Abstract
Single molecule observation of fluorescence resonance energy transfer can be used to provide insights into the structure and dynamics of proteins. Using a straightforward triple-colour labelling strategy, we present a measurement and analysis scheme that can simultaneously study multiple regions within single intrinsically disordered proteins.
Single molecule observation of fluorescence resonance energy transfer (smFRET) between a suitable dye pair has become a widely used tool in biology. Since smFRET can provide a dynamic distance readout on the molecular level, it has frequently been used to study intrinsically disordered proteins (IDPs).1–6 These proteins have recently gained much attention due to their involvement in many neurodegenerative diseases such as Alzheimer, Prion and Parkinson disease. Furthermore, IDPs are also enriched in many eukaryotic mechanisms, including nucleocytoplasmic transport, transcription and epigenetic regulation.7
In a classical dual-colour/single-pair smFRET experiment of a protein, a Donor (D) and an Acceptor (A) dye forming a FRET pair are site-specifically installed in the protein chain.8 Analysis of the resulting smFRET data reveals information about the structure and dynamics of the protein domain that is sandwiched between the two dyes. For a static protein, the FRET efficiency (EFRET) depends on the distance r between the two dyes according to
| (1) |
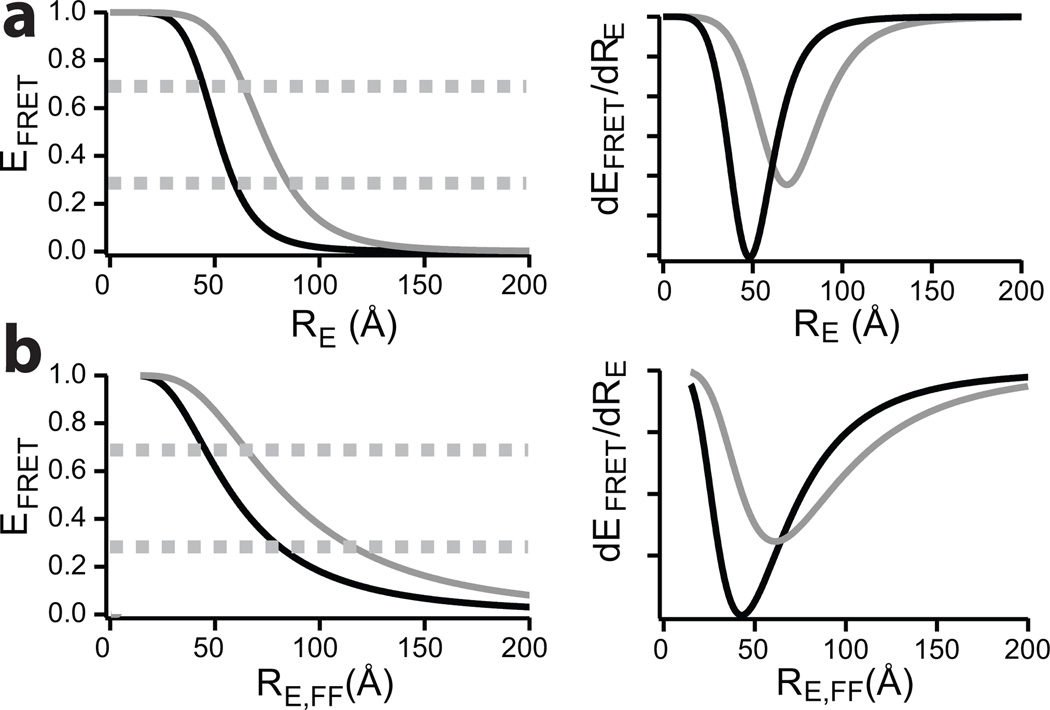
Where RE is the distance between the two dyes (defined as end-to-end distance, or more accurately in case of FRET, the dye-to-dye distance) and R0 is the Forster distance defined as the distance RE for which EFRET= 0.5. R0 is a characteristic of the dye pair under the given experimental conditions. Due to the power 6 dependence of EFRET on RE, FRET efficiency changes can be measured most accurately near R0, and thus long distances can be measured with higher accuracy using dye pairs with large R0 and vice versa for short distances (Figure 1). This effect is easily visualized when inspecting the first derivative of the EFRET vs RE distribution (Figure 1, right panels). For large proteins, a single dye pair can only be used to sample one part of the protein, making it very laborious to sample multiple regions with suitably accurate dye pairs. For example, for the approximately 600 amino acid long intrinsically disordered nucleoporin 153 phenylalanine-glycine (FG) repeat domain, 6 dual mutants are able to sample only 50% of the entire sequence.5
Figure 1.
Left panel shows the distance dependence of EFRET for the two dye pairs Alexa488-Alexa647 (black, R0 = 51 Å) and Alexa594-Alexa647 (grey, R0 = 73 Å) for static proteins in (a) and for dynamic proteins in (b). Right panel shows corresponding first derivative. Dashed lines show upper and lower boundaries for sensitive measurements in the left panel.
A key prerequisite for any smFRET study is the dual-labelling of the sample. In contrast to synthetically accessible DNA/RNA or small peptides, large proteins cannot be easily labelled site-specifically.6, 9, 10 When possible, the most common approach is to engineer two cysteines, which are then reacted with maleimide derivatives of single molecule suitable dyes. Since both cysteines offer the identical thiol chemistry, this usually leads to a random labelled sample containing DA, AD, DD and AA species, which can sometimes result in unwanted sample heterogeneities.11 A method bypassing this issue is the use of site-specific dual-labelling by introducing an orthogonal labelling site via a genetically encoded unnatural amino acid into a protein in addition to a specific single cysteine.6, 9, 10
It has been demonstrated that the concept of dual-colour FRET can also be extended to a three-colour dye system on the single molecule level. Here a second acceptor dye (A2) is introduced into the system, so that energy transfer can occur between D-A1, D-A2, but also from A1–A2, as A1 can now also function as a donor dye. The ability of three-colour single molecule FRET measurements to probe multiple proximities at the same time facilitates the study of more complex biological mechanisms. However, site-specific three-colour labelling of single proteins imposes additional difficulties. Consequently, most three-colour smFRET studies were performed intermolecularly and/or on nucleic acids.12–15 The same holds true for recent, still more complex, smFRET four-colour studies.16, 17
By using a combination of site-specific labelling via an unnatural amino acid and statistical labelling of two cysteine residues, we present here an extension of protein labelling to three-colour single molecule FRET measurements within a single protein. We show that this approach is applicable for studying IDPs with smFRET. Using a three-dye system with substantially different Forster distances in combination with multiplexed/alternating laser excitation,18, 19 we demonstrate that in fact the “random labelling” step can confer an advantage by offering an increased dynamic range of the smFRET measurement from a single sample preparation. We present a data analysis scheme that makes it possible to unambiguously identify properly labelled species that were prepared in two simple consecutive steps from a single sample preparation.
To develop a strategy to label a single protein with three dyes we choose the FG-rich domain of the yeast nucleoporin 49 (yNup49FG). These FG-rich proteins have previously been shown to be intrinsically disordered, i.e. no stable secondary structure can be observed in the native state.5, 6, 20 Such a protein can be described as a polymer chain that rapidly fluctuates in the buffer. To extract information about the disordered state of the protein, we aimed to label the protein with dye pairs that can undergo FRET if in sufficient proximity. To detect FRET on the single molecule level, we use confocal spot detection of freely diffusing molecules. In such a detection scheme, a single fluorescently labelled yNup49FG molecule will sample an ensemble of different states while diffusing through the laser focus for about 1 ms during which time it emits a fluorescence signal. In the case of such a dynamic protein, the EFRET term has to be extended to account for distance fluctuations between the two dyes that arise from the fast dynamics of the fluctuating chain.1, 4, 21, 22 As the protein explores a large conformational space in a short period, it basically behaves like a polymer in solution, and it has previously been described that this can be nicely approximated by a Gaussian chain model.1, 4, 21–23
Therefore the measured FRET efficiency can be described by
| (2) |
where the radial probability distribution of a Gaussian chain is
| (3) |
| (4) |
with the dye-to-dye distance for the fast fluctuating molecule being
| (5) |
Figure 1b illustrates that for dynamic proteins, the FRET efficiency has a shallower decay for long dye-to-dye distances compared to the static case (Figure 1b). As R0 is highly dependent on the overlap integral J(λ) (see Supplementary Methods) between the emission spectrum of the donor dye and the absorption spectrum of the acceptor dye, Alexa594-Alexa647 has a large R0 (~73 Å) and is thus well suited to measure long distances (Figure 1b for EFRET and first derivative plots). For measuring short distances Alexa488-Alexa647 is an ideal dye pair, since R0 is low (Figure 1b, ~51 Å). Furthermore, due to the Poisson nature of photon detection, the need to correct raw data for background, signal leakage, and the low signal, FRET efficiencies near 0 and 1 often suffer from numerical artefacts, so optimal measurements are performed in a FRET regime around EFRET= 50% (i.e ~30–70%).
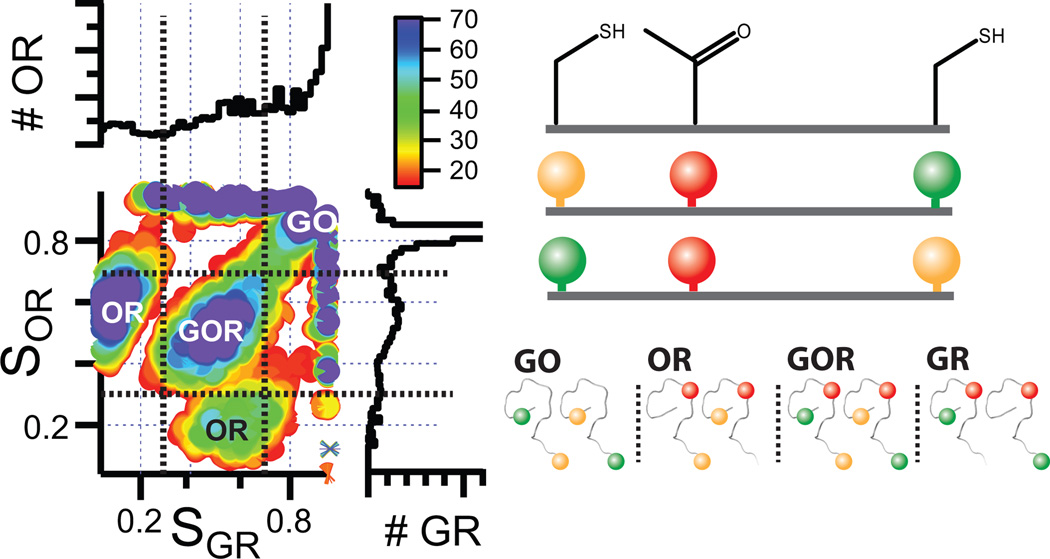
With a single dye pair, a single region of the protein can be studied. To extend beyond this, we label a protein with three dyes. To exploit the substantial difference of R0 for different dye pairs, we performed the following labelling strategy: We generated a yNup49FG mutant that contains two cysteines in the otherwise cysteine-free protein engineered at amino acid (AA) positions 120 and 250. In addition, the unnatural amino acid (UAA) p-acetylphenylalanine was introduced in response to suppression of the Amber nonsense mutation TAG by expressing the protein in an E. coli strain engineered with a suitable orthogonal tRNA/synthetase pair.9, 10 The Amber mutation TAG was purposely asymmetrically placed in between the two cysteines at position 155, such that the distance between one of the C and the TAG site is much shorter than the other (120C-155TAG= 35 AA vs. 155TAG-250C= 95 AA). Reaction with a hydroxylamine derivative of the red fluorescent Alexa647 (R) specifically labels this UAA position via oxime ligation.9, 10 The cysteines were reacted statistically in a 1:1 stoichiometry with green fluorescent Alexa488 (G) and orange fluorescent Alexa594 (O) maleimide dye derivatives. This creates more than eight different populations of labelled yNup49FG (Figure 2, right panel). To analyze this mixture without any further purification, we performed solution based measurements of single molecules using three multiplexing lasers. The first laser at λ= 488 nm excites Alexa488 and subsequent energy transfer to A1 and A2 are observed by detecting photon counts on three spectrally separated channels (green (G), orange (O) and red (R)). This laser pulse is followed by an orange pulse at λ= 570 nm, for detection of energy transfer from the orange (O, Alexa594) to the red dye (R, Alexa647). This is followed by another laser pulse at λ= 660 nm, which probes for the presence of red labelled species. Plotting the stoichiometry (S) of orange and red (SOR) vs. the S of green and red (SGR) yields a 2D plot showing several (Figure 2). We can then specifically select for those triple-labelled populations that contain one label each of Alexa488 (D, G), Alexa594 (A1, O) and Alexa647 (A2, R) (Figure 2). Due to the stochastic labelling of position 120C and 250C, the two different extracted species are 120C-G/155TAG-R/250C-O and 120C-O/155TAG-R/250C-G (shown in Figure 2).
Figure 2.
Stoichiometry analysis of sample containing up to 8 different labelled species. Plotting SOR vs SGR (left panel shows 2D plot as well projections on the individual axes) makes it possible to select the populations (black dashed lines) that contain each dye once (GOR labelled yNup49FG). The right panel shows a corresponding cartoon.
Figure 3 shows the experimental EFRET ratios measured for this mixture. In principle, in such a triple-colour labelled system FRET can occur between D-A1, D-A2 and A1–A2 thus creating intrinsically complex three dimensional data. However, our labelling scheme purposely places Alexa488 and Alexa594 far apart (total of 130 AA) so that no or only negligible energy transfer can occur between those two dyes (Figure 3a). This is easily achieved when labelling IDPs, as these proteins behave like a polymer in solution, i.e. the labelling positions spaced further apart in the protein sequence are also further apart in space. In contrast to a full three-colour analysis, which suffers from the complexity of simultaneously competing FRET pathways between the three dyes,12, 14 this simplifies our system to a regular dual-colour/single pair FRET analysis scheme.
Figure 1.
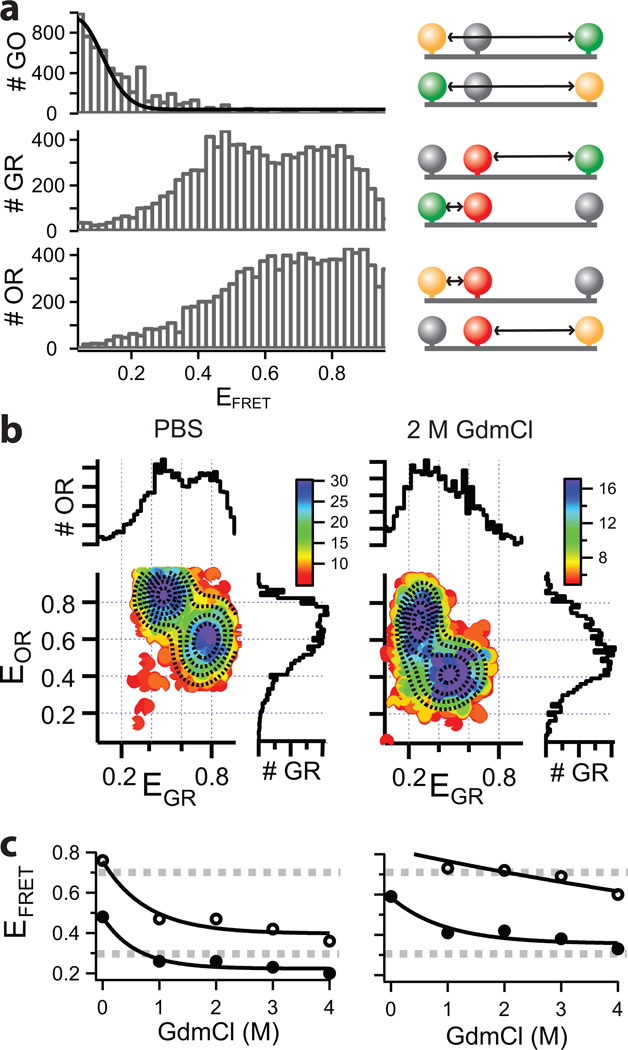
a) One dimensional FRET histograms of for the three dye combinations (EGO, EGR, EOR, left panel) are shown together with the corresponding labeling schemes (right panel). b) Shown is the 2D data of EOR vs. EGR for PBS and 4M GdmCl (left and right panel respectively) with their projections along both axes. The dashed lines are the result of a multivariate 2D Gaussian fit clearly identifying two species, one centered at (EOR, EGR)=(0.84,0.48) and one at (EOR, EGR)=(0.59,0.76) for PBS. Titration with increasing GdmCl decreases EFRET for all populations as summarized in (c). Open circles correspond to the short distance and filled circles to the long distance as indicated in the cartoon (a). Dashed lines indicate boundaries for more sensitive measurements (compare to Figure 1).
The EFRET analysis between D-A2 (EGR) and A1–A2 (EOR) does not show a simple Gaussian distribution (Figure 3a). As the FRET efficiencies are measured on a molecule by molecule basis (as they diffuse through the focal volume), it is possible to correlate differences in EOR and EGR for the sample species by plotting a 2D histogram. When EOR is plotted vs. EGR (Figure 3b), two populations can be clearly identified by a 2D double Gaussian fit. These populations correspond to the two sets of molecules that were differently labelled as also shown in Figure 3. One population shows the EFRET measurements for 120C-G/155TAG-R/250C-O species and the other population for the 120C-O/155TAG-R/250C-G species. Due to the different R0 of the two dye pairs, the EGR and EOR cannot probe the same amino acid spacing equally well. One EOR population is at 0.84, which is very close to 1 (were numerical artefacts can occur) and measures the short distance between 120C-O/155TAG-R, whereas this dye pair is instead rather suitable for longer distances. For this short distance the corresponding EGR species returns a value of EFRET= 0.76 and is thus nearer the sensitive detection regime (EFRET~ 0.3–0.7). When increasing the denaturant (guanidinium chloride, GdmCl) concentration, the protein further expands in solution as evident from a shift of all populations towards lower FRET efficiencies, further validating our experimental design (Figure 3b, right panel). Figure 3c summarizes the data from a titration experiment with increasing GdmCl concentrations illustrating that for the shorter distance the GR dye pair can follow the titration very well in contrast to the OR dye pair and vice versa for the longer distance.
Thus, within a single sample preparation, a short distance and a long distance can be measured reliably by using this labelling scheme with greatly different R0 for two dye pairs. It is important to note that multiple standard dual-colour measurements would be more laborious since they would require multiple independent sample preparations and measurements. Due to the different distances probed, a single dye pair cannot be used reliable in all cases and thus a longer amino acid distance would always need to be probed with a dye pair possessing a large R0, while a short amino acid distance requires a dye pair with small R0.
To conclude, we have achieved triple labelling of a single protein by combining maleimide chemistry with genetically encoding orthogonal chemical handles. We introduced an analysis scheme that, despite the partial random labelling, makes it possible to identify site-specifically labelled samples. Our labelling and analysis scheme requires that there be no FRET between the green and the orange dye. While future extensions of this method can in principle eliminate this requirement, we show that this can be used to our advantage because three-colour EFRET data is simplified to standard dual-colour data, and thus to data that can be easily and robustly analyzed. The approach is most useful when short and long distances occur simultaneously that cannot be well distinguished when using just a single dye pair. It enables us to obtain simultaneous readout of different regions from a single measurement and, more importantly, also from a single sample preparation, which greatly reduces the laborious and costly preparation of biological specimen for single molecule studies. Complex biological processes, where conformational changes occur across a broad distance range, such as those of intrinsically disordered proteins with high conformational flexibility, can therefore be studied faster and more easily.
Experimental procedures
Protein expression and purification
The yNup49FG containing cysteines at position 120 and 250 and an AMBER TAG mutation at site 155 was cloned into a pBAD expression vector. The vector contained an ampicillin resistance gene and was cotransformed with a plasmid harboring the tRNA/RS from M. jannaschii (under chloramphenicol resistance) for the selective expression of p-acetylphenylalanine in E. coli.24 Expression was done as previously described in terrific broth medium at 37°C.6 At OD600= 0.2–0.5, 1 mM of the p-acetylphenylalanine was added. The cells were induced at OD600= 0.4–1 with 0.02 % arabinose and incubated until harvesting. Ni-NTA resin (Qiagen, Dusseldorf, Germany) was used for the purification in phosphate buffered saline (PBS, pH 7.4, 2M urea). All purification buffers contained 0.2 mM tris(2-carboxyethyl)phosphine (TCEP) and 1 mM phenylmethane-sulfonylfluoride (PMSF).
Triple labelling and Three-colour single molecule fluorescence spectroscopy
A detailed procedure can be found in the Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Dr. Virginia VanDelinder for critical proofreading of this manuscript. SM acknowledges a fellowship from the Boehringer Ingelheim Fonds, EAL funding by the Emmy-Noether program of the Deutsche Forschungsgemeinschaft, and AAD grant GM066833 from the U.S. National Institutes of Health.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. Proc Natl Acad Sci U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. Proc Natl Acad Sci U S A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. Proc Natl Acad Sci U S A. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuler B, Eaton WA. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milles S, Lemke EA. Biophys J. 2011;101:1710–1719. doi: 10.1016/j.bpj.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milles S, Tyagi S, Banterle N, Koehler C, Vandelinder V, Plass T, Neal AP, Lemke EA. J Am Chem Soc. 2012 doi: 10.1021/ja210587q. [DOI] [PubMed] [Google Scholar]
- 7.Tompa P. Structure and Function of Intrinsiclaly Disordered Proteins. Boca Raton: CRC PRESS; 2010. [Google Scholar]
- 8.Deniz AA, Mukhopadhyay S, Lemke EA. J R Soc Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brustad EM, Lemke EA, Schultz PG, Deniz AA. J Am Chem Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemke EA, Gambin Y, Vandelinder V, Brustad EM, Liu HW, Schultz PG, Groisman A, Deniz AA. J Am Chem Soc. 2009;131:13610–13612. doi: 10.1021/ja9027023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo MH, Lee TS, Kim E, Cho YL, Park HS, Yoon TY, Kim HS. Anal Chem. 2011;83:8849–8854. doi: 10.1021/ac202096t. [DOI] [PubMed] [Google Scholar]
- 12.Clamme JP, Deniz AA. Chemphyschem. 2005;6:74–77. doi: 10.1002/cphc.200400261. [DOI] [PubMed] [Google Scholar]
- 13.Hohng S, Joo C, Ha T. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Biophys J. 2007;92:303–312. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratzke C, Berkemeier F, Hugel T. Proc Natl Acad Sci U S A. 2012;109:161–166. doi: 10.1073/pnas.1107930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Angew Chem Int Ed Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein IH, Steinhauer C, Tinnefeld P. J Am Chem Soc. 2011;133:4193–4195. doi: 10.1021/ja1105464. [DOI] [PubMed] [Google Scholar]
- 18.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Proc Natl Acad Sci U S A. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller BK, Zaychikov E, Brauchle C, Lamb DC. Biophys J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Proc Natl Acad Sci U S A. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nettels D, Gopich IV, Hoffmann A, Schuler B. Proc Natl Acad Sci U S A. 2007;104:2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nettels D, Muller-Spath S, Kuster F, Hofmann H, Haenni D, Ruegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, Best RB, Schuler B. Proc Natl Acad Sci U S A. 2009;106:20740–20745. doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopich IV, Szabo A. Journal of Physical Chemistry B. 2003;107:5058–5063. [Google Scholar]
- 24.Young TS, Ahmad I, Yin JA, Schultz PG. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.