Abstract
Escherichia coli O127:K63(B8) possesses high human blood group H (O) activity due to its O-antigen repeating unit structure. In this work, the wbiQ gene from E. coli O127:K63(B8) was expressed in E. coli BL21 (DE3) and purified as a fusion protein containing an N-terminal GST affinity tag. Using the GST-WbiQ fusion protein, the wbiQ gene was identified to encode an α1,2-fucosyltransferase using a radioactivity based assay, thin layer chromatography assay, as well confirming product formation by using mass spectrometry and NMR spectroscopy. The fused enzyme (GST-WbiQ) has an optimal pH range from pH 6.5 to pH 7.5 and does not require the presence of a divalent metal to be enzymatically active. WbiQ displays strict substrate specificity, displaying activity only towards acceptors that contain Gal-β1,3-GalNAc-α-OR linkages; indicating that both the Gal and GalNAc residues are vital for enzymatic activity. In addition, WbiQ was used to prepare the H-type 3 blood group antigen, Fuc-α1,2-Gal-β1,3-GalNAc-α-OMe, on a milligram scale.
Keywords: Escherichia coli, WbiQ, O-antigen, fucosyltransferase
1. Introduction
Fucosylation, the enzymatic transfer of L-fucose to either an oligosaccharide or a protein, is accomplished by a class of enzymes called fucosyltransferases (FucTs). FucTs are an important class of enzymes for both mammals as well as bacteria. In mammalian systems, fucose containing glycoconjugates are directly involved in many biological processes, such as fertilization, neuronal development, immune responses, cell adhesion, and in many human diseases [1–3]. For example, fucosylation occurs during the synthesis of the ABO(H) and Lewis antigens, which play important roles in human physiology [4, 5]. Contrastingly, fucosylation in prokaryotes is commonly observed in the O-antigens present in Gram-negative bacteria, the exposed portion of the lipopolysaccharides (LPS) [6]. Functions arising due to the O-antigens include but are not limited to virulence, molecular mimicry, clearance from the host’s immune system, cell adhesion, and localization [7, 8].
FucTs catalyze the transfer of one fucose residue from the donor, guanosine-5′-diphospho-β-L-fucose (GDP-Fuc), to a saccharide acceptor, forming a new glycosidic linkage. Based on the new glycosidic linkage formed (typically α1,2-, α1,3-, α1,4-, or α1,6-) FucTs can be classified into four different subfamilies. Among them, α1,2-FucTs belong to glycosyltransferase family 11 (http://www.cazy.org/fam/acc_GT.html) and are responsible for the transfer of fucose to galactose (gal) forming an α1,2-linkage. Belonging to this family are many other α1,2-FucTs from humans, other mammals, viruses, plants, and bacteria. Several of the genes responsible for eukaryotic α1,2-fucosyltransferases have been cloned and characterized, some of which have come from humans [9–12]. FUT1 and FUT2 are two human α1,2-FucTs that are responsible for the biosynthesis of different H-antigens [13]. Importantly, only a few α1,2-FucTs have been cloned from bacterial sources and subsequently characterized: WbsJ from E. coli O127, WbwK from E. coli O86, and α1,2-FucT from Helicobacter pylori [14–16]. Of these WbsJ and α1,2-FucT from H. pylori have unique substrate specificities and have demonstrated applicability in the synthesis of relevant fucose containing oligosaccharides [14, 16]. Regardless of the species that the α1,2- FucT was cloned from, no structural information is yet available for the α1,2-FucT subfamily (unlike the evolutionary related α1,6-FucT subfamily) [17]. As such, our understanding of the α1,2-FucT mechanism and roles of specific amino acid motifs are limited and are based off the available α1,6-FucT and α1,3-FucT crystal structures.
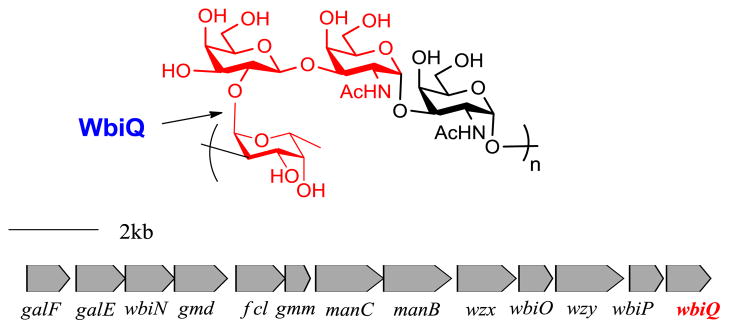
Enteropathogenic strain Escherichia coli O127:K63(B8) (EPEC) is associated with infantile diarrhea in developing countries and is an example of a pathogen that displays blood group antigens on its cell surface [18, 19]. E. coli O127’s O-antigen structure (Figure 1) expresses molecular mimicry of human blood group H-antigen and was reported to possess human blood group H (O) activity [20]. From the O-antigen biosynthetic gene cluster multiple genes were identified as glycosyltransferases involved in the assembly of the E. coli O127 polysaccharide, from which orf13 was identified as a putative α1,2-FucT [15, 21]. Thus, we propose that WbiQ encodes an α1,2-FucT that makes Fuc-α1,2-Gal-β-1,3-GalNAc (human blood group H-antigen mimic) present in the O-antigen repeating unit. Herein we describe the method for the overexpression, purification, and identification of the subcellular localization of GST-WbiQ. After overexpression in E. coli, the activity of WbiQ was optimized under different pH conditions and the influence of metal cations was tested. Furthermore, using a panel of acceptors, WbiQ showed strict acceptor substrate specificity and was only active toward acceptors that contained the Gal-β1,3-GalNAc-α-OR structure, forming Fuc-α1,2- Gal-β1,3-GalNAc-α-OR. Based on the acceptor substrate specificity, WbiQ was used in the preparative synthesis of H-type 3 blood group antigen (Fuc-α1,2- Gal-β1,3-GalNAc-α-OMe).
Figure 1.
O-antigen repeat unit structure of E. coli O127 and its biosynthetic gene cluster. The bond formed by WbiQ is indicated with an arrow, and the residues highlighted in red form the H-antigen mimic.
2. Materials and methods
2.1 Bacteria strains, plasmids, and reagents
E. coli competent cell DH5α [lacZΔM15 hsdR recA] was obtained from Invitrogen. E. coli competent cell BL21 (DE3) [F ompT hsdSB (rBmB) gal dcm (DE3)] was obtained from Stratagene. The plasmid, pGEX-4T-1 was obtained from GE Healthcare Life Sciences. Restriction enzymes were obtained from New England Biolabs. All reagents were from Sigma Aldrich unless otherwise noted.
2.2 Cloning and construction of wbiQ recombinant vector
The wbiQ gene was amplified by polymerase chain reaction (PCR) from chromosomal DNA of E. coli O127 with the forward primer 5′-ATGCGAATTCATGATGTATTGCTGTCTATCC (EcoRI restriction site underlined) and the reverse primer 5′-ATGCCTCGAGCTACATTGCTATCCAGTTT (XhoI restriction site underlined). The PCR product was digested with EcoRI and XhoI and inserted into the EcoRI/XhoI sites of plasmid pGEX-4T-1 such that the resulting expression plasmid, pGEX-wbiQ, has WbiQ fused to the gene encoding glutathione S-transferase (GST) in the same open reading frame. The constructs were transformed into E. coli DH5α cells, and the resulting recombinant plasmid was characterized by restriction mapping and DNA sequencing. The correct constructs were transformed into E. coli BL21 (DE3) for protein expression.
2.3 Overexpression and purification of WbiQ
E. coli BL21 (DE3) strain harboring the recombinant plasmid was grown in 1 L of LB medium at 37 °C. Once the OD600 reached 0.8, isopropyl-1-thio-β-D-galactosylpyranoside(IPTG) was added to a final concentration of 0.5 mM for induction. Protein expression proceeded for 15 hours at 16 °C. Cells were harvested by centrifugation (5000 g) and stored at −20 °C until needed. The cell pellet was resuspended in GST binding buffer (1x PBS, pH 7.4) and disrupted by sonication on ice (Branson Sonifier 450). The cell lysate was cleared by centrifugation (10000g, 45 min, 4 C) and the supernatant was loaded on to 4 mL of Glutathione Sepharose 4B slurry (GE Healthcare Life Sciences). The protein was subsequently eluted with GST elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione). Size exclusion chromatography was performed using Superdex 200 10/300 GL Column (GE Healthcare Life Sciences) equilibrated with 50 mM Tris-HCl, pH 7.5, 10% glycerol. Following the manufacturers’ protocol the column was calibrated using both the high and low molecular weight kits (GE Healthcare) and the molecular weight of the eluted GST-fusion protein was determined. The homogenous GST-WbiQ was stored at −80 °C in a buffer containing 50 mM Tris-HCL, pH 7.5, and 10% glycerol.
2.4 SDS-PAGE analysis, western blot analysis, and protein quantification
Protein expression and purification were analyzed by 12% SDS-PAGE and stained with Coomassie Brilliant Blue R250. For western blot analysis, GST-WbiQ was separated by 12% SDS-PAGE then electrophoretically transferred onto a Nitrocellulose membrane (Invitrogen), followed by blocking with 5% nonfat dry milk in 1x PBS buffer. All incubations were performed for 1 hour at room temperature, followed by 3 washings (10 min each) with 1x PBS-T. The GST tagged protein was first probed with mouse anti-GST polyclonal antibody (1:1000, Cell Signaling Technology). The blot was then probed with HRP-conjugated goat anti-mouse IgG (1:2000, GE Healthcare Life Sciences) and developed using either 3,3′,5,5′-Tetramethylbenzidine (TMB) or the ECL Western Blotting Detection Reagents (GE Healthcare). Protein concentration was determined by using the BCA Protein Assay Kit (Thermo Scientific).
2.5 Fucosyltransferase activity assay, effects of pH and metal cations
Enzyme activity was determined at 37 C for 1 hour in a final volume of 100 μL containing 20 mM Tris-HCl (pH 7.5), 0.3 mM GDP-β-L-fucose (supplemented with GDP- L-[U-14C]fucose (9000 cpm, American Radiolabeled Chemicals), 20 mM acceptor, and 10 μg GST-WbiQ. The acceptor was omitted in the control reaction. The reaction was terminated by adding 100 μL of ice cold water followed by addition of 800 μL (v/v = 1/1) Dowex 1×8 200–400 anion exchange resin. The mixture was centrifuged and the resulting supernatant was collected in a 20 mL plastic vial containing 10 mL of Scintiverse BD (Fisher Scientific). After thorough vortexing, the radioactivity of the mixture was counted in a Beckmann LS-3801 liquid scintillation counter. The activity of WbiQ under varying pH conditions was determined with 10 μg of GST-WbiQ in a 100 μL reaction mixture containing variable pH conditions (pH 5.0–9.5), 0.3 mM GDP-Fucose, and 20 mM Gal-β1,3-GalNAc-OMe for one hour. The activity of WbiQ in the presence of various divalent metal cations was determined in a 100 μL solution containing 10 μg GST-WbiQ, 20 mM Tris-HCl (pH 7.5) 0.3 mM GDP-Fucose, 20 mM Gal-β1,3-GalNAc-OMe, and 10 mM of a divalent metal, reacting for one hour.
2.6 Enzymatic synthesis of H-type 3 blood group
Using 1 mg of GST-WbiQ, milligram scale synthesis of Fuc-α1,2-Gal-β1,3-GalNAc-OMe was performed in a final volume of 3.0 mL at 37 C containing 20 mM Tris-HCl (pH 7.5), 10 mM Gal-β1,3-GalNAc-OMe (as prepared in [21]), and 15 mM GDP-fucose (as prepared in [22]). The reaction was monitored by thin-layer chromatography [i-PrOH/H2O/NH4OH = 7:3:2 (v/v/v)]. Products were stained with anisaldehyde/MeOH/H2SO4 = 1:15:2 (v/v/v). After complete conversion of acceptor to product, the protein was removed by boiling, followed by centrifugation (12000 g, 15 min). Excess GDP-fucose and the by product, GDP, were removed by anion exchange chromatography, and the final trisaccharide product was purified by Bio-Gel P-2 gel filtration (Bio-Rad) with a water mobile phase. The desired fractions were pooled, lyophilized, and stored at −20 °C.
2.7 Mass spectrometry and NMR
Electrospray ionization mass spectrometry (ESI-MS) assay was conducted using at The Ohio State University mass spectrometry facility on a Bruker micrOTOF Instrument provided by a grant from the Ohio BioProducts Innovation Center. 1H NMR and 13C NMR (Bruker Avance 500 MHz NMR spectrometer) were used for product confirmation. The trisaccharide product was dissolved in D2O and lyophilized before the NMR spectra were recorded at 303 K in a 5 mM tube.
3. Results and Discussion
3.1 Identification of wbiQ
From previous work, the O-antigen biosynthesis gene cluster was identified and several glycosyltransferases and processing enzymes were indentified from E. coli O127 (GenBank Accession no. AY493508). Among them, wbiQ, was identified as a putative α1,2-fucosyltranferase. After performing a BLAST search of WbiQ (Protein Accession no. AAR90894), it was identified to belong to glycosyltransferase family 11, characterized by a putative conserved domain. Glycosyltransferase family 11 contains α1,2-FucTs from all domains of life such as bacteria, virus, mammals, and humans [7]. The 299 amino acid WbiQ demonstrated high level amino acid identity towards several other bacterial α1,2-FucTs: WfbI from Salmonella enterica O13 (61%) and WbwK from E. coli O86 (48%). Also in this family, with lower sequence identity, are several other characterized fucosyltransferases such as WbsJ of E. coli O128:B12 (26%), FutC from H. pyolori (25%), and human FUT1 (15%). Similar to recently characterized WbsJ, WbiQ contains several conserved motifs, shown by the sequence alignment (Figure S1). The three motifs labeled I, II, and III are conserved across both bacterial and mammalian α1,2-FucTs, α1,6-FucTs, and O-FucTs. Motif I contains several basic residues, notably HxRRxD, which has been shown to be important for interacting with the donor GDP-fucose. The other two motifs, motif II and III, were observed in the crystal structure of human α1,6-FucT (FUT8), which may be involved with binding of GDP-fucose [23]. However, to fully elucidate the roles of these motifs in α1,2-FucTs, a three-dimensional structure would need to be determined.
Comparing WbiQ to recently characterized WbwK, they exhibit approximately 48% sequence similarity [15]. Similar to WbwK, WbiQ contains a putative transmembrane domain as identified by using TMpred (Prediction of Transmembrane Regions and Orientation), amino acids 246 to 264. Bacterial glycosyltransferases involved with the O-antigen biosynthesis are associated with the inner membrane facing the cytoplasmic side [24, 25].
3.2 Expression, purification, and subcellular localization of WbiQ from E. coli O127
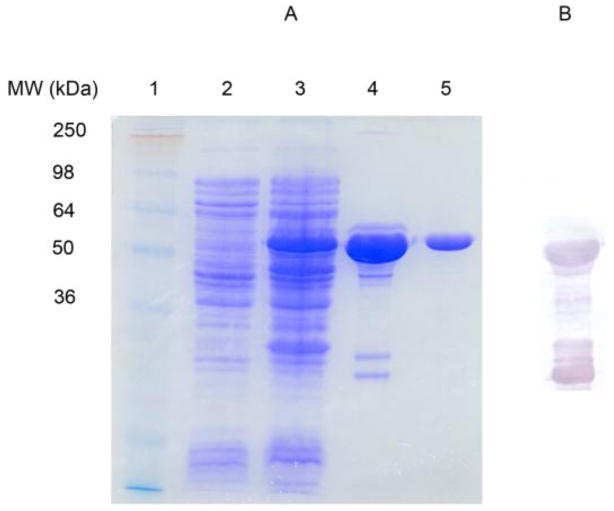
While previous attempts at expressing WbiQ with a His6 tag using the pET-15b produced large concentrations of enzyme (200 mg/mL), the purified protein was not enzymatically active. Thus pGEX-4T-1 was chosen to express WbiQ with a GST affinity tag in order to improve the enzyme’s solubility and stability. Expression of WbiQ with an N-terminal GST tag was carried out in 1 L of LB under induction of IPTG. The fusion protein GST-WbiQ was purified in one step GST-affinity chromatography, as shown by 12% SDS-PAGE (Figure 2A, Lane 4). The recombinant protein has an apparent molecular weight of 60 kD as estimated by the SDS-PAGE and anti-GST western blot (Figure 2B), which is similar to the theoretical molecular weight (61 kD), as calculated from its primary amino acid sequence. The major impurity from the SDS-PAGE (Lane 4) and anti-GST western blot appears to be soluble GST and/or truncated forms of GST-WbiQ. As such, we attempted to cleave the GST tag from the fusion protein by using thrombin; however, the cleavage efficiency was low and there appeared to be nonspecific cleavage. Thus, GST-WbiQ was further purified to near homogeneity using gel filtration chromatography (Figure 2A Lane 5). From the gel filtration profile (Figure S2), the molecular weight of GST-WbiQ is approximately 120 kDa, indicating dimerization, which may be due to the fusion tag, considering GST exists as a homodimer in nature. The GST-fusion protein purified using gel filtration was subsequently used for all experiments.
Figure 2.
Purification of GST-WbiQ fusion protein. (A) SDS-PAGE; Lane 1: Molecular weight marker; Lane 2: Pre-induction; Lane 3: Post-induction with IPTG; Lane 4: GST-WbiQ after elution from GST-resin.; Lane 5: GST-WbiQ after elution from Superdex 200 gel filtration. (B) Anti-GST western blot from sample in Figure 2A, Lane 4.
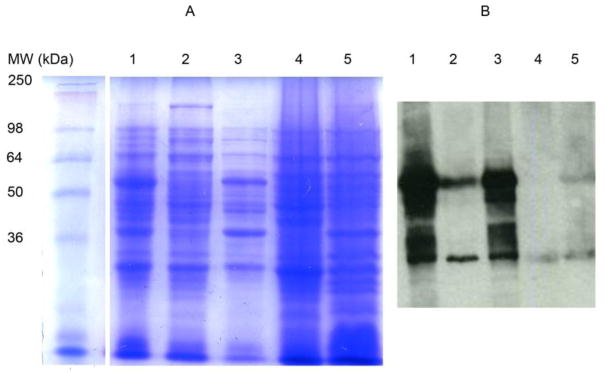
The subcellular localization of GST-WbiQ was investigated by using differential centrifugation methods, and was subsequently analyzed by SDS-PAGE and anti-GST western blot [26]. After lysis of the strain harboring pGEX-4T-1-wbiQ by sonication, the resulting lysate was subjected to centrifugation at 12000 g. Soluble GST-WbiQ (60 kDa) was present in the 12000 g centrifugation supernatant as shown by the anti-GST western blot in Figure 3B, Lane 2. The formation of the pellet after 12000 g centrifugation contained a significant amount of recombinant protein, Figure 3A and 3B Lane 3, suggesting that GST-WbiQ forms inclusion bodies. The supernatant after the 12000 g centrifugation was subjected to centrifugation at 50000 g for two hours, after which the supernatant was removed and centrifuged at 50000 g for another hour. The supernatant after 50000 g centrifugation contained soluble GST (22 kDa), as visualized by the anti-GST western blot, Figure 3 Lane 4. After purification of the membranes using ultracentrifugation, the resulting pellet contained both GST-WbiQ and GST, Figure 3B Lane 5, suggesting that GST-WbiQ is associated with the membrane in the E. coli host. This result is consistent with other bacterial glycosyltransferases, whereby the proteins are soluble but have some association with the inner membrane in their E. coli hosts [26]. While GST-WbiQ appears to associate with the inner membrane according to our studies, the roles of the GST-fusion tag and the putative transmembrane domain in this association are unknown.
Figure 3.
Subcellular localization of GST-WbiQ. (A) SDS-PAGE; Lane 1: Post-induction whole cell lysate; Lane 2: Supernatant after 12000 g centrifugation; Lane 3: Cell pellet formed after 12000 g centrifugation; Lane 4: Supernatant after 50000 g ultracentrifugation; Lane 5: Cell pellet formed after 50000 g ultracentrifugation. (B) Anti-GST western blot; Lane 1: Post-induction whole cell lysate; Lane 2: Supernatant after 12000 g centrifugation; Lane 3: Cell pellet formed after 12000 g centrifugation; Lane 4: Supernatant after 50000 g ultracentrifugation; Lane 5: Cell pellet formed after 50000 g ultracentrifugation.
3.3 Detection of α1,2-FucT activity and acceptor specificity
WbiQ belongs to glycosyltransferase family 11, and as such, is predicted to transfer L-fucose from GDP-β-L-fucose to β-D-Gal through an α1,2 linkage. Based on the O-antigen repeating unit of E. coli O127, a panel of acceptors were chosen to detect α1,2-FucT activity, as well as provide the relative activity for various acceptors. The results show that WbiQ is active with the Gal-β1,3-GalNAc-OR acceptors, which are derivatives of blood group T-antigen (Table 1). These acceptors are also structurally similar to the native O-antigen repeating unit. WbiQ exhibits strict acceptor substrate specificity, as it did not recognize any of the other disaccharides (lactose, lactulose, Gal-β1,4-glucitol) that contained the β-D-Gal residue at the nonreducing end. Acceptors Gb3 and α-Gal both have the β-D-Gal, not at the nonreducing end, and neither of these were suitable acceptors for WbiQ. Lastly, the monosaccharide, β-D-Gal, did not serve as a suitable acceptor for WbiQ. The inability to accept β-D-Gal contrasts many other α1,2-FucTs from family 11 that readily accept β-D-Gal as an acceptor [14, 16].
Table 1.
Acceptor substrate specificity of GST-WbiQ
| Acceptor | Specific Activity (nmol min−1 mg−1) |
|---|---|
| Gal-β1,3-GalNAc-O-Me (T-antigen) | 4.4 |
| Gal-β1,3-GalNAc-O-OH (T-antigen) | 3.9 |
| Gal-β1,4-Glc (Lactose) | ND* |
| Gal-β1,4-Fru (Lactulose) | ND |
| Gal-β1,4-glucitol | ND |
| Gal-β-OMe | ND |
| Gal-α1,3-Gal-β1,4Glc (Gb3) | ND |
| Gal-α1,4-Gal-β1,4Glc (α-Gal) | ND |
ND: not detectable.
Further verification of enzymatic activity was demonstrated by using TLC as the method of detection. A 100 μL reaction mixture was set up containing 10 μg of GST-WbiQ, with GDP-fucose as the donor and Gal-β1,3-GalNAc-OH as the acceptor. The trisaccharide product was visualized by TLC, whereby, after 12 hours of incubation at 37 °C a third spot is clearly visible, which runs slower than the acceptor (Figure 4 lane 4). After 48 hours, we observe complete consumption of GDP-fucose and formation of the trisaccharide product (Figure 4 lane 5).
Figure 4.
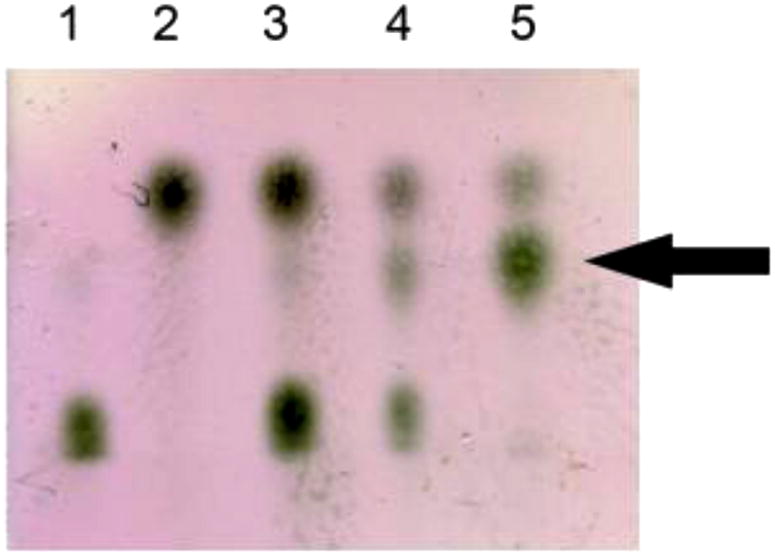
TLC demonstrating α1,2-FucT activity of GST-WbiQ. Lane 1: GDP-Fucose; Lane 2: Gal-β1,3-GalNAc-OH; Lane 3: Reaction mixture at time 1 hour; Lane 4: Reaction mixture after 12 hours; Lane 5: Reaction mixture after 48 hours; product formation indicated with an arrow.
3.4 Effects of pH and metal ions on α1,2-FucT activity
The effect of pH on α1,2-FucT activity was determined under varying pH conditions at 37 C using Gal-β1,3-GalNAc-OMe as the acceptor. Figure S3 shows the pH profile, which has an optimal pH range from 6.5 to 7.5. While the shape of the pH curve is the characteristic “bell shape,” there are two maximums at pH 6.5 and 7.5. This observed result may be due to the effect of the pH by ionizing a specific catalytic residue, affecting the binding affinity, affecting the stability of the protein, or a combination of all these effects [14].
WbiQ was labeled as a putative α1,2-fucosyltransferase belonging to glycosyltransferase family 11, which are characterized by a GT-B type fold. Glycosyltransferases characterized by a GT-B type fold typically exhibit activities independent of divalent metals. Contrastingly, GT-A type glycosyltransferases have a DXD motif, which coordinates a divalent metal, meaning a divalent metal is required for catalysis. WbiQ does not contain a DXD motif, and as such, it was expected not to require a metal for catalysis [27]. The effects of various divalent metal cations and EDTA on the α1,2-FucT activity of WbiQ were tested. From Figure S4 it showed that WbiQ does not require a divalent metal cation for catalysis, as it exhibited full activity when no divalent metal is added or when 10 mM EDTA is present. Upon adding 10 mM of any of the various divalent metal cations, inhibition of enzymatic activity is observed. These results are in agreement with other GT-B type fucosyltransferases whereby a metal binding site (DXD motif) is not present in the primary amino acid sequence and therefore enzymatic activity is independent of metal ions [14].
3.5 Preparative Enzymatic synthesis of H-type 3 blood group antigen
WbiQ was used to create the H-type 3 blood group antigen trisaccharide on a milligram scale. The reaction was carried out for 4 days and approximately 19 mg of Fuc-α1,2-Gal-β1,3-GalNAc-OMe was obtained from the reaction containing GDP-Fucose, Gal-β1,3-GalNAc-OMe, and purified GST-WbiQ. For confirmation of the correct linkage and structure, after gel filtration, the purified trisaccharide was analyzed by electrospray mass spectrometry and NMR. The assignment of 1H NMR and 13C NMR of Fuc-α1,2-Gal-β1,3-GalNAc-OMe are found in Figures S5 and S6, respectively, and are consistent with those reported previously [28].
4. Conclusions
In this work we identified, purified, characterized, and demonstrated the subcellular location of WbiQ, the putative α1,2-FucT from E. coli O127:K63(B8). The wbiQ gene was biochemically proven to encode an α1,2-FucT through a radioactivity based assay, TLC monitored assay, ESI/MS, and NMR. While the endogenous substrate (Gal-β1,3-GalNAc-α1,3-GalNAc-O-PP-Und) was not available for testing in our experiments, we demonstrate that WbiQ can efficiently recognize Gal-β1,3-GalNAc-α-OMe as a suitable substrate, possibly suggesting that the reducing end of the O-antigen repeating unit beyond this disaccharide may not be essential for WbiQ activity. Based on this substrate specificity, WbiQ could be characterized as a Family 4 α1,2-FucT whereby it recognizes Gal-β1,3-GalNAc-α acceptors but not other Gal-β containing acceptors; similar to WbwK from E. coli O86 and an α1,2-FucT from Caenorhabditis elegans [29]. Furthermore, we show that WbiQ can be used for the efficient synthesis of H-type 3 blood group antigen. WbiQ also appears to be a characteristic α1,2-FucT from glycosyltransferase family 11, containing several well conserved motifs as well as not containing a DxD metal cation binding motif. To further confirm the roles of the individual amino acids in motifs I-III within α1,2-FucTs, the three dimensional structure needs to be resolved. Due to the large concentration of soluble protein available from 1 L cultures (and unavailability of current α1,2-FucT crystal structures), crystallographic studies of WbiQ are currently underway.
Supplementary Material
Acknowledgments
P.G. Wang acknowledges the US National Institutes of Child Health & Human Development (R01 HD061935) and the US National Institutes of Genetic Medical Sciences (R01 GM085267) for financial support.
Abbreviations
- GT
glycosyltransferase
- Gal
galactose
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- Fuc
L-Fucose
- α1
2FucT, α1, 2-fucosyltransferase
- GDP-Fucose
guanosine-5′-diphospho-β-L-fucose
- IPTG
isopropyl-β-D-thiogalactopyranoside
- Me
methyl
- E. coli
Escherichia coli
- E. coli O127, Escherichia coli O127
K63(B8)
- GalNAc-O-PP-Und
Undecaprenyl linked GalNAc-pyrophosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES
- 1.Pang PC, Tissot B, Drobnis EZ, et al. Expression of bisecting type and Lewisx/Lewisy terminated N-glycans on human sperm. J Biol Chem. 2007;282:36593–602. doi: 10.1074/jbc.M705134200. [DOI] [PubMed] [Google Scholar]
- 2.Nishihara S, Iwasaki H, Nakajima K, et al. Alpha1,3-fucosyltransferase IX (Fut9) determines Lewis X expression in brain. Glycobiology. 2003;13:445–55. doi: 10.1093/glycob/cwg048. [DOI] [PubMed] [Google Scholar]
- 3.Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 4.Orntoft TF, Greenwell P, Clausen H, et al. Regulation of the oncodevelopmental expression of type 1 chain ABH and Lewis(b) blood group antigens in human colon by alpha-2-L-fucosylation. Gut. 1991;32:287–93. doi: 10.1136/gut.32.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oriol R, Samuelsson BE, Messeter L. ABO antibodies--serological behaviour and immuno-chemical characterization. J Immunogenet. 1990;17:279–99. doi: 10.1111/j.1744-313x.1990.tb00880.x. [DOI] [PubMed] [Google Scholar]
- 6.Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 7.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 8.Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr Res. 2008;343:1952–65. doi: 10.1016/j.carres.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Larsen RD, Ernst LK, Nair RP, et al. Molecular cloning, sequence, and expression of a human GDP-L-fucose:beta-D-galactoside 2-alpha-L-fucosyltransferase cDNA that can form the H blood group antigen. Proc Natl Acad Sci U S A. 1990;87:6674–8. doi: 10.1073/pnas.87.17.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarnesto A, Kohlin T, Hindsgaul O, et al. Purification of the secretor-type beta-galactoside alpha 1----2-fucosyltransferase from human serum. J Biol Chem. 1992;267:2737–44. [PubMed] [Google Scholar]
- 11.Hitoshi S, Kusunoki S, Kanazawa I, et al. Molecular cloning and expression of two types of rabbit beta-galactoside alpha 1,2-fucosyltransferase. J Biol Chem. 1995;270:8844–50. doi: 10.1074/jbc.270.15.8844. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Rasko DA, Sherburne R, et al. Molecular genetic basis for the variable expression of Lewis Y antigen in Helicobacter pylori: analysis of the alpha (1,2) fucosyltransferase gene. Mol Microbiol. 1999;31:1265–74. doi: 10.1046/j.1365-2958.1999.01268.x. [DOI] [PubMed] [Google Scholar]
- 13.Oriol R. Genetic control of the fucosylation of ABH precursor chains. Evidence for new epistatic interactions in different cells and tissues. J Immunogenet. 1990;17:235–45. doi: 10.1111/j.1744-313x.1990.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Liu XW, Shao J, et al. Characterization of a novel alpha1,2-fucosyltransferase of Escherichia coli O128:b12 and functional investigation of its common motif. Biochemistry. 2008;47:378–87. doi: 10.1021/bi701345v. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Shen J, Liu X, et al. Identification of a new alpha1,2-fucosyltransferase involved in O-antigen biosynthesis of Escherichia coli O86:B7 and formation of H-type 3 blood group antigen. Biochemistry. 2008;47:11590–7. doi: 10.1021/bi801067s. [DOI] [PubMed] [Google Scholar]
- 16.Daniel BS, Yu-Nong L, Chun-Hung L. Characterization of Helicobacter pylori alpha1,2-Fucosyltransferase for Enzymatic Synthesis of Tumor-Associated Antigens. Advanced Synthesis & Catalysis. 2008;350:2313–2321. [Google Scholar]
- 17.Chazalet V, Uehara K, Geremia RA, et al. Identification of essential amino acids in the Azorhizobium caulinodans fucosyltransferase NodZ. J Bacteriol. 2001;183:7067–75. doi: 10.1128/JB.183.24.7067-7075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivas M, Miliwebsky E, Balbi L, et al. Intestinal bleeding and occlusion associated with Shiga toxin-producing Escherichia coli O127:H21. Medicina (B Aires) 2000;60:249–52. [PubMed] [Google Scholar]
- 19.Stenutz R, Weintraub A, Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev. 2006;30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 20.Widmalm G, Leontein K. Structural studies of the Escherichia coli O127 O-antigen polysaccharide. Carbohydr Res. 1993;247:255–62. doi: 10.1016/0008-6215(93)84258-8. [DOI] [PubMed] [Google Scholar]
- 21.Yi W, Perali RS, Eguchi H, et al. Characterization of a bacterial beta-1,3-galactosyltransferase with application in the synthesis of tumor-associated T-antigen mimics. Biochemistry. 2008;47:1241–8. doi: 10.1021/bi7020712. [DOI] [PubMed] [Google Scholar]
- 22.Zhao G, Guan W, Cai L, et al. Enzymatic route to preparative-scale synthesis of UDP-GlcNAc/GalNAc, their analogues and GDP-fucose. Nat Protoc. 5:636–46. doi: 10.1038/nprot.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihara H, Ikeda Y, Toma S, et al. Crystal structure of mammalian alpha1,6-fucosyltransferase, FUT8. Glycobiology. 2007;17:455–66. doi: 10.1093/glycob/cwl079. [DOI] [PubMed] [Google Scholar]
- 24.Wang QM, Peery RB, Johnson RB, et al. Identification and characterization of a monofunctional glycosyltransferase from Staphylococcus aureus. J Bacteriol. 2001;183:4779–85. doi: 10.1128/JB.183.16.4779-4785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Boulton PG, Chan NW, et al. Novel Helicobacter pylori alpha1,2-fucosyltransferase, a key enzyme in the synthesis of Lewis antigens. Microbiology. 1999;145(Pt 11):3245–53. doi: 10.1099/00221287-145-11-3245. [DOI] [PubMed] [Google Scholar]
- 26.Saksouk N, Pelosi L, Colin-Morel P, et al. The capsular polysaccharide biosynthesis of Streptococcus pneumoniae serotype 8: functional identification of the glycosyltransferase WciS (Cap8H) Biochem J. 2005;389:63–72. doi: 10.1042/BJ20050217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lairson LL, Henrissat B, Davies GJ, et al. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–55. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 28.Yi W, Shao J, Zhu L, et al. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J Am Chem Soc. 2005;127:2040–1. doi: 10.1021/ja045021y. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Q, Van Die I, Cummings RD. A novel alpha1,2-fucosyltransferase (CE2FT-2) in Caenorhabditis elegans generates H-type 3 glycan structures. Glycobiology. 2008;18:290–302. doi: 10.1093/glycob/cwn007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.