Abstract
Recent findings indicate that in addition to its location in the peripheral plasma membrane, H-Ras is found in endomembranes like the endoplasmic reticulum and the Golgi complex. In these locations H-Ras is functional and can efficiently engage downstream effectors, but little is known about how its activation is regulated in these environments. Here we show that the RasGRF family exchange factors, both endogenous and ectopically expressed, are present in the endoplasmic reticulum but not in the Golgi complex. With the aid of H-Ras constructs specifically tethered to the plasma membrane, endoplasmic reticulum, and Golgi complex, we demonstrate that RasGRF1 and RasGRF2 can activate plasma membrane and reticular, but not Golgi-associated, H-Ras. We also show that RasGRF DH domain is required for the activation of H-Ras in the endoplasmic reticulum but not in the plasma membrane. Furthermore, we demonstrate that RasGRF mediation favors the activation of reticular H-Ras by lysophosphatidic acid treatment whereas plasma membrane H-Ras is made more responsive to stimulation by ionomycin. Overall, our results provide the initial insights into the regulation of H-Ras activation in the endoplasmic reticulum.
Ras family GTPases, H-Ras, N-Ras, and K-Ras 4B/4A, are key mediators in signaling pathways that convey extracellular signals from surface receptors to the interior of the cell, functioning as molecular switches in essential cellular processes (14, 33). It is well known that in order to be functional, Ras proteins must be attached to the inner leaflet of the plasma membrane (PM) (56). This is accomplished by posttranslational lipidic additions that take place in the C terminus, within a segment that has been termed the heterogeneous or hypervariable region (32). This region contains the essential signal for localizing Ras to membranes: the CAAX box (where C is cysteine, A is an aliphatic amino acid, and X is serine or methionine). The CAAX box undergoes a posttranslational modification that makes it more hydrophobic. The cysteine is farnesylated, the AAX sequence is proteolysed, and the newly C-terminal cysteine is carboxymethylated (32). A second signal is required for efficiently positioning Ras in the membrane. This is accomplished by palmitoylation of cysteine 181 in N-Ras and cysteines 181 and 184 in H-Ras. In K-Ras4B, the second signal consists of a polybasic motif of six lysines (positions 175 to 180) that is thought to interact electrostatically with the negatively charged membrane (24-26).
New findings indicate that the three Ras isoforms are located in distinct PM microdomains with different biochemical and physical-chemical characteristics (35, 42). Recently, it was demonstrated that the information required for accurate membrane localization is contained within the hypervariable region (29, 55), which also dictates how Ras proteins traffic to their destinations. Whereas H- and N-Ras traffic to the PM along the secretory pathway through the Golgi complex, K-Ras4B is directly routed from the endoplasmic reticulum (ER) to the PM by still unknown mechanisms (2, 12). Lately, a new twist has been provided by reports indicating that, in addition to its location in the PM, H-Ras is present in endomembrane systems such as endosomes, ER, and the Golgi complex (11, 44). The presence of H-Ras in these intracellular locations seems not to be a transient event associated with the transport and/or recycling of H-Ras proteins to and from the PM; instead, a pool of H-Ras appears to permanently reside in these organelles. More importantly, in these endomembranes H-Ras is active and can signal to downstream effectors like the Raf/ERKs pathway (11, 12, 44). However, to date, little is known about the mechanistics and the identity of the proteins involved in the regulation of H-Ras in endomembranes.
RasGRF1 and RasGRF2 are guanine nucleotide exchange factors (GEFs) that display a high selectivity for H-Ras in vivo (30, 35). In mammalians, RasGRF1 is expressed at high levels in the brain, in particular in the hippocampus (34, 54), although traces can also be detected in other tissues (23). RasGRF2, although expressed at high levels in the central nervous system, exhibits a more widespread expression pattern (18). The primary structures of these GEFs reveal a number of motifs presumably involved in diverse regulatory mechanisms. These include a Dbl homology (DH) domain, present mainly in GEFs for the Rho family GTPases. The DH domain is flanked by two pleckstrin homology domains of largely unknown function, although they are suggested to play a role in targeting mechanisms (46). Regarding their regulation, it has been shown that RasGRF1 and RasGRF2 are activated by G-protein-coupled receptors but are largely insensible to receptors of the tyrosine kinase type (18, 36, 48, 59). Augmentation of intracellular calcium levels by calcium ionophores can also bring about the activation of RasGRF1 and RasGRF2. This is achieved by a mechanism mediated through a calmodulin-binding isoleucine-glutamine (IQ) domain present in the N terminus of this GEF (7, 18-20; R. E. Cheney and M. S. Mooseker, Abstract, Mol. Biol. Cell 5:21a, 1994). Furthermore, we have recently demonstrated that the ability of RasGRF1 to activate Ras is regulated by the Rho family GTPase Cdc42, by a mechanism that entails the translocation of RasGRF1 to the cell particulate fraction (3, 4).
Since RasGRF family GEFs can be activated by stimuli mediated by the G-protein-coupled-type cell surface receptors and also by stimuli that provoke the release of calcium from intracellular stores like the ER, these GEFs possess the functional characteristics that turn them into ideal candidates for regulating the activation of H-Ras in this endomembrane. In this study we have addressed this hypothesis. Herein, we provide evidence indicating that RasGRF family GEFs colocalize with H-Ras in the ER but not in the Golgi complex. We demonstrate that Ras-GRF1 and RasGRF2 can efficiently induce nucleotide exchange on reticular H-Ras. We present data about differences in the role played by the RasGRF DH domain in the activation of H-Ras at the PM and at the ER. Finally, we show that RasGRF mediation primes membrane and reticular H-Ras to activation by distinct stimuli. Overall, our results provide the initial evidence for the regulation of H-Ras in the ER.
MATERIALS AND METHODS
Constructs.
The plasmid encoding HA-RasGRF1 was provided by R. R. Mattingly; those encoding FLAG-RasGRF2 wild-type, ΔDH, and ΔIQ were provided by M. F. Moran; that encoding avian infectious bronchitis virus M1 protein was provided by C. E. Machamer; that encoding KDELr-H-Ras SS was provided by X. Bustelo; that encoding HA-SOS1 was provided by J. M. Rojas; that encoding Ras-GRP1 was provided by by J. C. Stone. Plasmids encoding RasGRF1, RasGRF1 DH− and IQ− and GFP-H-Ras have been described previously (3, 22, 35). To generate M1-H-Ras SS, H-Ras SS (C181, 184S) was prepared by PCR-directed mutagenesis and verified by sequencing. It was then subcloned in-frame into pCEFL-HA (35). M1 codons 1 to 66 were amplified by PCR and cloned as a HindIII-BamHI fragment directly upstream of the HA tag. To generate CD8-H-Ras SS, HA-H-Ras SS was subcloned in pCDNACD8, directly downstream of the sequence coding for the transmembrane domain of the CD8 receptor (13). Sequences of the oligonucleotides used for the different constructs are available on request.
Cell culture.
COS-7 and HeLa cells were regularly grown in Dulbecco minimal essential medium supplemented with 10% fetal calf serum. COS-7 subconfluent cells were transfected with DEAE-dextran (3) or with LipofectAmine (for cells used for immunofluorescence studies). HeLa cells were transfected with Lipofectamine. Prior stimulation, cells were starved for 18 h. Lysophosphatide acid (LPA), brefeldin A, and ionomycin were from Sigma. Epidermal growth factor was from UBI.
Hippocampal and dorsal root ganglion neurons preparation.
Male 3-month-old Sprague-Dawley rats were housed and sacrificed according to European Union regulations. Rats were perfused with 3.7% formaldehyde in phosphate-buffered saline (PBS) (pH 7.4), for 15 min at room temperature. Tissue fragments of the hippocampal formation and dorsal root ganglia were dissected out of 500-μm-thick slices, washed in PBS for 1 h, transferred to a drop of PBS on a siliconized slide, covered with a coverslip, and squashed by percussion. The slides with adhered neurons were sequentially dehydrated in 96 and 70% ethanol at 4°C for 10 min, rinsed in PBS, and processed for immunofluorescence analysis.
Immunofluorescence.
Cultured cells were washed twice in PBS, fixed with 3.7% formaldehyde in PBS for 10 min at room temperature, and permeabilized with 0.5% Triton X-100-PBS for 20 min. Preparations were sequentially incubated with 0.5% Triton X-100-PBS for 15 min, and 0.1 M glycine-PBS for 30 min and blocked with 1% bovine serum albumin-0.01% Tween 20 in PBS for 5 min. They were then rinsed in PBS-0.05% Tween 20, incubated for 1 h with the primary antibodies, washed, and incubated for 45 min with the appropriate secondary antibodies conjugated to fluorescein isothiocyanate (FITC) or Texas Red. Coverslips were mounted in VectaShield and sealed with nail polish. Confocal microscopy was performed with a Bio-Rad MRC-1024 microscope, using excitation wavelengths of 488 nm (for FITC) and 543 nm (for Texas Red).
Antibodies.
Mouse monoclonal anti-HA was from Babco. Rabbit polyclonal anti-FLAG was from Invitrogen. Anti-RasGRF2 rabbit and goat polyclonal antibodies and anti-RasGRF1, anti-ERK2, anti-phospho-ERK, and anti-SOS rabbit polyclonal antibodies were from Santa Cruz Laboratories. Rabbit pan-RasGRF was provided by E. Santos (Salamanca, Spain). Rabbit polyclonal antitransferrin receptor was from Zymed. Mouse monoclonal anti-calnexin was from Becton Dickinson. Rabbit polyclonal anticalreticulin was from Calbiochem. Antigiantin mouse monoclonal antibodies were supplied by H. P. Hauri (Basel, Switzerland). Pan-histone mouse monoclonal antibody was from Boehringer Mannheim.
Immunoblotting.
Total lysates and immunoprecipitates were fractionated in sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose filters as described previously (1). Immunocomplexes were visualized by enhanced chemiluminescence detection (Amersham), using horseradish peroxidase-conjugated secondary antibodies (Cappel).
Ras GTP loading.
Ras GTP loading was performed basically as described previously (4). Ras-GTP was affinity sequestered using GST-Raf RBD (amino acids 1 to 149). Immunoblot analyses were performed as described above, using anti-HA antibody, and quantitated by densitometry with the program NIH Image 1.60. Ras-GTP levels were related to total Ras protein levels as determined by anti-HA immunoblotting in the corresponding total-cell lysates.
Kinase assays.
ERK2 kinase activities were determined as previously described (15), using anti-HA immunoprecipitates with myelin basic protein (MBP) as the substrate (Sigma).
Subcellular fractionations.
Briefly, previously minced and homogenized hippocampi, COS-7 cells, and HeLa cells were incubated in 20 mM HEPES (pH 7.4) for 20 min in ice and passed five times through 80- to 90- and 23- to 25-gauge syringes sequentially. Postnuclear supernatants were centrifuged at 100,000 × g for 30 min to yield the S100 (supernatant) and the P100 (pellet) fractions. P100 was resuspended in a volume of 20 mM HEPES (pH 7.4) equal to that of the S100 fraction. To separate the Triton-soluble and insoluble fractions, 1% Triton X-100 was added to the P100 fraction, which was then kept for 20 min in ice and centrifuged at 100,000 × g for 30 min. The supernatant (Triton-soluble fraction) was collected, and the pellet (Triton-insoluble fraction) was resuspended in a volume of 20 mM HEPES (pH 7.4) equal to that of the Triton-soluble fraction. Loading buffer was added to samples of the fractions containing equivalent amounts of protein, and they were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
RESULTS
RasGRFs colocalize with H-Ras in the ER.
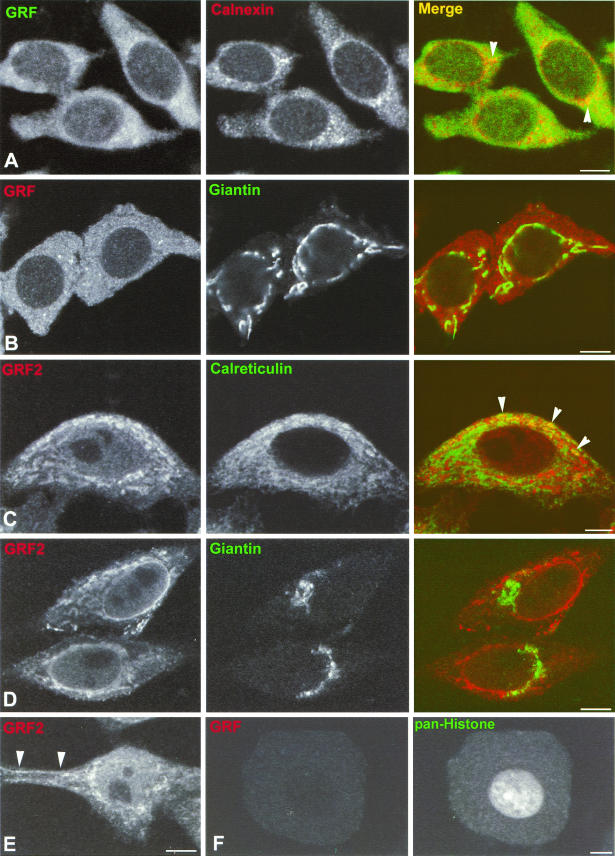
It is conceivable that GEFs must be found at some stage in organelles in which their cognate GTPases are located. Therefore, to study the involvement of RasGRF family GEFs in the regulation of H-Ras in endomembranes, we examined whether these GEFs were detectable in cellular locations, like the ER and the Golgi complex, where H-Ras seems to be active (11). First, we investigated the cellular distribution of RasGRF1 and RasGRF2 in their physiological environments. As such, we looked at hippocampal neurons, where RasGRF1 is naturally expressed (54). Hippocampal pyramidal neurons, exhibiting their characteristic cellular morphology and dispersed chromatin configuration within the nucleus, were immunostained with pan-RasGRF antibodies, and confocal microscopy revealed that RasGRF displayed a nonhomogeneous, diffuse cytoplasmic pattern, with a few small punctate structures of higher-intensity staining. These punctate structures were more abundant at the perinuclear region (Fig. 1A, left panel) and presumably correspond to small tubulovesicular elements of the ER, characteristic of these small pyramidal neurons of the hippocampal formation. Staining with anticalnexin, an ER marker, also disclosed this typical ER morphology of small vesicles, which were especially prominent around the nuclear envelope (Fig. 1A, middle panel). The overlay of RasGRF and calnexin staining revealed the colocalization of both molecules in the ER, specially in the punctate structures around the nucleus (Fig. 1A, right panel). Giantin, a Golgi marker, exhibited the typical perinuclear localization of the neuronal Golgi complex. However, the overlay of RasGRF and giantin demonstrated a conspicuous absence of RasGRF from the Golgi complex (Fig. 1B). To further substantiate our observations, we also investigated the subcellular distribution of RasGRF2, a GEF whose expression pattern is not restricted to the nervous system but can be detected in other tissues and cell lines (18). Immunofluorescence in HeLa cells with RasGRF2 antibodies revealed that this GEF displayed a reticular pattern, especially marked in the perinuclear region (Fig. 1C and D, left panels). Endogenous RasGRF2 was also evident in regions of the PM (Fig. 1E). Costaining with the ER marker calreticulin demonstrated a partial although clear colocalization with RasGRF2 in ER structures (Fig 1C., right panel). On the other hand, upon costaining with antigiantin, it was found that RasGRF2 was completely excluded from the Golgi complex (Fig. 1D). To ascertain that the signals yielded by the RasGRF-specific antibodies were indeed specific, we performed immunofluorescence analysis using the pan-RasGRF antibody in dorsal root ganglia neurons, which do not express RasGRF1 (58). As shown in Fig. 1F (left panel), no signal was evident in this type of neuron. Likewise, the anti-RasGRF2 antibody stained negatively in COS-7 cells that lack endogenous RasGRF2 (data not shown). These results demonstrated that RasGRF1 and RasGRF2 were present in the ER but not in the Golgi complex of cells naturally expressing these GEFs.
FIG.1.
Subcellular localization of endogenous RasGRFs. (A and B) Localization of RasGRF in hippocampal neurons. Shown are confocalmicrographs representative of hippocampal pyramidal neurons doubly immunostained with anti-panRasGRF and anticalnexin (red) (A) or giantin (green) (B). Intense colocalization in punctate structures around the nucleus is indicated by arrow heads. (C to E) Localization of RasGRF2 in HeLa cells immunostained with anti-RasGRF2 (red) and anti-calreticulin (green) (C) or giantin (green) (D). Localization of RasGRF2 in regions of the PM is indicated by arrow heads in panel E. Calreticulin costaining demonstrated colocalization with RasGRF2 in the ER, as indicated by arrowheads in panel C. (F) Confocal micrographs of dorsal root ganglion neurons doubly immunostained with anti-panRasGRF (left panel) and with anti-pan-histone (right panel). All panels show equatorial sections at the cell nucleus level, except for panel E, which is more tangential. Bars, 10 μm (5 μm for panels C and F).
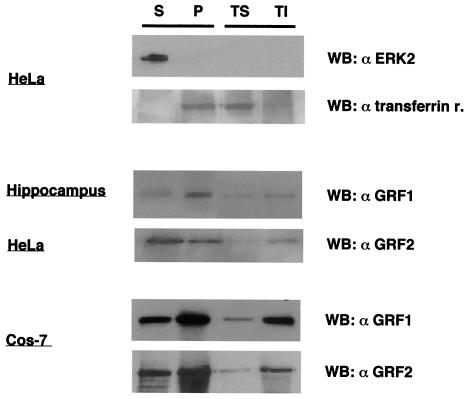
The cellular localization of endogenous RasGRF1 and RasGRF2, observed by immunofluorescence, was in agreement with the results obtained when their subcellular distribution was studied by cellular fractionation. This technique provided soluble and particulate fractions with minimal cross-contamination, as verified by immunoblotting using antibodies against ERK2 and transferrin receptor as markers for the cytoplasmic and membrane fractions, respectively (Fig. 2, top panels). RasGRF1 from hippocampal neurons was detected in the soluble fraction and, more prominently, in the particulate fraction, especially in the Triton-insoluble subfraction. RasGRF2 in HeLa cells exhibited a similar distribution, although it was slightly more prominent in the soluble fraction (Fig. 2, middle panels). Remarkably, the distribution of RasGRF1 and RasGRF2 in their physiological settings was largely mirrored in an ectopic expression system like COS-7 cells transfected with RasGRF1 or with RasGRF2. Even though these GEF expression levels were notably higher, ectopic RasGRF1 and RasGRF2 segregation was very similar to that observed for the endogenous GEFs (Fig. 2, bottom panels). In light of this data, we proceeded to determine the cellular localization of transfected RasGRF GEFs in COS-7 cells in further detail by immunofluorescence. This should enable us to validate this model for further experimentation, which otherwise would be complex in other cellular systems.
FIG. 2.
Analysis of RasGRF1 and RasGRF2 distribution by cellular fractionation in hippocampal neurons, HeLa cells, and RasGRF1- and RasGRF2-transfected (1 μg) COS-7 cells, revealed by immunoblotting with the indicated antibodies. ERK2 and transferrin receptor immunoblots served as controls for the soluble and particulate fractions, respectively. Lysates were fractionated as described in Materials and Methods. Lanes S, S100 soluble fraction; P, P100 particulate fraction. The particulate fraction was subsequently fractionated into Triton-soluble (TS) and Triton-insoluble (TI) fractions. WB, Western blot.
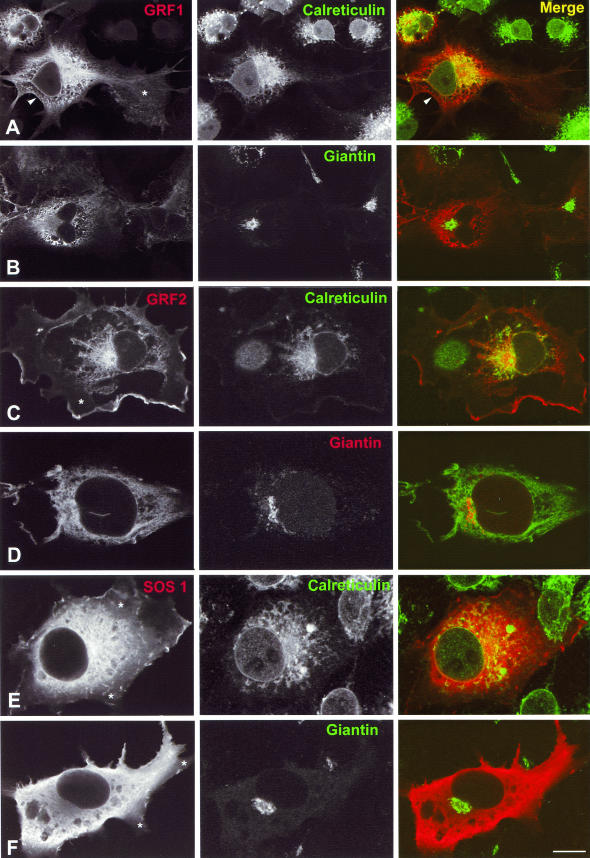
In COS-7 cells transfected with HA-tagged RasGRF1, RasGRF1 was visualized at the PM (Fig. 3A, left and right panels) and particularly in the perinuclear zone, including the nuclear envelope and a profuse ER network. Some diffuse labeling, characteristic of a cytosolic location, was also evident around the nucleus (Fig. 3A). The ER distribution was confirmed by double immunostaining with calreticulin. The overlay showed extensive colocalization of the two proteins in the ER. By contrast, costaining with giantin revealed a complete absence of RasGRF1 from the Golgi complex (Fig. 3B). Similar results were obtained for transfected FLAG-RasGRF2. RasGRF2 was observed in the nuclear envelope, ER, and, more prominently than RasGRF1, in some domains of the PM (Fig. 3C and D). Double-staining experiments showed that RasGRF2 also colocalized with calreticulin in the nuclear envelope and in the ER (Fig. 3C) but did not colocalize with giantin at the Golgi complex (Fig. 3D). For comparative purposes, we also studied the cellular distribution of SOS1. This GEF exhibited a more diffuse labeling than RasGRF1-2, indicative of a cytoplasmic distribution. SOS1 also localized to some domains of the PM (Fig. 3E). Double-labeling experiments also demonstrated its localization in the ER and the nuclear envelope (Fig. 3E) and, similarly to RasGRF1-2, a complete absence from the Golgi complex (Fig. 3F). Remarkably, while all GEFs displayed some degree of cytoplasmic staining, this was more marked in the cell body and GEFs were mainly absent from cytoplasmic prolongations like lamellipodia (Fig. 3A, C, E, and F). It is also noteworthy that the distribution of RasGRF1 and RasGRF2 within the ER was polarized, being most conspicuous at one of the nuclear poles. This phenomenon was evident both for ectropically expressed (Fig. 3A and C) and endogenous (Fig. 1A and C) GEFs.
FIG.3.
Subcellular localization of ectopic GEFs in COS-7 cells. Shown are confocal laser micrographs of COS-7 cells transfected with HA-RasGRF1 (A and B), Flag-RasGRF2 (C and D), or HA-SOS1 (E and F) (1 μg in each case). Cells expressing these different constructs were costained with antibodies directed against calreticulin (A, C, and E) or giantin (B, D, and F). RasGRF1 was revealed by anti-HA antibodies in panel A and by anti-RasGRF1 in panel B. RasGRF2 was revealed by anti-RasGRF2 antibodies in panel C and by anti-FLAG in panel D. SOS1 was revealed by anti-HA antibodies in panel E and by anti-SOS1 in panel F. Arrowheads in panel A indicate RasGRF1 at the PM. Asterisks in panels A, C, E, and F indicate an absence of GEFs from cytoplasmic extensions such as lamellipodia. Bars, 10 μm.
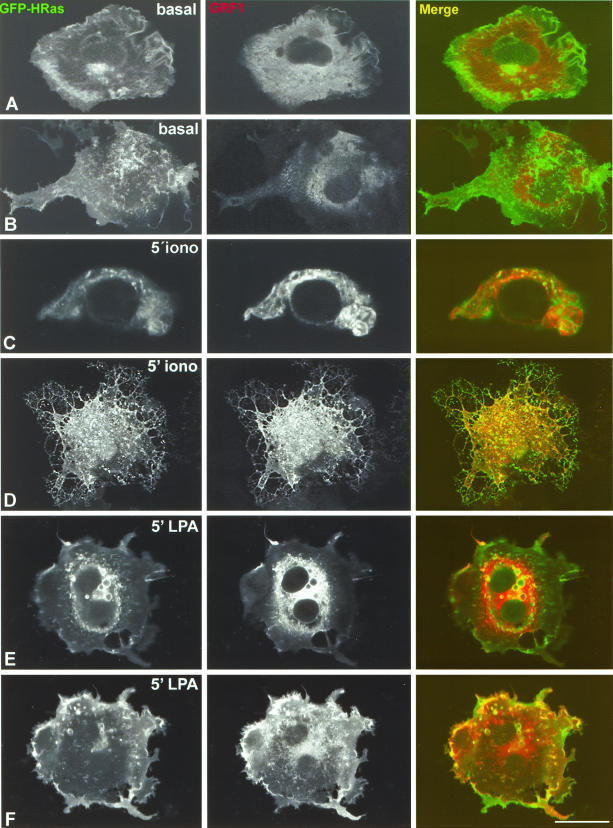
These results clearly indicated that RasGRF1 and RasGRF2, expressed at concentrations at which they effectively activate H-Ras at steady state, were present at the ER and at the PM, locations where H-Ras activation takes place (11). The next question we addressed was how RasGRF colocalized with H-Ras under acute stimulation by agents that unleash RasGRF exchange activity. For this purpose, their colocalization in COS-7 cells transfected with GFP-H-Ras in addition to suboptimal concentrations of RasGRF1 was analyzed by confocal microscopy. The ability of RasGRF1 at this concentration to activate H-Ras per se is greatly reduced; therefore, the basal activity levels of H-Ras are very low. This condition was indirectly ascertained by immunofluorescence analysis using antibodies against phosphorylated ERK (data not shown). Under unstimulated conditions, H-Ras was present at the PM, Golgi complex, and ER structures including the nuclear envelope and dispersed cisternae (Fig. 4A, left panel) whereas RasGRF1 was prominently detected at the ER, the cytoplasm, and the PM. (Fig. 4A, middle panel). Under these circumstances, there was little colocalization between RasGRF1 and H-Ras (Fig. 4A, right panel). This was shown in further detail by a confocal section extending through the PM, in which, even though the presence of GFP-H-Ras and RasGRF1 was remarkable (Fig. 4B, left and middle panels), the overlay was minimal (Fig. 4B, right panel). Upon stimulation for 5 min with the calcium ionophore ionomycin, a strong colocalization of the two proteins was observed in some ER elements, disclosed by an equatorial confocal section (Fig. 4C), and also in extensive domains of the PM, as demonstrated by a tangential section disclosing the cell surface (Fig. 4D). A similar situation was observed in cells treated with LPA, in which the two proteins colocalized at the perinuclear region, particularly at the limiting membrane of variable size vesicles and ER elements (Fig. 4E). A marked colocalization was also detected at the PM in a confocal section illustrating the cell surface (Fig. 4F). By contrast, we could not detect colocalization of H-Ras and RasGRF1 at the Golgi complex regardless of the circumstances (data not shown). On the other hand, a pool of RasGRF1 exhibiting a diffuse staining, typical of cytosolic localization, persisted even under LPA stimulation (Fig. 4E and F). Similar results were obtained with RasGRF2 (data not shown). These data demonstrated that under conditions at which RasGRF1 is active, this GEF is present and colocalizes with H-Ras at the PM and ER but not at the Golgi complex.
FIG. 4.
Colocalization of RasGRF1 and H-Ras under stimulation. COS-7 cells were cotransfected with suboptimal concentrations of RasGRF1 (0.1 μg) and with GFP-H-Ras (0.25 μg) and analyzed using confocal microscopy. (A and B) Basal conditions. An equatorial confocal section at the level of the cell nucleus (A) and a tangential section illustrating the PM (B) are shown. (C and D) Treatment with 1 μM ionomycin for 5 min. A confocal section at the cell nucleus level (C) and a confocal section sweeping through the cell surface (D) are shown. (E and F) Treatment with 10 μM LPA for 5 min. A confocal section at the cell nucleus level (E) and a confocal section illustrating the cell surface (F) are shown. Bars, 10 μm (5 μm for panel C).
RasGRF1 and RasGRF2 activate H-Ras in the ER but not in the Golgi complex.
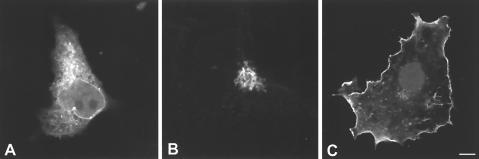
Our results showed a significant colocalization of RasGRF1 and RasGRF2 with H-Ras at the PM and the ER. However, the fact that two proteins share a location does not necessarily imply a functional relationship. Therefore, it was important to determine whether RasGRF GEFs could stimulate nucleotide exchange on H-Ras at the membrane systems where the two molecules coexisted. For this purpose, we engineered H-Ras constructs specifically tethered to defined subcellular sites. First, we generated a palmitoylation-deficient H-Ras by mutating cysteines 181 and 184 to serines (termed H-Ras SS). This mutant is not efficiently retained in the PM and exists in a dynamic equilibrium, shifting between reticular and cytoplasmic pools (11). Next, the signal provided by the palmitoylation was replaced by an alternative cue that would specifically direct H-Ras to the desired location. To deliver H-Ras SS to the ER, we fused to its N terminus amino acids 1 to 66 of the avian infectious bronchitis virus M protein (M1), shown to be restricted to reticular endomembranes (49). A PM-targeted H-Ras SS was generated by placing in its N terminus the transmembrane domain of the CD8 receptor, which we have previously shown to be an effective PM anchor (13). An HA tag was included to enable the detection of the expressed proteins. In addition, we assayed the activation of H-Ras at the Golgi complex, using an H-Ras SS N-terminally fused to the KDEL receptor (KDELr), a resident Golgi protein at steady state (10, 40). To verify whether our location-specific H-Ras proteins were correctly distributed, the constructs were transfected into COS-7 cells and their localization was visualized by immunofluorescence using anti-HA antibodies. As shown in Fig. 5A, M1-H-Ras SS displayed a typical reticular staining and was undetectable at the PM or at the Golgi. KDELr-H-Ras SS was restricted mainly to the Golgi apparatus, with very low reticular staining, and virtually absent from the PM (Fig. 5B). CD8-H-Ras SS exhibited a characteristic peripheral membrane staining, with only traces being detectable at the ER and Golgi (Fig. 5C). Therefore, these results validated our constructs for further experimentation.
FIG. 5.
Cellular localization of location-specific H-Ras constructs. COS-7 cells were transfected with the indicated constructs (0.25 μg in each case). (A) M1-H-Ras SS. (B) KDELr-H-Ras SS. (C) CD8-H-Ras SS. Immunofluorescence was performed using anti-HA antibodies. Bars, 10 μm.
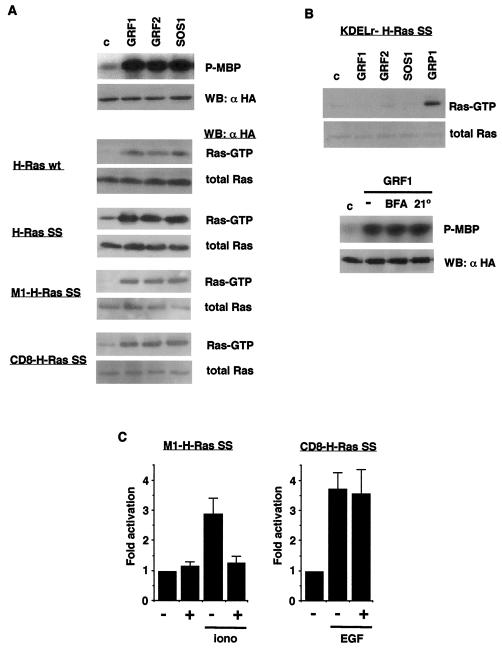
To examine whether RasGRF1 and RasGRF2 were capable of activating H-Ras in defined membrane systems, we transfected COS-7 cells with the location-specific H-Ras constructs and with RasGRF1 and RasGRF2. We also included SOS1 for comparisons. Ras activation was analyzed by pull-down assays using the GST-Raf Ras-binding domain (RBD) to affinity precipitate GTP-bound Ras (50), which was revealed by anti-HA immunoblotting. First, we checked that on cotransfection with HA-ERK2, as expected, all three GEFs could efficiently activate ERK2. (Fig. 6A, top panel). In the same fashion, we ascertained that these GEFs could induce exchange on wild-type H-Ras to a similar extent (Fig. 6A, upper middle panel). This was also the case for H-Ras SS. Since this mutant is found predominantly in the reticulum, this suggested that RasGRF1, RasGRF2, and SOS1 could stimulate GDP-GTP exchange on H-Ras ER pool. This was confirmed by probing the activation of M1-H-Ras SS, revealing that the three GEFs were capable of inducing a robust exchange on this reticulum-tethered H-Ras (Fig. 6A, lower middle panel). Likewise, RasGRF1, RasGRF2, and SOS1 also strongly activated CD8-H-Ras SS, indicating that these GEFs could stimulate PM H-Ras (Fig. 6A, bottom panel). On the other hand, neither RasGRF1, RasGRF2, nor SOS1 proved competent for catalyzing exchange on the Golgi-bound KDELr-H-Ras SS, while under the same experimental conditions RasGRP1 was capable of efficiently activating Golgi-targeted H-Ras (Fig. 6B, top), in agreement with recent reports (6, 10), suggesting that the H-Ras pool in this organelle was unreactive with or inaccessible to RasGRF and SOS GEFs. To verify this point further, we investigated the effects of interfering with Golgi functions on RasGRF1 signaling. In COS-7 cells cotransfected with RasGRF1 and HA-ERK2, it was found that neither treatment with brefeldin A, which causes a redistribution of Golgi proteins to the cytosol and cessation of vesicle formation (28), nor prolonged culture at 21°C, which blocks post-Golgi transport (45), had any effect on the activation of ERK2 induced by RasGRF1 (Fig. 6B, bottom), thereby demonstrating that the H-Ras Golgi pool is not a necessary mediator for the activation of the ERK pathway by RasGRF1.
FIG. 6.
Activation of H-Ras by GEFs in distinct membrane systems. (A) (Top) Activation of ERK2 by RasGRFs and SOS1. COS-7 cells were cotransfected with HA-ERK2 and the indicated GEFs (1 μg each). Kinase assays were performed in anti-HA immunoprecipitates, using MBP as substrate. (Middle) Levels of HA-ERK2 as determined by immunoblotting using anti-HA antibodies. (Bottom) Activation of H-Ras constructs tethered to defined locations. Ras GTP loading was determined, as described in Materials and Methods, in COS-7 cells transfected with the different GEFs (1 μg) in addition to the site-specific H-Ras constructs (0.25 μg), as indicated. H-Ras-GTP levels present in affinity precipitates using glutathione S-transferase (GST)-Raf RBD as bait and the total H-Ras levels in the corresponding total lysates were detected by anti-HA immunoblotting. WB, Western blotting; wt, wild type. (B) Activation of H-Ras in the Golgi complex. (Top) Ras GTP loading was determined, as described in Materials and Methods, with COS-7 cells transfected with the different GEFs (1 μg) as indicated, in addition to the Golgi-tethered KDELr-H-Ras SS construct (0.25 μg). (Bottom) Activation of ERK2 by RasGRF1 after treatments that affect the Golgi complex. COS-7 cells were cotransfected with HA-ERK2 and RasGRF1 (1 μg each), treated with 5 μg of BFA/ml for 30 min, or subjected to 21°C for 2 h. Kinase assays were performed with anti-HA immunoprecipitates, using MBP as the substrate. (Lower panel). Protein levels of HA-ERK2 as determined by immunoblotting using anti-HA antibodies. (C). Activation of reticular H-Ras by endogenous RasGRF. HeLa cells were transfected with M1-H-Ras SS or with CD8-H-Ras SS constructs (0.25 μg) as indicated, in addition to empty vector (−) or RasGRF1 ΔCdc25 (0.5 μg) (+), and stimulated with 1 μM ionomycin or with 100 ng of EGF/ml for 5 min where indicated. Ras-GTP loading was determined as previously described. Data show the mean of at least three independent experiments relative to the Ras-GTP levels detected in control cells. Error bars indicate standard error of the mean (SEM).
It was of interest to examine, whether endogenous RasGRF was capable of activating the H-Ras pool present in the ER, thereby corroborating the physiological relevance of our findings using ectopic RasGRF. To this end, HeLa cells were transfected with ER-tethered M1-H-Ras SS and subsequently starved and subjected to stimulation with ionomycin, a well-known activator of RasGRF GEFs. This treatment resulted in almost a threefold increase on M1-H-Ras SS GTP levels (Fig. 6C). Since ionomycin could affect Ras GTP levels by different mechanisms, it was crucial to determine to what extent this effect of ionomycin on ER H-Ras was attributable to RasGRF. For this purpose, we used a construct encoding RasGRF1 devoid of its Cdc25 domain (FLAG-GRF1 ΔCdc25) (3). This protein acts as a dominant inhibitory mutant by sequestering upstream activators away from endogenous RasGRF in an unproductive fashion, since it cannot itself induce nucleotide exchange. On cotransfecting GRF1 ΔCdc25 into HeLa cells, it was found that this construct could diminish ionomycin-induced M1-H-Ras SS GTP levels to almost basal levels. On the other hand, in spite of being well expressed (data not shown), GRF1 ΔCdc25 had no effect on the nucleotide exchange induced by EGF over PM-tethered CD8-H-Ras SS (Fig. 6C), a process known to be independent of RasGRF, thereby demonstrating the specificity of the inhibitory effects of this construct. These results indicated that endogenous RasGRF was capable of activating the H-Ras pool present in the ER upon stimulation with ionomycin.
The RasGRF1 DH domain is required for H-Ras activation in the ER but not in the PM.
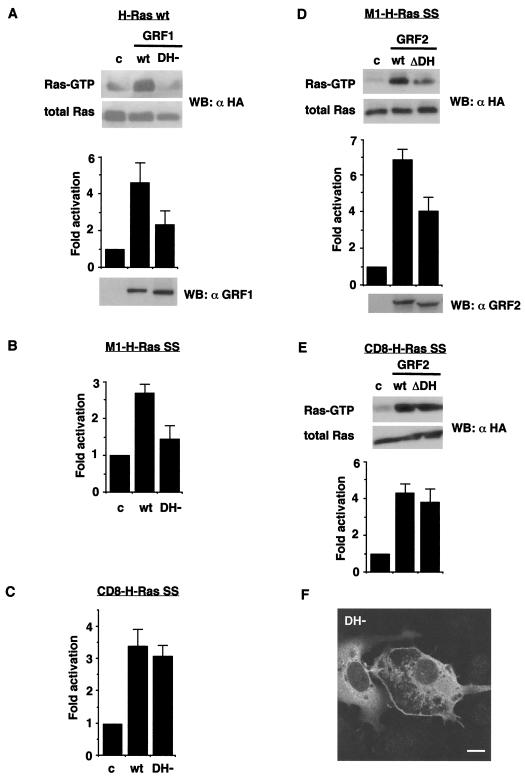
Our recent results have demonstrated that curtailing its DH domain precludes RasGRF1 from activating Ras/ERK (3). Therefore, it was of interest to determine whether the DH domain was required for activating H-Ras regardless of its cellular location, or, alternatively, was necessary only for stimulating a subset of H-Ras in a definite site. To test this hypothesis, we investigated if a RasGRF1 DH domain mutant (DH−) with a diminished ability for activating Ras/ERK (3, 22) was competent for activating the reticular and PM H-Ras pools in COS-7 cells. In agreement with our previous results, Ras-GTP pull-down assays revealed that the ability of RasGRF1 DH- to activate total H-Ras experienced a ≈60% reduction in comparison to the wild-type GEF (Fig. 7A). Interestingly, the DH- mutant activated M1-H-Ras SS only up to 50% of the level induced by wild-type RasGRF1 (Fig. 7B). However, this mutant was just as efficient as wild-type RasGRF1 for activating CD8-H-Ras SS (Fig. 7C). RasGRF2 behaved in an identical fashion: a RasGRF2 DH domain deletion mutant (ΔDH) (19) was impaired in its ability to induce exchange on M1-H-Ras SS (Fig. 7D) but was perfectly capable of augmenting CD8-H-Ras SS GTP levels (Fig. 7E). These results suggested that the RasGRF DH domain was required for catalyzing nucleotide exchange in the reticular fraction but not in the PM H-Ras fraction. We then asked if these results bore some relationship to the cellular distribution of RasGRF on curbing the DH domain. To investigate this, we transfected RasGRF1 DH- into COS-7 cells and examined its localization. As shown in Fig. 7F, RasGRF1 DH- was markedly present in the peripheral membrane. However, on the perinuclear zone, the reticular pattern characteristic of wild-type RasGRF1 (Fig. 3) was largely lost and the DH- mutant presented a more diffuse distribution (as shown in the cell on the left) reminiscent of soluble, cytoplasmic proteins. This result suggested that the diminished ability of DH domain mutant forms for activating the reticular H-Ras pool was a consequence of their loss of localization to the ER.
FIG. 7.
Role of RasGRF1 DH domain on the activation of H-Ras in distinct cellular locations. (A) Activation of total H-Ras by RasGRF1 DH−. H-Ras wild-type (wt) (0.25 μg) was transfected in COS-7 cells in addition to RasGRF1 wild type and the DH− mutant (1 μg) as indicated. (Upper panel) H-Ras-GTP levels from a representative experiment; Ras-GTP loading was determined as described in Materials and Methods. Data show means and SEM of at least five independent experiments relative to the Ras-GTP levels detected in control cells. (Lower panel) RasGRF1 expression levels as determined by immunoblotting using anti-RasGRF1 antibodies. (B) Activation of ER-bound M1-H-Ras by RasGRF1 DH−. Data show means and SEM of at least five independent experiments. (C) Activation of PM-tethered CD8-H-Ras by RasGRF1 DH−. Data show means and SEM of at least five independent experiments. (D) Activation of ER-tethered M1-H-Ras by RasGRF2 ΔDH. M1-H-Ras SS (0.25 μg) was transfected in COS-7 cells in addition to wild-type RasGRF2 and the ΔDH mutant (1 μg) as indicated. (Upper panel) H-Ras-GTP levels from a representative experiment. Data show means and SEM of at least five independent experiments relative to the Ras-GTP levels detected in control cells. (Lower panel) RasGRF2 expression levels as determined by immunoblotting using anti-RasGRF2 antibodies. (E) Activation of membrane-tethered CD8-H-Ras by RasGRF2 ΔDH. Data show means and SEM of at least five independent experiments. (F) Cellular localization of RasGRF1 DH− (1 μg) transfected in COS-7 cells. Bar, 10 μm.
Distinct pools of H-Ras are differently induced by RasGRF-activating stimuli.
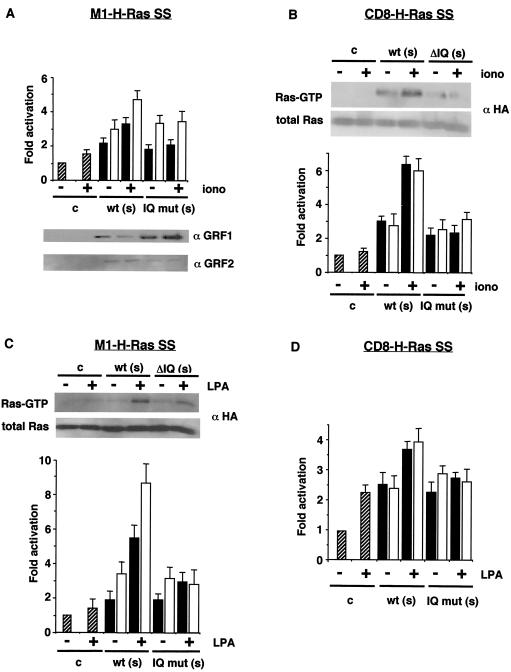
Finally, since RasGRF1 and RasGRF2 are activated by two types of stimuli, mediated by either heterotrimeric G proteins or calcium-calmodulin (43), we investigated whether, on mediation by RasGRF, these stimuli preferentially activated H-Ras within a defined location. COS-7 cells were cotransfected with suboptimal concentrations of RasGRF1 and RasGRF2 in addition to ER- or PM-targeted H-Ras, and, after starvation, Ras activation was assayed on stimulation with ionomycin or LPA. As shown in Fig. 8A, in RasGRF1- and RasGRF2-transfected cells, ionomycin induced only a moderate increase on Ras-GTP levels on the ER-targeted M1-Ras SS, not even reaching a twofold increase in either case. As a control, we included mutants defective for IQ domain functions, ΔIQ for RasGRF2 (19) and IQ− for RasGRF1 (22), that, as expected, were unreactive with ionomycin. Notably, the RasGRF-mediated response to ionomycin was more pronounced on PM-tethered CD8-H-Ras SS (Fig. 8B), suggesting that RasGRF1 and RasGRF2 preferentially primed the H-Ras PM pool to induction by calcium ionophores. Interestingly, the outcome was the opposite for the effect of LPA. LPA induced almost a threefold increase in H-Ras activation at the ER mediated by both RasGRF1 and RasGRF2 (Fig. 8C), but only a small increase in the activation of the peripheral-membrane H-Ras pool was evident (Fig. 8D). As previously shown for RasGRF1 (27) deletion of the IQ domain also rendered RasGRF2 unreactive with LPA. These results demonstrated that depending on its localization, H-Ras was distinctively sensitive to RasGRF-mediated activation triggered by different types of stimuli.
FIG. 8.
Distinct pools of H-Ras are differentially induced by RasGRF-activating stimuli. (A) Activation of ER-bound H-Ras by ionomycin. COS-7 cells were cotransfected with M1-H-Ras SS (0.25 μg) in addition to vector (hatched bars) or suboptimal concentrations (s = 0.1 μg) of constructs encoding RasGRF1 (open bars) or RasGRF2 (solid bars), in their wild-type (wt) versions and IQ domain mutant forms (IQ− for RasGRF1, ΔIQ for RasGRF2). After starvation, the cells were treated with 1 μM ionomycin for 5 min where indicated (+) and Ras-GTP levels were determined as described above. Data show means and SEM of at least five independent experiments relative to the Ras-GTP levels detected in control cells. (Lower panel) RasGRF1 and RasGRF2 expression levels as determined by immunoblotting using specific antibodies. (B) Activation of PM-bound CD8-H-Ras SS by ionomycin. (Upper panel) H-Ras-GTP levels from a representative experiment with RasGRF2. Data show means and SEM of at least five independent experiments relative to the Ras-GTP levels detected in control cells. (C) Activation of ER-tethered M1-H-Ras SS by LPA. (Upper panel) H-Ras-GTP levels from a representative experiment with RasGRF2 on activation with 5 μM LPA for 5 min where indicated (+). Data show means and SEM of at least five independent experiments relative to the Ras-GTP levels detected in control cells. (D) Activation of PM-bound CD8-H-Ras SS by LPA. Data show means and SEM of at least five independent experiments relative to the Ras-GTP levels detected in control cells.
DISCUSSION
In an attempt to examine the regulation of Ras in endomembranes, we have investigated the involvement of RasGRF family GEFs as regulators of the activation of H-Ras in the ER and Golgi complex. Identifying the cellular sites to which a GEF localizes can give important clues to its functions. Previous studies undertaken with different cell types probing the distribution of ectopically expressed RasGRF1 by simple fractionation have yielded similar results. RasGRF1 can be found in the soluble and, more prominently, the particulate fractions of NIH 3T3 (47), HEK293T (7) and COS-7 (reference 3 and this study) cells. Using the same method, we show here that endogenous RasGRF1 from hippocampal neurons fractionates in a similar fashion. RasGRF1 is also predominantly particulate in fractionated rat brain (53). RasGRF2 from HeLa cells is also markedly detected in the particulate fraction. Separation of the particulate fraction into Triton-soluble and Triton-insoluble subfractions revealed that RasGRF1 and RasGRF2 are found in both fractions although mainly in the Triton-insoluble phase. It is not known to what cellular structure(s) these fractions correspond. The Triton-soluble fraction probably represents the bulk membranes, irrespective of their origin. It is conceivable that the Triton-insoluble fraction may be partly constituted by PM lipid rafts and caveolae, where H-Ras is markedly located (42). Indeed, recent results from our laboratory suggest that H-Ras activation by RasGRF1 and RasGRF2 takes place in these compartments (35). Another possibility that we cannot presently rule out is that a fraction of the Triton-insoluble RasGRF1 and RasGRF2 could be associated with microtubules. This would not be unprecedented, since another RasGEF, RasGRP, is partly associated with microtubules in certain cell types (41). Even though, thus far, no structures resembling lipid rafts have been described within the ER, it is becoming clear that this organelle is not a uniform compartment but, rather, a spatially and functionally heterogeneous structure (39). Therefore, it cannot be ruled out that hitherto unidentified ER-derived structures could also contribute to the RasGRF1 and RasGRF2 Triton-insoluble pool.
We have extended these biochemical observations with detailed analyses of RasGRF1 and RasGRF2 distribution by immunofluorescence that reveal that RasGRF1 and RasGRF2 localize strongly to the ER but are completely excluded from the Golgi complex. This is the case for the endogenous RasGRF expressed in hippocampal neurons, mainly RasGRF1, since RasGRF2 expression is almost undetectable in hippocampus (21), and for endogenous RasGRF2 present in HeLa cells. Interestingly, RasGRF distribution in the ER exhibits a polarized pattern, being most prominent at one of the nuclear poles. The significance of this phenomenon is currently unknown. Remarkably, a localization almost identical to that found in physiological environments is observed in COS-7 cells expressing ectopical RasGRF1 or RasGRF2, an observation that suggests that this cellular system should be used for further experimentation.
Our results demonstrate that on treatment with stimuli that trigger the enzymatic activity of RasGRF1, there is a marked augmentation in the colocalization of this GEF with H-Ras at the PM and at the ER but not at the Golgi complex, in comparison to basal, unstimulated conditions under which only scarce colocalization between RasGRF1 and H-Ras can be observed at the PM and ER. Seemingly, this increase in colocalization between RasGRF1 and H-Ras does not require major changes in RasGRF distribution within the cell, since the gross cellular localization of RasGRF1 and RasGRF2 does not experience obvious changes under stimulation or as a result of massive overexpression (our unpublished results). Cellular fractionation has also failed to identify major changes in RasGRF1 distribution as a consequence of stimulation (reference 7 and our unpublished results). This is in sharp contrast to SOS, which is markedly cytoplasmic under basal conditions but undergoes an abrupt redistribution to the particulate fraction as a result of EGF stimulation. This is visualized by a conspicuous recruitment to the peripheral membrane and the perinuclear region (references 8 and 31 and our unpublished results). As such, our results suggest that RasGRF activation does not imply massive translocations from one cellular compartment to another. If any, there would be subtle changes, since they cannot be identified by immunofluorescence or by fractionation. Overall, the system could be envisioned as a RasGRF cytoplasmic pool serving as a dormant reserve, readily available whenever necessary to replenish the RasGRF activatable pool at confined locations within the targeted organelle. Under stimulation, RasGRF would experience a short shuttle to H-Ras compartments within the same organelle, where Ras activation would ensure. This model reconciles our observations showing an increase in the colocalization between RasGRF1 and H-Ras on stimulation with those demonstrating that the gross subcellular distribution of RasGRF1 is mostly unaltered under stimulation. In this perspective, spillage of excess RasGRF to nearby H-Ras compartments could explain the activation of H-Ras by overexpressed RasGRFs under basal conditions.
Colocalization at the PM and the ER would just be circumstancial evidence in the absence of functional proof that RasGRF1 and RasGRF2 can activate H-Ras therein. With the use of H-Ras constructs specifically tethered to the PM, ER, and Golgi, we have demonstrated that RasGRF1, RasGRF2, and SOS1 can effectively induce GDP-GTP exchange at the PM and ER, but not Golgi-associated H-Ras. Furthermore, treatments that markedly alter Golgi functions do not affect the output signal of RasGRF1. These functional pieces of evidence are in full agreement with our data on subcellular localization, which demonstrate that RasGRF1, RasGRF2, and SOS1 are present at the PM and ER but not at the Golgi complex. In this respect, recent data demonstrate that H-Ras activation at the Golgi is mediated mainly by RasGRP1 (6, 10). The molecular basis of the attraction toward different endomembranes exhibited by distinct Ras GEFs is not known. Overall, our results demonstrate that RasGRF1, RasGRF2, and SOS1 mediate in the activation of H-Ras at the ER while RasGRPs would activate H-Ras at the Golgi complex (6, 10).
These results seem opposed to those in a recent report, utilizing the “Raichu” construct (37), suggesting that GEFs activate Ras at the PM but not at the central region of the cell. As an explanation, this study proposes the interesting hypothesis that Ras GAP activity would exhibit a gradient, with the highest levels at the central region of the cell, decreasing toward the periphery (38). However, other possibilities are open. It is possible that Raichu-Ras may not totally reflect the mechanism by which endogenous Ras is switched off. Under stimulation, Raichu-Ras will emit a fluorescent signal only when bound to its own Raf RBD. The likely event of binding to an endogenous Raf protein would prevent the Raichu system from emitting an “on” signal. Therefore, Raichu-Ras binding to any endogenous effector molecule would have a similar effect to binding to a GAP molecule. Therefore, in addition to a putative GAP gradient, local increases in Ras effector concentrations could contribute to quenching the Raichu-Ras signal.
Past results from our laboratory have demonstrated that deleting its DH domain results in a defective RasGRF1, with a diminished capacity for catalyzing GTP incorporation into Ras (3). Herein, we have extended this observation by investigating the role of the DH domain on the activation of H-Ras ER and PM fractions. We demonstrate that RasGRF1 and RasGRF2 DH mutant forms are impaired in their ability to activate reticular H-Ras but that their capacity to induce GDP-GTP exchange on the H-Ras PM fraction is unchanged. Interestingly, the reduced capacity of the RasGRF1 DH mutant for activating H-Ras in the ER correlates with the loss of its localization to the perinuclear ER, while its presence at the PM is unaltered. These results strongly suggest that the DH domain harbors some targeting signal necessary to adequately position RasGRF1 in the ER. This signal would be dispensable for its interaction with the PM. Consistently, we have previously shown that ablation of the DH domain markedly diminished the amount of RasGRF1 in the particulate fraction and that the addition of an H-Ras CAAX box restored the localization of the DH mutant to this subcellular fraction and its capability to activate Ras (3). Nevertheless, our results indicate that even the DH− mutant retains some activity over the H-Ras pool in the ER. It is therefore conceivable that other signals within RasGRF would also contribute to some extent to the adequate positioning of the GEF and the activation of H-Ras in this compartment.
Our final goal has been to evaluate whether distinct types of stimuli that unleash the catalytic activity of RasGRF will stimulate different H-Ras pools. We demonstrate that under RasGRF mediation, ionomycin preferentially activates H-Ras at the PM. This was somewhat unexpected, since we reasoned that because the ER is the main calcium intracellular reservoir, calcium release would preferentially activate the nearby H-Ras ER pool. Surprisingly, this was not the case. Calcium-induced RasGRF activation requires the participation of calmodulin (20) a protein that is ubiquitous within the cell (51) and has been shown to intervene in the control of the Ras/ERK pathway at the PM on calcium efflux (17). Furthermore, calcium-calmodulin has been recently implicated in the regulation of K-Ras (52), an isoform that, thus far, has been shown to be functional only at the PM. Thus, calcium-calmodulin-mediated mechanisms regulating Ras functions at the PM seem to be widespread. In contrast to the effect of ionomycin, we have observed that LPA is most stimulatory over the H-Ras ER fraction, and this effect was also dependent on the RasGRF2 IQ domain. Other studies indicate that calcium chelators, calmodulin inhibitors, or the deletion of the RasGRF1 IQ domain preclude LPA-mediated activation of the Ras pathway (27, 57), consistent with our results. It is intriguing how two stimuli that switch on the same regulatory mechanism will activate H-Ras differently depending on its location, but our results suggest that the differences must be in mechanisms other than calmodulin binding.
Compared to the wild-type protein, RasGRF1 and RasGRF2DH domain mutant forms are impaired over 50% in their ability to activate total H-Ras (references 3, 19, and 22 and this study). This would imply that the H-Ras ER fraction is the main mediator of RasGRF output signal, as opposed to the H-Ras PM pool. However, we demonstrate that RasGRF can potently activate PM H-Ras, and the fact that stimuli like ionomycin lead to a preferential RasGRF-mediated activation of H-Ras PM pool clearly indicates that RasGRF GEFs are competent stimulators of PM H-Ras signals under certain circumstances. The molecular determinants and the cues that dictate the selectivity of RasGRF toward the ER and PM H-Ras pools under different stimuli are thus far unknown and are under investigation. With our current method, the use of different H-Ras constructs precludes an accurate quantitative comparison of H-Ras activation in the PM and the ER. We are currently designing a system that will enable us to undertake this task.
Lastly, our study suggests that the presence of H-Ras in the ER is not simply a temporal event reflecting the transient passage of this protein toward its final destination at the PM. In the absence of studies addressing the comparative stabilities of H-Ras pools at the different cellular locations in relation to their functions, it is becoming clear that the ER H-Ras pool possesses remarkable biochemical and biological activities. As such, non palmitoylated H-RasV12, which accumulates predominantly in the ER (2, 12), activates Raf, phosphoinositide-3-kinase, and Jun N-terminal kinase less efficiently that palmitoylated H-RasV12 but sufficiently well to retain up to 75% of its ability to transform NIH 3T3 fibroblasts (9, 11, 25, 26). In agreement with these data, we demonstrate here that within this location, H-Ras is fully responsive to GEFs. This may seem in conflict with a recent study in which no activation of Ras was detected on the ER, as opposed to the Golgi, upon mitogenic stimulation (11). However, it should be noticed that that study was undertaken with COS-1 cells, which do not express RasGRF1 or RasGRF2 (our unpublished results). Furthermore, the mitogenic stimulus used therein was epidermal growth factor, to which any trace of RasGRF would be entirely unresponsive (18, 36, 48, 59). Therefore, rather than contradicting our present results, it adds further support to the notion that RasGRF family GEFs would be the main mediators of Ras activation in the ER and suggests that SOS, which we demonstrate herein to be capable of activating the H-Ras ER pool, would play a more restricted role in the activation of Ras at this location.
From our data, it can be concluded that the PM H-Ras pool is a nonspecific target for all RasGEF families, since RasGRFs, SOSs, and RasGRPs (10) are all capable of catalyzing nucleotide exchange therein. H-Ras activation in endomembranes appears to be a more selective process, where different RasGEF families display marked preferences. The explanation of this phenomenon is not known but may indicate some relationship with the tissue-specific roles that Ras-GRPs and RasGRFs possess (43). In this respect, it is well known that RasGRFs are preferentially expressed in the nervous system (18, 21, 34, 54), in particular in mature neurons (58), basically non-proliferative tissue. Therefore, it could be hypothesized that the role of RasGRF could be that of a GEF specialized in regulating Ras signaling under nonmitogenic stimulation. Although highly speculative at this stage, evidence is mounting that spatial and temporal variations in calcium signals can dramatically affect biological outcomes (16). For example, calcium triggers exocytosis within microseconds at the synaptic junctions but calcium signals that last over minutes to hours are required to drive events such as gene transcription or cell proliferation (5). Therefore, it is tempting to speculate that at the ER, the main source of internal calcium, the function of RasGRF would be that of a GEF especially sensitive to acute, short-duration calcium peaks, thereby modulating the activation of Ras in a nonproliferative fashion. As of today, the biological significance of the activation of H-Ras in endomembranes is still unclear. Among the plethora of H-Ras biochemical and biological functions, we ignore those that are being specifically regulated from the different, confined platforms where Ras activation ensues. Without any doubt, this will be a hot topic of research in the near future and will surely provide valuable clues to explain the specific roles of GEFs in distinct endomembrane systems.
Acknowledgments
We thank X Bustelo, L. A. Feig, R. R. Mattingly, C. E. Machamer, M. F. Moran, H. P. Hauri, J. M. Rojas, J. C. Stone, and E. Santos for providing us with reagents.
The laboratory of P.C. is supported by grant BMC2002-01021 from the Spanish Ministry of Education and grant 01-087 from the Association for International Cancer Research (AICR). M.L. and M.T.B. are supported by grant BFI2002-00454 and G.E. is supported by grant SAF 2000-0042 from the Spanish Ministry of Education. I.A., D.M., and F.C. are predoctoral fellows of the Spanish Ministry of Education. V.S. is a Lady Tata Memorial Trust postdoctoral fellow.
REFERENCES
- 1.Ajenjo, N., D. S. Aaronson, E. Ceballos, C. Richard, J. León, and P. Crespo. 2000. Myeloid leukemia cell growth and differentiation are independent of mitogen-activated protein kinases ERK1/2 activation. J. Biol. Chem 275:7189-7197. [DOI] [PubMed] [Google Scholar]
- 2.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arozarena, I., D. S. Aaronson, D. Matallanas, V. Sanz, N. Ajenjo, S. Tenbaum, H. Teramoto, T. Ighishi, J. C. Zabala, J. S. Gutkind, and P. Crespo. 2000. The Rho family GTPase Cdc42 regulates the activation of Ras/MAPK pathway by the exchange factor Ras-GRF. J. Biol. Chem. 275:26441-26448. [DOI] [PubMed] [Google Scholar]
- 4.Arozarena, I., D. Matallanas, and P. Crespo. 2001. Maintenance of Cdc42 GDP-bound state by Rho-GDI inhibits MAP kinase activation by the exchange factor Ras-GRF. J. Biol. Chem. 276:21878-21884. [DOI] [PubMed] [Google Scholar]
- 5.Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signaling: dynamics homeostasis and remodeling. Nat. Rev. Mol. Cell Biol. 4:517-529. [DOI] [PubMed] [Google Scholar]
- 6.Bivona, T. G., I. Perez de Castro, I. M. Ahearn, T. M. Grana, V. K. Chiu, P. J. Lockyer, P. J. Cullen, A. Pellicer, A. D. Cox, and M. R. Philips. 2003. Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424:694-698. [DOI] [PubMed] [Google Scholar]
- 7.Buchsbaum, R., J. B. Telliez, S. Goonesekera, and L. A. Feig. 1996. The N-terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor Ras-GRF act cooperatively to facilitate activation by calcium. Mol. Cell. Biol. 16:4888-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buday, L., and J. Downward. 1993. Epidermal growth factor regulates p21 Ras through the formation of a complex of receptor, Grb2 adaptor protein and SOS nucleotide exchange factor. Cell 73:611-620. [DOI] [PubMed] [Google Scholar]
- 9.Cadwallader, K. A., H. Paterson, S. G. MacDonald, and J. F. Hancock. 1994. N-terminal myristoylated ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol. Cell. Biol. 14:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caloca, M. J., J. L. Zugaza, and X. R. Bustelo. 2003. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 278:33465-33473. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson, A. D. Cox, and M. R. Philips. 2002. Ras signaling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 12.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 13.Crespo, P., T. G. Cachero, N. Xu, and J. S. Gutkind. 1995. Dual effect of β-adrenergic receptors on mitogen-activated protein kinase: evidence for a βγ-dependent activation and Gαs-cAMP-mediated inhibition. J. Biol. Chem. 270:25259-25265. [DOI] [PubMed] [Google Scholar]
- 14.Crespo, P., and J. Leon. 2000. Ras proteins in the control of the cell cycle and cell differentiation. Cell. Mol. Life Sci. 57:1613-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crespo, P., N. Xu, W. F. Simonds, and J. S. Gutkind. 1994. Ras-dependent activation of MAP kinase pathway mediated by G-protein βγ subunits. Nature 369:418-420. [DOI] [PubMed] [Google Scholar]
- 16.Cullen, P. J., and P. J. Lockyer. 2002. Integration of calcium and Ras signaling. Nat. Rev. Mol. Cell Biol. 3:339-348. [DOI] [PubMed] [Google Scholar]
- 17.Egea, J., C. Espinet, R. M. Soler, S. Peiro, N. Rocamora, and J. X. Comella. 2000. Nerve growth factor activation of the extracellular signal-regulated kinase pathway is modulated by Ca2+ and calmodulin. Mol. Cell. Biol. 20:1931-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fam, N. P., W. Fan, Z. Wang, L. Zhang, H. Chen, and M. F. Moran. 1997. Cloning and characterization of Ras-GRF2, a novel nucleotide exchange factor of Ras. Mol. Cell. Biol. 17:1396-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, W., C. A. Koch, C. L. de Hoog, N. P. Fam, and M. F. Moran. 1998. The exchange factor Ras-GRF2 activates Ras-dependent and Rac-dependent mitogen activated protein kinase pathways. Curr. Biol. 8:935-938. [DOI] [PubMed] [Google Scholar]
- 20.Farnsworth, C. L., N. W. Freshney, L. B. Rosen, A. Ghosh, M. E. Greenberg, and L. A. Feig. 1995. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature 376:524-527. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Medarde, A., L. M. Esteban, A. Nuñez, A. Porteros, L. Tessarollo, and E. Santos. 2002. Targeted disruption of Ras-GRF2 shows its dispensability for mouse growth and development. Mol. Cell. Biol. 22:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freshney, N. W., S. D. Goonesekera, and L. A. Feig. 1997. Activation of the exchange factor Ras-GRF by calcium requires an intact Db1 homology domain. FEBS Lett. 407:111-115. [DOI] [PubMed] [Google Scholar]
- 23.Guerrero, C., J. M. Rojas, M. Chedid, Esteban. L. M., D. Zimonjic, N. Popescu, J. Font de Mora, and E. Santos. 1996. Expression of alternative forms of Ras exchange factors GRF and SOS1 in different human tissues and cell lines. Oncogene 12:1097-1107. [PubMed] [Google Scholar]
- 24.Hancock, J. F., K. A Cadwallader, H. Paterson, and C. J. Marshall. 1991. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targetting of Ras proteins. EMBO J. 10:4033-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock, J. F., A. I. Magee, J. E. Childs, and C. J. Marshall. 1989. All Ras proteins are polyisoprenylated but only some are palmytoylated. Cell 57:1167-1177. [DOI] [PubMed] [Google Scholar]
- 26.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmytoilation is required in addition to the CAAX motif to localize p21Ras to the membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 27.Innocenti, M., R. Zippel, R. Brambrilla, and E. Sturani. 1999. CDC25 Mm/Ras-GRF1 regulates both Ras and Rac signaling pathways. FEBS Lett. 460:357-362. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, C. L. 2000. Brefeldin A revealing the fundamental principles governing membrane dynamics and protein transport. Subcell. Biochem. 34:233-272. [DOI] [PubMed] [Google Scholar]
- 29.Jaumot, M., J. Yan, J. Clyde-Smith, J. Sluimer, and J. F. Hancock. 2002. The linker domain of the Ha-Ras hypervariable region regulates interactions with exchange factors, Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 277:272-278. [DOI] [PubMed] [Google Scholar]
- 30.Jones, M. K., and J. H. Jackson. 1998. Ras-GRF activates Ha-Ras but not N-Ras or K-Ras4B protein in vivo. J. Biol. Chem. 273:1782-1787. [DOI] [PubMed] [Google Scholar]
- 31.Li, N., A. Batzer, R. Daly, V. Yajnik, E. Skolnick, P. Chardin, D. Bar-Sagi, B. Margolis, and J. Schlessinger. 1993. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signaling. Nature 363:85-88. [DOI] [PubMed] [Google Scholar]
- 32.Lowy, D. R., and B. M. Willumsen. 1993. Function and regulation of Ras. Annu. Rev. Biochem. 62:851-891. [DOI] [PubMed] [Google Scholar]
- 33.Malumbres, M., and A. Pellicer. 1998. Ras pathways to cell cycle control and cell transformation. Front. Biosci. 3: d887-d912. [DOI] [PubMed]
- 34.Martegani, E., M. Vanoni, R. Zippel, P. Coccetti, R. Brambilla, C. Ferrari, E. Sturani, and L. Alberghina. 1992. Cloning by functional complementation of a mouse cDNA encoding a homologue of CDC25, a Saccharomyces cerevisiae Ras activator. EMBO J. 11:2151-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matallanas, D., I. Arozarena, M. T. Berciano, D. S. Aaronson, A. Pellicer, M. Lafarga, and P. Crespo. 2003. Differences in the inhibitory specificities of H-Ras, K-Ras and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J. Biol. Chem. 278:4572-4581. [DOI] [PubMed] [Google Scholar]
- 36.Mattingly, R. R., and I. G. Macara. 1996. Phosphorylation-dependent activation of the Ras-GRF exchange factor by muscarinic receptors and G-protein βγ subunits. Nature 382:268-272. [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki, N., S. Yamashita, K. Kurokawa, Y. Ohba, T. Nagai, A. Miyakawi, and M. Matsuda. 2001. Spatio-temporal images of growth factor induced activation of Ras and Rap1. Nature 411:1065-1068. [DOI] [PubMed] [Google Scholar]
- 38.Ohba, Y., K. Kurokawa, and M. Matsuda. 2003. Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 22:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papp, S., E. Dziak, M. Michalak, and M. Opas. 2003. Is all the endoplasmic reticulum created equal? The effects of the heterogeneous distribution of endoplasmic reticulum Ca2+ handling proteins. J. Cell Biol. 160:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelham, H. R. 1996. The dynamic organisation of the secretory pathway. Cell Struct. Funct. 21:413-419. [DOI] [PubMed] [Google Scholar]
- 41.Pierret, P., A. Vallee, N. Mechawar, N. A. Dower, J. C. Stone, P. M. Richardson, and R. J. Dunn. 2001. Cellular and subcellular localization of Ras guanyl nucleotide-releasing protein in the rat hippocampus. Neuroscience 108:381-390. [DOI] [PubMed] [Google Scholar]
- 42.Prior, I. A., and J. F. Hancock. 2001. Compartmentalization of Ras proteins. J. Cell Sci. 114:1603-1608. [DOI] [PubMed] [Google Scholar]
- 43.Quilliam, L. A., J. F. Rebhun, and A.F. Castro. 2002. A growing family of guanine nucleotide exchange factors is responsible for activation of ras family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 71:391-444. [DOI] [PubMed] [Google Scholar]
- 44.Roy, S., B. Wyse, and J. F. Hancock. 2002. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol. Cell. Biol. 22:5128-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saraste, J., and E. Kuismanen. 1984. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell 38:535-549. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, A., and A. Hall. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587-1609. [DOI] [PubMed] [Google Scholar]
- 47.Shou, C., C. L. Farnsworth, B. G. Neel, and L. A. Feig. 1992. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature 358:351-354. [DOI] [PubMed] [Google Scholar]
- 48.Shou, C., A. Wurmser, K. Ling, M. Barbacid, and L. A. Feig. 1995. Differential response of the Ras exchange factor, Ras-GRF to tyrosine kinase and G protein mediated signals. Oncogene 10:1887-1893. [PubMed] [Google Scholar]
- 49.Swift, A. M., and C. E. Machamer. 1991. A Golgi retention signal in a membrane spanning domain of coronavirus E1 protein. J. Cell Biol. 115:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, S. J., and D. Shalloway. 1996. Cell-cycle dependent activation of Ras. Curr. Biol. 6:1621-1627. [DOI] [PubMed] [Google Scholar]
- 51.Vetter, S. W., and E. Leclere. 2003. Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 270:404-414. [DOI] [PubMed] [Google Scholar]
- 52.Villalonga, P., C. Lopez-Alcala, M. Bosch, A. Chiloeches, N. Rocamora, J. Gil, R. Marais, C. J. Marshall, O. Bachs, and N. Agell. 2001. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 21:7345-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei, W., B. Das, W. Park, and D. Broek. 1994. Cloning and analysis of human cDNA encoding a 140 kDa brain guanine nucleotide exchange factor Cdc25 GEF, which regulates the function of Ras. Gene 151:279-284. [DOI] [PubMed] [Google Scholar]
- 54.Wei, W., S. S. Schereiber, M. Baudy, G. Tocco, and D. Broek. 1993. Localization of the cellular expression pattern of cdc25 NEF and Ras in the juvenile rat brain. Mol. Brain Res. 19:339-344. [DOI] [PubMed] [Google Scholar]
- 55.Welman, A., and M. M. Burger. 2000. Structure and function of the C-terminal hypervariable region of K-Ras4B in plasma membrane targeting and transformation. Oncogene 19:4582-4591. [DOI] [PubMed] [Google Scholar]
- 56.Willumsen, B. M., A. Christensen, N. L. Hubbert, A. G. Papageorge, and D. R. Lowy. 1984. The p21 Ras c-terminus is required for transformation and membrane association. Nature 310:583-586. [DOI] [PubMed] [Google Scholar]
- 57.Zippel, R., M. Balestrini, M. Lomazzi, and E. Sturani. 2000. Calcium and calmodulin are essential for Ras-GRF1-mediated activation of the Ras pathway by lysophosphatidic acid. Exp. Cell Res. 258:403-408. [DOI] [PubMed] [Google Scholar]
- 58.Zippel, R., N. Gnesuta, N. Matus-Leibovitch, E. Mancinelli, D. Saya, Z. Vogel, and E. Sturani. 1997. Ras-GRF the activator of Ras is preferentially expressed in mature neurons of the central nervous system. Mol. Brain Res. 48:140-144. [DOI] [PubMed] [Google Scholar]
- 59.Zippel, R., S. Orecchia, E. Sturani, and E. Martegani. 1996. The brain specific Ras exchange factor CDC25Mm: modulation of its activity through Gi mediated signals. Oncogene 12:2697-2703. [PubMed] [Google Scholar]