Abstract
Three cyclin-dependent kinases, CDK7, -8, and -9, are specifically involved in transcription by RNA polymerase II (Pol II) and target the Pol II C-terminal domain (CTD). The role of CDK7 and CDK8 kinase activity in transcription has been unclear, with CDK7 shown to have variable effects on transcription and CDK8 suggested to repress transcription and/or to target other gene-specific factors. Using a chemical genetics approach, the Saccharomyces cerevisiae homologs of these kinases, Kin28 and Srb10, were engineered to respond to a specific inhibitor and the inhibitor was used to test the role of these kinases in transcription in vivo and in vitro. In vitro, these kinases can both promote transcription, with up to 70% of transcription abolished when both kinases are inhibited together. Similarly, in vivo inhibition of both kinases together gives the strongest decrease in transcription, as measured by chromatin immunoprecipitation of Pol II. Kin28 and Srb10 also have overlapping roles in promoting ATP-dependent dissociation of the preinitiation complex (PIC) into the Scaffold complex. Using the engineered kinases and an ATP analog, specific kinase substrates within the PIC were identified. In addition to the previously known substrate, the Pol II CTD, it was found that Kin28 phosphorylates two subunits of Mediator and Srb10 targets two subunits of TFIID for phosphorylation.
An initial step in transcription by RNA polymerase II (Pol II) is the formation of a preinitiation complex (PIC), in which Pol II and the general transcription factors are stably bound at the promoter but Pol II is not yet in an active state to begin RNA synthesis (23, 29). In the next step, the DNA helicase XPB promotes ATP-dependent isomerization of the PIC into the Open complex. In this state, a single-stranded DNA bubble is formed spanning the transcription start site, and the template DNA strand is pulled into the active site of Pol II. Upon addition of the remaining nucleotides, polymerase initiates transcription. In concert with these events, serine 5 in the C-terminal domain (CTD) of Pol II becomes phosphorylated independently of Open complex formation (17, 32, 43). In two cases, this was shown to promote escape of Pol II from the promoter (2, 18). In addition to Pol II, two general transcription factors, TFIIB and TFIIF, dissociate from the promoter during the initiation process, leaving the remaining general factors at the promoter in the Scaffold complex (49). In vitro, this complex can serve as an intermediate in transcription reinitiation.
Genetic and biochemical approaches have identified four Saccharomyces cerevisiae cyclin-dependent kinases specifically involved in transcription: Kin28 (CDK7), Srb10 (CDK8), Ctk1, and Bur1/Sgv1. The latter two kinases are related to mammalian CDK9 (32). All four of these kinases are known to phosphorylate the Pol II CTD, but each plays a different role in gene expression. Kin28 is an essential gene and is a subunit of the general factor TFIIH, but the role of Kin28/CDK7 kinase activity in transcription is controversial. Northern and genome-wide expression analyses have shown that Kin28 is required for normal levels of Pol II transcripts (16, 45). Kin28 activity is also required for binding of capping enzymes to the phosphorylated CTD (21, 38). However, studies examining the effect of Kin28 on transcription using chromatin immunoprecipitation (IP) have given contradictory results as to the importance of Kin28 (21, 38). Likewise, in vitro studies using the kinase inhibitor H8 or mutations in Kin28 or human CDK7 that reduce kinase activity have shown effects on transcription ranging from none to strong dependence (2, 17, 18, 20, 25, 39).
Srb10, originally identified as a suppressor of CTD truncations, is a nonessential subunit of the Mediator complex. Mediator binds RNA Pol II and is required for yeast transcription in vivo and in vitro in cellular extracts (23). Genetically, Srb10 has been found to act both positively and negatively in gene expression. On a genome-wide scale, deletion of Srb10 derepressed expression of 173 genes in rich glucose medium (16). In other studies, mutation of Srb10 was found to induce expression of genes repressed by glucose, mating type-specific genes, and genes involved in stress response and in nutrient foraging (9). Consistent with a repressive function, it was found that Srb10 could phosphorylate and inactivate Pol II in vitro prior to PIC formation (14). CDK8, the mammalian homolog of Srb10 in the Mediator complex NAT, was found to repress transcription in vitro by phosphorylation of cyclin C, the cofactor for CDK7 (1). In contrast, Srb10 is required for efficient activation of transcription by both Gal4 and Sip4 (15, 46). Finally, it was found that Srb10 phosphorylation of the activators Gcn4 and Ste12 destabilizes these proteins (10, 27).
Both yeast Ctk1 and Bur1/Sgv1 are related to mammalian CDK9 (32). CDK9 is a subunit of the factor P-TEFb that stimulates Pol II elongation by counteracting the action of negative factors NELF and DSIF (30). Genetically, Ctk1 and Bur1 are suggested to be elongation factors, since mutations in both cause sensitivity to 6-azauracil and each shows genetic interactions with known Pol II elongation factors (32). However, these two kinases may have different targets, as BUR1 is an essential gene whereas CTK1 is not.
In contrast to the very stable PIC, the Open complex is unstable. In the human system, purified PICs rapidly lose activity when treated with ATP (8). In the yeast system, PICs incubated with ATP rapidly dissociate into the Scaffold complex, which contains all PIC components except Pol II, TFIIB, and TFIIF (49). Scaffold complexes formed by addition of ATP alone or by addition of all four nucleotides appear identical (35, 49). This destabilization of Pol II may be required for productive transcription, allowing Pol II to escape the promoter. Scaffold complex formation requires a hydrolyzable form of ATP; therefore, it is possible that one or more of the kinases identified above are involved in PIC dissociation to the Scaffold complex. To investigate the role of kinase activities in transcription and Scaffold complex formation, we have used a chemical genetics approach (4, 5) to specifically inhibit individual kinases in vivo and in vitro. Surprisingly, we found that Kin28 and Srb10 have overlapping roles in promoting transcription and in formation of the Scaffold complex. We have also identified new unexpected targets of these kinases within the transcription machinery.
MATERIALS AND METHODS
Yeast strains.
Strains with triple Flag epitope tags at the C terminus of coding regions were derivatives of BY4705 (7) or BJ5460 (19). These strains were constructed using the vector p3FLAG-KanMX as described previously (13) or p3FLAG-Hyg (a gift from T. Tsukiyama).
The Kin28 L83G allele was generated by in vitro mutagenesis, and the gene was cloned into the integration vector pRS306 (42) to generate plasmid pSH573. This plasmid was cut in the Kin28 coding sequence with HindIII and transformed to strain BY4705, selecting for Ura+ integrants. Strains which had lost the wild-type KIN28 gene, leaving kin28 L83G at the KIN28 chromosomal locus, were selected on 5-fluorouracil plates, screened for sensitivity to NA-PP1, and confirmed by PCR amplification and sequencing of the KIN28 locus. This strain was triple Flag epitope-tagged at Kin28 as above to create strain SHY483. Western analysis showed that the nontagged kin28 L83G strain (SHY473) had two- to threefold lower levels of Kin28 protein than wild-type cells, probably due to lower stability of the mutant protein. To compensate for this, the strain was transformed with plasmid pSH579 containing the kin28 L83G allele on an ARS CEN URA3 vector. The resulting strain, SHY508, contained levels of Kin28 L83G protein comparable to Kin28 in wild-type strains. SHY508 was used for the biochemical studies described in this paper.
To create the Srb10 Y236G strain, the SRB10 open reading frame was first replaced by the KanMX gene in strain BY4705. This strain was transformed with plasmid pSH599 (ARS CEN srb10 Y236G) to create strain SHY543. To create the kin28 srb10 double analog-sensitive mutants, the SRB10 gene was deleted from SHY508 and then transformed with pSH599 to create strain SHY549.
For purification of the CAK complex containing Tfb3, Ccl1, and Kin28, the TFB3 gene was tandem affinity purification (TAP) tagged in either wild-type or Kin28 L83G strains as described previously (36) to create strains SHY405 and SHY532. To purify Srb10 kinase by IP, in vitro mutagenesis was used to triple Flag tag the C terminus of the SRB10 and srb10 Y236G genes on an ARS CEN plasmid. These plasmids were used to transform a Δsrb10 strain to create strains SHY607 (SRB10-Flag3) and SHY608 (srb10 Y236G-Flag3).
Yeast growth assays.
To test the sensitivity of strains to PP1 derivatives, 12 μl of a saturated yeast culture grown in yeast extract-peptose-dextrose (YPD) medium was diluted in 2.5 ml of YPD top agar (1% agar) and plated to YPD plates. A 6-mm disk of 3MM paper (Whatman) was placed on top of the agar and spotted with 3 μl of 1 or 10 mM NA-PP1 or NM-PP1 in dimethyl sulfoxide. Plates were incubated 14 to 24 h at 30°C.
Preparation of kinase complexes and in vitro kinase assay.
Kin28 complexes (CAK) were purified by the TAP-tag purification method (36) with the following modifications. Whole-cell extracts (WCE) (∼300 mg of protein) made from SHY405 and SHY532 were loaded onto a 5-ml immunoglobulin G-Sepharose column (Amersham Biosciences). After extensive washing with buffer A (20 mM HEPES [pH 7.9], 300 mM potassium acetate [KOAc], 0.5 mM EDTA, 10% glycerol, 0.05% NP-40, 1 mM dithiothreitol [DTT], and protease inhibitors), the resin was incubated with 6 U of TEV-protease per mg of WCE at 16°C for 4 h. The supernatant was made 2 mM in CaCl2 and diluted four times with buffer B (20 mM Tris [pH 8.0], 300 mM KOAc, 1 mM MgOAc, 1 mM imidazole, 2 mM CaCl2, 10% glycerol, 0.01% NP-40, 2 mM DTT, and protease inhibitors). It was then loaded onto a 0.8-ml calmodulin affinity column (Stratagene), washed with buffer B, and eluted with buffer C (20 mM Tris [pH 8.0], 300 mM KOAc, 1 mM MgOAc, 1 mM imidazole, 3 mM EGTA, 10% glycerol, 0.01% NP-40, 2 mM DTT, and protease inhibitors). Fractions containing CAK were collected and concentrated by using Centricon 30 (Millipore). Srb10 complexes were immune precipitated from nuclear extracts made from SHY607 (SRB10-Flag3) and SHY608 (srb10 Y236G-Flag3) strains. Typically, 0.5 mg of nuclear extract was preincubated with 10 μl of protein G beads for 1 h. After brief centrifugation, the supernatant was incubated with 3 μl of anti-Flag M2 antibody for 2 h at 4°C, and then 10 μl of fresh protein G beads was added for an additional 1 h. The beads were then washed extensively with buffer D (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% NP-40, 2 mM DTT, and protease inhibitors) and resuspended in transcription buffer for kinase assay.
The substrate glutathione S-transferase (GST)-CTD, used in the kinase assay, contained the entire Rpb1 CTD from amino acids 1534 to 1733 fused to GST, cloned in the vector pGEX-5X-1 (Amersham), and was purified from Escherichia coli using glutathione-Sepharose. For both Kin28 and Srb10 kinase assays, 1 μg of GST-CTD was added to kinase complex in a 20-μl reaction volume containing transcription buffer plus phosphatase inhibitors (1 mM NaN3, 1 mM NaF, 0.4 mM NaVO3, and 0.4 mM Na3VO4). Various amounts of NA-PP1 immediately followed by 600 μM ATP-5 μCi of [γ-32P]ATP was then added for 4 min (for Kin28 assay) or 30 min (for Srb10 assay) at room temperature. The reactions were stopped by adding NuPAGE LDS sample buffer (Invitrogen), resolved by 4-to-12% NuPAGE gel, and quantitated by PhosphorImager (Molecular Dynamics).
Transcription.
Plasmid template transcription was carried out as described previously (35) except that phosphocreatine and creatine phosphokinase were omitted from transcription reaction mixtures and another transcription buffer (20 mM HEPES [pH 7.6], 100 mM KOAc, 0.05 mM EDTA, 3.5% glycerol, 2.5 mM DTT) was used. After PICs were formed by incubating nuclear extract, activator Gal4-AH, and plasmid pSH515 for 40 min, various concentrations of NA-PP1 immediately followed by 250 μM nucleoside triphosphates (NTPs) were added for 4 min to give a single round of initiation.
Immobilized template transcription was performed as described previously (35) with the following modifications. PICs were assembled by incubating nuclear extract, activator Gal4-AH, and the immobilized template for 40 min and then washed with transcription buffer plus 0.05% NP-40 three times. To initiate transcription, various concentrations of NA-PP1 immediately followed by 600 μM NTP were added for 4 min at room temperature.
Immobilized template assay.
PIC formation was performed as described previously (49) except for the modifications described above. PIC dissociation experiments were performed as described elsewhere (49) with the following modifications. After washing, PICs were resuspended in 50 μl of transcription buffer containing 0.025% NP-40 and 1 μg of HaeIII-digested E. coli DNA competitor. Various concentrations of NA-PP1 immediately followed by 600 μM ATP-10 μCi of [γ-32P]ATP were added for 4 min at room temperature. The supernatants were removed and precipitated with trichloroacetic acid. The templates were washed once and digested with PstI to isolate the Scaffold complexes. All samples were analyzed by a 4 to 12% NuPAGE or a 3 to 8% Tris-acetate gel (Invitrogen) and electroblotted to polyvinylidene difluoride membranes. Phosphorimager and Western blot analysis were performed on the same membrane using the methods described previously (35).
32P labeling of N6-benzyl-ATP.
[γ-32P]N6-benzyl-ATP was produced by a method described previously (31). Briefly, 10-His-tagged nucleoside-diphosphate kinase (NDPK) was overexpressed and purified from E. coli cells by using a HIS-Select HC nickel affinity gel (Sigma). NDPK was then bound to iminodiacetic acid-Co2+ Sepharose beads (Sigma) in a Bio-Spin column (Bio-Rad). After washing, [γ-32P]ATP was loaded to the column to produce a population of autophosphorylated NDPK. The column was then washed with buffer to remove residual [γ-32P]ATP and then washed with N6-benzyl-ADP to yield an eluted mixture of N6-benzyl-ADP and [γ-32P]N6-benzyl-ATP. Following quantitation, the product was used in substrate labeling experiments.
Substrate labeling by [γ-32P]N6-benzyl-ATP.
The substrate labeling experiments were performed the same as the immobilized template assay described above, except that 1 to 5 μCi of [γ-32P]N6-benzyl-ATP was used instead of [γ-32P]ATP.
Chromatin IP analysis.
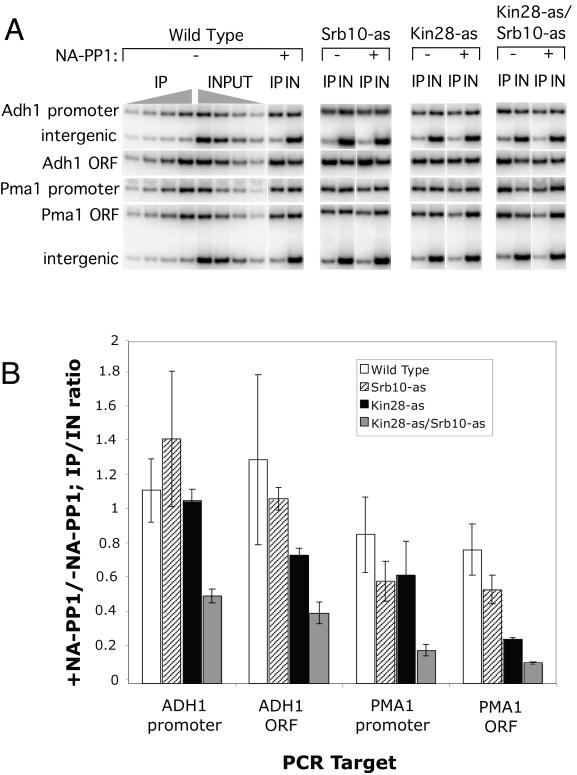
Strains containing the indicated wild-type or analog-sensitive kinase (as-kinase) mutations were grown at 30°C in synthetic glucose medium containing only required amino acids. At an A600 of ∼1.5, cultures were split into two and NA-PP1 was added to one culture at a final concentration of 6 μM. Strains were incubated at 30°C with shaking for an additional 12 min. Formaldehyde was added to each culture to a final concentration of 1% and incubated for 10 min at room temperature. Cross-linking was stopped by the addition of 125 mM glycine followed by a 5-min incubation. Cells were harvested and lysed, and extracts were made essentially as described previously (22). DNA was sonicated to obtain an average size of ∼300 bp. IPs used 1 mg of protein extract and 5 μl of polyclonal αRpb3, or no antibody, as a control. After incubation of antibody with extract at 4°C for 3 h, protein A-Sepharose was added for an additional 1 h. Beads were collected and washed a total of eight times and eluted twice with buffer containing 1% sodium dodecyl sulfate (SDS) at 65°C. Cross-linking was reversed overnight at 65°C, and DNA was isolated using the QIAquick PCR purification kit (Qiagen). Input DNA was purified from 0.01 mg of protein extract. PCRs used the ADH1 (−255, −13; 844, 1013) and PMA1 (−370; 2018, 2290) primers described in reference 21. Primer PMA1 −370 was paired with primer PMA1_M47, CAATGATTTTCTTTAACTAGCTGGGG. The intergenic primers were complementary to a sequence beyond the end of the Gal1 gene: GCTTTCAACCGCTGCGTTTTGG and CTGCATCTCGTCAGTTGGCAAC. A 20-μl PCR volume included 2 μCi of [α-32P]dCTP, and amplification conditions were 95°C for 2 min; 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min for 26 cycles; and 72°C for 3 min. The IP/input (IN) ratio of DNA used in PCR for each replicate was 1:2. All samples were assayed as a titration of four replicates to confirm linearity and quantified by phosphorimager. The ratio of IP/IN was calculated for each replicate and adjusted for background by subtracting the corresponding intergenic IP/IN ratio. These results were used to determine the means from two independent experiments. The averages and standard deviations for those experiments are shown below graphically in Fig. 4B.
FIG. 4.
Inhibition of Kin28 and Srb10 in vivo. (A) Representative chromatin IP data from one experiment. The indicated strains were either treated or not treated with 6 μM NA-PP1 for 12 min and cross-linked with formaldehyde, and sheared DNA was isolated. Pol II cross-linking was assayed by IP with Rpb3 antisera followed by quantitative PCR. For all samples, a series of four different DNA concentrations were used for PCR in the linear range of the assay. Shown are PCR products of representative IN or IP samples. (B) Quantitation of results from two separate chromatin IP experiments. Results are plotted as the ratio of signals seen with and without NA-PP1 treatment.
Extracts and antibodies.
Nuclear and small-scale WCE were prepared as described on the Hahn Laboratory website (www.fhcrc.org/labs/hahn). Monoclonal Rpb1 antibodies 8WG16 and H14 were purchased from Covance Co., and YN-18 was obtained from Santa Cruz Biotechnology. Anti-TAF2 antibody was a gift from R. Tjian. Other antibodies used in this study have been described previously (24, 35).
RESULTS
Kin28 and Srb10, but not Bur1 or Ctk1, are stably associated with PICs.
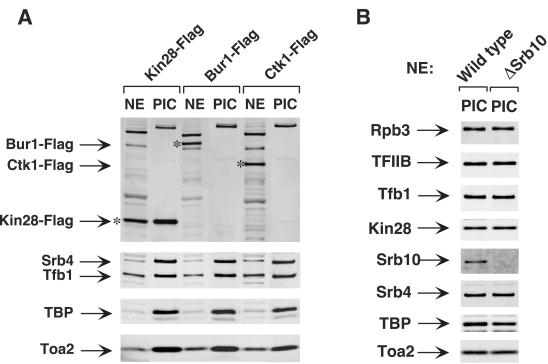
The four kinases specifically implicated in transcription, Kin28, Srb10, Bur1, and Ctk1, are candidates for promoting the Scaffold complex formation. Among them, Kin28 and Srb10 were shown to be PIC components (Fig. 1) (24, 29, 34). Using chromatin IP, Ctk1 has been localized near promoter regions, but it was not distinguished if Ctk1 is a stable PIC component or associates with newly elongating RNA Pol II (11). To investigate whether Ctk1 and Bur1 are stable PIC components, the immobilized template system was used to form PICs with extracts made from strains containing C-terminal Flag-tagged Bur1, Ctk1, or Kin28. Although similar levels of the three Flag-tagged kinases were present in the nuclear extracts, Bur1 and Ctk1 were not detectable in the PICs by Western blotting (Fig. 1). In contrast, Kin28 was clearly enriched in the PIC compared to nuclear extract. Other known PIC components, such as Srb4, Tfb1, TBP, and Toa2, were also probed by Western analysis and shown to be equivalent in PICs formed by all three Flag-tagged extracts. These results show that Ctk1 and Bur1 are not stable PIC components and suggest that either Kin28 and/or Srb10 could be the kinase responsible for PIC dissociation. Mass spectrometry analysis of yeast PICs formed by the same methods used here has not detected the presence of any other kinases in yeast PICs (33).
FIG. 1.
Bur1 and Ctk1 are not PIC components. (A) Nuclear extracts (NE) made from Kin28-Flag, Bur1-Flag, and Ctk1-Flag strains were incubated with the immobilized template for 40 min. PICs were isolated and analyzed by Western blotting. The top panel was probed with the anti-Flag M2 antibody. The asterisk indicates the position of Kin28-Flag, Bur1-Flag, or Ctk1-Flag protein. The lower three panels were probed with antibodies directed against known PIC components. The experiment shown in panel B is the same as that in panel A but demonstrates that Srb10 is a stable PIC component (24).
Specific inhibition of Kin28 and Srb10 kinases.
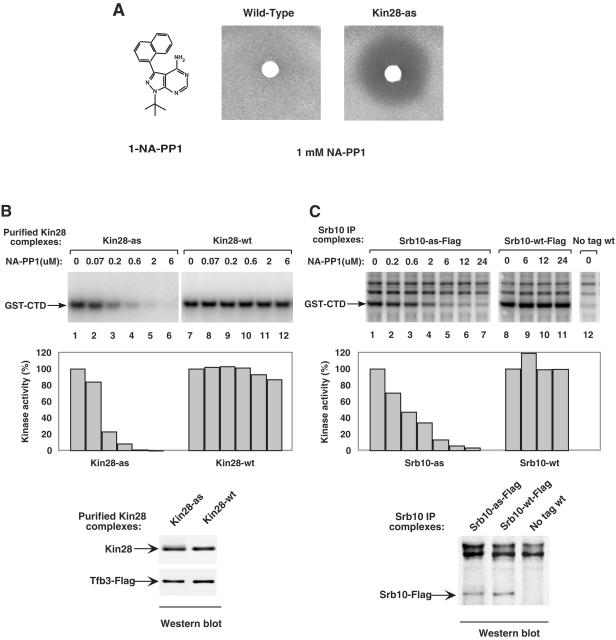
To investigate the roles of Kin28 and Srb10 in transcription, we first attempted to use several nonspecific kinase inhibitors, including H8, which has been previously used to test the role of mammalian CDK7. However, none of the inhibitors tested reduced either transcription or CTD phosphorylation in the yeast nuclear extract system (N. Yudkovsky, personal communication). We also attempted to express a dominant-negative Kin28 mutant in yeast in order to purify the mutant kinase. Unfortunately, this lethal mutation severely inhibited cell growth in heterozygous diploids (N. Yudkovsky, personal communication). As an alternative, we used a chemical genetics strategy that allowed specific inhibition of individual kinases by a small molecule inhibitor (4, 5). In this approach, a specific conserved bulky residue in the kinase ATP binding pocket is mutated to either glycine or alanine, resulting in an enlarged pocket that is not present in other endogenous kinases. Chemically modified PP1 derivatives, such as NA-PP1 (Fig. 2A), can specifically fit into the enlarged nucleotide binding pocket and inhibit kinase activity of these analog-sensitive (as) mutants. For the c-Src kinase, enlarging the ATP binding pocket by this method affected the structure of the nucleotide binding pocket without affecting the phospho-acceptor binding site (47). Further, the peptide substrate specificities of the wild type and as-kinases were identical, as determined using a degenerate peptide library, consistent with the identical structures of the phospho-acceptor binding site in both enzymes (47).
FIG. 2.
Inhibition of Kin28-as and Srb10-as activities by NA-PP1. (A) Growth inhibition of either wild-type or Kin28-as strains. Three microliters of 1 mM NA-PP1 was spotted on a 0.6-cm filter disk placed on a soft agar plate containing the indicated strain and incubated for 16 h at 30°C. The structure of NA-PP1 is shown. (B) Kin28-as or Kin28 wild-type (wt) complexes were purified from yeast WCE as described in Materials and Methods. The effects of NA-PP1 on the kinase activities of these complexes were assayed using GST-CTD as a substrate. Reactions were performed in transcription buffer containing 600 μM ATP-5 μCi of [γ-32P]ATP. After SDS-PAGE, the phosphorylated GST-CTD was quantitated by phosphorimaging. The activities of Kin28-as (lane 1) and Kin28-wt (lane 7) were normalized to 100%. Shown below is Western analysis of equal amounts of the two Kin28-purified complexes. (C) Immune precipitates from Srb10-as-Flag, Srb10-wt-Flag, and untagged strains were assayed for kinase activities in the presence of increasing amounts of NA-PP1. Reactions and quantitation were performed as described for panel B. Immune precipitate from an untagged strain was used as a control. The lower panel is a Western analysis of equal amounts of the anti-Flag immune precipitates.
Using this strategy to create kinase-as mutations, yeast strains containing Kin28 L83G, Srb10 Y236G, or Kin28 L83G/Srb10 Y236G double mutants were constructed and tested for growth inhibition by the PP1 derivatives. NA-PP1 had no effect on growth of the wild-type strain but strongly and equivalently inhibited growth of the Kin28-as and Kin28-as/Srb10-as double mutant (Fig. 2A and data not shown). In contrast, NA-PP1 added to the Srb10-as strain had no detectable effect in this assay, since Srb10 is not an essential gene (data not shown). All strains were engineered to express the as-kinase mutants at levels equivalent to the endogenous wild-type kinases (see Materials and Methods). None of these strains showed any obvious growth phenotypes, such as heat or cold sensitivity.
To further characterize the inhibitor sensitivity of the mutant kinases, in vitro kinase assays were performed using GST-CTD as a substrate. To mimic the experimental conditions of the Scaffold formation and transcription assays, the kinase assay was conducted in transcription buffer and 600 μM ATP/[γ-32P]ATP. Purified yeast CAK complexes containing Ccl1, TAP-tagged Tfb3, and either Kin28 or Kin28-as were used in the GST-CTD kinase assay (Fig. 2B, upper panel). Under these reaction conditions, 6 μM NA-PP1 almost completely inhibited Kin28-as activity but had little effect on wild-type Kin28 activity. Western analysis of the CAK complexes (Fig. 2B, lower panel) confirmed that equivalent amounts of Kin28 and Kin28-as were used in the kinase assay. To test the sensitivity of Srb10-as kinase to NA-PP1 inhibitor, immune precipitates from Srb10-Flag and Srb10-as-Flag extracts were tested (Fig. 2C, upper panel). Again, equivalent amounts of wild-type and mutant kinases were used in the in vitro assay (Fig. 2C, lower panel). The activity associated with Srb10-as kinase decreased dramatically with increasing NA-PP1 concentrations. The 6 μM concentration of NA-PP1 inhibited kinase activity about 90%, whereas the activity of Srb10 wild type was not affected. Two other phosphorylated polypeptides were observed in this assay (Fig. 2C, upper panel) and were likely due to background phosphorylation of nonspecifically precipitated proteins by other kinases in the immune precipitates, since they were also seen in control reactions using strains lacking any Flag tag (lane 12).
Both Kin28 and Srb10 can promote transcription in vitro.
The Kin28-as and Srb10-as strains were next used to investigate the role of the kinases in transcription. These experiments were performed using PICs isolated on immobilized promoter templates (Fig. 3). Nuclear extracts and the activator Gal4-AH were incubated with the immobilized HIS4 promoter containing a single upstream Gal4 binding site (34). PIC formation was allowed to proceed for 40 min, and then PICs were washed and resuspended in transcription buffer. NA-PP1 was added, immediately followed by all four NTPs, and the reactions were then incubated for 4 min, giving approximately a single round of initiation. Primer extension assay of RNA synthesized is shown in the top panel of Fig. 3A, and the results are quantitated in the lower panel. A 12 μM concentration of NA-PP1, which completely inhibited Kin28-as kinase activity (Fig. 2), reduced transcription only 40% (lane 7). Under the same conditions which inhibit Srb10 kinase activity ∼95% (Fig. 2), inhibition of Srb10 alone did not significantly reduce transcription. In contrast, inhibition of both kinases together gave a 70% reduction of transcription (lane 14). Similar results were seen using plasmid promoter templates where the transcription reaction was done in the presence of nuclear extract instead of using purified PICs (data not shown). To compare this unexpected result with the effect of genetic mutations in the two kinases, extracts were made from strains containing either a Kin28 ts mutant (45) or a Kin28-as Δsrb10 double mutation. Consistent with the above results, addition of 24 μM NA-PP1 inhibited transcription by 80% in the Kin28-as Δsrb10 extract (Fig. 3B, lanes 5 and 6). In contrast, transcription was inhibited only 50% in the kin28 ts extract when compared to addition of saturating amounts of purified wild-type CAK complex to the mutant extract (Fig. 3B, lanes 9 and 10). Taken together, our results demonstrate that both Kin28 and Srb10 can stimulate transcription with Kin28, the dominant kinase, able to promote normal levels of transcription in the absence of Srb10. However, when Kin28 is inhibited, Srb10 can promote significant levels of transcription. This is in contrast to previous models for Srb10 function where it was thought to have either no role or an inhibitory function in general transcription. The low level of transcription remaining when both Srb10 and Kin28 were inhibited may be due to either kinase-independent transcription, incomplete inhibition of Kin28 and/or Srb10, or transcription dependent on other kinase activities in the reaction. As described below, other kinase activities that are not sensitive to NA-PP1 are present in the purified PICs.
FIG. 3.
Both Kin28 and Srb10 can promote transcription. (A) Nuclear extracts (NE) made from wild-type, Kin28-as, Srb10-as, or Kin28-as Srb10-as strains were used as indicated. The transcription reactions were performed as described in Materials and Methods. Quantitation of the results is shown in the lower panel. Transcription activity of each nuclear extract without inhibitor was normalized to 100%. (B) Transcription reaction performed as described for panel A using immobilized templates and NA-PP1 with extracts from the indicated strains. In lane 9, a saturating amount of purified wild-type CAK was added.
Both Kin28 and Srb10 can promote transcription in vivo.
To examine the role of these kinases in vivo, we analyzed strains containing the kinase-as mutations for Pol II transcription with or without NA-PP1 addition. As described above, it is likely that Kin28 kinase activity is required for mRNA stability. Therefore, when Kin28 is inhibited, the association of Pol II with coding regions of transcribed genes measured by chromatin IP is likely a better measure of transcription (21, 38). NA-PP1 added to yeast at concentrations below 10 μM was previously shown to specifically inhibit as-kinase activity (5). Exponentially growing cells were incubated with 6 μM NA-PP1 for 12 min and cross-linked with formaldehyde for 10 min, and then chromatin was isolated and analyzed using chromatin IP. Western analysis of total cell protein (data not shown) demonstrated that this brief treatment gave a significant decrease in the level of the Pol IIo form in both the Kin28-as and Srb10-as/Kin28-as double mutant but not in wild-type or Srb10-as strains. To measure association of Pol II cross-linked to DNA, antiserum against the Pol II subunit Rpb3 was used in the IPs, as binding of this antibody is independent of the phosphorylation state of the CTD. Comparisons below are made for Pol II cross-linking in cultures either with or without the 12-min drug treatment.
Strikingly, we found that the in vivo cross-linking results paralleled those seen in vitro for the positive role of the two kinases in transcription (Fig. 4). In Fig. 4B, the chromatin IP/IN DNA signal was plotted as a ratio of the signals observed with or without addition of NA-PP1. Addition of inhibitor to the Kin28-as strain gave a 2.6-fold decrease in Pol II cross-linking at the PMA1 open reading frame. In agreement with the in vitro results, addition of inhibitor to the Kin28-as Srb10-as double mutant strain gave a stronger reduction in Pol II cross-linking, with a 5.7-fold decrease compared to the wild-type strain. Similar results were seen at the ADH1 open reading frame. Inhibition of both kinases together caused nearly a twofold greater decrease in Pol II cross-linking compared to inhibition of Kin28 alone. In contrast, inhibition of Srb10 alone did not give a significant decrease in Pol II cross-linking at either ADH1 or PMA1. We also probed for Pol II cross-linking using PMA1 and ADH1 probes which detect Pol II at both the promoter and the 5′ end of the coding sequence. Interestingly, inhibition of Kin28 or Srb10 alone did not cause a significant decrease in Pol II cross-linking at either promoter. In contrast, inhibition of Kin28 and Srb10 together caused a significant decrease in Pol II cross-linking. This decrease could be due to either instability of the PIC and/or a decrease in Pol II at the 5′ end of the coding sequence.
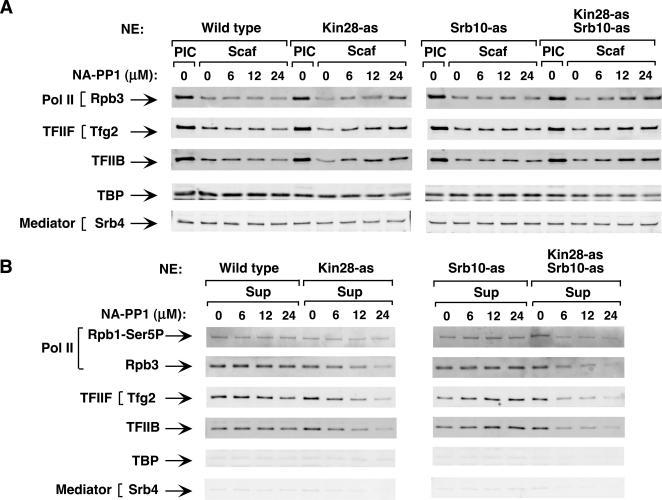
Kin28 and Srb10 possess the activity responsible for PIC dissociation.
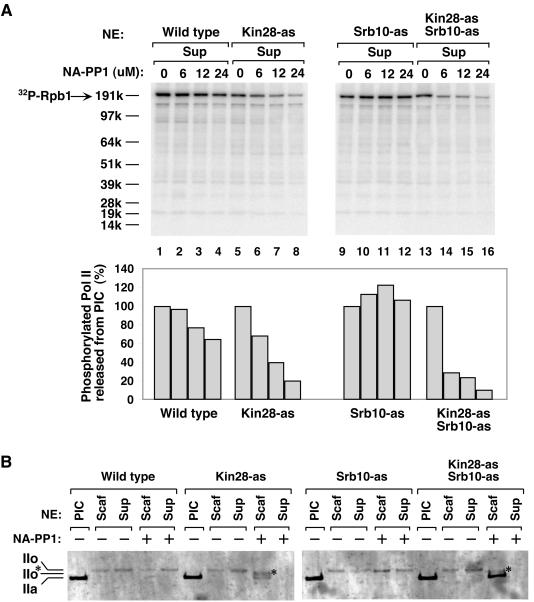
To investigate the roles of Kin28 and Srb10 in PIC dissociation, PICs were formed on immobilized templates; various concentrations of NA-PP1 were added and immediately followed by ATP/[γ-32P]ATP addition. After incubation for 4 min, factors released into the supernatant and factors remaining on the promoter template were analyzed by Western blotting (Fig. 5). [γ-32P]ATP was added to detect phosphorylation and will be addressed below. Little effect of NA-PP1 was observed on the dissociation of PICs containing wild-type kinases or the Srb10-as kinase. In contrast, PIC dissociation was reduced when Kin28 alone or both Kin28 and Srb10 were inhibited. Increasing levels of NA-PP1 blocked the release of factors (Pol II, IIF, and IIB) into the supernatant and increased the level of factors remaining on the promoter (Fig. 5). PIC components that do not normally dissociate were not affected (TBP and Srb4). The inhibition of PIC dissociation was more complete when both Kin28 and Srb10 were inhibited compared to inhibiting Kin28 alone. Comparing Western blotting signals when 6 μM NA-PP1 inhibitor was used (Fig. 5B), up to 70% inhibition of Pol II, TFIIB, and TFIIF release was observed in the double mutant strain compared to at most 30% inhibition in the Kin28-as single mutant. From these results, we conclude that Kin28 and Srb10 kinase activities both promote PIC dissociation into the Scaffold complex.
FIG. 5.
Inhibition of Kin28 and Srb10 kinases inhibits PIC dissociation. PICs were assembled on the immobilized template using nuclear extracts (NE) made from wild-type, Kin28-as, Srb10-as, and Kin28-as Srb10-as strains. Variable amounts of NA-PP1 along with 600 μM ATP-10 μCi of [γ-32P]ATP were added for 4 min. (A) Scaffold complexes (Scaf) were washed, isolated by PstI digestion, and analyzed by Western blotting for the indicated factors. Factors in the PIC without ATP addition (PIC) are shown for comparison. (B) Factors released from the PIC upon ATP addition into the supernatant (Sup) (Pol II, IIB, and IIF) were recovered and analyzed by Western blotting.
PIC dissociation is accompanied by phosphorylation of the Pol II CTD by Kin28 and Srb10.
To investigate the activity of Kin28 and Srb10 kinases on Pol II CTD phosphorylation during Pol II dissociation, the membrane from the experiment of Fig. 5 was analyzed for 32P incorporation to examine the effect of inhibiting Kin28 and Srb10 on Pol II phosphorylation. Figure 6A shows the phosphorimage of the membrane containing factors released from the template upon ATP addition. Inhibition of Srb10 alone had little effect on the amount of phosphorylated Pol II released (Fig. 6A, lanes 9 to 12). When Kin28 was inhibited, the phosphorylated Pol II released was reduced by 30% at 6 μM and by 80% at 24 μM NA-PP1 (Fig. 6A, lanes 5 to 8). In contrast, when both Kin28 and Srb10 were inhibited, the amount of phosphorylated Pol II released was reduced by 70% at 6 μM and by 90% at 24 μM NA-PP1 (lanes 13 to 16).
FIG. 6.
Inhibition of Kin28 and Srb10 kinases inhibits phosphorylation of the Pol II CTD. (A) Phosphorylated Pol II released into the supernatant (Sup) during Scaffold complex formation. Phosphorimager analysis was performed on the membrane containing the supernatants (Fig. 5), and the quantitation of the phosphorylated Pol II CTD is shown below. The phosphorylation signal of each extract without inhibitor (lanes 1, 5, 9 and 13) was normalized to 100% (lower panel). (B) Both Kin28 and Srb10 contribute to hyperphosphorylation of the CTD during Scaffold complex formation. PIC and Scaffold complexes formed from the indicated extracts were fractionated on a 3 to 8% Tris-acetate gel and analyzed by Western blotting using antibody YN-18, which detects Rpb1 independent of the phosphorylation state. IIo* represents a partially phosphorylated Pol II form.
To examine this in more detail, samples from a PIC dissociation experiment were analyzed for the state of Pol II phosphorylation by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a 3-to-8% Tris-acetate gel to separate different phosphorylated forms of Rpb1 (Fig. 6B). For this analysis, the Western blot was probed with antisera to the N terminus of Rpb1. As expected, PICs contained the nonphosphorylated IIa form of Pol II. Without NA-PP1, addition of ATP to all extracts led to multiple phosphorylation of the CTD, giving the slower-mobility IIo form. This result showed that the wild-type and mutant kinases were equally capable of processive CTD phosphorylation. In the Kin28-as mutant extract, NA-PP1 inhibited this processive phosphorylation, and Pol II remaining on the template was found equally in the IIa form and in a form of intermediate mobility, labeled IIo*. In contrast, in a PIC with both Kin28-as and Srb10-as enzymes, NA-PP1 almost completely inhibited the shift of Pol II into any slower-mobility forms. Together, the results of Fig. 6 demonstrate that both Srb10 and Kin28 are the primary kinases responsible for CTD phosphorylation upon formation of the Scaffold complex.
Identification of other specific targets of Kin28 and Srb10 kinases.
From the above results and from previous studies, it is clear that both Kin28 and Srb10 phosphorylate the CTD in the PIC. However, whether these kinases have any specific targets in other general factors or coactivators when present in the PIC has not been examined. To determine if Kin28 and Srb10 target any other PIC components for phosphorylation, the labeling of proteins in the Scaffold complex and supernatant by [γ-32P]ATP was examined (Fig. 7A, lanes 1 to 6). From this, it was apparent that many PIC components and probably other contaminating factors on the immobilized templates become phosphorylated upon addition of ATP. When these reactions were performed using the Kin28-as Srb10-as double mutant extract with added NA-PP1, phosphorylation of these factors apart from Rpb1 was not obviously inhibited, strongly suggesting the presence of other kinases on the immobilized templates (data not shown). Since these other kinases were not detected by mass spectrometry analysis, they are more likely present at substoichiometric levels compared to authentic PIC components. To test for specific phosphorylation of PIC components by Kin28 or Srb10 that may have been masked by this background phosphorylation and to separately identify the targets of the two kinases, we again used the as-kinases. It was previously shown that as-kinases with the enlarged ATP binding pocket can often use the modified ATP analog N6-benzyl-ATP much more efficiently than other wild-type kinases, allowing direct identification of substrates for the engineered kinase (31, 40, 41). In addition, it was shown that an as-kinase utilizing N6-benzyl-ATP had the same polypeptide substrate specificity as a wild-type kinase with ATP substrate (47).
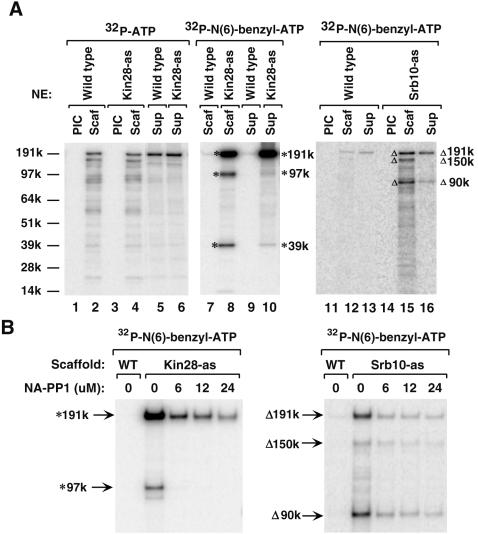
FIG. 7.
Identification of Kin28 and Srb10 substrates in the PIC. (A) The immobilized template assay was performed as described in the legend for Fig. 6, using the indicated nuclear extracts (NE). After PICs were assembled, 600 μM ATP/[γ-32P]ATP or 600 μM ATP/[γ-32P]N6-benzyl-ATP was used to form Scaffolds. Scaffold complexes (Scaf) and supernatants (Sup) were analyzed on a 4 to 12% NuPAGE gel and visualized by phosphorimaging. Lanes 1 to 6, PIC phosphorylation after adding ATP/[γ-32P]ATP; lanes 7 to 16, specific labeling of Kin28 or Srb10 substrates after adding ATP/[γ-32P]N6-benzyl-ATP. * and Δ indicate the three major substrates of Kin28 and Srb10, respectively. (B) NA-PP1 inhibited specific labeling of the Kin28 and Srb10 substrates. Wild-type (WT), Kin28-as, and Srb10-as Scaffolds were formed using 600 μM ATP/[γ-32P]N6-benzyl-ATP in the absence or presence of an increasing amount of NA-PP1. Samples were resolved on a 3 to 8% Tris-acetate gel. The positions of Kin28 and Srb10 substrates are indicated by the * and Δ symbols, respectively.
PIC dissociation was performed as before except that ATP/[γ-32P]N6-benzyl-ATP was added. For Kin28-as, three major polypeptides of 191, 97, and 39 kDa were phosphorylated (Fig. 7A, lanes 8 and 10). The majority of the 191-kDa protein dissociated into the supernatant, whereas the 97- and 39-kDa proteins mostly remained in the Scaffold. The total phosphorylation signal of the 191-kDa protein was about 20-fold stronger than that observed in the 97- and 39-kDa polypeptides. For Srb10-as, many polypeptides were phosphorylated at a low level; however, the phosphorylation levels of three polypeptides of 191, 150, and 90 kDa were strongest (Fig. 7A, lanes 15 and 16). In contrast to the Kin28 substrates, the phosphorylation levels of all three Srb10 substrates were similar. In both the Kin28-as and Srb10-as labeling experiments, wild-type PICs were assayed as controls, and the N6-benzyl-ATP-phosphorylated proteins could barely be observed (lanes 7, 9, 12, and 13), confirming the specificity of these assays.
Labeling of Kin28 and Srb10 substrates is inhibited by NA-PP1.
Earlier, it was shown that the phosphorylation of GST-CTD by the purified as-kinases was inhibited by NA-PP1 (Fig. 2). However, this did not directly address whether the as-kinases could be inhibited when assembled into the PIC and their natural substrates were modified. To examine this, we tested if phosphorylation of the specific substrates identified above using N6-benzyl-ATP could be inhibited by NA-PP1. Kin28-as and Srb10-as Scaffolds were formed as before by adding ATP/[γ-32P]N6-benzyl-ATP in the presence of increasing amounts of NA-PP1. Samples were resolved by SDS-PAGE. The labeling of both Kin28 and Srb10 substrates was greatly reduced with increasing NA-PP1 concentration (Fig. 7B). Specifically, the 191-kDa Kin28 substrate showed 95% inhibition and the 191-kDa Srb10 substrate showed 90% inhibition by 24 μM NA-PP1. The 39-kDa substrate was not examined in this experiment, as it was run off the bottom of the gel. These results, together with the in vitro kinase assay, confirmed the NA-PP1 sensitivity of the as-kinases when assembled into PICs.
Identification of Mediator and TFIID subunits as kinase targets.
To identify the kinase targets seen above, genes encoding candidate PIC components were C-terminally epitope tagged using three copies of the Flag epitope. Since Flag alters the mobility of polypeptides on SDS-PAGE, comparison of phosphorylated proteins in tagged and untagged extracts will identify the target. To simplify this approach, nuclear extracts from Kin28-as or Srb10-as (1st NE) were mixed with equal amounts of an extract from a Flag-tagged strain (2nd NE), and PICs were formed. Scaffolds were generated using ATP/[γ-32P]N6-benzyl-ATP and analyzed by phosphorimaging and Western blotting. It was expected that one half of the PICs would contain the as-kinase. If the kinase and the Flag-tagged protein are not tightly associated in the starting extracts, PICs containing the kinase-as should contain a mixture of tagged and untagged PIC component. Thus, if the specific target is tagged, a doublet of phosphorylated polypeptides arising from the tagged and untagged polypeptides will be observed.
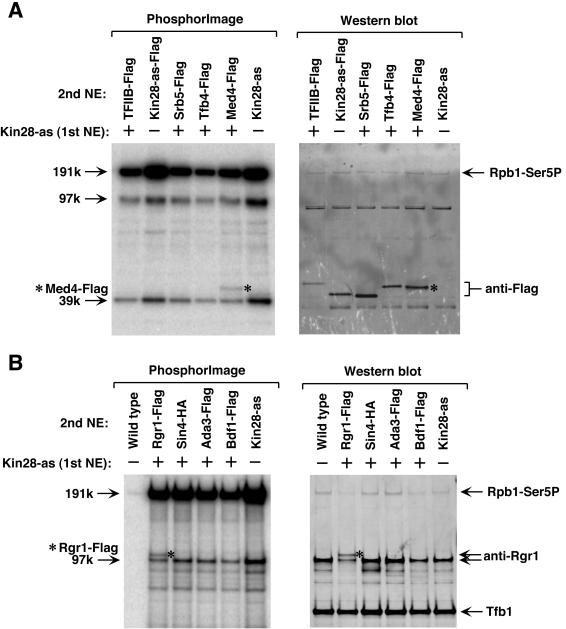
For the 39-kDa Kin28 substrate, TFIIB, Kin28, Srb5, Tfb4, and Med4 were chosen as the most likely candidates, considering factors presumed to be PIC components in the range of 39 kDa. The genes encoding these factors were Flag tagged, and extracts were made from the tagged strains. PICs were formed on immobilized templates using the strategy above, except for Kin28-as-Flag, which was used directly. Only the Scaffold that formed with Med4-Flag plus Kin28-as extracts showed a doublet of phosphorylated polypeptides (Fig. 8A, left panel). Western blotting using anti-Flag antibody confirmed that the top band was Med4-Flag (Fig. 8A, right panel). Similarly, Rgr1, Sin4, Ada3, and Bdf1 were chosen as the 97-kDa Kin28 substrate candidates, and doublet bands were seen only in the Scaffold formed with Rgr1-Flag plus Kin28-as extract. Western blotting using anti-Rgr1 and anti-Flag antibody confirmed that the doublet was Rgr1-Flag and Rgr1 (Fig. 8B and data not shown). Thus, we conclude that the 39-kDa and 97-kDa substrates of Kin28 are Med4 and Rgr1. Med4 and Rgr1 are two subunits of the Mediator complex. A phosphorylated species of Med4 was identified in yeast extract, but the corresponding kinase was not determined (3). As expected, the 191-kDa Kin28 substrate was Rpb1, as shown by H14 antibody (Fig. 8).
FIG. 8.
Identification of Med4 and Rgr1 as Kin28 substrates in the PIC. (A) Identification of Med4 as the 39-kDa Kin28 substrate. Substrate candidates were Flag tagged in a wild-type strain background (except Kin28, which directly used Kin28-as-Flag). Nuclear extract made from a Flag-tagged strain (2nd NE) was mixed with Kin28-as nuclear extract (1st NE) in equal amounts. Scaffolds were formed as described in the legend for Fig. 7, loaded onto a 4 to 12% NuPAGE gel, and electroblotted to a polyvinylidene difluoride membrane. Phosphorimaging and Western blotting were performed on the same membrane. The * indicates the position of Med4-Flag on both phosphorimaging and Western blotting. (B) Identification of Rgr1 as the 97-kDa Kin28 substrate. The procedure was the same as for panel A, except samples were separated on a 3 to 8% Tris-acetate gel. The * indicates the position of Rgr1-Flag on both phosphorimaging and Western blotting. In addition, Rpb1 was identified as the 191-kDa Kin28 substrate by H14 antibody in both panels A and B.
A similar method was used to identify Srb10 substrates in the PIC. For the 90-kDa substrate, Scaffold formed with Bdf1-Flag and Srb10-as extracts showed doublet bands, suggesting Bdf1 was the 90-kDa Srb10 substrate (Fig. 9, lane 6). This result was further confirmed by directly Flag tagging Bdf1 in the Srb10-as strain background. Scaffold formed from this extract displayed a slower-migrating phosphorylated polypeptide than the 90-kDa substrate (Fig. 9, lane 5). Western blotting with anti-Flag antibody confirmed the slower-migrating band was Bdf1-Flag (Fig. 9, lanes 11 and 12). We also directly Flag tagged TAF2 in the Srb10-as strain, and a slower migrating band was displayed in the Scaffold, thus confirming TAF2 was the 150-kDa Srb10 substrate (Fig. 9, lanes 1 and 3). TAF2 is a subunit of the TFIID complex (44). Interestingly, Bdf1 was also reported to be a factor associated with TFIID and is the yeast counterpart to a TAF1 domain in larger eukaryotes (26). Thus, we identified Bdf1 and TAF2 as two novel substrates of Srb10.
FIG. 9.
Identification of Bdf1 and TAF2 as Srb10 substrates. The same methods were used as for Fig. 8, except that some of the strains contained both a Flag-tagged gene as well as the Srb10-as mutation. In lane 6, Srb10-as extract was mixed with a Bdf1-Flag extract. The Δ indicates the position of Bdf1-Flag, and the “o” indicates the position of TAF2-Flag. For Western blotting, anti-Flag M2 and the H14 anti-Ser5 antibodies were used as indicated.
DISCUSSION
Previous studies have given contradictory results on the role of CDK7/Kin28 in transcription. In vitro studies using either the nonspecific kinase inhibitor H8 or a kinase-defective mutant have shown transcription dependence on CDK7/Kin28 ranging from no effect to strong dependence. At the CTD-dependent dihydrofolate reductase and AdE4 promoters, dependence was attributed to polymerase escape, because kinase activity was not required for synthesis of the first phosphodiester bond during abortive initiation or Open complex formation (2, 18). In vivo, temperature-sensitive mutants of Kin28 have been used in mRNA analysis. However, general mRNA stability is likely affected in these mutants, since mRNA capping enzyme is not recruited to Pol II after Kin28 inactivation (21, 38). In more direct studies using chromatin IP to examine Pol II located at promoters and coding sequences, one study found a dramatic decrease in the level of Pol II associated with both promoter and gene sequences after heat shock of a ts kin28 mutant (38). In contrast, another study using a partially defective Kin28 kinase mutant found little difference in Pol II occupancy despite the fact that these strains had much lower levels of the hyperphosphorylated IIo form of Pol II (21).
Our original goal in examining the two kinases was to discover the mechanism of PIC dissociation to the Scaffold complex. Since inhibitors such as H8 and kinase-defective mutations were difficult to use in our system, we used as-kinase mutations for both Kin28 and Srb10 (4, 5). When mutated, both kinases retained activity but were specifically inhibited by NA-PP1, allowing us to test the role of these kinases in transcription and PIC dissociation. An advantage of using this method is that the inhibitor is specific for only the mutated kinase and the inhibitor is cell permeable, allowing tests of kinase function both in vivo and in vitro.
Surprisingly, we found that both kinases could contribute to transcription in vitro. Addition of 12 μM NA-PP1 had no significant effect on transcription using extracts from either wild-type or Srb10-as strains. In contrast, a 40% decrease in transcription was seen when Kin28 alone was inhibited. However, in a strain where both kinases contained analog-sensitive mutations, transcription was inhibited up to 70%. These results were also consistent with those seen in extracts made from strains containing a Kin28 ts mutation or a Kin28-as Δsrb10 double mutation. Strikingly, the positive function of Kin28 and Srb10 was also observed using chromatin IP to examine Pol II associated in vivo with either the coding sequence or 5′ end of two genes. At the two promoters tested, inhibition of both kinases together gave a two- to threefold greater decrease in Pol II cross-linking compared to inhibition of Kin28 alone.
Combined, our in vivo and in vitro studies demonstrate that both Kin28 and Srb10 have a positive role in promoting transcription by Pol II, with Kin28 being the dominant kinase. The result that Srb10 can promote significant levels of transcription in the absence of Kin28 contradicts previous models where Srb10 had either no role or an inhibitory role in general transcription. As yet, we cannot determine which step in transcription requires the action of the two kinases. In the yeast nuclear extract system, we have been unable to observe promoter-specific abortive initiation, so we cannot test whether initiation or promoter escape is a kinase-requiring step in yeast. Previous studies have shown only gene-specific negative and positive functions for Srb10 in transcription. However, in light of these new studies, a general role for Srb10 in initiation would have been masked by the function of Kin28. Genetic analysis has shown that Kin28 and Srb10 are not redundant, since only Kin28 is essential for growth. From Western analysis of drug-treated cells (data not shown), it is clear that Kin28 is primarily responsible for the highly phosphorylated form of Pol II. Previous studies have suggested that this highly phosphorylated form is required for recruitment of capping and other RNA processing factors (30). Thus, one essential role of Kin28 kinase activity not fulfilled by Srb10 is stimulation of RNA processing, since Srb10 is much less processive in phosphorylation than Kin28 (Fig. 6) (6). In contrast, we propose that transcription does not require the highly phosphorylated IIo form of Pol II and that this function can be fulfilled to a significant extent by a lesser phosphorylated state generated by the Srb10 kinase.
When both kinases are inhibited in vitro or in vivo, a fraction of normal transcription is still observed. Whether this remaining transcription is due to incomplete inhibition of Srb10 and Kin28, is kinase independent, or is promoted by one or more other kinases is not yet known. Of the four S. cerevisiae cyclin-dependent kinases known to be involved in transcription, we have shown that only Kin28 and Srb10 are stable components of the PIC. Although we found evidence for other kinase activities in purified PICs, these enzymes are likely very substoichiometric with other PIC components, since they were not observed in mass spectrometry analysis of PICs purified using the immobilized template method (33).
Blocking Kin28 and Srb10 kinase activities was also found to inhibit PIC dissociation into the Scaffold complex upon ATP addition. This was demonstrated both by a decrease in the amount of phosphorylated Pol II released as well as by a decrease in the amount of total Pol II, TFIIB, and TFIIF released upon ATP addition. The inhibition of Scaffold complex formation correlated very closely with the decrease in transcription seen, suggesting that these two processes are closely related. Similar to the transcription results, blocking both kinases inhibited PIC dissociation to the greatest extent.
Since altered-specificity kinases can preferentially use ATP analogs such as N6-benzyl-ATP, we tested directly whether either Kin28 or Srb10 had any relevant targets in the PIC apart from the Pol II CTD. For Kin28, it was clear that two polypeptides in the PIC apart from the CTD were specific targets for phosphorylation. These additional polypeptides were identified as the Mediator subunits Med4 and Rgr1. From the electron microscopy structure of the Pol II-Mediator complex, both of these subunits are proposed to be located in the middle or tail region of Mediator. This region is proposed to be near the base of the CTD on Pol II (12). Therefore, it is reasonable that one kinase could target the CTD as well as these two Mediator subunits for phosphorylation in the PIC. Previously observed in vitro CDK7 targets seen in isolated kinase assays (TBP, TFIIFα, and TFIIEα) (28, 37, 48) were not phosphorylated in the PIC, suggesting that phosphorylation of these polypeptides does not normally occur during the transcription reaction.
In contrast to the high specificity seen with Kin28, Srb10 phosphorylated many more PIC polypeptides at a low level. Despite this, we found three polypeptides were phosphorylated by Srb10 at a higher level than others. These were identified as the Pol II CTD and two TFIID subunits, Bdf1 and TAF2. Srb10 is known to both repress and activate a subset of genes; however, the direct target is unknown except in a few cases. It is possible that phosphorylation of TFIID contributes to gene-specific regulation at some of these Srb10-regulated promoters, since these subunits appear to be required at only a subset of all yeast genes.
From the results presented here, it is clear that PIC dissociation is dependent on the kinase activities of Kin28 and Srb10. The simplest model for this dissociation is that phosphorylation of the Pol II CTD leads to instability of Pol II in the Open Complex, and dissociation of Pol II, TFIIB, and TFIIF follows. The CTD is the most likely target for this reaction, since both Kin28 and Srb10 have overlapping activities in PIC dissociation and the CTD is the only common target of these kinases. However, until the sites of phosphorylation on the TFIID and Mediator subunits are identified and mutated, we cannot rule out a role for these modifications in PIC dissociation and transcription. Possible roles for these modifications could be stabilization of the Scaffold to facilitate transcription reinitiation, promoter escape, or initiation. Testing these models will require mutation of these target polypeptides to eliminate the modification and measurement of genome-wide effects as well as specific effects on transcription initiation and reinitiation in vitro.
Acknowledgments
We are grateful to N. Yudkovsky for important initial work, S. Biggins and E. T. Young for advice, S. Tatsutani for excellent technical help, and J. Ranish for sharing information prior to publication. We thank H.-T. Chen, N. Mohibullah, L. Warfield, W. Reeves, and G. Rani for generous gifts of Flag-tagged nuclear extracts and proteins, T. Tsukiyama for p3FLAG-Hyg plasmid, and R. Tjian for the TAF2 antibody. We also thank W. Reeves and B. Moorefield for comments on the manuscript.
This work was supported by a grant from the National Institutes of Health to S.H. and K.S. (CA70331 and AI44009). S. Hahn is an Associate Investigator and Y. Liu is a Postdoctoral Associate of the Howard Hughes Medical Institute.
REFERENCES
- 1.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 2.Akoulitchev, S., T. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. [DOI] [PubMed] [Google Scholar]
- 3.Balciunas, D., M. Hallberg, S. Bjorklund, and H. Ronne. 2003. Functional interactions within yeast mediator and evidence of differential subunit modifications. J. Biol. Chem. 278:3831-3839. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, A. C., O. Buzko, and K. M. Shokat. 2001. Magic bullets for protein kinases. Trends Cell Biol. 11:167-172. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray, J. Blethrow, E. Shimizu, J. Z. Tsien, P. G. Schultz, M. D. Rose, J. L. Wood, D. O. Morgan, and K. M. Shokat. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. [DOI] [PubMed] [Google Scholar]
- 6.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Cai, H., and D. S. Luse. 1987. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 262:298-304. [PubMed] [Google Scholar]
- 9.Carlson, M. 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13:1-23. [DOI] [PubMed] [Google Scholar]
- 10.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, J. A., Y. Takagi, R. D. Kornberg, and F. A. Asturias. 2002. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10:409-415. [DOI] [PubMed] [Google Scholar]
- 13.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 15.Hirst, M., M. S. Kobor, N. Kuriakose, J. Greenblatt, and I. Sadowski. 1999. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 3:673-678. [DOI] [PubMed] [Google Scholar]
- 16.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, Y., and J. D. Gralla. 1994. RNA polymerase II phosphorylation: uncoupling from GAL4-VP16 directed Open complex formation and transcription in a reconstituted system. Nucleic Acids Res. 22:4958-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, Y., M. Yan, and J. D. Gralla. 1996. A three-step pathway of transcription initiation leading to promoter clearance at an activation RNA polymerase II promoter. Mol. Cell. Biol. 16:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194:428-453. [DOI] [PubMed] [Google Scholar]
- 20.Keogh, M. C., E. J. Cho, V. Podolny, and S. Buratowski. 2002. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 23.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y., J. A. Ranish, R. Aebersold, and S. Hahn. 2001. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 276:7169-7175. [PubMed] [Google Scholar]
- 25.Makela, T. P., J. D. Parvin, J. Kim, L. J. Huber, P. A. Sharp, and R. A. Weinberg. 1995. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc. Natl. Acad. Sci. USA 92:5174-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, C., S. Goto, K. Lund, W. Hung, and I. Sadowski. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187-190. [DOI] [PubMed] [Google Scholar]
- 28.Ohkuma, Y., and R. G. Roeder. 1994. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature 368:160-163. [DOI] [PubMed] [Google Scholar]
- 29.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 30.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 31.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell 1:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranish, J. A., E. C. Yi, D. M. Leslie, S. O. Purvine, D. R. Goodlett, J. Eng, and R. Aebersold. 2003. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33:349-355. [DOI] [PubMed] [Google Scholar]
- 34.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves, W. M., and S. Hahn. 2003. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 23:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 37.Rossignol, M., I. Kolb-Cheynel, and J. M. Egly. 1997. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 16:1628-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serizawa, H., J. W. Conaway, and R. C. Conaway. 1993. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature 363:371-374. [DOI] [PubMed] [Google Scholar]
- 40.Shah, K., Y. Liu, C. Deirmengian, and K. M. Shokat. 1997. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. USA 94:3565-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah, K., and K. M. Shokat. 2002. A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 9:35-47. [DOI] [PubMed] [Google Scholar]
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirode, F., D. Busso, F. Coin, and J. M. Egly. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3:87-95. [DOI] [PubMed] [Google Scholar]
- 44.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 45.Valay, J.-G., M. Simon, M.-F. Dubois, O. Bensaude, C. Facca, and G. Faye. 1995. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 249:535-544. [DOI] [PubMed] [Google Scholar]
- 46.Vincent, O., S. Kuchin, S. P. Hong, R. Townley, V. K. Vyas, and M. Carlson. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witucki, L. A., X. Huang, K. Shah, Y. Liu, S. Kyin, M. J. Eck, and K. M. Shokat. 2002. Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme. Chem. Biol. 9:25-33. [DOI] [PubMed] [Google Scholar]
- 48.Yankulov, K. Y., and D. L. Bentley. 1997. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 16:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]