Abstract
Previous studies of human hepatitis B virus (HBV) transcription revealed the requirement of two enhancer elements. Enhancer I (EnhI) is located upstream of the X promoter and is targeted by multiple activators, including basic leucine zipper proteins, and enhancer II (EnhII) is located upstream to the PreCore promoter and is targeted mainly by nuclear receptors (NRs). The mode of interplay between these enhancers and their unique contributions in regulating HBV transcription remained obscure. By using time course analysis we revealed that the HBV transcripts are categorized into early and late groups. Chang (CCL-13) cells are impaired in expression of the late transcripts. This could be corrected by overexpressing EnhII activators, such as hepatocyte nuclear factor 4α, the retinoid X receptor α, and the peroxisome proliferator-activated receptor α, suggesting that in Chang cells EnhI but not EnhII is active. Replacing the 5′-end EnhI sequence with a synthetic Gal4 response (UAS) DNA fragment ceased the production of the early transcripts. Under this condition NR overexpression poorly activated EnhII. However, activation of the UAS by Gal4-p53 restored both the expression of the early transcripts and the EnhII response to NRs. Thus, a functional EnhI is required for activation of EnhII. We found a major difference between Gal4-p53 and Gal4-VP16 behavior. Gal4-p53 activated the early transcripts, while Gal4-VP16 inhibited the early transcripts but activated the late transcripts. These findings indicate that the composition of the EnhI binding proteins may play a role in early to late switching. Our data provides strong evidence for the role of EnhI in regulating global and temporal HBV gene expression.
Hepatitis B virus (HBV) is the prototype of the hepadnaviridae, a family of small hepatotropic enveloped viruses. The HBV genome consists of a partially double-stranded 3.2-kb DNA with four major open reading frames (ORFs). These ORFs encode the reverse transcriptase (Pol protein), Core proteins (preCore and Core), three surface antigen proteins (preS1, preS2, and S), and the X protein (pX). Upon infection the viral genome targets the host nucleus, where it detaches from the viral polymerase and is repaired, acquiring a covalently closed circular DNA (cccDNA) configuration. cccDNA serves as a template for mRNA synthesis by the host polymerase II. At least five promoters control the synthesis of the six major viral transcripts. Two distinct transcripts are initiated at the X-gene promoter (12, 20, 28), both encoding the X protein (12). One is a short 0.7-kb transcript named short-X RNA (sxRNA), and the other is a 3.9-kb transcript named long-X RNA (lxRNA). The preCore and Core promoters are about 30 bp apart and initiate the synthesis of the pregenomic and preCore RNA species, designated pg/pcRNA. The pgRNA has a dual function: it is used as a template for viral replication and is translated into the Core protein. This transcript is assumed to translate the Pol protein. Other major HBV transcripts initiate either at the preS1 or preS2/S gene promoters. Their corresponding transcripts are named preS1 and preS2/S RNA, respectively. These are the major known HBV transcripts that escape splicing.
Two enhancers, designated enhancer I (EnhI) and enhancer II (EnhII), have been identified in the HBV genome. Both enhancers exhibit greater activity in cell lines of hepatic origin, and they also function in conjunction with heterologous promoters (3, 17, 23, 25, 46, 53, 57). EnhI regulates not only the juxtapositioned X promoter (16) but also all the other viral promoters (1, 11, 23, 24). EnhI is essential for HBV transcription and can be partially replaced by the simian virus 40 enhancer (24). HBV-transgenic mice lacking EnhI at the 5′ end of the inserted DNA are defective in virion production and poorly supported liver-specific HBV expression (19). A region within EnhI binds multiple transcription activators of the basic leucine zipper family, including C/EBP (9), the AP-1 complex (13), and ATFs (35). This region possesses an intrinsic enhancer activity in a variety of hepatic cell lines (13, 29, 48). In addition, cellular factors involved in cell cycle control and apoptosis, including the tumor suppressor protein p53 (39), its homologue p73 (11), the proto-oncoprotein c-Abl (10), and RFX-1 (26, 45), specifically bind and regulate EnhI activity. Recently, in vivo footprinting analysis has demonstrated that the EnhI region is occupied by the aforementioned cell cycle control proteins (43). EnhII is situated immediately upstream to the pg/pc promoter and has been implicated in regulating its transcription as well as the transcription of the preS2/S promoters (30, 56).
Nuclear receptors (NRs) are a superfamily of over 150 different intracellular proteins that directly control the activity of target genes through interaction with small lipophilic signaling molecules (reviewed in reference 36). There is a substantial body of evidence indicating that NRs have an effect on HBV gene expression through their nuclear receptor responding elements (NRREs) that are found in both EnhI and EnhII/pre-C regions (25, 42, 47, 54). The HBV NRREs mainly bind the hepatocyte nuclear factor 4α (HNF4α), the retinoid X receptor α (RXRα), and the peroxisome proliferator-activated receptor α (PPARα). Recent studies suggest that while the preCore NRRE is important for viral pgRNA and DNA synthesis, the NRRE of EnhI plays a more global role and can be compensated for by other binding factors (53). The HBV NRREs play an important role in the HBV life cycle, as has been demonstrated by mutational analysis (46, 53, 55). Recently it has been reported that overexpressing HNF4α and RXRα-PPARα support HBV expression and replication in a heterologous nonpermissive cell line (46). This suggests that these NRs are not only required but are also sufficient in supporting HBV transcription. However, the obtained level of expression under this condition was very low, implying a more complex regulation mechanism of HBV gene expression.
Temporal HBV gene expression has been poorly investigated due to the lack of efficient infectious culture systems. It has been reported that the expression of HBV sxRNA is transient, and its level drops to minimal levels much before the other transcripts (51). The question of whether the X promoter is the first to be activated has not been addressed. We found this to be a possible explanation for the requirement of two enhancers. It is possible that the two enhancers, each regulating a different set of transcripts, are functional at different stages of the HBV life cycle. EnhI is active early upon transfection, while EnhII is activated later, concomitant with the silencing of EnhI. Our data provides strong evidence in support of this model. Early-late switching of transcription of many DNA viruses is executed by virally encoded proteins. We show that the early-late switching in HBV transcription can be determined by an alternative mechanism. This switching mechanism involves the transcription activation domain of the activators together with EnhI. The molecular basis of this alternative mechanism remained to be resolved.
MATERIALS AND METHODS
Plasmid constructions.
HBV DNA was inserted into pGEM-3Z (Promega), an inert vector containing no eukaryotic regulatory elements. A 2xHBV plasmid containing two head-to-tail copies of HBV full-length DNA (subtype adw) was ligated into pGEM-3Z by using the unique EcoRI sites. The 1.3xHBV DNA construct contains an overlength HBV genome of 4,195 bp. It has a unique EcoRV site (nucleotide 1043) at the 5′ terminus and a unique TaqI site (nucleotide 2017) at the 3′ terminus. This EcoRV-TaqI HBV fragment was inserted between the SmaI and AccI unique sites of pGEM-3Z as described previously (12). The 1.3xHBV-Luciferase plasmid was constructed by substitution of the HBV sequences between the BglII and SpeI restriction sites with the Luciferase ORF. The functional ATG of the inserted luciferase gene is of the Core ORF, and therefore it is produced by the pgRNA under the regulation of the pgRNA promoter. To obtain a circular genomic DNA, 200 μg of linear HBV DNA was excised from the 2xHBV plasmid by EcoRI and was religated in a diluted (100 ml) ligation mix to encourage intramolecular self ligation (12). The construction of the 1.3xX-Core and the 1.3xXKO-Core HBV DNA was previously described (12). For production of the Gal4-1.3xHBV construct, the DNA fragment of the 5′ EnhI-X gene copy between the unique EcoRV and SphI restriction sites has been replaced with four copies of a Gal4-responsive sequence (5′ CGGAGTACTGTCCTCCGAG 3′).
Cell culture, DNA transfection, and RNA analysis.
Huh7 and Chang cells were cultured in Dulbecco's modified Eagle's minimal essential medium (D5796; Sigma) containing 100 U of penicillin and 100 μg of streptomycin per ml, supplemented with 8% fetal bovine serum (GIBCO Laboratories). Transfection was carried out by the CaPi method as previously described (12). Cells were seeded in 9-cm-diameter plates 6 to 8 h prior to transfection and reached 60% confluence at the time of transfection. The transfection was carried out with 15 μg of HBV DNA and 10 μg of carrier pGEM-3Z plasmid DNA per 10-cm dish. Other expression plasmids were used in amounts as indicated in the figure legends, but the total DNA amount was kept constant. In each case a green fluorescent protein expression plasmid was used to monitor transfection efficiency.
When NR ligands were used, cells were plated in medium supplemented with 8% charcoal-treated fetal bovine serum starting 24 h before transfection. About 16 h after transfection, cells were washed twice with a solution of phosphate-buffered saline (PBS) plus Ca2+ and Mg2+ and were incubated with fresh medium for an additional 30 h before harvesting.
For RNA analysis, total RNA was extracted and analyzed by Northern blotting as previously described (12). HBV radioactive probes were prepared by using the DNA of the X-gene region. For a GAPDH probe, 1.3 kb of GAPDH cDNA was used. Probes were labeled by using a random priming protocol with the desired DNA template and [α-32P]dCTP (3,000 ci/mmol; Amersham). About 106 cpm (10 ng of DNA) labeled DNA fragments were used per 1 ml of hybridization buffer. After hybridization the membrane was washed for 20 min at 65°C in a 0.1% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate buffer and was exposed to an X-ray film for autoradiography.
Protein analysis.
Proteins were extracted from cells by TRI-REAGENT (MRC, Inc.) according to the manufacturer's instructions and were solubilized in Laemmli sample buffer containing 4 M urea by incubation for 30 min at 50°C. Soluble proteins were boiled for 10 min and subsequently were fractionated on sodium dodecyl sulfate-10% polyacrylamide gels. For Western blot analysis, gels were electroblotted to a nitrocellulose membrane for 1 h at 200 mA. Membrane filters were stained after blotting by ponceau S and were soaked for 2 h at room temperature in a blocking solution (PBS containing 10% nonfat milk and 0.01% [vol/vol] Tween-20 [Sigma]). All further incubation steps were performed with the same solution. Filters were incubated for 1 to 2 h at room temperature in the presence of either monoclonal mouse anti-HBcAg (mAb 22), generated as previously described (40), polyclonal (immunoglobulin G purified) rabbit anti-Gal4 generated in our laboratory (22), or anti-β-tubulin (clone no. TUB2.1; Sigma) antibodies, and washed three times with PBS plus 0.01% Tween-20. Goat anti-rabbit or goat anti-mouse antibody conjugated with horseradish peroxidase (ICN laboratories) was added (diluted 1:10,000 in blocking solution) and was incubated for an additional 1 h, followed by three rounds of washes. Antibody-antigen complexes were visualized by the ECL chemiluminescent detection system (Pierce) according to the manufacturer's instructions.
RESULTS
Early and late HBV transcripts.
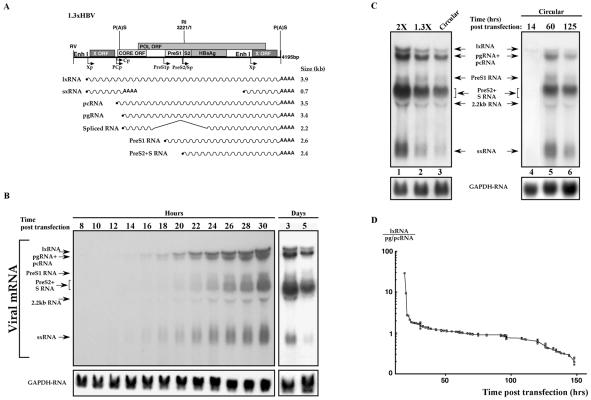
To study the temporal aspect of HBV transcription, Huh7, a highly differentiated hepatocellular carcinoma cell line, was transfected with HBV DNA and was harvested for RNA analysis every 2 h starting at 8 h posttransfection. Given the circular nature of the HBV 3.2-kb DNA, the cloned genome does not contain a contiguous viral genome. We generated and used a construct containing a 1.3X genome in tandem and in head-to-tail configuration (Fig. 1A). The 1.3xHBV 4.2-kb DNA construct is capable of programming the expression of all the HBV mRNA species and supports viral replication in hepatoma cells and hepatocytes of transgenic mice (19). About 16 h after transfection the two X RNA species were the first to be detected (Fig. 1B). These RNA bands include the long 3.9-kb transcript, named lxRNA, that is active in expression of the X protein (12), and the short 0.7-kb transcript, referred to as sxRNA. The other HBV transcripts, namely pg/pcRNA and the PreS2/S RNA species, began to accumulate about 4 h later. The 2.2-kb transcript is a spliced variant and is reactive to a Core gene-specific probe (data not shown). The HBV lxRNA and sxRNA levels were significantly reduced at longer time points and were hardly detected 5 days posttransfection. These results are in agreement with previous reports on the temporal program of HBV transcription (51). To rule out the possibility that this pattern of gene expression is unique to the employed DNA configuration, we transfected cells with an HBV dimer DNA and a circularized monomer genome that was generated by in vitro self ligation (12). The pattern of transcription is identical in all three DNA configurations (Fig. 1C, lanes 1 to 3). Furthermore, the circularized HBV DNA monomer shows the same temporal transcription program, with the X transcripts being the first to be detected (Fig. 1C, lanes 4 to 6). To quantify the difference in the relative amounts of the various transcripts over time, the levels of the lxRNA and pg/pcRNA from 14 different experiments were determined by phosphorimager. The ratio between the level of these early (lxRNA) and late (pg/pcRNA) transcripts was calculated and plotted versus time (hours) in a semilogarithmic scale. At the early time points there is over 20 times more early RNA, whereas at the late time points the ratio drops to less than 0.2 (Fig. 1D). The differences between the stability of these transcripts cannot account for these kinetics as previously reported (12). These data indicate that HBV transcripts are categorized into two groups based on their timing of expression. One, the early transcripts, initiate at the X-gene promoter, and the other, the late transcripts, initiate from all the other known HBV promoters. Given the fact that the X-gene promoter is under regulation of EnhI, the early transcripts are expected to be regulated by this enhancer (see below).
FIG. 1.
Temporal HBV gene expression. (A) Schematic illustration of the 1.3xHBV DNA construct used in this study and the expected mRNA species. The different HBV ORFs and promoters (arrows) are shown. P(A)S indicates the position of the polyadenylation signal. The EcoRV (RV) and EcoRI (RI) unique sites are indicated. (B) Huh7 cells were transfected with plasmids containing 1.3 copies of HBV DNA and harvested at the indicated time points (hours/days) posttransfection. RNA was extracted, separated on a formaldehyde-agarose gel, and analyzed by using a 32P-X gene DNA probe. A GAPDH probe was used to quantify RNA in each lane. Arrows indicate the position of the known viral transcripts. The 2.2-kb transcript is an HBV spliced RNA (data not shown). Note that lx- and sxRNA are the first to be visible (16 h posttransfection) and are the first to disappear at later time points. (C) The pattern of HBV transcription is not template dependent. The transcription pattern obtained by three different HBV DNA configurations is shown in lanes 1 to 3. At lane 1 two tandem copies of complete HBV genome ligated at the unique EcoRI site was used (2X). For lane 2 the construct shown in panel A was used. For lane 3 HBV DNA was linearized at the EcoRI site and was self ligated as described previously (12) to obtain a circular intact HBV genome. The circular HBV DNA template was used in a time course experiment (lanes 4 to 6). (D) Time-dependent ratio between the lxRNA and the pg/pcRNA level. The levels of lxRNA and the pg/pcRNA were measured by phosphorimager at different time points and were plotted in a semilogarithmic scale versus time (hours). The data summarize 14 different experiments, each with about 10 different time points (n = 153).
Chang cells are defective in supporting expression of the HBV late transcripts.
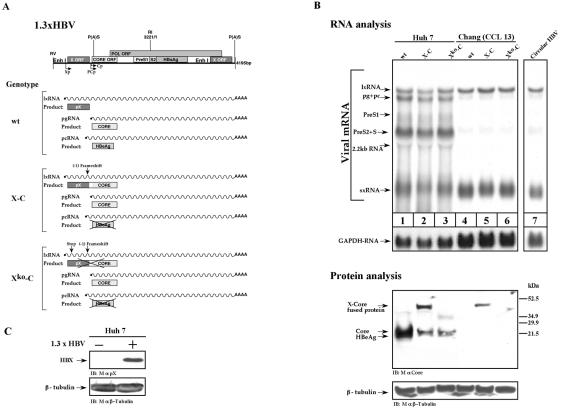
A number of liver-derived cell lines are commonly used to study HBV transcription. Among the employed lines, Chang cells (CCL-13) are exceptional, as their liver origin has not been fully validated. Huh7 and Chang cells were transfected with either wild-type or mutant HBV DNA (Fig. 2A), and the transcription pattern was determined and compared. In the Chang cells, the early lx- and sxRNA species were the major transcripts detected, along with very poor expression of the other transcripts (Fig. 2B, compare lanes 1 and 4). Protein analysis revealed that Chang cells are inefficient in the production of the Core protein. This finding is in agreement with the low level of the pgRNA that encodes the Core protein (Fig. 2B, lanes 1 and 4). Both sxRNA and lxRNA encode the X protein (Fig. 2C), in agreement with a previous report (12). However, lxRNA also contains a complete CORE ORF (Fig. 1A). An HBV DNA mutant (construct X-C), whose X ORF was fused to that of CORE, can express the X-Core chimera 40-kDa protein, provided that lxRNA is efficiently produced (12). Protein analysis revealed that Chang cells efficiently express the X-Core chimera protein (Fig. 2B, lanes 2 and 5), further reinforcing the fact that Chang cells express lxRNA. As expected, a double HBV DNA mutant containing a stop codon in the X ORF did not support the production of the X-Core fusion protein (Fig. 2B, construct Xko-Core). These data suggest that Chang cells favorably support the transcription from the X-gene promoter (see below). Given the fact that activation of the X-gene promoter is an early event, we concluded that Chang cells are impaired in synthesis of the HBV late transcripts.
FIG. 2.
Chang cells are defective in late gene expression. (A) Schematic drawing of HBV transcripts and proteins produced by the three HBV constructs. The wild-type (wt) 1.3xHBV construct programs the synthesis of the X, Core, and HBeAg proteins by the lxRNA, pgRNA, and pcRNA, respectively. The X-C construct, harboring a fused X-Core gene, directs the synthesis of a 40-kDa X-Core fused product by the lxRNA. The X-C mutant directs the synthesis of Core protein by the pcRNA but is incapable of producing the HBeAg by the pcRNA, since the HBeAg gene resides upstream of the frameshift mutation site (12). The Xko-C mutant harbors an additional stop mutation at position 27 of the 5′-end X gene and is therefore incapable of producing both the X-Core fused product and the HBeAg. (B) Huh7 and Chang cells were transfected with either the wt 1.3xHBV DNA (lanes 1 and 4), the X-C mutant (lanes 2 and 5), or the Xko-C mutant (lanes 3 and 6). Lane 7 shows the RNA pattern obtained upon transfecting Chang cells with a circular HBV DNA. RNA was analyzed as described in the legend to Fig. 1. For Northern blotting hybridization the X-gene DNA probe was used. For protein analysis, monoclonal mouse anti-Core (mAb 22) and a monoclonal mouse anti-β-tubulin antibody were used. (C) The Huh7-transfected cells with wt HBV DNA were analyzed for pX production by using anti-pX-specific monoclonal antibody generated in our laboratory. IB, immunoblot.
Regulation of HBV transcription by hepatocyte nuclear factors.
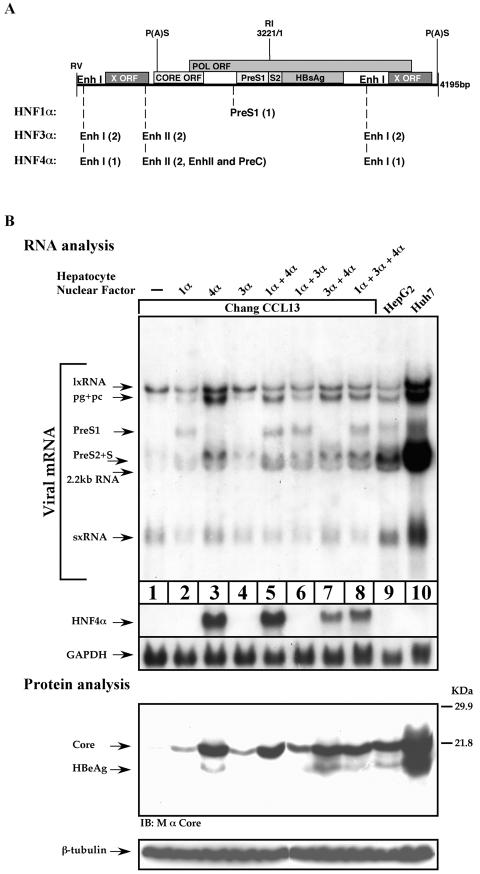
Two distinct enhancers regulate HBV transcription. EnhI is likely to be responsible for the activation of the early transcripts, and EnhII is likely to be responsible for the late transcripts. If true, the unique HBV transcription pattern in Chang cells may result from a lack of EnhII activity. Hepatocyte nuclear factors, such as HNF1, HNF3, and HNF4, play an important role in HBV transcription, and several associated binding sites were mapped within the HBV genome at both enhancers (Fig. 3A). One of the nuclear factors, HNF4α, preferentially activates EnhII (42, 53).
FIG. 3.
Overexpression of HNF4α is sufficient to restore HBV gene expression. (A) A schematic drawing depicting the different hepatocyte nuclear factor-responsive elements situated along the 1.3xHBV DNA. Each element is depicted by its position within the HBV DNA, and the number of binding sites is shown in parenthesis. (B) Chang cells were transfected with wild-type HBV DNA alone or together with 200 ng of different combinations of plasmids expressing HNF1α, HNF3α, or HNF4α proteins. As a positive control highly differentiated HepG2 and Huh7 cells were transfected only with HBV DNA. Cells were harvested 40 h posttransfection for total RNA and protein analysis. The same blotting membrane was subsequently used for hybridization with the X-gene DNA, HNF4α DNA, and GAPDH DNA probes. For protein analysis monoclonal mouse anti-Core or anti-β-tubulin antibody was used. IB, immunoblot
Chang cells were transfected with an HBV plasmid together with the hepatocyte nuclear factors in different combinations, and the levels of HBV transcripts were determined by Northern blotting. The data shows that HNF4α increased the expression of both the pc/pgRNA and the preS2/S RNA (Fig. 3B). Elevated amounts of the Core and HBeAg proteins were shown to be directly related to the pg/pcRNA levels (Fig. 3B, protein analysis). HNF1α gave rise to a high level of the preS1 RNA (Fig. 3, compare lanes 1 and 2), which is consistent with the fact that the preS1 promoter contains an active HNF1α binding site (58). Expression of HNF3α alone did not significantly affect the HBV transcription pattern (Fig. 3B, compare lanes 1 and 4) but supported accumulation of the late transcripts when expressed together with HNF1α. Coexpression of two activators in different combinations or all three together did not further improve the level of the HBV transcripts above that obtained with HNF4α alone. These results indicate that the hepatocyte nuclear factors support HBV transcription, and in Chang cells expression of the late HBV transcripts can be improved by overexpression of HNF4α.
Induction of HBV late transcription by NRs.
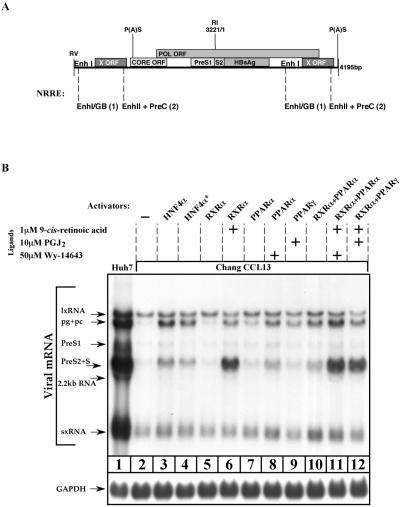
HBV transcription is regulated by a number of NRs apart from HNF4α. RXRα heterodimers, together with either PPΑRα or PPΑRγ, bind to NRREs found within the HBV genome (Fig. 4A). Different NRs compete in binding to a given NRRE. Some NRs cause repression activity (42, 55). It has been reported that overexpression of PPARα-RXRα in the presence of their associated ligands leads to an increase in pgRNA synthesis with little or no effect on other HBV transcripts (53). This is possibly done via preferential activation of EnhII. To examine the involvement of NRs in transcription of the late transcripts, Chang cells were cotransfected with HBV DNA in various combinations in the presence or absence of their respective ligands. RXRα alone did not improve HBV transcription, but a significant induction in the level of the late transcripts was obtained in the presence of retinoic acid, its cognate ligand (Fig. 4B, lanes 5 and 6). PPARα, in the presence of its ligand, PGJ2, increased the level of the late transcripts, although to a much lower extent than RXRα, with no significant effect on the sx- and lxRNA levels (Fig. 4B, lanes 7 and 8). The effect of PPARγ overexpression on pg/pcRNA synthesis was only moderate when transfected alone or together with RXRα. The highest induction in the level of late transcripts was obtained when RXRα and PPARα were cotransfected (Fig. 4B, lane 11). Collectively these results suggest that in Chang cells the late pg/pc and PreS2/S promoters are poorly functional, possibly due to the inefficient activity of EnhII.
FIG. 4.
Induction of the HBV late transcripts by overexpression of NRs. (A) Schematic presentation of the different NRREs situated along the 1.3xHBV DNA. Each element is depicted by its position within the HBV DNA, and the number of binding sites is indicated in parenthesis. (B) Chang cells were transfected with wild-type HBV DNA alone or together with 200 ng of different combinations of plasmids that express HNF4α, RXRα, PPARα, and PPARγ proteins. As a positive control, highly differentiated Huh7 cells were transfected only with HBV DNA. Cells (except control cells of line 4*) were plated in medium supplemented with 8% charcoal-treated fetal bovine serum from 24 h prior to transfection up to harvesting. After removal of transfected excess DNA, 9-cis-retinoic acid, prostaglandin J2 (PGJ2), and Wy-14643 were added to the cell media at the amounts indicated in the final concentrations. These are the ligands of RXRα, PPARγ, and PPARα, respectively. All cells were harvested 40 h posttransfection for total RNA analysis. For Northern blotting hybridization the X-gene DNA probe was used. GAPDH probe was used to quantify RNA in each lane.
An active upstream enhancer is required for efficient HBV early and late transcription.
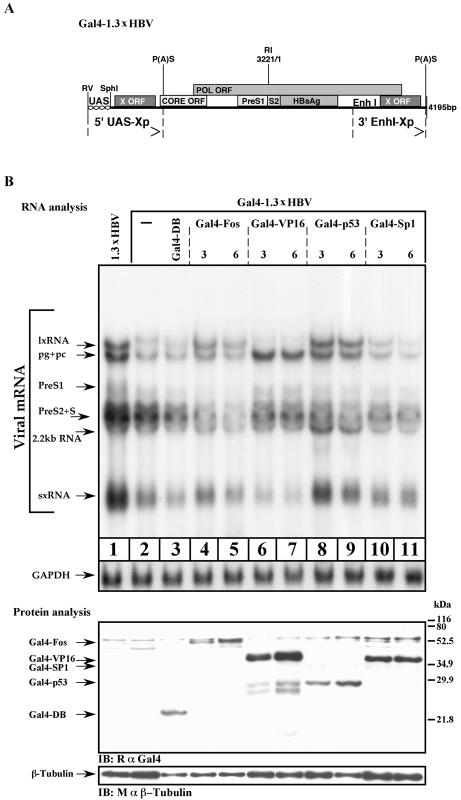
Having demonstrated that in Chang cells accumulation of the late transcripts can take place by overexpression of a set of specific NRs, we next examined the role of EnhI in this process. To this end, we have replaced the upstream (5′) EnhI element of the 1.3xHBV construct with Gal4-responsive sequences (Fig. 5A). This artificial enhancer is inactive in animal cells unless strong activators in the form of Gal4-chimera proteins are provided. The chimera proteins contain the Gal4 DNA binding domain that is fused to desired activation domains (ADs). This construct, designated Gal4-1.3xHBV, contains a wild-type EnhI at its 3′ end.
FIG. 5.
EnhI regulates HBV transcription. (A) Schematic illustration of the Gal4-1.3xHBV construct. The 5′ copy of EnhI has been replaced by four repeats of a synthetic Gal4-responsive element (UAS). EcoRV and SphI sites are the unique restriction sites flanking the UAS region. Arrows indicate the redundant termini. (B) Huh7 cells were transfected with Gal4-1.3xHBV DNA alone or together with 3 or 6 μg of Gal4-Fos, Gal4-VP16, Gal4-p53, and Gal4-Sp1 expression plasmids as indicated. Cells were harvested 40 h posttransfection for total RNA and protein analysis. For Northern blotting hybridization, the X-gene DNA probe was used. GAPDH probe was used to quantify RNA in each lane. For protein analysis, polyclonal rabbit anti-Gal4 (RαGal4) or anti-β-tubulin antibodies were used. The position of the different Gal4 chimera is shown. IB, immunoblot.
Huh7 cells were transfected with Gal4-1.3xHBV and with the wild-type 1.3xHBV DNA as a positive control. Removal of the 5′ end of EnhI resulted in reduced levels of all the transcripts (Fig. 5B), suggesting that EnhI supports the activity of all the HBV promoters. Next the Gal4 HBV plasmid was transfected together with a plasmid expressing one of the following chimera activators: Gal4-DBD, Gal4-Fos, Gal4-VP16, Gal4-p53, or Gal4-Sp1. To confirm the expression of the Gal4-fused proteins, Western blot analysis was performed by using anti-Gal4 antibodies (Fig. 5B, protein analysis panel). Gal4-VP16 activator exclusively supported the accumulation of the late transcripts with concomitant repression of the early transcripts (Fig. 5B, lanes 6 and 7). This may be the result of the known VP16 acidic AD activity that supports the far downstream promoter while repressing the proximal one (2, 18, 31, 49). In contrast to VP16, the Gal4-p53 activator supported the expression of all the HBV promoters (Fig. 5B, compare lane 8 to lane 1). These data indicate that EnhI and the nature of the associated transcription activators determine the pattern of HBV gene expression.
Domination of EnhI in HBV transcription.
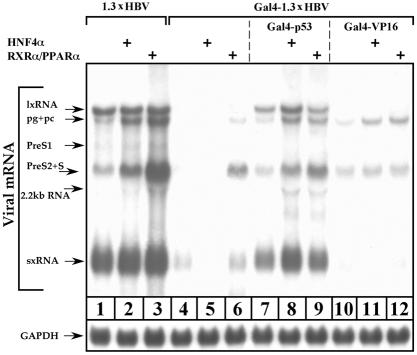
To further examine the overall role that EnhI plays in regulation of HBV transcription, we used Chang cells that are inefficient in late gene transcription. Chang cells transfected with Gal4-1.3xHBV DNA showed no lxRNA and a very low level of sxRNA (Fig. 6, lane 4). The absence of lxRNA was expected. However, the fact that sxRNA was poorly expressed despite the fact that the HBV sequence contains an intact X gene, including its promoter and EnhI at the 3′ end, was rather surprising. These findings strongly indicate that the 5′ region of EnhI is functionally more important and that the majority of the sxRNA are transcribed from the 5′ end of the X gene.
FIG. 6.
EnhI predominates HBV gene expression; analysis by Northern blotting. Chang cells were transfected with a wild-type 1.3xHBV DNA or were cotransfected with a mutant Gal4-1.3xHBV DNA together with 3 μg of Gal4-p53 or Gal4-VP16 expression plasmids as indicated. In addition, cells were cotransfected with 200 ng of HNF4α or 150 ng of RXRα and PPARα expression plasmids. Cells were plated in medium supplemented with 8% charcoal-treated fetal bovine serum from 24 h prior to transfection until harvesting. After removal of transfected excess DNA, 1 μM 9-cis-retinoic acid and 50 μM Wy-14643 were added to the RXRα-PPARα-transfected cells. Cells were harvested 40 h posttransfection for total RNA analysis as performed above.
As described above, the Chang cells' missing function can be complemented by overexpression of either HNF4α or RXRα-PPARα (Fig. 3 and 4). Unexpectedly, HNF4α does not support late gene expression in the absence of a 5′-end functional EnhI (Fig. 6, lane 5). Thus, the capacity of HNF4α to support late gene expression depends on a functional EnhI. To a lesser extent, late gene expression can be obtained with RXRα-PPARα as well (Fig. 6, lane 6).
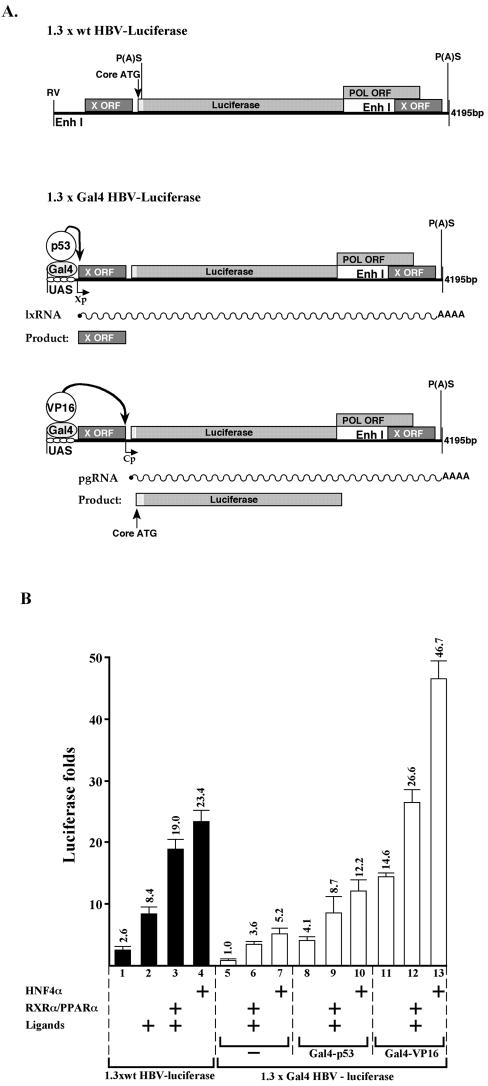
Activation of the artificial UAS enhancer by overexpressing Gal4-p53 and Gal4-VP16 chimera proteins is in agreement with the data obtained from Huh7 cells. The former supported transcription of all the HBV RNA, whereas the latter supports only the late RNA species (Fig. 6, lanes 7 and 10). Overexpression of HNF4α and RXRα-PPARα further supported HBV transcription in Gal4-p53 but not in the Gal4-VP16-activated enhancer. Similar results were obtained in the context of reporter plasmids. The region between nucleotides 1987 and 682 (BglII-SpeI sites) was removed from the HBV genome to insert the Luciferase gene (Fig. 7A, 1.3x wt HBV-Luciferase). This HBV-based reporter plasmid is activated by overexpression of HNF4α and RXRα-PPARα (Fig. 7B, lanes 1 to 4). However, in the context of 1.3xGal4 HBV-Luciferase, where the 5′-end EnhI was replaced by the Gal4 binding site, the activity of the reporter plasmid was much weaker (lanes 5 to 7). The activity was partially improved by the cotransfected Gal4-p53 that activates the X promoter and hence is expected to mainly support the production of the X protein and not the Luciferase (lanes 8 to 10). In contrast, substantial activation was obtained by cotransfected Gal4-VP16 (lanes 11 to 13), in agreement with its capacity to stimulate the promoter of the Core gene (Fig. 7A). These findings indicate that the HBV promoters are distinct and are differentially regulated in an EnhI-dependent manner. Furthermore, our data revealed a hierarchical regulation of HBV gene expression, whereby the activity of the EnhII depends on the prior activation of EnhI.
FIG. 7.
EnhI predominates HBV gene expression; analysis by HBV-based reporter plasmids. (A) The structure of the constructed HBV-based reporter plasmids is shown. Also indicated are the predicted modes of activity of the different Gal4-chimera activators and the resultant transcripts and proteins. (B) Chang cells were transfected with the plasmids indicated in panel A, and level of Luciferase activity was measured. The reporter plasmids were cotransfected with the indicated plasmids and were treated with ligands (1 μM 9-cis-retinoic acid and 50 μM Wy-14643).
DISCUSSION
In this paper we have elucidated the intradynamic mechanisms that govern HBV gene expression. We show that the HBV transcripts can be categorized into two distinct groups. One group encompasses the sxRNA and lxRNA mRNA species expressing the X protein. These are accumulated early after transfection and disappear soon after. This is the characteristic of the viral early genes, and therefore these transcripts are regarded in this study as the HBV early transcripts. All the other HBV RNA species are collectively referred to as the late transcripts. In this study we show that the expression of these two sets of RNA species can be uncoupled. Previously, many researchers have only been able to detect the late transcripts and therefore questioned the existence of the X-gene-specific mRNA species. One possibility is that these studies measured the level of HBV transcription during the late stages of viral gene expression.
In this study, functional dissection of HBV gene expression has been achieved by using an undifferentiated hepatocyte cell line called Chang (32). These cells support EnhI activity and selectively produce the early transcripts. The defect in programming the synthesis of the late genes correlates with the absence of expression of the major activators of EnhII, namely HNF4α and RXRα-PPARα (data not shown). We have shown that ectopically expressed HNF4α and RXRα-PPARα in Chang cells was sufficient to restore the transcription of the late genes while hardly affecting early transcription. This finding is in agreement with the fact that these activators bind the EnhII NRREs with higher affinity than that of EnhI (53). The emerging picture of the HBV transcription program is that each of the two HBV enhancers regulates a distinct phase of the viral life cycle. EnhI is responsible for the expression of the early transcripts, whereas EnhII regulates the late ones. The study of the Chang cells revealed that EnhI is active under conditions whereby no EnhII activity is detected, suggesting that EnhI is functionally autonomous.
Unlike EnhI, in the context of the HBV genome EnhII is functionally not autonomous, and its activity depends on the presence of an active EnhI. Substitution of a 5′-end EnhI sequence with UAS sequence resulted in a sharp reduction in the expression of the late genes. This is despite the fact that the construct contained an intact EnhI at its 3′ end. Substitution of the two copies of EnhI resulted in a complete abolishment in the HBV gene expression (data not shown). This was the case for Huh7 cells expressing HNF4α and RXRα-PPARα, the EnhII activators. Furthermore, in Chang cells where the activity of EnhII is induced by overexpression of HNF4α and RXRα-PPARα, no EnhII activation was obtained with a construct lacking the 5′-end EnhI sequences. The response to the activators can be reestablished by artificial activation of the 5′ end with the Gal4 chimera proteins, such as Gal4-p53 and Gal4-VP16. These findings indicate that a functional EnhI is required for EnhII to be active. Thus, there is a hierarchical relationship between the two HBV enhancers that may determine the sequential pattern of HBV transcription. Expression of the late genes, those that are EnhII dependent, begins only after activation of EnhI and early gene expression.
The described pattern of temporal gene expression is characteristic of many DNA viruses and enables developmental programs in the viral life cycle. In general, viral regulatory proteins like the papovavirus large T antigen (27, 50), adenovirus E1a (21, 33), and herpes VP16 and Epstein Barr Virus Zta proteins (2, 7, 18) are made during the initial period of infection and induce the expression of the late structural genes. The X regulatory protein of HBV that is expressed by the early transcripts may play a similar role. However, no role was assigned to pX in EnhII activation to support the expression of the late genes. It was previously reported that the effect of X protein on EnhI is dose dependent (15). At low levels pX positively regulates EnhI activity and supports transcription from the X promoter, whereas at high levels it plays an opposite role and represses the activity of the X promoter. Although not shown, it is very likely that repression of EnhI permits activation of EnhII. According to this regulatory loop pX functions as an early-late switch. However, the fact that HBV late transcripts could be detected under environments where no pX is produced suggests that although pX might be needed, it is not essential for switching the transcription program from early to late stage.
The PreS2/S promoter that controls transcription of the small- and middle-surface RNAs is TATA-less and shares extensive homologies with the simian virus 40 late promoter (5, 8, 14). As such, it was shown that in contrast to other HBV control elements the factors that bind this promoter are not hepatocyte specific (4, 34, 37). In this study we show that the pc/pg and PreS2/S promoters are coregulated by EnhII. Turning on EnhII, either indirectly by Gal4-p53 or directly by Gal4-VP16, provoked PreS2/S promoter activity. These data, as well as the fact that EnhII specific induction by Gal4-VP16 stimulates the PreS2/S promoter, strongly argue that an active EnhII is both required and sufficient for PreS2/S promoter function. This is rather surprising considering the fact that enhancers preferentially activate the most immediate promoter, rarely acting upon the downstream ones.
In contrast, the PreS1 promoter that controls transcription of the large surface antigen RNA is independently regulated. This is the only HBV promoter containing a classical TATA box sequence (44). For its activation, the binding of both the ubiquitous Oct-1 and the liver-specific factor HNF1 is required (6, 56, 58). In contrast to the PreS2/S promoter, HNF1α is sufficient to stimulate the PreS1 promoter, regardless of EnhII activity (data not shown). Occupation of two independent regulatory mechanisms for PreS1 and PreS2/S RNA synthesis emphasizes the demand for their precise relative ratio during HBV life cycle. Indeed, this ratio has been shown to be crucial for HBV virion production and maturation and nuclear cccDNA synthesis (37, 38, 41, 52).
While attempting to activate the UAS enhancer through expression of the chimera Gal4-based activators we found a major difference between two very close ADs, namely p53 and VP16. The p53 AD supported expression of the early transcripts and restored response to NRs. This implies that p53 activity is similar to that obtained with the authentic EnhI-protein complex. This conclusion may explain our previous finding, that the p53 binding upstream to EnhI interferes with the EnhI activity (11, 39). The interference is simply the result of competition between p53 and the EnhI complex for the same set of coactivators. In sharp contrast to p53, the VP16 AD repressed the early promoters and supported the expression of the late ones. This happened in the absence of transfected NRs, suggesting direct recognition of the late promoters by the VP16 AD. Understanding the molecular basis of this differential regulation by two very close transcription ADs is an interesting issue that should be addressed in the future. This may provide a mechanistic insight into how the HBV transcription switching is ensured. Soon after infection, EnhI is occupied by a set of proteins with p53 AD-like activity that supports early transcription. This complex is transient and is replaced by a second complex that has a VP16-like activity to support late gene transcription. This novel dynamic mechanism of enhancer activity that we found in HBV may have genome-wide implications in cell regulation and development as well.
Acknowledgments
We thank A. Cooper for the analysis of pX expression and S. Budilovsky for excellent technical assistance.
Y. Shaul holds the Oscar and Emma Getz professorial chair.
REFERENCES
- 1.Antonucci, T. K., and W. J. Rutter. 1989. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J. Virol. 63:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Levy, R., O. Faktor, I. Berger, and Y. Shaul. 1989. Cellular factors that interact with the hepatitis B virus enhancer. Mol. Cell. Biol. 9:1804-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock, C. T., S. Kubicka, M. P. Manns, and C. Trautwein. 1999. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology 29:1236-1247. [DOI] [PubMed] [Google Scholar]
- 5.Cattaneo, R., H. Will, N. Hernandez, and H. Schaller. 1983. Signals regulating hepatitis B surface antigen transcription. Nature 305:336-338. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H. K., B. Y. Wang, C. H. Yuh, C. L. Wei, and L. P. Ting. 1989. A liver-specific nuclear factor interacts with the promoter region of the large surface protein gene of human hepatitis B virus. Mol. Cell. Biol. 9:5189-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, T., and M. Carey. 1993. The ZEBRA activation domain: modular organization and mechanism of action. Mol. Cell. Biol. 13:7045-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De-Medina, T., O. Faktor, and Y. Shaul. 1988. The S promoter of hepatitis B virus is regulated by positive and negative elements. Mol. Cell. Biol. 8:2449-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dikstein, R., O. Faktor, and Y. Shaul. 1990. Hierarchic and cooperative binding of the rat liver nuclear protein C/EBP at the hepatitis B virus enhancer. Mol. Cell. Biol. 10:4427-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dikstein, R., D. Heffetz, Y. Ben-Neriah, and Y. Shaul. 1992. c-abl has a sequence-specific enhancer binding activity. Cell 69:751-757. [DOI] [PubMed] [Google Scholar]
- 11.Doitsh, G., and Y. Shaul. 1999. HBV transcription repression in response to genotoxic stress is p53-dependent and abrogated by pX. Oncogene 18:7506-7513. [DOI] [PubMed] [Google Scholar]
- 12.Doitsh, G., and Y. Shaul. 2003. A long HBV transcript encoding pX is inefficiently exported from the nucleus. Virology 309:339-349. [DOI] [PubMed] [Google Scholar]
- 13.Faktor, O., S. Budlovsky, R. Ben-Levy, and Y. Shaul. 1990. A single element within the hepatitis B virus enhancer binds multiple proteins and responds to multiple stimuli. J. Virol. 64:1861-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faktor, O., T. De-Medina, and Y. Shaul. 1988. Regulation of hepatitis B virus S gene promoter in transfected cell lines. Virology 162:362-368. [DOI] [PubMed] [Google Scholar]
- 15.Faktor, O., and Y. Shaul. 1990. The identification of hepatitis B virus X gene responsive elements reveals functional similarity of X and HTLV-I tax. Oncogene 5:867-872. [PubMed] [Google Scholar]
- 16.Fukai, K., S. Takada, O. Yokosuka, H. Saisho, M. Omata, and K. Koike. 1997. Characterization of a specific region in the hepatitis B virus enhancer I for the efficient expression of X gene in the hepatic cell. Virology 236:279-287. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, A. D., P. Ostapchuk, and P. Hearing. 1993. Functional interaction of nuclear factors EF-C, HNF-4, and RXR alpha with hepatitis B virus enhancer I. J. Virol. 67:3940-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerster, T., and R. G. Roeder. 1988. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc. Natl. Acad. Sci. USA 85:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, W. T., J. Wang, G. Tam, T. S. Yen, and J. S. Ou. 1991. Leaky transcription termination produces larger and smaller than genome size hepatitis B virus X gene transcripts. Virology 181:630-636. [DOI] [PubMed] [Google Scholar]
- 21.Hardy, S., D. A. Engel, and T. Shenk. 1989. An adenovirus early region 4 gene product is required for induction of the infection-specific form of cellular E2F activity. Genes Dev. 3:1062-1074. [DOI] [PubMed] [Google Scholar]
- 22.Haviv, I., D. Vaizel, and Y. Shaul. 1995. The X protein of hepatitis B virus coactivates potent activation domains. Mol. Cell. Biol. 15:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honigwachs, J., O. Faktor, R. Dikstein, Y. Shaul, and O. Laub. 1989. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J. Virol. 63:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, K. Q., and A. Siddiqui. 1991. Regulation of the hepatitis B virus gene expression by the enhancer element I. Virology 181:721-726. [DOI] [PubMed] [Google Scholar]
- 25.Huan, B., M. J. Kosovsky, and A. Siddiqui. 1995. Retinoid X receptor alpha transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J. Virol. 69:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katan, Y., R. Agami, and Y. Shaul. 1997. The transcriptional activation and repression domains of RFX1, a context-dependent regulator, can mutually neutralize their activities. Nucleic Acids Res. 25:3621-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller, J. M., and J. C. Alwine. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36:381-389. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S. H., S. P. Hong, S. K. Kim, W. S. Lee, and H. M. Rho. 1992. Replication of a mutant hepatitis B virus with a fused X-C reading frame in hepatoma cells. J. Gen. Virol. 73:2421-2424. [DOI] [PubMed] [Google Scholar]
- 29.Kosovsky, M. J., V. I. Khaoustov, M. Rushton, and B. Yoffe. 2000. Induction of hepatitis B virus gene expression at low temperature. Biochim. Biophys. Acta 1490:63-73. [DOI] [PubMed] [Google Scholar]
- 30.Kramvis, A., and M. C. Kew. 1999. The core promoter of hepatitis B virus. J. Viral Hepatol. 6:415-427. [DOI] [PubMed] [Google Scholar]
- 31.Lai, J. S., M. A. Cleary, and W. Herr. 1992. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 6:2058-2065. [DOI] [PubMed] [Google Scholar]
- 32.Lee, J. S., and S. S. Thorgeirsson. 2002. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology 35:1134-1143. [DOI] [PubMed] [Google Scholar]
- 33.Lillie, J. W., and M. R. Green. 1989. Transcription activation by the adenovirus E1a protein. Nature 338:39-44. [DOI] [PubMed] [Google Scholar]
- 34.Lu, C. C., and T. S. Yen. 1996. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology 225:387-394. [DOI] [PubMed] [Google Scholar]
- 35.Maguire, H. F., J. P. Hoeffler, and A. Siddiqui. 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252:842-844. [DOI] [PubMed] [Google Scholar]
- 36.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 37.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1997. The small envelope protein is required for secretion of a naturally occurring hepatitis B virus mutant with pre-S1 deleted. J. Virol. 71:5449-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nassal, M. 1999. Hepatitis B virus replication: novel roles for virus-host interactions. Intervirology 42:100-116. [DOI] [PubMed] [Google Scholar]
- 39.Ori, A., A. Zauberman, G. Doitsh, N. Paran, M. Oren, and Y. Shaul. 1998. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 17:544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paran, N., B. Geiger, and Y. Shaul. 2001. HBV infection of cell culture: evidence for multivalent and cooperative attachment. EMBO J. 20:4443-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raney, A. K., C. M. Eggers, E. F. Kline, L. G. Guidotti, M. Pontoglio, M. Yaniv, and A. McLachlan. 2001. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J. Virol. 75:2900-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raney, A. K., J. L. Johnson, C. N. Palmer, and A. McLachlan. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71:1058-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shamay, M., R. Agami, and Y. Shaul. 2001. HBV integrants of hepatocellular carcinoma cell lines contain an active enhancer. Oncogene 20:6811-6819. [DOI] [PubMed] [Google Scholar]
- 44.Siddiqui, A., S. Jameel, and J. Mapoles. 1986. Transcriptional control elements of hepatitis B surface antigen gene. Proc. Natl. Acad. Sci. USA 83:566-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegrist, C. A., B. Durand, P. Emery, E. David, P. Hearing, B. Mach, and W. Reith. 1993. RFX1 is identical to enhancer factor C and functions as a transactivator of the hepatitis B virus enhancer. Mol. Cell. Biol. 13:6375-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, H., A. K. Raney, and A. McLachlan. 2001. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J. Virol. 75:3937-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vannice, J. L., and A. D. Levinson. 1988. Properties of the human hepatitis B virus enhancer: position effects and cell-type nonspecificity. J. Virol. 62:1305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, S., S. Hayes, and P. O'Hare. 1994. Site-specific conformational alteration of the Oct-1 POU domain-DNA complex as the basis for differential recognition by Vmw65 (VP16). Cell 79:841-852. [DOI] [PubMed] [Google Scholar]
- 50.Wiley, S. R., R. J. Kraus, F. Zuo, E. E. Murray, K. Loritz, and J. E. Mertz. 1993. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 7:2206-2219. [DOI] [PubMed] [Google Scholar]
- 51.Wu, H. L., P. J. Chen, M. H. Lin, and D. S. Chen. 1991. Temporal aspects of major viral transcript expression in Hep G2 cells transfected with cloned hepatitis B virus DNA: with emphasis on the X transcript. Virology 185:644-651. [DOI] [PubMed] [Google Scholar]
- 52.Xu, Z., G. Jensen, and T. S. Yen. 1997. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J. Virol. 71:7387-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, X., and J. E. Mertz. 2001. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J. Virol. 75:11354-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, X., and J. E. Mertz. 1997. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J. Virol. 71:9366-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, X., and J. E. Mertz. 2003. Distinct modes of regulation of transcription of hepatitis B virus by the nuclear receptors HNF4α and COUP-TF1. J. Virol. 77:2489-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuh, C. H., and L. P. Ting. 1990. The genome of hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J. Virol. 64:4281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, P., A. K. Raney, and A. McLachlan. 1992. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology 191:31-41. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, D. X., and T. S. Yen. 1991. The ubiquitous transcription factor Oct-1 and the liver-specific factor HNF-1 are both required to activate transcription of a hepatitis B virus promoter. Mol. Cell. Biol. 11:1353-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]