changes in methylation patterns imposed on the genome during embryogenesis can be not only maintained in the adult but also transmitted to the next generation. It has been shown that maternal diet can affect the methylation state of the IAP element and can cause a shift in coat color distribution in progeny from Avy mothers (4, 44). This is an interesting result that emphasizes that environmental stimuli such as diet can profoundly affect the methylation state of the genome and result in gene expression states and phenotypic changes that are stable throughout life (16).
It has been established that methylation can suppress the expression of IAP elements (41), but the role of the specific Dnmt1 isoforms in this suppression has not yet been addressed. We tested the hypothesis that Dnmt1 plays a crucial role in the establishment and successful somatic propagation of these epigenetic states during the earliest stages of embryogenesis. For this, we examined the effect of Dnmt1 mutant alleles that generate lower Dnmt1 levels in the embryo on the expression of Aiapy in the adult. Our results suggest a role for the shorter oocyte-specific Dnmt1 isoform in maintaining IAP methylation in the cleavage-stage embryo, whereas the longer isoform found in the male gametes and somatic cells is important only after implantation. Our results further suggest that interference with the methylation machinery in early embryogenesis results in stable changes of IAP methylation and altered gene expression in the adult.
MATERIALS AND METHODS
Genotypes and breeding.
Four mutant alleles of Dnmt1 were used: the null allele (c allele [20]), the hypomorphic chip allele (39), and the 2lox and 1lox alleles (15). The Msx2Cre transgene contains the Cre recombinase cDNA under the control of the Msx2 regulatory sequences (37) and was always transmitted through the female germ line. The Aiapy allele contains an IAP insertion upstream of the agouti gene (28) and was always transmitted through the male germ line from pseudoagouti or near-pseudoagouti males.
Phenotype classification and statistics.
The overall degree of yellow fur contribution in Aiapy mice was determined visually. In all cases, the examiner was blind to the genotype. The mice were classified in four different groups: 0 to 10% yellow contribution (all or almost all brown), 15 to 50% contribution, 55 to 85% contribution, and 90 to 100% contribution (all or almost all yellow). The P values were obtained using the Mann-Whitney rank sum test.
DNA preparation and methylation assay.
Tissues were digested in lysis buffer (100 mM Tris HCl [pH 8.5], 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl, plus 100 μg of proteinase K/ml) overnight (o/n) at 55°C, phenol-chloroform extracted, and precipitated with an equal volume of isopropanol. A 10-μg aliquot of DNA was digested with the stated restriction endonuclease for 12 to 16 h. The methyl-sensitive restriction enzyme HpaII is blocked by methylation of its recognition sequence. The enzyme only cuts when the site is demethylated, yielding smaller DNA products (see Fig. 3A). The products were resolved on an agarose gel, transferred to nylon membranes (Genescreen), and hybridized in Church buffer (0.5 M NaPO4 [pH 7.5], 7% SDS, 2 mM EDTA) with a radioactively labeled probe at 65°C o/n. Probe 1 was generated by PCR amplification from mouse genomic DNA using the following primers (5′ to 3′): F, GCTTCTCAGGATGGATGTCA; R, GCCCCAGTTTCATCACTTGT, yielding a 736-bp fragment. The probe was synthesized with random hexamers. Final wash was done with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS at 65°C.
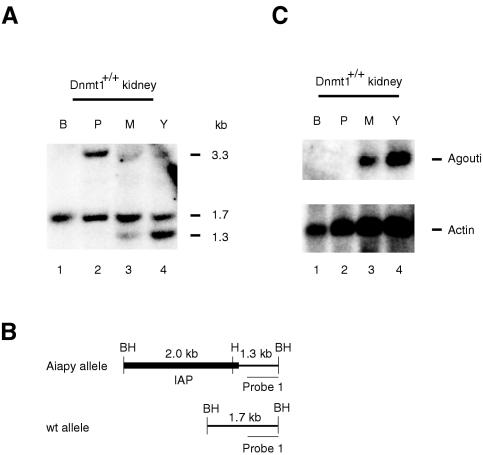
FIG. 3.
Methylation status of the IAP allele correlates with agouti expression and coat color. (A) Southern blot analysis of kidney tissues from black (B), pseudoagouti (P), mottled (M), and yellow (Y) animals. The methylation-sensitive restriction enzyme HpaII (H) in combination with BamHI (BH) was used to detect the methylation status of the 5′ LTR of the IAP. The wild-type allele does not contain a HpaII site yielding a fragment of 1.7 kb (see schematic diagram in panel B). Depending on the methylation status of the 5′ LTR, the Aiapy allele will yield a 1.3-kb fragment if unmethylated or a 3.3-kb fragment if methylated. Because the mice were heterozygous for the Aiapy allele, a wild-type 1.7-kb fragment was observed in all samples. All three fragments are indicated on the right side of the panel. A clear correlation between Aiapy hypomethylation and coat color can be observed. (B) Schematic diagram of the fragment sizes predicted from the Aiapy and wild-type alleles. (C) Northern blot analysis of total RNA from black (B), pseudoagouti (P), mottled (M), and yellow (Y) mice hybridized to an agouti probe. A clear correlation between methylation (panel A) and expression at the 5′ LTR of the IAP can be seen. The agouti mRNA is 692 bases. As a loading control, an actin probe was used on the same blot (lower panel).
Northern analysis.
RNA was isolated using TRIzol (Gibco), separated on a formaldehyde-agarose gel, and transferred to a nylon membrane (Genescreen). Hybridization was carried out at 65°C o/n in Church buffer. The agouti probe (500 bp) was generated by PCR using the primers (5′ to 3′) F3 (CCACCCTAGTGAGCTTCCTG) and R2 (GCCCACAAGTCACAACCAC) and reverse-transcribed liver RNA from yellow mice. The agouti probe was radioactively labeled and synthesized with random hexamers.
Western analysis.
Female mice were hormone primed with 5 IU of pregnant mare serum gonadotropin (Calbiochem) intraperitoneally followed 48 h later by 5 IU of recombinant human chorionic gonadotropin (Calbiochem) intraperitoneally. Eggs were harvested from the oviducts 24 h after human chorionic gonadotropin treatment. Ten eggs of each genotype were lysed in sample buffer (2% SDS, 100 mM dithiothreitol, 60 mM Tris [pH 6.8], 0.01% bromophenol blue), boiled, and loaded on an SDS-8% polyacrylamide electrophoresis gel. The gel was blotted on a nitrocellulose membrane (catalog no. RPN2020D; Amersham Pharmacia Biotech). A chicken C-terminal Dnmt1 antibody was used at 1/1,000 dilution (8), and detection was performed with the chemiluminescence method (ECL; Amersham Pharmacia Biotech).
RESULTS
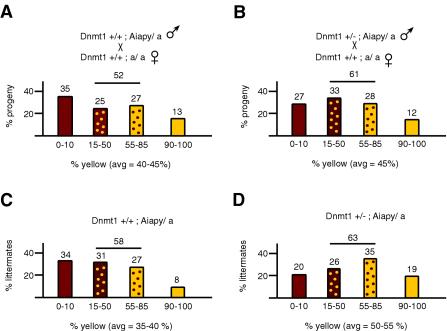
To investigate the effect of reduced Dnmt1 levels in embryos on the methylation of paternally transmitted Aiapy alleles, we determined by visual examination the coat color distribution in offspring from Dnmt1+/+; Aiapy/a and Dnmt1 +/−; Aiapy/a males bred to wild-type Dnmt1+/+; a/a females. A mutation in the agouti locus results in no detectable agouti expression and a recessive black hair phenotype. Expression of agouti from the Aiapy allele is dominant and gives coat colors ranging from brown (agouti) to yellow, depending on the level of agouti expression. To exclude transgenerational inheritance at the agouti locus in our experiments, we transmitted the Aiapy allele through the paternal germ line and always used males that were pseudoagouti or with a very high pseudoagouti contribution. Coat colors were broken down into four categories: agouti (0 to 10% yellow coat color), mottled (15 to 50% and 55 to 85% yellow coat color), and yellow (90 to 100% yellow coat color) (Fig. 1 and 2). The distribution of coat color in the progeny from Dnmt1+/+ fathers was 35% agouti and 13% yellow (the remainder being mottled) (Fig. 2A). The distribution of coat color in progeny from Dnmt1+/− fathers containing 50% less Dnmt1 than wild type (20) showed that 27% of the progeny was agouti and 12% was yellow (Fig. 2B). Thus, reduction of Dnmt1 levels by half in the male germ line did not significantly (P = 0.8) affect the overall coat color distribution in the offspring. However, when the coat color of the offspring was separated by genotype, we noted that 20% of the Dnmt1+/− animals were agouti and 19% were yellow (average yellow color = 50 to 55%) (Fig. 2D) compared to Dnmt1+/+ animals, which were 34% agouti and 8% yellow, a difference that was significant (average yellow color = 35 to 40%; P = 0.02) (Fig. 2C). Specifically, the Dnmt1+/− progeny were about half as likely to be agouti and twice as likely to be yellow compared to Dnmt1+/+ siblings. The coat color of Dnmt1+/+ progeny from Dnmt1+/− fathers (Fig. 2C) showed a similar distribution as progeny from Dnmt1+/+ fathers (Fig. 2A), suggesting that reducing the levels of Dnmt1 in the male differentiating gametes did not affect coat color distribution in the progeny. Thus, the apparent shift of the Dnmt1+/− progeny average contribution toward yellow likely reflected the loss of methylation in somatic cells. To confirm that the coat color of our animals correlated with the methylation status of the agouti IAP LTR, as would be expected (28), we determined the extent of methylation at that locus in organs from Aiapy agouti, mottled, and yellow animals. We found that the methylation of the Aiapy insertion correlated with coat color and IAP-driven agouti RNA expression (Fig. 3).
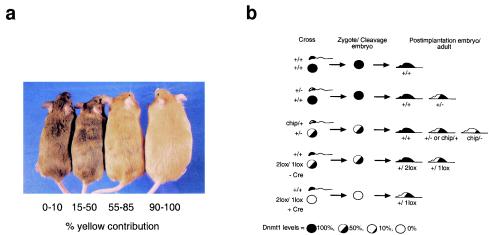
FIG. 1.
Coat color of Aiapy mice. (a) Aiapy siblings show a wide degree of expressivity of the agouti gene, resulting in coat colors ranging from pseudoagouti (left) to completely yellow (right). Constitutive expression of the agouti gene also correlates with an obesity phenotype, which can be seen in the solid yellow mouse (28). (b) Relative levels of Dnmt1 in parental gametes and progeny from the crosses performed. The strength of the Dnmt1 mutant alleles relative to wild type (set at 100%) is as follows: Dnmt1+/2lox (100%), Dnmt1chip/+ (60%), Dnmt1+/− (50%), Dnmt1+/1lox (50%), Dnmt12lox/1lox (50%), and Dnmt1chip/− (10%). Wild-type levels of Dnmt1 are indicated by a solid black color. For example, Dnmt1+/− mice, which contain only 50% of wild-type Dnmt1 levels, are depicted as 50% black. The genotypes of the gametes represent the genotype of the parental animals, as gametes are haploid and can carry either allele. The levels of Dnmt1 depicted in the sperm represent expected premeiosis levels, since mature sperm does not contain detectable Dnmt1. Those levels indicate the degree of Dnmt1 levels that the male gamete eventually contributes to the implantation embryo and adult when Dnmt1 expression is turned on. Dnmt1+/− and Dnmt1chip/+ mice are depicted by a 50% black mouse for simplicity but contain, respectively, 50 and 60% of wild-type levels of Dnmt1.
FIG. 2.
Distribution of coat color of Aiapy progeny. All results are expressed in terms of the percentage of yellow coat color (x axis). The degree of yellow expression was scored in four different ranges of yellow contribution: 0 to 10%, 15 to 50%, 55 to 85%, and 90 to 100%. The y axis represents the percentage of the progeny which fell in a specific range and is indicated above each group. The total percentage of mottled contribution is indicated over the mottled ranges and represents the percentage of progeny falling in the 15 to 85% range. The average yellow contribution of each panel is indicated and is labeled “avg.” The average determined was the arithmetic average, specifically, the sum of the estimated yellow contribution of each mouse divided by the number of mice. Coat color distribution are shown for progeny from fathers containing wild-type levels of Dnmt1 (A) or heterozygous levels of Dnmt1 (B) bred to C57BL/6 female mice. A wide range of coat color contribution is evident. The distribution in panel B was broken down according to whether the mice were Dnmt1+/+ (C) or Dnmt1+/− (D). Sample sizes (n) were 96 (A), 133 (B), 60 (C), and 73 (D). The genetic background of all mice was C57BL/6.
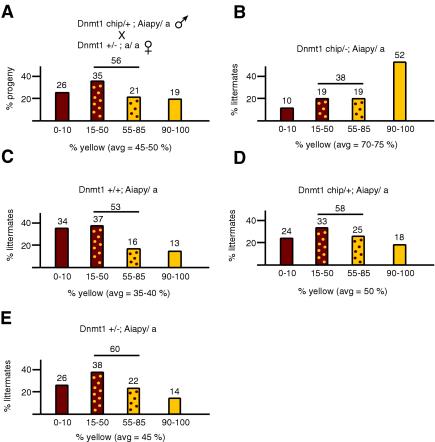
If the loss of methylation at the Aiapy locus in somatic cells were directly linked to the level of Dnmt1, further reduction of Dnmt1 levels would be predicted to result in a more significant increase in yellow progeny. To test this hypothesis, we crossed males containing a hypomorphic allele of Dnmt1 (Dnmt1chip) and the Aiapy allele with C57BL/6 females heterozygous for the Dnmt1 null allele to generate compound heterozygous Dnmt1chip/− mice, which express 10% of wild-type levels of Dnmt1 and are globally hypomethylated (7). Progeny from this cross had a coat color distribution of 26% agouti and 19% yellow (average yellow contribution = 45 to 50%) (Fig. 4A), a result similar to what was observed with Dnmt1+/− fathers and wild-type mothers (Fig. 2B). However, when the genotype of the offspring was taken into account, the distribution of coat color showed that the hypomethylated Dnmt1chip/− animals were on average more yellow than their wild-type or Dnmt1 heterozygous siblings (Fig. 4B to E), consistent with Dnmt1chip/− mice being more hypomethylated. Specifically, 10% of Dnmt1chip/− animals were in the 0 to 10% agouti group and 52% were 90 to 100% yellow (average yellow contribution = 70 to 75%; P = 0.001). This cross yielded three additional genotypes, Dnmt1+/+, Dnmt1chip/+, and Dnmt1+/−. As shown in Fig. 4C, the wild-type progeny had a coat color distribution similar to that in the control experiment shown in Fig. 2A, suggesting that halving the maternal store of Dnmt1 in early embryos did not have an effect on the paternally transmitted Aiapy. The Dnmt1chip/+ and Dnmt1+/− mice had a slight bias towards the yellow color in comparison to wild-type progeny (50 and 45% average yellow color, respectively; Fig. 4D and E). These results are reminiscent of the Dnmt1 heterozygotes in Fig. 2D, which also displayed a slight yellow bias. Thus, our results suggest that reduction of the longer Dnmt1 isoform levels affects the methylation of the agouti locus in embryonic cells during postimplantation and leads to a loss of Aiapy methylation.
FIG. 4.
Coat color distribution from parents containing low levels of Dnmt1. The progeny resulting from the cross in panel A was plotted according to the respective genotypes (B to E). Sample sizes (n) were 155 (A), 21 (B), 38 (C), 51 (D), and 45 (E). A clear switch to yellow coat color can be seen in Dnmt1chip/− animals (B). The genetic background of the mice was mostly C57BL/6 but contained small amounts of 129 and BALB/c, which came from the Dnmt1chip allele.
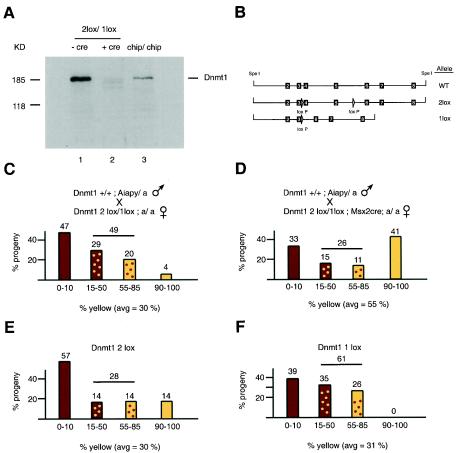
The oocyte contains a large store of a shorter Dnmt1 isoform which is present during the cleavage stage of embryogenesis. To test whether this isoform plays a role in maintaining IAP methylation during that stage, we sought to reduce the levels of maternal Dnmt1 in oocytes to ensure minimal contribution of that protein to the early embryo. To do this, we used females containing a conditional allele of Dnmt1 (Dnmt12lox [15]) combined with a null allele (Dnmt11lox) and a Cre recombinase transgene driven by the Msx2 promoter (37, 23), which expresses Cre in the oocytes. The Dnmt12lox allele functions normally in vivo and produces wild-type levels of Dnmt1 (15). The Cre-mediated recombination of this allele in oocytes deletes exons 4 and 5, which contain the translational start site of the oocyte-specific form of Dnmt1. The effect of this rearrangement of the Dnmt1 locus in reducing Dnmt1 levels in oocyte was measured by immunoblotting of oocytes from hormone-primed Dnmt12lox/1lox Msx2Cre females (Fig. 5A). Levels of the oocyte-specific Dnmt1 isoform were found to be much reduced in Dnmt12lox/1lox oocytes containing Msx2Cre compared to Dnmt12lox/1lox oocytes that did not contain Msx2Cre. Lower levels of smaller proteins were also detected in the Msx2Cre oocytes and may have represented products made from downstream translational initiation. Similar products were also observed in cells containing an N-terminal knockout of the Dnmt1 gene in which the translational start site for the oocyte-specific form was deleted and resulted in an embryonic lethal phenotype in homozygous mutant animals (Dnmt1n allele) (22).
FIG. 5.
Coat color distribution of progeny from mothers with reduced Dnmt1o. (A) Immunoblot of 10 oocytes from Dnmt12lox/1lox; a/a females without Msx2Cre (lane 1) or with Msx2Cre (lane 2) and from females homozygous for the hypomorphic Dnmt1chip allele (lane 3). A C-terminal Dnmt1 antibody was used. The predicted size of the oocyte form is 170 kDa. (B) Schematic diagram of the Dnmt12lox (2lox) and Dnmt11lox (1lox) alleles compared to the wild-type allele (WT). The exons are indicated by the black boxes and corresponding exon numbers, and the loxP sites are indicated by arrowheads. The 1lox allele contains a deletion of genomic sequences that includes exons 4 and 5. (C and D) Coat color distribution of progeny from Dnmt1+/+; Aiapy/a fathers and Dnmt12lox/1lox; a/a mothers without Msx2Cre (C) or heterozygous for Msx2Cre (D). Sample sizes (n) were 45 (C) and 27 (D); P = 0.004. The background of the mice was mostly C57BL/6 but contained small contributions from FVB and 129, which came from the 2lox, 1lox, and Msx2Cre alleles. (E and F) The progeny in panel C were broken down according to whether they were Dnmt12lox (E) or Dnmt11lox (F).
To test the effect of low maternal Dnmt1 levels on agouti expression in Aiapy progeny, we crossed Dnmt1+/+; Aiapy/a males with black Dnmt12lox/1 lox; a/a females carrying or not carrying the Msx2Cre transgene (Fig. 5C and D). All Dnmt12lox/1lox females containing Cre transgenes transmitted a recombined 1lox allele, showing that the Cre-mediated recombination in the oocyte was very efficient (data not shown). The litter size from the Msx2Cre-carrying mothers was normal (eight on average). These results contrasted with those of the knockout of the maternal store of Dnmt1 by deletion of the oocyte-specific promoter, which resulted in embryonic death in most fetuses during the last third of gestation (13), a difference which could be explained by the presence of low levels of truncated Dnmt1 in our experiment that might have provided residual activity and rescued the null phenotype. Interestingly, Msx2Cre transgenic females generated litters that were more yellow on average than the litters from females that didn't carry the Msx2Cre transgene. Specifically, progeny from Msx2Cre mothers were 33% agouti and 41% yellow (Fig. 5D; average yellow contribution = 55%) compared to 47% agouti and 4% yellow (Fig. 5C; average yellow contribution = 30%) when the mothers did not contain the Msx2Cre allele, a difference that was statistically significant (P = 0.004). To confirm that the results obtained in Fig. 5C were not biased by the Dnmt1 genotype of the progeny, we plotted the pups in Fig. 5C according to whether they were Dnmt12lox/+ or Dnmt11lox/+ (Fig. 5E and F). The results showed that the color distribution between these two genotypes was similar (P = 0.9) and suggested that the effect seen in Fig. 5D was not simply the result of lower Dnmt1 levels in somatic cells (Dnmt11lox/+). Thus, in addition to maintaining imprinted gene methylation in early embryos (13), the maternal Dnmt1 may also play a role in maintaining IAP methylation.
DISCUSSION
In this work we have studied the role of Dnmt1 in the methylation and expression of IAP elements. In normal development, high levels of Dnmt1 are present during embryogenesis. We have investigated whether the level of Dnmt1 expressed during cleavage and after implantation is important for the maintenance of IAP methylation. Using coat color as the readout, we found that the level of Dnmt1 expression was important for the maintenance of IAP methylation during cleavage and early developmental stages. IAP methylation patterns established during early development are maintained throughout life and can profoundly affect gene expression and phenotype in the postnatal animal.
The most profound shift in coat color was seen when Dnmt1o expression was inhibited during oogenesis. This suggests that the maternal store of Dnmt1o not only maintains monoallelic methylation of imprinted genes but is also crucial for the maintenance of IAP methylation. It has been shown that Dnmt1o is present mostly in the cytoplasm during early embryogenesis but enters the nucleus at the eight-cell stage and immediately exits (2). Thus, Dnmt1o may play a role in keeping IAPs silent solely during this stage. Alternatively, it is possible that Dnmt1o is present in the nucleus throughout early development in levels that are below detection by fluorescence in situ hybridization but sufficient to maintain IAP silencing. Large amounts of Dnmt1o may be briefly ferried to the nucleus to resupply existing Dnmt1o levels. A previous study where Dnmt1o was deleted by genetic inactivation of the Dnmt1o promoter showed that demethylation did not occur at IAPs (13). However, in contrast to our experiments which analyzed a single IAP element, that previous study investigated the large number of IAP elements carried in the mouse genome. It is possible that Dnmt1o maintains methylation levels of only some but not most IAP elements. Additionally, the progeny described here is heterozygous for Dnmt1 (Dnmt1+/1lox), in contrast with previous experiments in which the mice contained wild-type levels of somatic Dnmt1 (13). The lower expression of the somatic form of Dnmt1 in Dnmt1+/− pups resulted in a slight increase in the fraction of mice with a yellow coat, whereas the reduction of the Dnmt1s level to 10%, as in Dnmt1chip/− mice, substantially increased the shift to yellow coat color. These observations suggest that the level of somatic Dnmt1 is also important for the maintenance of IAP methylation. Because Dnmt1s is not translated during cleavage (27), it likely functions after implantation, when Dnmt1o levels decrease. Although reduction of Dnmt1 levels in embryos resulted in yellower mice on average, some mice with Dnmt1o deleted or with 10% somatic Dnmt1 were completely agouti, indicating methylation at Aiapy in all cells of the animal. Thus, reduction of Dnmt1 levels in embryos does not always cause IAP demethylation and suggests that the loss of methylation at Aiapy is a stochastic event. The final color or degree of mottledness of the coat is likely a result of the existing IAP methylation state combined with the probabilistic event of losing additional CpG methylation during cleavage or after implantation as a result of lowered Dnmt1 levels.
In wild-type mice, IAPs are resistant to demethylation throughout embryonic development (19). Methylation of IAP in cleavage embryos is probably important in preventing IAP expression, which could lead to retrotransposition and insertional mutagenesis (41). Our results show that the shorter Dnmt1 isoform is important in keeping the IAPs silent during cleavage development. Changes of IAP methylation during embryogenesis are faithfully maintained in tissues of the adult, as evidenced by the direct correlation between coat color and the overall level of Aiapy methylation in the liver and kidney (Fig. 3 and data not shown). Proper establishment of these embryonic patterns of methylation are crucial in setting up chromatin states following remethylation of DNA during postimplantation development (11). Because structural patterns of chromatin established during this embryonic stage are maintained in the adult, changes of methylation prior to chromatin assembly may have long-lasting effects impacting all subsequent cell lineages.
Stochastic changes in methylation patterns in early embryogenesis induced by dietary or other factors can have far-reaching effects in adulthood or in the next generation, such as the determination of coat color of Avy mice or the kinked-tail phenotype of AxinFu mice (29, 42, 44). Such modifications in genomic methylation patterns may lead to stable changes in gene expression that may have phenotypic consequences. For instance, the methylation and expression of the reelin gene, whose disruption leads to a phenotype similar to schizophrenia, has been shown to be influenced by an l-methionine-supplemented diet (38). It has also been suggested that cancer and a number of diseases such as multiple sclerosis, rheumatoid arthritis, and diabetes may have an epigenetic basis (32). The expression of disease genes could be affected by methylation directly, like the reelin gene, or indirectly by interference from nearby retroviral-like elements, such as the Aiapy allele in mice. Sequencing of the mouse and human genomes has revealed that a large proportion of their sequences, 37 and 42%, respectively, is derived from retrotransposable elements (18, 43) which are normally silenced by methylation. In humans, long interspersed transposable elements can alter the expression of neighboring genes (36) and short interspersed transposable elements are excluded from imprinted regions in the genome (9), suggesting that monoallelic expression of imprinted genes may be more vulnerable to methylation-mediated chromatin silencing occurring in short interspersed transposable elements. Our observations are consistent with the notion that changes of the epigenetic state of the genome can be induced early in development by genetic and environmental conditions. Such changes can have profound consequences for gene expression in the adult and for disease manifestation (16).
Acknowledgments
We thank R. Flannery for help with the mouse colony and K. Hong, J. Gribnau, and K. Eggan for helpful discussion. We also wish to thank E. Michaud for providing the Aiapy mouse strain and G. Martin for providing the Msx2Cre mice.
This work was supported by grant no. 5-R01-CA87869 from the National Institutes of Health.
REFERENCES
- 1.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 2.Carlson, L. L., A. W. Page, and T. H. Bestor. 1992. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 6:2536-2541. [DOI] [PubMed] [Google Scholar]
- 3.Chen, R. Z., U. Pettersson, C. Beard, L. Jackson-Grusby, and R. Jaenisch. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395:89-93. [DOI] [PubMed] [Google Scholar]
- 4.Cooney, C. A., A. A. Dave, and G. L. Wolff. 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132:2393S-2400S. [DOI] [PubMed] [Google Scholar]
- 5.Duhl, D. M., H. Vrieling, K. A. Miller, G. L. Wolff, and G. S. Barsh. 1994. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8:59-65. [DOI] [PubMed] [Google Scholar]
- 6.Eden, A., F. Gaudet, A. Waghmare, and R. Jaenisch. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:455. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet, F., J. G. Hodgson, A. Eden, L. Jackson-Grusby, J. Dausman, J. W. Gray, H. Leonhardt, and R. Jaenisch. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489-492. [DOI] [PubMed] [Google Scholar]
- 8.Gaudet, F., D. Talbot, H. Leonhardt, and R. Jaenisch. 1998. A short DNA methyltransferase isoform restores methylation in vivo. J. Biol. Chem. 273:32725-32729. [DOI] [PubMed] [Google Scholar]
- 9.Greally, J. M. 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. USA 99:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gwynn, B., K. Lueders, M. S. Sands, and E. H. Birkenmeier. 1998. Intracisternal A-particle element transposition into the murine β-glucuronidase gene correlates with loss of enzyme activity: a new model for β-glucuronidase deficiency in the C3H mouse. Mol. Cell. Biol. 18:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimshony, T., J. Zhang, I. Keshet, M. Bustin, and H. Cedar. 2003. The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 34:187-192. [DOI] [PubMed] [Google Scholar]
- 12.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 13.Howell, C. Y., T. H. Bestor, F. Ding, K. E. Latham, C. Mertineit, J. M. Trasler, and J. R. Chaillet. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104:829-838. [DOI] [PubMed] [Google Scholar]
- 14.Howlett, S. K., and W. Reik. 1991. Methylation levels of maternal and paternal genomes during preimplantation development. Development 113:119-127. [DOI] [PubMed] [Google Scholar]
- 15.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 16.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.):245-254. [DOI] [PubMed] [Google Scholar]
- 17.Kafri, T., M. Ariel, M. Brandeis, R. Shemer, L. Urven, J. McCarrey, H. Cedar, and A. Razin. 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 6:705-714. [DOI] [PubMed] [Google Scholar]
- 18.Landers, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 19.Lane, N., W. Dean, S. Erhardt, P. Hajkova, A. Surani, J. Walter, and W. Reik. 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35:88-93. [DOI] [PubMed] [Google Scholar]
- 20.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 21.Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3:662-673. [DOI] [PubMed] [Google Scholar]
- 22.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y. H., L. Ma, L. Y. Wu, W. Luo, R. Kundu, F. Sangiorgi, M. L. Snead, and R. Maxson. 1994. Regulation of the Msx2 homeobox gene during mouse embryogenesis: a transgene with 439 bp of 5′ flanking sequence is expressed exclusively in the apical ectodermal ridge of the developing limb. Mech. Dev. 48:187-197. [DOI] [PubMed] [Google Scholar]
- 24.Lorincz, M. C., D. Schubeler, S. R. Hutchinson, D. R. Dickerson, and M. Groudine. 2002. DNA methylation density influences the stability of an epigenetic imprint and Dnmt3a/b-independent de novo methylation. Mol. Cell. Biol. 22:7572-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyko, F., B. H. Ramsahoye, H. Kashevsky, M. Tudor, M. A. Mastrangelo, T. L. Orr-Weaver, and R. Jaenisch. 1999. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 23:363-366. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, W., A. Niveleau, J. Walter, R. Fundele, and T. Haaf. 2000. Demethylation of the zygotic paternal genome. Nature 403:501-502. [DOI] [PubMed] [Google Scholar]
- 27.Mertineit, C., J. A. Yoder, T. Taketo, D. W. Laird, J. M. Trasler, and T. H. Bestor. 1998. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development 125:889-897. [DOI] [PubMed] [Google Scholar]
- 28.Michaud, E. J., M. J. van Vugt, S. J. Bultman, H. O. Sweet, M. T. Davisson, and R. P. Woychik. 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 8:1463-1472. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, H. D., H. G. Sutherland, D. I. Martin, and E. Whitelaw. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23:314-318. [DOI] [PubMed] [Google Scholar]
- 30.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 31.Oswald, J., S. Engemann, N. Lane, W. Mayer, A. Olek, R. Fundele, W. Dean, W. Reik, and J. Walter. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10:475-478. [DOI] [PubMed] [Google Scholar]
- 32.Petronis, A. 2001. Human morbid genetics revisited: relevance of epigenetics. Trends Genet. 17:142-146. [DOI] [PubMed] [Google Scholar]
- 33.Rakyan, V. K., S. Chong, M. E. Champ, P. C. Cuthbert, H. D. Morgan, K. V. Luu, and E. Whitelaw. 2003. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 100:2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reik, W., and J. Walter. 2001. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2:21-32. [DOI] [PubMed] [Google Scholar]
- 35.Rougier, N., D. Bourc'his, D. M. Gomes, A. Niveleau, M. Plachot, A. Paldi, and E. Viegas-Pequignot. 1998. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 12:2108-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speek, M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21:1973-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, X., M. Lewandoski, E. N. Meyers, Y. H. Liu, R. E. Maxson, Jr., and G. R. Martin. 2000. Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat. Genet. 25:83-86. [DOI] [PubMed] [Google Scholar]
- 38.Tremolizzo, L., G. Carboni, W. B. Ruzicka, C. P. Mitchell, I. Sugaya, P. Tueting, R. Sharma, D. R. Grayson, E. Costa, and A. Guidotti. 2002. An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc. Natl. Acad. Sci. USA 99:17095-17100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker, K. L., C. Beard, J. Dausmann, L. Jackson-Grusby, P. W. Laird, H. Lei, E. Li, and R. Jaenisch. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10:1008-1020. [DOI] [PubMed] [Google Scholar]
- 40.Ukai, H., H. Ishii-Oba, M. Ukai-Tadenuma, T. Ogiu, and H. Tsuji. 2003. Formation of an active form of the interleukin-2/15 receptor beta-chain by insertion of the intracisternal A particle in a radiation-induced mouse thymic lymphoma and its role in tumorigenesis. Mol. Carcinog. 37:110-119. [DOI] [PubMed] [Google Scholar]
- 41.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 42.Waterland, R. A., and R. L. Jirtle. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterston, R. H., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 44.Wolff, G. L., R. L. Kodell, S. R. Moore, and C. A. Cooney. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12:949-957. [PubMed] [Google Scholar]
- 45.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]