Abstract
We devised a sensitive method for the site-specific detection of rare meiotic DNA strand breaks in germ cell-enriched testicular cell populations from mice that possess or lack an active recombination hot spot at the H2-Ea gene. Using germ cells from adult animals, we found an excellent correlation between the frequency of DNA breaks in the 418-bp H2-Ea hot spot and crossover activity. The temporal appearance of DNA breaks was also studied in 7- to 18-day-old mice with an active hot spot during the first waves of spermatogenesis. The number of DNA breaks detected rose as leptotene and zygotene spermatocytes populate the testis with a peak at day 14 postpartum, when leptotene, zygotene, and early pachytene spermatocytes are the most common meiotic prophase I cell types. The number of DNA breaks drops precipitously 1 day later, when middle to late pachytene spermatocytes become the dominant subtype. The recombination-related breaks in the hot spot likely reflect SPO11-induced double-strand breaks and/or recombination intermediates containing free 3′ hydroxyl groups.
Meiotic recombination leads to crossover events critical for the proper segregation of homologous chromosomes. It has been known for decades that recombination events may not be randomly distributed along each chromosome. The result is that some regions exhibit a recombination frequency greater than that expected based on the genome average relationship between physical distance and crossing over (reviewed in references 2, 31, 37, 38, and 54). These so-called recombination hot spots have been found in every organism examined carefully. A number of crossover hot spots exist in the mouse major histocompatibility complex (MHC; also called H-2) (see Fig. 1), a region rich in genes that control immune function (reviewed in references 14, 31, 38, 45, 49, and 54). Among the best characterized are hot spots associated with loci located in the class II region of the MHC: Eb1 (7, 25, 57, 62), Ea (19, 24, 27, 28), Pb (20, 45, 59), and Lmp-2 (now called Psmb9) (17, 44, 46). At each locus, the bulk of the crossover events localize to regions of a few hundred to a few thousand base pairs and recombination rates per unit of physical distance (centimorgan [cM] per megabase) vary from ∼175 to ∼2,600 times that expected for DNA segments of the same size based on the mouse genome average rate (0.5 cM/Mb).
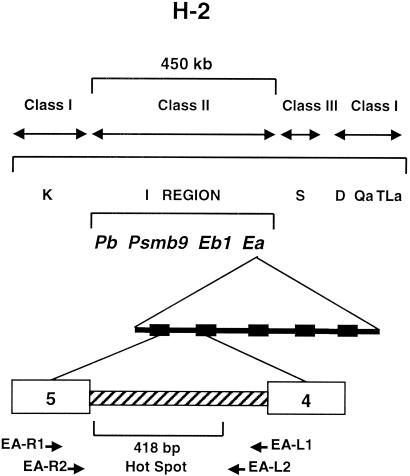
FIG. 1.
Location of the recombination hot spot in the Ea gene within the context of the mouse H-2 region on chromosome 17. The organization of the class I, II, and III genes is shown. The Ea gene is contained in the I region. The K, D, and S regions include other gene families involved in immune function. Exons 4 and 5 of the Ea gene (numbered boxes) are indicated, and the intron between the two exons contains the hot spot. The approximate locations of the other well-characterized hot spots, Eb, Pb, and Lmp2 (or Psmb9), are also indicated. The positions of the first- and second-round Ea primers used to identify breaks in the hot spot are shown from the transcriptional perspective of either the sense (EA-L primers) or template (EA-R primers) DNA strands.
One unusual feature of recombination hot spots in the mouse H-2 region is the dependence on genetic background for activity (reviewed in references 14, 31, 38, 45, 49, and 54). Like the MHC of most animals, the H-2 region contains many highly polymorphic genes. H-2 regions from different mouse strains have different constellations of alleles at many of these loci. For any one inbred strain, the particular allelic constellation in this region defines its haplotype. Only mice carrying certain combinations of haplotypes in the H-2 region exhibit recombination hot spot activity at a particular hot spot. A hot spot region active in the context of one pair of haplotypes may be virtually inactive (a cold spot) in another combination. Thus, strains with an active Ea hot spot show no crossing over at the Eb1 hot spot (19, 24).
The H2-Ea (designated Ea here) gene (Fig. 1) contains a haplotype-specific hot spot in intron 4 (19, 24, 27-29). In a backcross between B10 mice and F1 animals derived from crossing the strains B10.F(13R) and B10.S(9R), all crossover events observed in the ∼450-kb H-2 interval (Fig. 1) mapped to a 418-bp region in intron 4 of the Ea gene (19, 24). The 13R and 9R strains were derived using C57BL/10 mice and are genetically identical except in the H-2 region (congenic strains). At the Ea gene, the 13R strain is homozygous for the p haplotype, whereas 9R is homozygous for the k haplotype. The hot spot at Ea is detected in F1 animals heterozygous for these two haplotypes (p/k), but when a chromosome with a p haplotype Ea gene is combined with an Ea gene of the d or s haplotype (p/d or p/s), crossing over is not detected at Ea (19). Estimates of the frequency of recombination at the Ea hot spot lie between 1/50 and 1/250 (24).
There is considerable evidence indicating that the formation and processing of double-strand breaks (DSBs) (53) constitute the major pathway leading to crossing over (reviewed in references 31, 36, and 37). DSBs originating early in meiosis are processed to yield 3′ single-strand tails that can initiate recombination through invasion of the other homolog. Following DNA repair synthesis, a double Holliday junction can be formed. Depending on how these junctions are resolved, flanking marker exchange typical of crossing over may be observed. Meiotically induced DSBs have been detected by direct molecular experiments in both budding yeast (51, 52) and fission yeast (8). In addition, a strong correlation exists between the location of DSBs in budding and fission yeast and known recombination hot spots (reviewed in references 31, 36, and 37).
Emerging molecular evidence suggests that the relationship between DNA breaks and recombination in mammalian meiosis is similar to that in yeast meiosis. There is evidence for the involvement of DSBs in mouse recombination based on studies of animals deficient in Spo11 gene function (4, 42), immunocytochemical analysis using antibodies against the phosphorylated form of histone H2AX (γ-H2AX) that accumulates in chromatin at the sites of DSBs (18, 32, 40, 41) and the demonstration of DNA strands with 3′ overhangs using an in situ DNA labeling protocol on sectioned seminiferous tubules (61).
However, direct biochemical evidence for meiotic DSBs at a recombination hot spot is more difficult to obtain in mice than in yeasts. First, although there can be more than 200 RAD51/DMC1 foci per spermatocyte nucleus (33), any one mouse meiotic cell undergoes only ∼25 meiotic exchanges, detected as foci of MLH1 protein (1, 15, 26). These few events are distributed across the total haploid genome length of ∼3,000 Mb, and on average, a genetic distance of 1 cM corresponds to a physical distance of 2 Mb. Per unit of physical distance, mice have ∼650 times fewer meiotic exchange events than budding yeast, where a genetic distance of 1 cM corresponds to a physical distance of ∼3 kb (37). Second, yeast cells can be synchronized to enter meiosis so a relatively large proportion of the cells will have a break at a particular recombination hot spot. In the adult mammalian testis, primary spermatocytes account for only ∼20% of the germ cell population (5), and only a small proportion of these cells can possibly have an unligated DNA break at any one site. These factors dramatically reduce the chance that a locus-specific DNA strand break could be directly detected in a population of adult testis cells.
To provide some insight into the molecular basis for haplotype-specific hot spot activity, we examined the Ea hot spot region in meiotic cells for the presence of DNA strand breaks using a sensitive PCR assay. The assay is capable of detecting not only DSBs but also any recombination intermediate with a free 3′ OH group. We studied testis cell populations enriched in germ cells from 13R and 9R mouse strains and their F1 progeny as well as individuals with a p/d haplotype in the H-2 region. We found an excellent correlation between the frequency of DNA breaks in the intron 4 hot spot of the Ea gene and those Ea haplotype combinations known to be associated with high or low frequencies of meiotic crossing over. We also exploited the first wave of spermatogenesis in prepubertal mouse testes to obtain germ cells at progressively more advanced stages of meiotic prophase I (MPI) in order to follow the developmental timing of meiotic DNA breaks.
MATERIALS AND METHODS
Animals.
Mice of the B10.S(9R) and B10.F(13R) strains (referred to hereafter as 9R and 13R, respectively) and their F1 progeny were used for both measurement of strand breaks and determination of the temporal appearance of the stage of MPI. Breeding colonies were maintained in the Animal Resource Facilities at the University of Southern California (USC), Los Angeles, and the University of Tennessee, Knoxville. F1 mice heterozygous at the Ea gene for a p haplotype (from P/J females) and a d haplotype (from B10 · D2-H2dH2-T18cHco/oSnJ males) were generated at USC. The two parental strains were purchased from The Jackson Laboratory (Bar Harbor, Maine).
Cell preparation.
A cell population enriched in germ cells was obtained from adult testes (2- to 2.5-month-old mice) by enzymatic digestion with collagenase followed by trypsin and DNase I according to published procedures (5). To correlate DNA breaks with the stage of meiosis, separate germ cell preparations were isolated from two different sets of litters. Animals were sacrificed at daily intervals over a period from days 7 to day 18 postpartum (pp). Testes from two to five mice of each age were pooled for germ cell isolation.
Plug formation.
Cells were suspended in enriched Krebs Ringer buffer (5) at a concentration of 2 × 107 cells/ml. The solution containing 2.0% InCert (BioWhittaker Molecular Applications, Rockland, Maine) low-melting-point agarose was made with enriched Krebs Ringer buffer. Equal volumes of cell suspension and agarose solution (cooled to ∼40°C) were mixed to produce a final concentration of 104 cells/μl in 1.0% agarose. After the solution was pipetted into 75-mm3 plug molds (Bio-Rad, Hercules, Calif.), the plugs were cooled at 4°C for 5 min and incubated at 50°C for 40 to 48 h in lysis solution (0.5 M EDTA [pH 7.9], 1.0% Sarkosyl, 100 μg of proteinase K per ml; Sigma, St. Louis, Mo.). Plugs were washed 15 times with Tris-EDTA buffer at room temperature for 10 min each time and used immediately in the next step.
TdT tailing.
Plugs were incubated in tailing buffer (20 mM Tris acetate [pH 7.9], 10 mM magnesium acetate, 50 mM potassium acetate, 1 mM dithiothreitol, 0.25 mM CoCl2, 0.8 mM dGTP) at 4°C for 2 to 3 h. The plugs were transferred to 300 μl of fresh tailing buffer containing 15 μl (20 U/μl) of terminal transferase (TdT) (New England Biolabs, Beverly, Mass.), incubated on ice for ∼15 h, and then incubated at 37°C for 2 h. The plugs were then washed once with Tris-EDTA buffer, and DNA was extracted by a standard method for isolating DNA from low-melting-temperature agarose (43). The DNA was dissolved in 75 μl of deionized water (Sigma) so that 1 μl contained DNA from the equivalent of 104 cells (∼60 ng) and stored at 4°C.
Primer extension and first-round PCR of dGTP-tailed DNA fragments.
The sequences of the primers used in these experiments are shown in Table 1. A single primer extension was performed in a 50-μl reaction mixture containing 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 2.0 mM MgCl2, 0.01% gelatin, 50 μM concentrations of dATP, dGTP, dCTP, and dTTP (Amersham Pharmacia Biotech, Piscataway, N.J.), 1 μl of DNA, 1 U of Taq DNA polymerase (Promega, Madison, Wis.), and 0.2 μM primer C+P containing C10 at the 3′ end. The DNA was denatured at 94°C for 4 min and incubated at 50°C for 3 min and then at 72°C for 3 min. The reaction mixture was heated at 94°C, and PCR was started after the addition of 0.2 μM primer P and 0.2 μM first-round gene-specific primer (EA-L1, EA-R1, or EB-L1; see Results). The cycling conditions were as follows: denaturation at 94°C for 45 s, followed by annealing and extension at 66°C for 3 min for 30 cycles. The final extension step was for 5 min at 72°C.
TABLE 1.
Oligonucleotide primers for primer extension and PCR
| Oligonucleotide primer | Sequence |
|---|---|
| (C + P) | 5′ GTTAACCGCAACGTACCGTTGTTTGAGCAGGCCCCCCCCCC 3′ |
| P | 5′ GTTAACCGCAACGTACCGTTGTTTGAGCAGG 3′ |
| EA-L1 | 5′ GCGTTAAATGTGCTCAGAGACTGACAGATGTGTG 3′ |
| EA-L2 | 5′ CTGAGGGAGGAAAGCACGAGTG 3′ |
| EA-R1 | 5′ GATGAGGTCTCCGCTGAGATGAACAACTGAAAGG 3′ |
| EA-R2 | 5′ GCTGTCACAGCATGTTTCTTCACTG 3′ |
| EB-L1 | 5′ GGACAGAAACTCCAGCAGGGCAGGAACTTG 3′ |
| EB-L2 | 5′ AGGCCATGGAGAGTGGGTTG 3′ |
Second-round PCR.
The concentrations of the components in the buffer and Taq polymerase were identical to those used in the first round of PCR. Primer P was used at a concentration of 0.2 μM. A new nested gene-specific primer (EA-L2, EA-R2, or EB-L2) was used after 32P labeling in a 10-μl reaction mixture containing 2 μl of 20 μM primer, 1 μl (10 U) of T4 polynucleotide kinase, 5 μl of [γ-32P]ATP (4,500 Ci/mmol, 10 μCi/μl; ICN, Irvine, Calif.), 1 μl of 10× kinase buffer, and 1 μl of H2O and incubated for 2 h at 37°C. The labeling mixture plus 3 μl (20 μM) of the same unlabeled primer was added to a 480-μl PCR master mix. One microliter of first-round PCR product was added to 24 μl of the master mix. The final concentration of the 32P-labeled gene-specific primer was 0.08 μM, and the final concentration of unlabeled gene-specific primer was 0.12 μM. The cycling conditions included an initial denaturation step of 4 min at 94°C and 30 PCR cycles. One PCR cycle consisted of 45 s at 94°C and 1.5 min at 63°C (EA-L2) or 60°C (EA-R2 and EB-L2). The final extension step was 5 min at 72°C.
Restriction enzyme digestion and electrophoresis.
Second-round PCR products were analyzed with and without digestion by restriction enzyme BslI (EA-L and EB-L products) or BanII (EA-R products). Both enzymes were obtained from New England Biolabs. Two microliters of the second-round product was mixed with 5 to 10 U of enzyme and incubated at 55°C (BslI) or 37°C (BanII) for 2 h. Digested or undigested PCR product (0.25 μl) was mixed with 7.75 μl of sequencing buffer (98% formamide, 10 mM EDTA [pH 8.0], 0.25% xylene cyanol FF, 0.25% bromophenol blue) and denatured at 95°C for 10 min before loading on 6% polyacrylamide sequencing gel (7.5 M urea) and subjected first to electrophoresis and then autoradiography. A 32P-labeled MspI digest of pBR322 was included as a molecular size marker.
Immunostaining.
To assess the MPI stage, a small aliquot of cells from the preparations used for the strand break assay was fixed by the method of Moens et al. (33). Fixed cells were spread on slides (ThermoShandon, Pittsburgh, Pa.) at a concentration of approximately 6,000 cells/well. The cells were incubated simultaneously with antibodies to the synaptonemal complex protein SYCP3 and the testis-specific histone H1t (13). After overnight incubation, the slides were washed and incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit and rhodamine-conjugated goat anti-rat secondary antibodies (Pierce, Rockford, Ill.), washed and counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes, Eugene, Oreg.). Coverslips were mounted with Prolong Antifade (Molecular Probes). Immunofluorescent staining was observed with an epifluorescence microscope (Olympus America, Melville, N.Y.). Images were captured to Adobe Photoshop (Adobe Systems, Inc., San Jose, Calif.) with a C5810 color chilled 3CCD camera (Hamamatsu, Hamamatsu City, Japan).
Cell counts.
The frequency of cells at each stage of MPI through pachynema was determined on the basis of the staining pattern observed with anti-SYCP3 and anti-histone H1t. The criteria for prophase substage identification are described in Results and in the legend to Fig. 8. A minimum of 200 DAPI-stained cells were counted for each age on two replicate slides. The proportion of cells at each stage of MPI was calculated as a percentage of the total cell population or as a percentage of the total number of MPI cells present at each age.
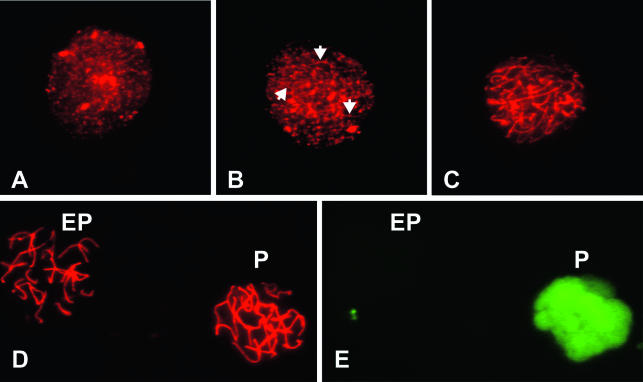
FIG. 8.
Immunostaining of spermatocytes. Spermatocytes were double labeled with antibodies to SYCP3 and histone H1t to determine the stage of MPI. (A) Leptonema was characterized by a punctate appearance of SYCP3. (B and C) Cells with patterns ranging from short linear patches (arrowheads in panel B) to staining along the full length of the incompletely paired chromosomes (C) were classified as zygonema. (D and E) Early pachytene spermatocytes (EP) are stained with SYCP3, but not histone H1t. Mid- to late pachytene spermatocytes (P) are positive for both SYCP3 and histone H1t. Panels D and E show the same field.
Statistics.
Statistical analysis was performed using the SPSS statistical software package (SPSS Inc., Chicago, Ill.) or Minitab (Minitab, Inc., State College, Pa.). Differences among groups in the number, frequency, or distribution of strand breaks were determined using either a one-way or two-way analysis of variance (ANOVA), two proportion comparisons, or chi-square analysis. When appropriate, we applied the Bonferroni or Tukey's multiple-comparison correction. Significance was assessed at the 0.05 level.
RESULTS
Analysis of breaks at the Ea locus.
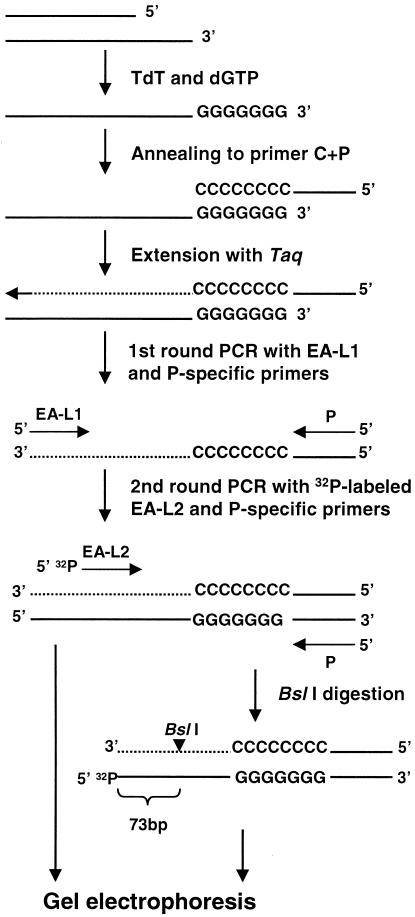
Our method is similar to that used previously to detect DNA breaks in yeast (56), except that modifications were made to enhance the specificity and sensitivity of the assay to allow its application to more-complex genomes. The experimental procedures were developed using reconstruction experiments where I-SceI-digested DNA from a CHO cell line containing a single recognition site for the enzyme (30) was mixed with live cells not treated with I-SceI (data not shown). An outline of the protocol used to detect rare DNA breaks in mouse testis cells is shown in Fig. 2. To detect rare breaks due to recombination, those that result from shearing by mechanical forces during conventional DNA purification must be avoided. To achieve this, a cell preparation enriched in germ cells was dissociated from the seminiferous tubules of testes of 2- to 2.5-month-old mice and embedded in low-melting-point agarose to form plugs. The cells were lysed by detergent and proteolytic enzyme digestion. To mark the sites of strand breaks, the plugs were dGTP tailed with TdT. Sequencing revealed a range of 8 to 13 guanines incorporated into the final PCR product (data not shown). TdT is capable of extending 3′ OH groups found at 3′ overhangs, internal nicks, blunt ends, and when the 3′ end is recessed with an efficiency that depends on the base at the 3′ end, the length of the single strand, and the divalent cation used (10, 11, 22). Importantly, once the G tails are added to the DNA in the plug, any breaks incurred thereafter are not detected by our assay. Following tailing, phenol-chloroform extraction and ethanol precipitation, the G-tailed fragments were extended with an oligonucleotide containing both a 3′ poly(C) region (C) and a 5′ unique (P) sequence (C+P). Two rounds of PCR using the P- and Ea-specific primers (EA-L1 for the first round and 32P-labeled EA-L2 for the second round) followed. BslI-treated and untreated second-round PCR products were sized on DNA sequencing gels. The BslI site is located 73 bp from the 5′ end of the second-round Ea PCR primer. Thus, all PCR products arising from Ea templates with DNA breaks anywhere in the hot spot should be cut down to a 32P-labeled 73-bp fragment after BslI digestion. The Ea origin of a few PCR products was confirmed by DNA sequencing of reamplified, second-round PCR products extracted from a gel (Fig. 3).
FIG. 2.
Outline of the method to detect rare DNA breaks. A DNA fragment with a 3′ overhang is used in this example. The method is applicable to any DNA that had an available 3′ OH group. For an explanation of the method and details, see Materials and Methods and Results.
FIG. 3.
Locations of meiotic DNA breakpoints in the Ea gene. The sequence of intron 4 of the Ea gene is shown beginning with the splice site adjacent to exon 4 (nucleotide 2160 in National Center for Biotechnology Information [NCBI] accession no. V00834). The uppercase letters identify nucleotides of the hot spot region. The positions of six breaks identified by DNA sequencing (see Results) are indicated by arrows. The underlined region of the intron shows the sequence of the 32P-labeled second-round PCR primer EA-L2 that locates DNA breaks on the sense strand. The bold uppercase letters denote the 52- to 105-bp bin with the greatest number of breaks (see Fig. 7). The first uppercase letter is the nucleotide at position 2307 in NCBI accession no. V00834.
DNA breaks in 9R and 13R parental strains.
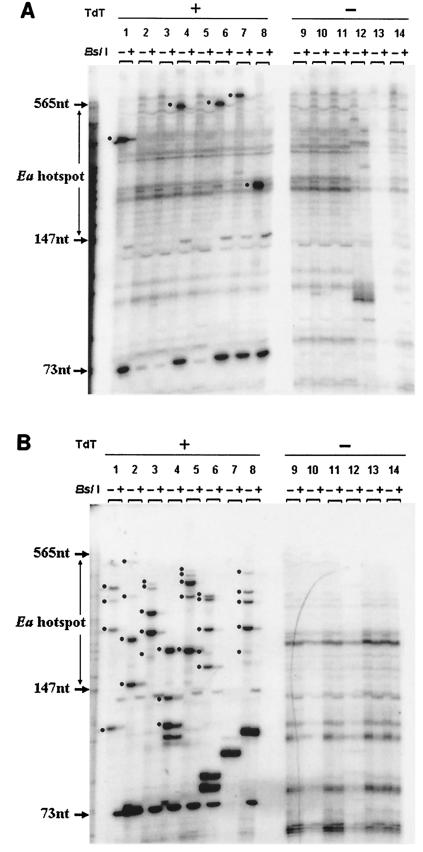
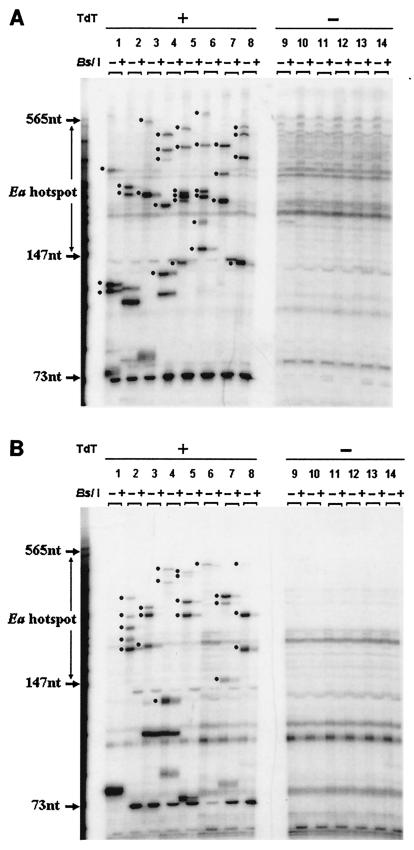
Eight aliquots from a TdT-treated plug and six aliquots from an untreated plug (each aliquot containing DNA from 10,000 testis cells) were examined for each of six 9R animals (homozygous for the k haplotype at Ea). After two rounds of PCR, the product from each aliquot was divided into two fractions, one of which was digested with BslI. Results from one animal are shown in Fig. 4A. Without BslI digestion (− lanes), bands can be seen throughout an approximately 700-bp molecular size range (the largest bands were of weaker intensity and are seen more clearly on the original autoradiographs than in the figures prepared for publication; data not shown). Some bands are artifacts of amplification, since they appear in both the TdT-treated and untreated plugs. In the TdT-treated aliquots, a number of bands appear that are absent from the untreated control. Each band in a lane labeled − (uncut) whose intensity is reduced by BslI digestion as shown in the + (cut) lane represents PCR product of a single Ea gene segment that contained a specific DNA break at the time of plug formation. The molecular size region bounded by the arrows is the 418-bp Ea recombination hot spot. On the basis of gel analysis of eight 10,000 cell aliquots from each of six 9R animals (480,000 cells in total), we counted 48 bands that could be digested by BslI. Thus, we estimate the frequency of specific Ea DNA breaks as one break per 10,000 cells. Table 2 shows that approximately one-half of the breaks (0.52 break/10,000 cells) were within the 418-bp hot spot region, while the other half (0.48 break/10,000 cells) lay outside the hot spot region (in the latter case, ∼300 bp can be resolved using the original autoradiographs).
FIG. 4.
Detection of DNA breaks in testis cells from strains 9R (A) and 13R (B). Representative data from a single experiment on a single mouse is shown in each case along with the no-TdT control (−). In each autoradiograph, the boundaries of the recombination hot spot are indicated between two molecular size markers. The lanes (each loaded with PCR product made using 10,000 cell equivalents of template) are arranged in pairs. The first member of the pair (−) contains second-round PCR products undigested by BslI. The second member of the pair (+) contains the same amplification product but after digestion with BslI. The 73-nucleotide (73nt) molecular size marker shows the position of the labeled BslI digestion product derived from all DNA breaks at the Ea hot spot. The thickness of individual bands reflects not only the amount of PCR product produced but also the fact that DNA slippage can occur in the TdT-added G/C mononucleotide repeat tract. In both panels, a black dot is positioned next to each band that we consider an Ea-specific break because the band in the − lane is cut down by BslI (see + lane). Bands not labeled by a dot either are not cut by the enzyme or are found in the no-TdT control. Bands more than 565 nucleotides long were evaluated using the original autoradiographs (not shown) and not the figures prepared for publication.
TABLE 2.
Number of DNA breaks in the 418-bp Ea hot spot region
| Animal | Avg no. of DNA breaks/10,000 adult testis cellsa
|
||||
|---|---|---|---|---|---|
| 9R (k/k) | 13R (p/p) | A cross (k/p) | B cross (p/k) | P/J × B10.D2 (p/d) | |
| 1 | 0.25 | 1.62 | 3.00 | 2.13 | 0.63 |
| 2 | 1.13 | 3.00 | 2.88 | 2.50 | 0.63 |
| 3 | 0.50 | 3.00 | 2.50 | 1.50 | 0.38 |
| 4 | 0.38 | 2.13 | 1.88 | 3.13 | 0.38 |
| 5 | 0.13 | 2.00 | 2.50 | ||
| 6 | 0.75 | 2.00 | 2.00 | ||
| Avgb | 0.52 (0.48) | 2.44 (0.69) | 2.40 (0.60) | 2.30 (0.84) | 0.51 (0.10) |
For each animal, a total of 80,000 cells were examined.
The average number of breaks outside the hot spot per 10,000 cells is shown in parentheses.
Many more DNA breaks were found in 13R mice (homozygous at Ea for the p haplotype) than in 9R mice. A typical result from one 13R animal is shown in Fig. 4B. On the basis of gel analysis of 320,000 cells (eight 10,000 cell aliquots from each of four animals), we counted 78 breaks within the hot spot region or an average of 2.44 DNA breaks/10,000 cells (Table 2). Outside the hot spot region, the average was 0.69 breaks/10,000 cells, a result similar to that found in 9R animals. Given the low frequency and large size range of the observed DNA fragments, it is unlikely that two independent Ea fragments would give rise to identically sized PCR products in a single 10,000-cell aliquot.
DNA breaks in the F1 offspring of a 9R × 13R cross.
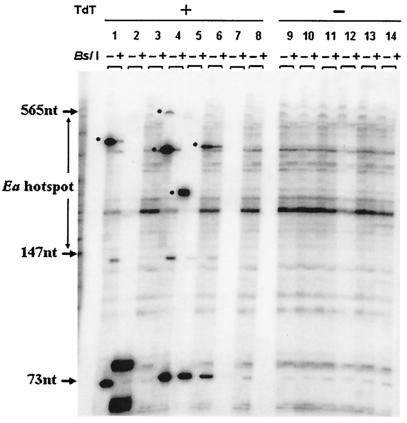
One type of F1 progeny (A cross) results from mating 13R males and 9R females to produce animals heterozygous for the p and k haplotypes at Ea. The other type (B cross) comes from the reciprocal mating. Figure 5 gives an example of the DNA break data for one of each of these mice, respectively. The results (based on 480,000 cells for each F1 progeny type) are summarized in Table 2. The A and B cross animals, like their 13R parent, had a greater average number of breaks per 10,000 cells within the hot spot region than the 9R parent (2.40 and 2.30 breaks/10,000 cells, respectively, for A and B cross offspring). Only average numbers of 0.60 (A cross) and 0.84 (B cross) breaks/10,000 cells were detected outside the hot spot, a number not very different from the numbers for the 9R and 13R strains.
FIG. 5.
Detection of DNA breaks in testis cells from the A cross (A) and B cross (B) progeny of matings between 13R and 9R mice. For details, see the legend to Fig. 4.
DNA strand breaks in mice heterozygous at Ea for the p and d haplotypes.
The same protocol was performed on testis cells from p/d male mice derived from crossing P/J mice (homozygous for the p haplotype at Ea) with the B10 · D2 strain (homozygous for the d haplotype at Ea). Genetic studies have shown a lack of Ea hot spot activity in p/d mice (19). An example of the data from one animal is shown in Fig. 6. Based on the study of 320,000 testis cells, the average number of breaks/10,000 cells in the hot spot region (0.50) is almost identical to the number in 9R animals (Table 2).
FIG. 6.
Detection of DNA breaks in testis cells from the progeny of crosses between P/J and B10 · D2 animals (p/d). For details, see the legend to Fig. 4.
DNA strand breaks depend on genotype.
When 9R, 13R, A cross, B cross, and p/d mice are compared (Table 2), the difference in DNA strand break frequency among them is highly significant (P ≪ 0.001). The frequency of breaks in the Ea hot spot region in 13R mice and its F1 progeny with 9R (A cross and B cross mice) is, on average, at least 4.6-fold greater than the frequency found in 9R and p/d haplotype mice. We note that the break frequencies in the hot spot region of the 9R and p/d mice are similar to the frequencies outside the hot spot in the hot 13R, A cross, and B cross animals, suggesting that these breaks may not be related to recombination (see Discussion).
Testing for strand bias in DNA breaks.
In a separate experiment, we compared the number of breaks within the hot spot region as a function of which strand contained the DNA break using two sets of Ea-specific nested primers (Fig. 1). One pair (EA-L1 and EA-L2) was identical to that used in the experiments described above and is located close to exon 4 and upstream of the hot spot relative to the direction of transcription. The second pair (EA-R1 and EA-R2) is located downstream of the hot spot. DNA plugs from 13R, 9R, A cross, and B cross mice (two animals for each of the four types) and p/d mice (four animals) were analyzed in parallel from both directions (Table 3). Considering the five strains together, the frequency of breaks per 10,000 cells did not differ significantly (P = 0.36) when counted using primers upstream compared to those downstream of the hot spot. This experiment independently confirmed that the 13R, A cross, and B cross animals have more breaks than the 9R parent and p/d mice (P ≪ 0.001).
TABLE 3.
Number of breaks in the 418-bp Ea hot spot region detected from upstream or downstream of the hot spot
| Strain (total no. of cells) | Avg no. of DNA breaks/10,000 adult testis cells
|
|
|---|---|---|
| EA-L1 + EA-L2 (left) | EA-R1 + EA-R2 (right) | |
| 9R (160,000) | 0.44 | 0.69 |
| 13R (160,000) | 2.56 | 1.88 |
| A cross (160,000) | 2.00 | 1.56 |
| B cross (160,000) | 2.25 | 1.88 |
| p/d (320,000) | 0.50 | 0.75 |
When the break data from 13R, A cross, and B cross animals were pooled however, 27% fewer breaks were found with the downstream Ea primer (mean, 1.7 breaks/10,000 cells) than with the upstream Ea primer (mean, 2.3 breaks/10,000 cells). This small difference in break number was significant (P = 0.03).
Distribution of DNA strand breaks in the hot spot.
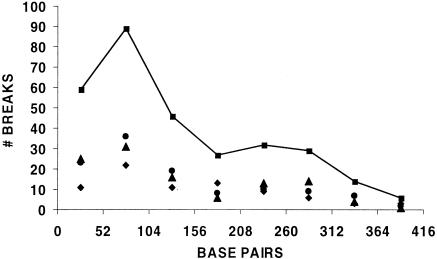
We determined the distribution of breaks within the hot spot of the 13R and A and B cross individuals (Fig. 7) using all of the data obtained with the upstream (EA-L) primers. The 418-bp hot spot region was divided into eight ∼52-bp bins, and the breaks were assigned to the bins on the basis of the molecular size markers run on each sequencing gel. Bin 1 was closest to the EA-L2 primer (close to exon 4). A total of 302 DNA strand breaks from 1.28 × 106 testis cells were analyzed. Overall, break position is not compatible with a random distribution throughout the hot spot (P ≪ 0.001). The greater concentration of breaks closer to the position of the EA-L2 primer could be due to more efficient amplification of short versus long PCR products, thereby making it easier to detect breaks close to this primer. If this were the only explanation however, break number might be expected to decrease monotonically with bin number. The significant peak detected in bin two does not support the idea of a monotonic function.
FIG. 7.
Distribution of DNA breaks in the Ea hot spot region. Break numbers are indicated for 13R (diamond), A cross (circle), B cross (triangle), and the pooled data set (squares). Although each bin of the 418-bp hot spot is actually 52.25 bp in width, the plot is made with 52.00-bp bins. Nucleotide 1 on the abscissa is closest to exon 4 and at position 2307 in NCBI accession no. V00834.
Results for the other strand using the EA-R primers (data not shown) provided no evidence for a monotonic decrease in break number as a function of bin number, although the data are not conclusive due to insufficient sample size (80 compared to 302 breaks). However, by pooling adjacent bins to reduce the number of bins from eight to four, we found a highly significant difference (P ≪ 0.001) between the break distributions determined using the upstream versus downstream Ea primer sets.
Analysis of the H2-Eb1 hot spot region.
Genetic studies indicate that the Eb1 hot spot is inactive in the A cross, B cross, and p/d haplotype combinations (19, 24). Using the equivalent of 80,000 cells for each strain, we examined the DNA sequences at the center of the Eb1 hot spot (57) for strand breaks in 9R and 13R mice, their F1 offspring, and the p/d haplotype mice (data not shown). Using P, C+P, EB-L1, and EB-L2 primers and the restriction enzyme BslI to cut the second-round PCR products, we observed no significant difference in the average number of Eb1 breaks (mean, 0.35 break/10,000 cells; 95% confidence interval, 0.12 and 0.58 break/10,000 cells) among the five strains (all P values > 0.1). However, an active Ea hot spot is 10 times “hotter” with respect to crossing over than an active Eb1 hot spot (7, 19, 24), so it is possible that recombination-specific DNA breaks even at an active Eb1 hot spot might not be detected above background levels using our present assay.
Temporal appearance of strand breaks during MPI.
The developmental appearance of germ cell DNA strand breaks in A cross and B cross offspring was correlated with different substages of MPI. The method of obtaining germ cell-enriched preparations and the strand break assay were described above. However, small aliquots of cells from each preparation were also stained to detect meiosis-related proteins (the synaptonemal complex protein SYCP3 and the testis-specific histone H1t) in order to identify substages of MPI. The staining patterns characteristic for each substage of MPI are shown in Fig. 8. The leptotene spermatocytes were identified as those with a punctate pattern of SYCP3 staining, localizing the emergence of lateral chromosomal axes (Fig. 8A). Zygotene spermatocytes were characterized by longer axes and some focal patches of chromosome pairing (Fig. 8B and C). Spermatocytes with thicker and somewhat shorter SYCP3-staining axes were defined as pachytene, and 20 pairs of chromosomes could be identified (Fig. 8D). Staining for histone H1t protein appears at mid-pachynema (9), so spermatocytes that were positive only for SYCP3 were identified as being in early pachynema, while those staining for both SYCP3 and histone H1t were identified as being in middle to late pachynema (Fig. 8).
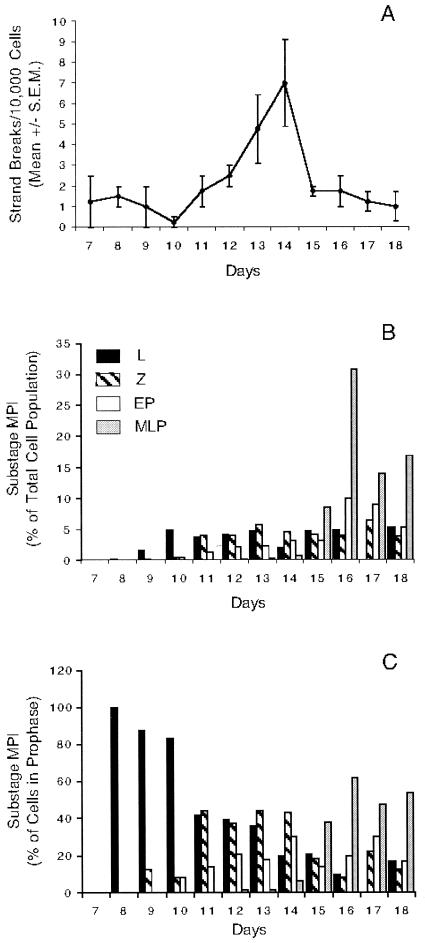
The proportion of each type of spermatocyte was determined at daily intervals for days 7 to 18 pp. The cells in each substage of meiotic prophase present on each day are depicted as a proportion of either the total population of cells in the preparation (Fig. 9B) or of all the MPI spermatocytes (Fig. 9C). No spermatocytes were present on day 7 pp, and leptotene spermatocytes appeared on day 8 pp, followed by zygotene spermatocytes on day 9 pp. Comparing Fig. 9B and C, it can be seen that while leptotene spermatocytes make up a significant proportion of prophase cells during this time, their absolute number in the total population is small. Early pachytene spermatocytes appeared on day 10 pp, and mid- to late-pachytene spermatocytes emerged on day 12 pp. The dynamic nature of the meiotic cell population during the first waves of spermatogenesis can be appreciated in Fig. 9C. Throughout the period, the proportion of developmentally earlier cells declined as more advanced cell stages appeared. This is especially noticeable on days 8 to 11 pp as the proportion of leptotene spermatocytes decreases (Fig. 9C), but it is also seen in the proportion of zygotene spermatocytes on days 14 to 16 pp. Expressed as a proportion of those cells in MPI (Fig. 9C), the leptotene and zygotene spermatocytes are present at similar levels on days 11 through 13 pp. As the pachytene spermatocytes become the predominant cell type in the population, the proportion of leptotene spermatocytes drops further on day 14, and the following day, the proportion of zygotene spermatocytes declines. This developmental pattern of sequential appearance of more advanced MPI substages correlates well with previous data (9, 16, 34).
FIG. 9.
Temporal appearance of DNA strand breaks and the stages of MPI. (A) Temporal appearance of mean number of DNA strand breaks/10,000 cells (± standard error of the mean [S.E.M.]). The break numbers are the mean values of four replicate measurements. (B and C) Temporal appearance of the stages of MPI shown as the proportion of the spermatocyte substages on days 7 to 18 pp among all testis cells (B) or among those cells in MPI (C). For the criteria used in identifying the stages of MPI, see the legend to Fig. 8. Bars represent the stages (L, leptonema; Z, zygonema; EP, early pachynema; MLP, mid- to late pachynema).
At daily intervals, the average number of Ea hot spot DNA strand breaks in the germ cell population was determined (Fig. 9A). For each time point, the equivalent of 40,000 testis cells was examined. On day 7 pp, before the appearance of the earliest meiotic spermatocytes, low levels of DNA strand breaks unrelated to crossing over were detected in spermatogonia and/or Sertoli cells, thereby providing an estimate of the background level of breaks for these nonmeiotic cells. The average number of Ea strand breaks began to increase on days 11 and 12 pp and reached a peak on day 14 pp, correlating with the presence of leptotene, zygotene, and early pachytene spermatocytes among the MPI cells over this period (Fig. 9C). The peak number of seven breaks per 10,000 cells in the hot spot region on day 14 pp was ∼5.4-fold higher than the number found in germ cells on day 7 pp and was significantly different than the numbers found on days 7 to 11 pp or days 17 and 18 pp. The average number of strand breaks in the adult A and B cross progeny (2.35 breaks/10,000 cells) was slightly lower than the numbers found in A and B cross animals 15 to 18 days pp.
DISCUSSION
Correlation between hot spot activity and DNA breaks.
Using a PCR-based assay designed to detect rare DNA strand breaks, we assessed meiosis-related breaks in mouse testicular germ cells. To enhance the detection of meiotic breaks, we studied the recombination hot spot in intron 4 of the H2-Ea gene in the mouse MHC complex. We showed that the number of DNA strand breaks in an adult testis cell population enriched for germ cells is highly correlated with genetic data on crossing over in that intron. We observed (Table 2) four- to fivefold more DNA strand breaks in the p/k haplotype animals than in p/d haplotype mice that lack crossover events at the Ea hot spot (19, 24, 27, 28).
At the Eb1 hot spot, the p/d mice, the 9R and 13R parents and their A cross and B cross progeny did not differ significantly from one another (P > 0.10) in having a very low number of DNA breaks (mean, 0.35 break/10,000 cells; 95% confidence interval, 0.12 and 0.58 break/10,000 cells). These results are expected, given that recombination at Eb1 has not been detected in mice carrying the p/k or p/d haplotypes at this gene (19, 24).
Classical linkage analysis (24) of the 418-bp Ea hot spot using the same strains we studied estimated that the crossover rate averaged ∼1 cM (based on 500 progeny), indicating that recombination is thousands of times greater than would be expected for a DNA segment of this size given the genome average rate in the mouse (0.5 cM/Mb). Using our assay, we detected a 4.6-fold-greater number of DNA breaks in the Ea hot spot than in flanking regions.
To reconcile the crossover data with the DNA break data, we must calculate the ratio of recombination-related breaks inside, compared to outside, the hot spot rather than using the total number of breaks. To do this, we need to know how the total number of observed breaks in any region are partitioned between those that are recombination related and those that are due to other causes. Endogenous physiological processes or even possibly the DNase I treatment used for the isolation of testis cells could be generating low levels of breaks detected by our assay. We can estimate the fraction of breaks that are unassociated with recombination by assuming that all breaks found (i) flanking the hot spot region in both the “cold” (9R and p/d) and “hot” (13R, A cross, and B cross) Ea mice and (ii) inside the hot spot region of the “cold” Ea mice do not contribute to crossing over. This is a reasonable assumption, since crossing over at Ea has not been detected outside the hot spot in “hot” mice or inside the hot spot in “cold” mice (19, 24, 27, 28). In addition, the DNA break frequencies are quite consistent (an average of 0.54 break per 10,000 cells) in all the above circumstances. Using this average break frequency as an estimate of the background level of our assay, we can calculate, for example, that if 99.9% of the breaks detected outside (or in inactive) hot spots were not related to crossover events, the ratio of DNA breaks related to recombination in the hot spot would be more than 4,000 times greater than recombination-related breaks outside it, assuming equal-size regions are being compared (for the A cross, [2.4 − (0.54 × 0.001)]/(0.54 × 0.001) = 4,443). The average number of breaks at the inactive Eb1 hot spot in the five mouse strains we studied (0.35 break/10,000 cells) provides an independent confirmation of the background frequency used in the above estimate.
We should also consider that the absolute number of Ea DNA strand breaks per 10,000 testis cells detected in the hot spot by our procedure may be lower than the numbers present in the whole testis. One mechanism that could bias against the detection of DNA breaks could involve a low efficiency of TdT tailing and amplification. However, earlier trials with the CHO cell system (mentioned in Results) showed that plugs containing 10,000 cells not cut by I-SceI and 5 genome equivalents of purified target DNA molecules with a cleaved I-SceI site (at exactly the same position), gave a single intense product after tailing and PCR, which suggests that even a single input molecule would be easily detected. A difficulty in detecting DNA breaks would also result if there were an experimental bias against recovering DNA fragments covalently attached at the 5′ end to SPO11 protein. This is an unlikely scenario, since our cells were first digested with an ionic detergent and proteinase K, as is the custom in studying enzymes that form covalent linkages with DNA (6, 50).
Molecular nature of the DNA breaks.
Our results on the Ea locus provide direct biochemical evidence for meiotic DNA breaks at a recombination hot spot without being able to specify the exact molecular nature of the breaks. Our data are consistent with DNA breaks having either blunt ends or 3′ or 5′ overhangs, as well as single-strand nicks or gaps, since TdT can extend 3′ OH groups in all these cases (10, 11, 22), albeit at different efficiencies.
The breaks detected at the Ea hot spot in the mixed cell population from adult mice could be DSBs induced, for example, by SPO11 protein (4, 32, 42), and/or DNA nicks or gaps with 3′ OH groups present in intermediates of the recombination process according to the classical (53) DSB model (including breaks related to DNA mismatch repair). The classical DSB model of recombination predicts that the frequency of DNA breaks detected on each of the two strands should be identical, and consistent with this, we observed only a small difference (27%). This difference might be due, in part, to different PCR efficiencies associated with the upstream versus downstream Ea primers, leading to the detection of more breaks using the upstream primer pair. In addition, random DNA breaks correlated with transcription could also contribute to the slightly greater number of breaks observed using the upstream primers, since these primers detect breaks on the nontemplate (and presumably transiently single-stranded) sense strand during Ea transcription.
Our results also show that the 302 DNA breaks detected using primers (EA-L) upstream of the hot spot were not distributed randomly within the hot spot but were concentrated in the first three bins and appeared to peak in the second bin (52 to 105 bp) (Fig. 7). Neither the G+C content (49 versus 46%) nor any remarkable DNA sequence or structural feature distinguishes the first three bins from the remainder of the region.
Only 80 DNA breaks were available from studies using the downstream primers (EA-R). However, comparison of the distribution of these breaks (data not shown) along the hot spot with the breaks detected by the upstream primers (EA-L; Fig. 7) indicates a significant difference (P ≪ 0.001). Additional experiments using the downstream primers are needed to determine the true nature of this distribution within the hot spot so that the reason for the difference in the distribution of breaks between the two strands can be better understood. Finally, regardless of their exact molecular nature, the correlation between DNA breaks in the Ea hot spot region and recombination activity is highly significant.
Temporal appearance of DNA breaks.
We also studied the temporal appearance of DNA breaks during the first wave of spermatogenesis as progressively more advanced germ cells emerge sequentially on successive days of postnatal development. In Fig. 9, the temporal appearance of DNA strand breaks is compared to the developmental pattern of emergence of different substages of MPI within the germ cell population. Data for budding yeast (35) and studies on immunocytochemical localization of SPO11, RAD51, and γ-H2AX in mouse spermatocytes (32, 34, 39) suggest that meiotic DSBs first arise during leptonema. However, our data show that the frequency of Ea DNA breaks from the cell populations isolated from mice 8 and 9 days pp (when spermatocytes in leptonema first appear and account for 80 to 100% of the MPI cell population; Fig. 9C) is not significantly different from the frequency observed in testis cells taken before meiotic cells appeared (day 7). This could be due to the fact that although leptotene spermatocytes make up more than 80% of all the MPI cells on days 8 to 10 pp (but drops thereafter), their absolute frequency among total testis tubule cells, including a majority of somatic cells (Fig. 9B), is quite low (0.3, 1.0, and 5.0%, for these three days, respectively). When the crossover frequency at Ea is also taken into consideration (24) (1/50 − 1/250), it becomes uncertain whether Ea-specific DNA breaks could be detected above the background level of breaks at the earliest stages of the developmental time course experiment when leptotene spermatocytes are so few in number.
As the first wave of spermatogenesis progresses simultaneously with the initiation of new waves, identifying which cell types contain the DNA breaks at the Ea hot spot is also difficult but for different reasons. First, the proportion of cells in the different substages of MPI are changing with respect to one another between days 11 and 15 pp, reflecting the asynchrony that exists in the onset of spermatogenesis (i.e., the subsequent waves of spermatogenesis are initiated as the first progresses). Second, the length of time a meiotic DNA strand break with a 3′ OH group can exist in spermatocytes before it is ligated to an adjacent 5′ phosphate is not known. As a result, the cell type (leptotene, zygotene, or early pachytene) harboring the strand break cannot be precisely determined. It is possible that all three may have recombination-associated strand breaks. Thus, for example, we could be measuring initiating DSBs in either leptotene or zygotene spermatocytes. This interpretation is consistent with the fact that Spo11−/− spermatocytes, which cannot form DSBs (4, 42), arrest in zygonema and undergo apoptosis, evidence that it is unlikely that initiating DSBs occur in the early pachytene spermatocytes. Likewise, MLH1 localization and data from the Mlh1 null mice (3, 39) indicate that chiasmata are formed in early to mid-pachynema. Therefore, it is reasonable to suggest that the breaks measured could also reflect other types of recombination intermediates present in at least the early pachytene spermatocytes at the time chiasmata are beginning to be established.
On day 14 pp, leptotene, zygotene, and early pachytene spermatocytes account for more than 90% of the MPI germ cells (Fig. 9C). In adult testis, leptotene, zygotene, and early pachytene spermatocytes represent 47% of the MPI germ cell population (data not shown). The number of breaks per 10,000 cells is 2 to 3 times higher on day 14 pp: ∼7 versus ∼2.5 breaks/10,000 cells in 2- to 2.5-month-old adults. It is noteworthy that the most precipitous transition in DNA break frequency we detected during early postnatal development was the drop between days 14 and 15 pp that was accompanied by a dramatic rise in the proportion (Fig. 9C) of cells in mid- to late pachynema with a corresponding drop in the frequency of zygotene and early pachytene spermatocytes. The association between the rise in mid- to late pachytene spermatocytes and a drop in DNA break number could be explained by ligation events associated with the final molecular steps of DSB repair. For example, recent studies (17) showed that crossover DNA molecules at the mouse Lmp2 recombination hot spot arise at some unknown time between postnatal days 11 and 16 after which the frequency does not change appreciably. Since unligated intermediates of DSB-induced crossover products at Lmp2 would not have been detected by the PCR assay used in that study, it suggests that the resolution of Holliday junctions reaches a steady-state level among the mixed population of primary spermatocytes sometime during this period. Once spermatogenesis is fully established in the 2- to 2.5-month-old adult, with the full complement of MPI germ cells present, the number of unligated strand breaks is also expected to reach a plateau, reflecting a steady-state frequency of germ cells initiating and progressing through recombination.
The temporal pattern of our detection of DNA strand breaks during meiosis in mice raises two interesting possibilities. First, it is possible that the earliest breaks we detected, SPO11 localization, and γ-H2AX modification of chromatin all exist before complete synapsis in the mouse (23, 32; this report). At least some of these breaks could be newly initiated SPO11-induced DSBs consistent with molecular and genetic data in yeast that indicate that DSB formation precedes full synapsis (35). Interestingly, there are species-specific differences in the timing of the DSBs that initiate recombination and synapsis; for example, DSB formation appears to occur after synapsis in Drosophila (21). Initiation of recombination is also dispensable for synapsis in Caenorhabditis elegans (12). However, until now, the timing of DSB formation in the mouse has been assessed by genomewide SPO11 localization and chromatin modifications, such as phosphorylation of histone H2AX. Clearly, more information will come from the direct detection of rare DSBs at specific sites in complex genomes when specific methods for distinguishing among DSBs and other rare recombination intermediates become available.
The second interesting possibility emerging from these results concerns the nature of Ea hot spot DNA strand breaks and their timing relative to the timing of SPO11 and γ-H2AX staining of spermatocytes. The appearance of these two proteins in leptonema is thought to reflect DSB formation. If our inability to specifically detect crossover-related DNA breaks in leptotene spermatocytes at days 8 to 10 is not simply due to the low numbers of this cell type in testes in mice of this age, our results could suggest that the positions of some DSBs that eventually lead to crossovers in the Ea hot spot region may lie outside the Ea hot spot itself (47, 48, 60). Thus, some DNA regions may be hot spots of genetically detected crossovers, because they tend to promote Holliday junction resolution leading to the exchange of flanking markers. The 3′ OH groups we detected could be present on the extension products of the original 3′ overhangs not yet ligated to an adjacent strand or could have arisen during the process of Holliday junction resolution (55) or DNA mismatch repair. Ultimately, the precise nature of the breaks and elucidation of the order of events will require coupling assays such as the one presented here with new methods of localization of proteins catalyzing specific steps in recombination at specific sites. Also required will be the development of methods that can yield large numbers of highly pure germ cells at specific meiotic substages under conditions where the background level of DNA breaks unrelated to recombination is minimal.
Haplotype specificity of recombination hot spots.
Genetic studies using mice heterozygous at Ea for the p haplotype and another (non-p) haplotype have shown that hot spot activity is not transmitted in a dominant fashion (19, 24). Although it is impossible to gather genetic mapping information by crossing male and female members of the same inbred mouse strain, our assay showed that the number of DNA breaks (Table 2) and their distribution (Fig. 7) at the Ea hot spot in the inbred 13R strain were similar to those for the A and B cross animals. This observation would naturally lend support to the simple idea that there is an association between the presence of DNA breaks at Ea and the presence of an Ea gene with the p haplotype. However, we found that break frequency in the p/d haplotype combination was more similar to the value for the inbred 9R strain (homozygous at Ea for the k haplotype). There are several possible explanations for the lack of a clear dominant effect of the p haplotype on DNA strand breaks. Allelic differences in the hot spot region itself cannot be responsible for the differences in recombination hot spot activity (58), since at least a 644-bp segment of the p haplotype Ea gene (including the hot spot) is the same in both the p/d and 13R parental strains (24). Another possibility stems from the fact that the 13R H-2 region is the product of a single crossover between two different strains somewhere between Ea and the S region (Fig. 1), a distance of ∼300 kb (24), whereas the chromosome carrying the p haplotype Ea gene in the p/d mice has an uninterrupted H-2 region. Perhaps the recombination hot spot activity found to be associated with the 13R H-2 region is associated with both the presence of an Ea p haplotype and cis sequences derived from another genetic background (19, 24). Genetic studies of the Lmp2 hot spot have revealed two regions that can enhance or suppress recombination; one is proximal and one is distal to the hot spot (46). That the initiation of DNA breaks may take place at a considerable distance from the site of a crossover in fission yeast (48, 60) may also be relevant.
Another possible explanation for the lack of a dominant transmission of p haplotype recombination hot spot potential at Ea is that sequences unlinked to the H-2 region may be playing a role in hot spot activity. Thus, the genetic background of the 13R, A cross, and B cross animals is pure C57BL/10 (except for the H-2 region), whereas the genetic backgrounds of the p/d and p/s mice that lack Ea hot spot activity are heterogeneous (P/J and C57BL/10 mice and P/J and A.SW mice, respectively). However, it is known that homozygosity for C57BL/10 outside the H-2 region is not required for hot spot activity (24).
Understanding the haplotype specificity of H-2 recombination hot spots in the future will require additional genetic experiments. The assay we describe can be used not only to determine the developmental timing of meiotic DNA breaks but also to estimate the number of breaks at specific sites. This makes it possible to relate DNA breaks to crossover events in genomes significantly larger than those of budding yeast and fission yeast. Ultimately, correlating strand breaks with localization of proteins mediating steps in recombination will resolve the temporal order of events.
Acknowledgments
We thank Mike Lichten, Tom Petes, and three anonymous reviewers for helpful comments on the manuscript. We are grateful to Peter Calabrese and Simon Tavaré for statistical advice and Cynthia Park and Debby Andreadis for caring for the animals.
This work was supported in part by NIH grants from the NIGMS (GM36745) (N.A.) and NICHD (HD31376 and HD33816) (M.A.H.).
REFERENCES
- 1.Anderson, L. K., A. Reeves, L. M. Webb, and T. Ashley. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnheim, N., P. Calabrese, and M. Nordborg. 2003. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am. J. Hum. Genet. 73:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336-342. [DOI] [PubMed] [Google Scholar]
- 4.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 5.Bellve, A. R. 1993. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225:84-113. [DOI] [PubMed] [Google Scholar]
- 6.Borgnetto, M. E., S. Tinelli, L. Carminati, and G. Capranico. 1999. Genomic sites of topoisomerase II activity determined by comparing DNA breakage enhanced by three distinct poisons. J. Mol. Biol. 285:545-554. [DOI] [PubMed] [Google Scholar]
- 7.Bryda, E. C., J. A. DePari, D. B. Sant'Angelo, D. B. Murphy, and H. C. Passmore. 1992. Multiple sites of crossing over within the Eb recombinational hotspot in the mouse. Mamm. Genome 2:123-129. [DOI] [PubMed] [Google Scholar]
- 8.Cervantes, M. D., J. A. Farah, and G. R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5:883-888. [DOI] [PubMed] [Google Scholar]
- 9.Cobb, J., B. Cargile, and M. A. Handel. 1999. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev. Biol. 205:49-64. [DOI] [PubMed] [Google Scholar]
- 10.Deininger, P. L. 1987. Full-length cDNA clones: vector-primed cDNA synthesis. Methods Enzymol. 152:371-389. [DOI] [PubMed] [Google Scholar]
- 11.Deng, G., and R. Wu. 1981. An improved procedure for utilizing terminal transferase to add homopolymers to the 3′ termini of DNA. Nucleic Acids Res. 9:4173-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser, and A. M. Villeneuve. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94:387-398. [DOI] [PubMed] [Google Scholar]
- 13.Eaker, S., A. Pyle, J. Cobb, and M. A. Handel. 2001. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J. Cell Sci. 114:2953-2965. [DOI] [PubMed] [Google Scholar]
- 14.Fischer Lindahl, K. 1991. His and hers recombinational hotspots. Trends Genet. 7:273-276. [DOI] [PubMed] [Google Scholar]
- 15.Froenicke, L., L. K. Anderson, J. Wienberg, and T. Ashley. 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71:1353-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetz, P., A. C. Chandley, and R. M. Speed. 1984. Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. J. Cell Sci. 65:249-263. [DOI] [PubMed] [Google Scholar]
- 17.Guillon, H., and B. de Massy. 2002. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat. Genet. 32:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Hamer, G., H. L. Roepers-Gajadien, A. van Duyn-Goedhart, I. S. Gademan, H. B. Kal, P. P. van Buul, and D. G. de Rooij. 2003. DNA double-strand breaks and gamma-H2AX signaling in the testis. Biol. Reprod. 68:628-634. [DOI] [PubMed] [Google Scholar]
- 19.Heine, D., S. Khambata, K. S. Wydner, and H. C. Passmore. 1994. Analysis of recombinational hot spots associated with the p haplotype of the mouse MHC. Genomics 23:168-177. [DOI] [PubMed] [Google Scholar]
- 20.Isobe, T., M. Yoshino, K. Mizuno, K. F. Lindahl, T. Koide, S. Gaudieri, T. Gojobori, and T. Shiroishi. 2002. Molecular characterization of the Pb recombination hotspot in the mouse major histocompatibility complex class II region. Genomics 80:229-235. [DOI] [PubMed] [Google Scholar]
- 21.Jang, J. K., D. E. Sherizen, R. Bhagat, E. A. Manheim, and K. S. McKim. 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116:3069-3077. [DOI] [PubMed] [Google Scholar]
- 22.Kato, K. I., J. M. Goncalves, G. E. Houts, and F. J. Bollum. 1967. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J. Biol. Chem. 242:2780-2789. [PubMed] [Google Scholar]
- 23.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 24.Khambata, S., J. Mody, A. Modzelewski, D. Heine, and H. C. Passmore. 1996. Ea recombinational hot spot in the mouse major histocompatibility complex maps to the fourth intron of the Ea gene. Genome Res. 6:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Kobori, J. A., E. Strauss, K. Minard, and L. Hood. 1986. Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science 234:173-179. [DOI] [PubMed] [Google Scholar]
- 26.Koehler, K. E., J. P. Cherry, A. Lynn, P. A. Hunt, and T. J. Hassold. 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafuse, W. P., N. Berg, S. Savarirayan, and C. S. David. 1986. Mapping of a second recombination hot spot within the I-E region of the mouse H-2 gene complex. J. Exp. Med. 163:1518-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafuse, W. P., and C. S. David. 1986. Recombination hot spots within the I region of the mouse H-2 complex map to the E beta and E alpha genes. Immunogenetics 24:352-360. [DOI] [PubMed] [Google Scholar]
- 29.Lafuse, W. P., S. T. Lee, and C. S. David. 1990. Molecular analysis of two recombinant mouse strains with crossovers in the E alpha recombination hot spot. J. Immunogenet. 17:169-176. [DOI] [PubMed] [Google Scholar]
- 30.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichten, M., and A. S. Goldman. 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29:423-444. [DOI] [PubMed] [Google Scholar]
- 32.Mahadevaiah, S. K., J. M. Turner, F. Baudat, E. P. Rogakou, P. de Boer, J. Blanco-Rodriguez, M. Jasin, S. Keeney, W. M. Bonner, and P. S. Burgoyne. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27:271-276. [DOI] [PubMed] [Google Scholar]
- 33.Moens, P. B., D. J. Chen, Z. Shen, N. Kolas, M. Tarsounas, H. H. Heng, and B. Spyropoulos. 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106:207-215. [DOI] [PubMed] [Google Scholar]
- 34.Moens, P. B., N. K. Kolas, M. Tarsounas, E. Marcon, P. E. Cohen, and B. Spyropoulos. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115:1611-1622. [DOI] [PubMed] [Google Scholar]
- 35.Padmore, R., L. Cao, and N. Kleckner. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66:1239-1256. [DOI] [PubMed] [Google Scholar]
- 36.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petes, T. D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2:360-369. [DOI] [PubMed] [Google Scholar]
- 38.Pittman, D. L., and J. C. Schimenti. 1998. Recombination in the mammalian germ line. Curr. Top. Dev. Biol. 37:1-35. [DOI] [PubMed] [Google Scholar]
- 39.Plug, A. W., A. H. Peters, K. S. Keegan, M. F. Hoekstra, P. de Boer, and T. Ashley. 1998. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 111:413-423. [DOI] [PubMed] [Google Scholar]
- 40.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 42.Romanienko, P. J., and R. D. Camerini-Otero. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6:975-987. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Shiroishi, T., N. Hanzawa, T. Sagai, M. Ishiura, T. Gojobori, M. Steinmetz, and K. Moriwaki. 1990. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics 31:79-88. [DOI] [PubMed] [Google Scholar]
- 45.Shiroishi, T., T. Koide, M. Yoshino, T. Sagai, and K. Moriwaki. 1995. Hotspots of homologous recombination in mouse meiosis. Adv. Biophys. 31:119-132. [DOI] [PubMed] [Google Scholar]
- 46.Shiroishi, T., T. Sagai, N. Hanzawa, H. Gotoh, and K. Moriwaki. 1991. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 10:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35:243-274. [DOI] [PubMed] [Google Scholar]
- 48.Steiner, W. W., R. W. Schreckhise, and G. R. Smith. 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9:847-855. [DOI] [PubMed] [Google Scholar]
- 49.Steinmetz, M., D. Stephan, and K. Fischer Lindahl. 1986. Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell 44:895-904. [DOI] [PubMed] [Google Scholar]
- 50.Strick, R., P. L. Strissel, S. Borgers, S. L. Smith, and J. D. Rowley. 2000. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc. Natl. Acad. Sci. USA 97:4790-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338:87-90. [DOI] [PubMed] [Google Scholar]
- 52.Sun, H., D. Treco, and J. W. Szostak. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64:1155-1161. [DOI] [PubMed] [Google Scholar]
- 53.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 54.Wahls, W. P. 1998. Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 37:37-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 56.Xu, F., and T. D. Petes. 1996. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics 143:1115-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yauk, C. L., P. R. Bois, and A. J. Jeffreys. 2003. High-resolution sperm typing of meiotic recombination in the mouse MHC Eβ gene. EMBO J. 22:1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshino, M., T. Sagai, K. F. Lindahl, Y. Toyoda, K. Moriwaki, and T. Shiroishi. 1995. Allele-dependent recombination frequency: homology requirement in meiotic recombination at the hot spot in the mouse major histocompatibility complex. Genomics 27:298-305. [DOI] [PubMed] [Google Scholar]
- 59.Yoshino, M., T. Sagai, K. F. Lindahl, Y. Toyoda, T. Shiroishi, and K. Moriwaki. 1994. No dosage effect of recombinational hotspots in the mouse major histocompatibility complex. Immunogenetics 39:381-389. [DOI] [PubMed] [Google Scholar]
- 60.Young, J. A., R. W. Schreckhise, W. W. Steiner, and G. R. Smith. 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9:253-263. [DOI] [PubMed] [Google Scholar]
- 61.Zenvirth, D., C. Richler, A. Bardhan, F. Baudat, A. Barzilai, J. Wahrman, and G. Simchen. 2003. Mammalian meiosis involves DNA double-strand breaks with 3′ overhangs. Chromosoma 111:369-376. [DOI] [PubMed] [Google Scholar]
- 62.Zimmerer, E. J., and H. C. Passmore. 1991. Structural and genetic properties of the Eb recombinational hotspot in the mouse. Immunogenetics 33:132-140. [DOI] [PubMed] [Google Scholar]