Abstract
Myc oncoproteins are overexpressed in most cancers and are sufficient to accelerate cell proliferation and provoke transformation. However, in normal cells Myc also triggers apoptosis. All of the effects of Myc require its function as a transcription factor that dimerizes with Max. This complex induces genes containing CACGTG E-boxes, such as Ornithine decarboxylase (Odc), which harbors two of these elements. Here we report that in quiescent cells the Odc E-boxes are occupied by Max and Mnt, a putative Myc antagonist, and that this complex is displaced by Myc-Max complexes in proliferating cells. Knockdown of Mnt expression by stable retroviral RNA interference triggers many targets typical of the “Myc” response and provokes accelerated proliferation and apoptosis. Strikingly, these effects of Mnt knockdown are even manifest in cells lacking c-myc. Moreover, Mnt knockdown is sufficient to transform primary fibroblasts in conjunction with Ras. Therefore, Mnt behaves as a tumor suppressor. These findings support a model where Mnt represses Myc target genes and Myc functions as an oncogene by relieving Mnt-mediated repression.
In normal cells Myc activation increases proliferative rates and provokes transformation by regulating genes that drive metabolism and the cell cycle and by regulating the angiogenic switch (reviewed in reference 33). However, Myc also triggers the apoptotic response (1, 14), which includes activation of the Arf-p53 tumor suppressor pathway (40), as well as suppression of the bcl-X and bcl-2 antiapoptotic genes (13).
Precisely how Myc induces such a wide array of biological responses is unclear, but all of the effects of Myc require its function as a basic helix-loop-helix-leucine zipper (b-HLH-Zip) transcription factor. The ability of Myc to bind to its recognition element, a CACGTG E-box (8), and to induce transcription requires dimerization with Max, a small b-HLH-Zip protein (9). The activity of the Myc-Max complex is thought to be harnessed by other b-HLH-Zip factors, the Mad proteins (2) and Mnt (or “Rox” [20, 29]), which also dimerize with Max and bind to these elements yet repress transcription through their associations with the general corepressors Sin3a and Sin3b and associated histone deacetylases (3). These findings, and the expression of some Mad proteins during differentiation (2), have suggested a model whereby activating Myc-Max complexes are antagonized by repressive Mad-Max or Mnt-Max complexes and the equilibrium between these complexes regulates cell fate. However, other alternatives are equally plausible; for example, Myc could function as an antagonist of Mad and/or Mnt and activate gene expression through the relief of active repression (4).
If Mad family members functioned to antagonize Myc, then one prediction was that their targeted deletion should release Myc activity to provoke inappropriate cell proliferation, apoptosis and, ultimately, transformation. Surprisingly, the knockouts of Mad1, Mxi1 (Mad2), and Mad3 revealed only modest and selective effects on cell differentiation rather than on cell growth or transformation (15, 35, 36). By contrast, little is known about the physiological role of Mnt, which, in contrast to Mad factors, is expressed in proliferating cells (20).
To evaluate the interplay of Myc, Mad, and Mnt in regulating transcription and cell fate, we reevaluated the mechanism by which c-Myc induces the transcription of the Ornithine decarboxylase (Odc) gene, which encodes the rate-limiting enzyme in polyamine biosynthesis. These studies pinpointed Mnt as a key regulator of Odc expression and demonstrated that Myc induces Odc transcription by relief of Mnt-mediated repression. Strikingly, knockdown of Mnt by stable RNA interference established that Mnt loss provokes the typical “Myc” response, even in cells lacking c-myc, and that Mnt behaves as a tumor suppressor.
MATERIALS AND METHODS
Cells and viral infections.
Wild-type and E2f1−/− mouse embryonic fibroblasts (MEFs) (5, 40), BALB/c-3T3 fibroblasts (30), HO.15 c-myc−/− Rat1a fibroblasts (27), wild-type and c-Myc-deficient embryonic stem (ES) cells (6), and interleukin-3 dependent FDC-P.1 myeloid cells (32) were cultured as previously described. Retroviral infections were performed as previously described (40). Ha-RasV12 and pBabe-Puro retroviral supernatants were a kind gift from Robert Lewis (Eppley Cancer Center).
Plasmids, reagents, and antibodies.
The vectors pRc-CMV-HA-Mnt and MSCV-IRES-GFP were kind gifts from Peter Hurlin and Elio Vanin, respectively. MSCV-Myc-ER-IRES-GFP was generated by cloning a cDNA encoding Myc-ER (from Michael Bishop) into the EcoRI site of MSCV-IRES-GFP. The MntRNAi 1-3 or control RNAi (directed against pBluescript) retroviruses were designed by PCR-SHAGging amplification and the RNAi oligo retriever program (see www.cshl.org/public/SCIENCE/hannon.html). The sequences were as follows: Mnt1, AAAAAAGCCACCCCCAGTAGCATCCCTATGCTCATCAAGCTTCATGAGCATAGAGACGCTACTGGAGGCGGCGGTGTTTCGTCCTTTCCACAA; Mnt2, AAAAAAGGAAGGCATAGTAACCATAGTCACAGGATCAAGCTTCACCCTGTGACTATGGTCACCATGCCCTCCGGTGTTTCGTCCTTTCCACAA; Mnt3, AAAAAAGCCCGGTCGCCCTCAAGCTGGCCTGTCCGCAAGCTTCCAGACAGGCCAGCCTGAGGACGACCAGGCGGTGTTTCGTCCTTTCCACAA; and pBluescript; AAAAAAAGAACAACACTCAACCCTATCCCGATCCACAAGCTTCTAGACCGAGATAGGGTTGAGTGTTGTTCCGGTGTTTCGTCCTTTCCACAA. The PCR product was cloned into the NotI site of MSCV-IRES-GFP. 4-Hydroxytamoxifen (4-HT [Sigma]) was used at 1 μM. Antibodies against c-Myc (Santa Cruz Biotechnologies), Max (Transduction Labs), Mnt (from Peter Hurlin and Robert Eisenman), Mad1 (Santa Cruz), and USF-1 (Upstate) were used for supershift analyses in electrophoretic mobility shift assays (EMSAs). Antibodies against Mnt (Santa Cruz), E2f1 (Transduction Labs), p27Kip1 (Transduction Labs), ODC (from Anthony Pegg), p53 (Calbiochem), p19Arf (Upstate), caspase-9 (Cell Signaling), and Bcl-X1. (Transduction Labs) were used for immunoblotting.
Viability and apoptosis assays.
To assess the effects of Mnt knockdown on cell survival, green fluorescent protein (GFP)-only- and MntRNAi-expressing cells were seeded in 100-mm plates with 5 × 105 cells. After 24 h, the cells were shifted to medium containing 0.1% fetal calf serum and harvested at specific intervals, and their viability was determined by trypan blue dye exclusion. Apoptosis was determined by propidium iodide staining and annexin V-Fluorescein isothiocyanate staining, which was performed as described by the manufacturer (BioWhittaker Inc. Walkersville, Md.). Briefly, 2 × 105 to 5 × 105 cells were washed in phosphate-buffered saline and resuspended in 190 μl of binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, 2.5 mM CaCl). A 10-μl portion of annexin V-fluorescein isothiocyanate was added, the mixture was incubated for 10 min, and the cells were washed once and resuspended in 190 μl of binding buffer. To this was added 10 μl of 20-μg/ml propidium iodide stock solution. The cells were then analyzed for the presence of apoptotic cells by fluorescence-activated cell sorter analysis.
RNA analysis.
RNA was extracted using Trizol (Invitrogen). Northern blot analysis was performed as described previously (31). Real-time PCR was performed using the Taq-Man EZ reverse transcription-PCR core kit as described by the manufacturer (Applera, Inc.). Primer and probe sequences are available on request.
Odc promoter analyses.
EMSAs were performed using extracts as described (24). Chromatin immunoprecipitations (ChIP) were performed on serum-starved and -stimulated BALB/c-3T3 fibroblasts by using a kit from Upstate and the antibodies described above. The primers used encompassed both E-boxes of the Odc gene. Primer sequences are available on request. Transient transfections of BALB/c-3T3 cells were performed using Lipofectamine 2000 (Invitrogen). The Odc promoter-reporter constructs (ODCΔLuc and ODCΔLuc5A) driving the expression of firefly luciferase have been described previously (32).
RESULTS
Mnt repression of Odc is relieved by c-Myc.
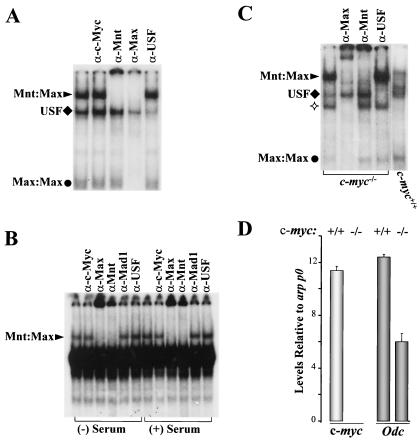
To initially assess which complexes of the Max network might contribute to Odc regulation, we performed EMSAs using antibodies that can supershift in vitro-generated Mad-Max, Myc-Max, and Mnt-Max complexes bound to probes containing the Odc E-boxes (data not shown). As observed in other systems (24), we failed to detect a Myc-Max complex or any Mad-Max complexes which bound to these elements in cell extracts from proliferating FDC-P1 myeloid cells (Fig. 1A), from proliferating or quiescent BALB/c-3T3 fibroblasts (Fig. 1B), or from wild-type or c-myc−/− ES cells (Fig. 1C). By contrast, a Mnt-Max complex was readily detected in extracts derived from all cell types (Fig. 1A to C). Moreover, the binding of the Mnt-Max complex was greatly augmented in extracts of c-myc−/− ES cells over wild-type ES cells (Fig. 1C), despite an equivalent expression of Mnt (data not shown). Therefore, loss of c-Myc increases the DNA binding activity of the Mnt-Max complex, and real-time PCR analyses established that this was associated with reductions in the level of Odc mRNA in c-myc−/− compared with wild-type ES cells (Fig. 1D).
FIG. 1.
(A and B) Mnt-Max is the predominant complex detected in EMSA using either Odc E-box1 (A) or E-box2 (B and C) probes, using extracts from proliferating FDC-P1 myeloid cells (A) growing (+ Serum) or quiescent (− Serum) BALB/c 3T3 fibroblasts (B). Arrowheads, diamonds, and circles indicate Mnt-Max, USF, and Max-Max complexes, respectively, as established by supershift analyses. Results using E-box probe 1 or 2 were comparable for all cell extracts (data not shown). (C) c-myc loss enhances Mnt-Max DNA binding activity in ES cells. EMSA was performed with extracts from the indicated ES cells. ✧ indicates a complex of unknown origin that is augmented in c-myc−/− ES cells and which contains Max, since it is supershifted by an antiserum against Max. (D). c-myc loss compromises Odc expression in ES cells. RNA was prepared from exponentially growing ES cell cultures, and levels of c-myc and Odc transcripts were determined by real-time PCR. Oligonucleotides and probes for the detection of acidic ribosomal protein P0 mRNA (arp p0) were used as an internal control, since this gene is not regulated by Myc (17).
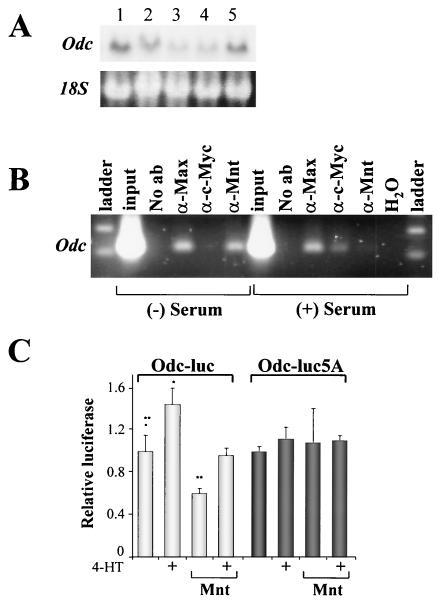
The binding of c-Myc to its response elements has been detected in some targets, such as Cyclin D2, using ChIP assays (10, 16). We therefore evaluated the occupation of the Odc E-boxes in quiescent and serum-stimulated BALB/c-3T3 fibroblasts (which express Odc at low or high levels [Fig. 2A, lanes 4 and 5, respectively). As expected, Max bound to these elements irrespective of the growth state of the cells (Fig. 2B). Interestingly, ChIP assays demonstrated that Mnt occupied the Odc E-boxes in quiescent cells (Fig. 2B) but that its binding was abolished in serum-stimulated fibroblasts, despite equivalent levels of the Mnt-Max complex (as determined by EMSA) in proliferating and quiescent cells (Fig. 1B). Rather, in proliferating fibroblasts ChIP analyses established that Mnt was effectively replaced by c-Myc (Fig. 2B). Therefore, the induction of Odc by growth factors is associated with the ability of Myc to displace Mnt-Max complexes.
FIG. 2.
Myc activates Odc transcription by relieving Mnt-mediated repression. (A) Odc expression in BALb/c-3T3 cells is serum dependent. Fibroblasts were cultured in the presence (lane 1) or absence of serum for 1, 2, and 3 days (lanes 2 to 4). After 3 days of starvation, fresh medium containing serum was added for 3 h (lane 5). Northern blot analysis was performed using a probe directed against Odc. 18S rRNA served as loading control. (B) Mnt-Max complexes occupy the Odc E-boxes in quiescent BALB/c-3T3 fibroblasts and are displaced by c-Myc-Max complexes in serum-stimulated cells. ChIP assays of serum-starved (2 days) or restimulated (for 3 h) BALB/c-3T3 cells were performed using antibodies directed against Max, c-Myc, or Mnt. The primers used for the PCR encompass the intronic E-boxes of the Odc gene. (C) Mnt represses the Odc promoter, and Mnt repression is relieved by Myc. BALB/c-3T3 cells infected with a Myc-ER-expressing retrovirus (MSCV-Myc-ER-IRES-GFP) were cotransfected with Odc promoter-luciferase constructs (containing intact [Odc-Luc] or mutated [Odc-luc5A] E-boxes) together with a Renilla luciferase control plasmid (pRL-SV40 [Promega]) and, where indicated, a Mnt expression vector (pRc-CMV-HA-Mnt). Transfections were balanced for DNA and promoter content by using the empty pRc vector plasmid. Myc-ER was activated by adding 4-HT for 24 h. Luciferase activity was determined using a luminometer. Results shown are the mean of triplicate assays and are representative of three independent experiments.
The induction of the Odc promoter by c-Myc (7) and the binding of the Mnt-Max complex present in cells (data not shown) are abolished by CACGTG-to-CACCTG point mutations of these E-boxes. To address whether Mnt would repress the Odc transcription and whether this suppression could be relieved by Myc activation, BALB/c-3T3 fibroblasts engineered to express the tamoxifen-inducible Myc-ER construct (25) were transiently transfected with Odc promoter-reporters and a Mnt expression construct. As expected, Mnt repressed Odc promoter activity, and this was dependent on the E-boxes. Moreover, activation of Myc-ER with 4-HT effectively relieved Mnt-mediated repression of the Odc promoter (Fig. 2C). Therefore, Myc antagonizes Mnt-mediated repression of Odc transcription.
Knockdown of Mnt expression accelerates growth, triggers apoptosis, and provokes transformation.
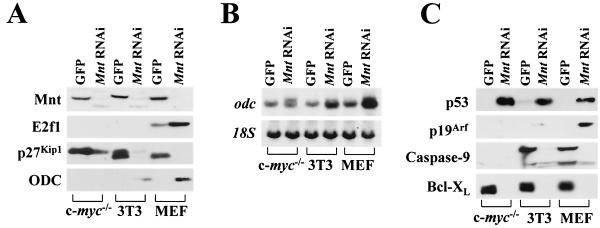
Collectively, these data were consistent with a model whereby Myc activates Odc through the displacement of Mnt, which binds to Odc E-boxes to repress transcription. If this scenario was generally true, then the effects of Mnt loss should essentially mimic those of Myc overexpression. To address this issue, we generated a retroviral vector (MSCV-MntRNAi-IRES-GFP) that expresses a short hairpin RNA (shRNA) that targets both mouse and rat Mnt mRNA, driven by the U6 RNA polymerase III promoter (19). Two fibroblast cell lines, mouse BALB/c-3T3 (30) and rat HO.15 c-myc−/− cells (27), and early-passage (passage 3) mouse embryonic fibroblasts (MEFs) were transduced with MSCV-MntRNAi-IRES-GFP virus, which also expresses the gene for GFP by virtue of an internal ribosome entry site (IRES). As a control, these cells were also infected in parallel with a MSCV-IRES-GFP virus and infected cells were sorted by flow cytometry for GFP. Using this strategy, there was a specific ablation of Mnt protein levels in all three MntRNAi-expressing fibroblast strains (see Fig. 7A).
FIG. 7.
Mnt knockdown triggers “Myc” transcriptional programs. (A) Immunoblot analyses of the indicated fibroblast strains infected with control (GFP) or MntRNAi-expressing retroviruses. The anti-Mnt and anti-ODC immunoblots demonstrate that the MntRNAi indeed ablates Mnt expression and up-regulates ODC. The absence of ODC and E2f1 signals in the c-myc−/− cells appears to reflect the inability of these antibodies to recognize rat ODC and E2f1. In addition, Mnt loss was associated with a marked down-regulation in levels of p27KipI and in MEFs with an up-regulation of E2f1. (B) Northern blot analysis demonstrates that Odc is up-regulated by MntRNAi in all three fibroblast strains. Evaluation of 18S rRNA levels served as a loading control. (C) The p19Arf-p53 apoptotic pathway and the Bcl-XL apoptotic pathway are triggered by Mnt loss. Knockdown of Mnt was associated with an increase in p53 levels in all fibroblasts and with the induction of Arf in MEFs. The loss of Mnt was also associated with the activation of caspase-9, since larger (inactive) proenzyme form of caspase-9 was lost in MntRNAi-expressing fibroblasts. Again, the absence of caspase-9 protein in the c-myc−/− cells may reflect the inability of this antibody to recognize rat caspase-9. Bcl-XL protein levels were dramatically reduced in all MntRNAi-expressing fibroblast strains.
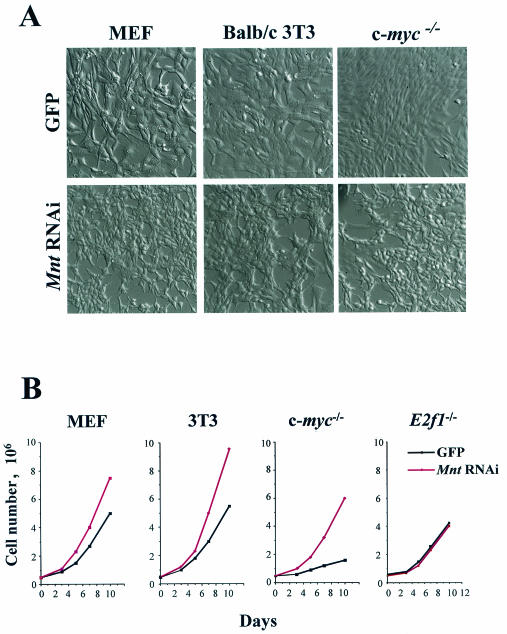
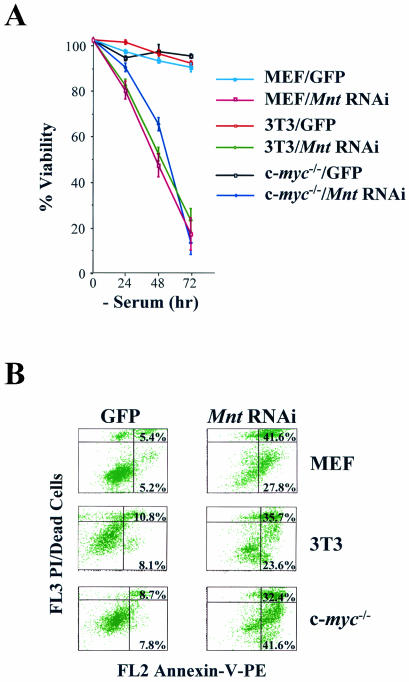
The hallmarks of Myc-overexpressing fibroblasts include accelerated rates of cell proliferation and changes in morphology to more refractile cells (27, 40). Strikingly, all cells expressing MntRNAi showed immediate and obvious effects of Mnt knockdown, since these fibroblasts were more refractile and proliferated at an accelerated rate (Fig. 3). Most notably, these changes were particularly evident in c-myc−/− fibroblasts, which also fail to express N-Myc or L-Myc (27); therefore, Mnt knockdown is sufficient to provoke the accelerated growth of fibroblasts independently of Myc.
FIG. 3.
Knockdown of Mnt by stable RNAi accelerates cell proliferation. (A) Morphological changes induced by MntRNAi expression in MEFs, BALB/c-3T3 cells, and c-myc−/− HO.15 fibroblasts are shown. (B) Acceleration of cell proliferation by ablation of Mnt is independent of c-myc but requires E2f1. Growth rates of the indicated MntRNAi-expressing cells were compared to GFP-only expressing fibroblasts.
Myc overexpression in fibroblasts also triggers apoptosis when cells are deprived of the survival factors present in serum (14, 18). As judged by their ability to exclude the vital dye trypan blue (Fig. 4A), by the appearance of annexin V-positive cells by FACS (Fig. 4B), and by the cleavage of pro-caspase-9 (Fig. 7C), all MntRNAi-expressing fibroblasts underwent rapid apoptosis when deprived of serum whereas GFP-only-expressing cells survived the withdrawal of serum. Again, since effects were obvious in c-myc−/− fibroblasts, apoptosis triggered by Mnt knockdown can also be Myc independent.
FIG. 4.
Mnt knockdown triggers apoptosis. (A) The indicated cells were shifted to medium containing 0.1% fetal calf serum, and their viability was determined by trypan blue dye exclusion. A representative experiment performed in triplicate is shown. (B) Apoptosis was determined by FACS analysis of annexin V-positive, propidium iodide-stained cells 48 h after serum depletion. The percentages of annexin V-positive cells are given in the quadrants (early apoptotic, lower right panels; late apoptotic, upper right panels). PE, phycoerythrin.
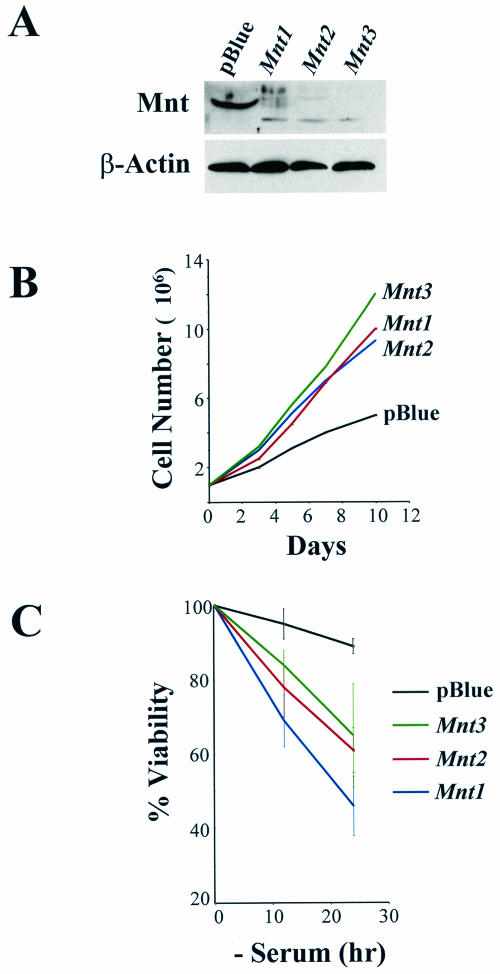
To rule out the possibility that the effects on cell proliferation and apoptosis were not in fact due to the expression of a short hairpin RNA, we designed three additional shRNAi-encoding retroviruses, one encoding an shRNAi targeting a sequence from pBluescript (which, according to BLAST searches, is not present in the mouse genome) and two additional shRNAi targeting Mnt. Together with the original MSCV-MntRNAi-IRES-GFP virus, these retroviruses were introduced into MEFs and assayed for cell proliferation and apoptosis. As is evident in Fig. 5A, all three MntRNAi constructs were very efficient in knocking down Mnt expression whereas cells expressing pBlueRNAi still expressed Mnt. Again, the effects of Mnt knockdown were remarkable, with changes in morphology (data not shown), increased cell proliferation (Fig. 5B), and induction of cell death following serum starvation (Fig. 5C).
FIG. 5.
Phenotypes observed in MEFs on Mnt knockdown are not due to general effects of expressing shRNAi. (A) Immunoblot analyses for Mnt of MEFs infected with control (MSCV-pBlueRNAi-IRES-GFP) or three different MSCV-MntRNAi-IRES-GFP-expressing retroviruses. β-Actin levels were assessed as a control. (B) MEFs expressing any of the three MSCV-MntRNAi-IRES-GFP retroviruses displayed accelerated rates of proliferation compared to MEFs expressing the MSCV-pBlueRNAi-IRES-GFP retrovirus. (C) MEFs expressing any of the three MSCV-MntRNAi-IRES-GFP retroviruses, but not those infected with the retrovirus encoding MSCV-pBlueRNAi-IRES-GFP, undergo rapid apoptosis following serum withdrawal.
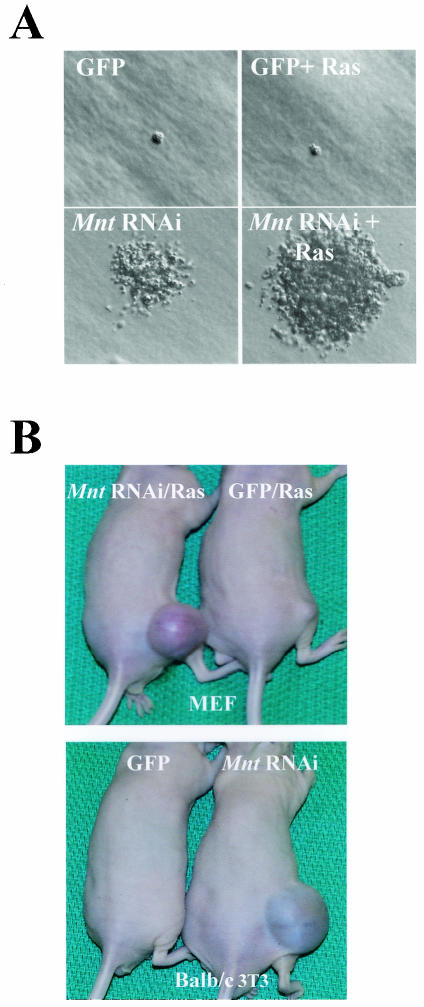
Myc also provokes transformation of primary fibroblasts in conjunction with activated Ha-Ras (22), and we therefore also assessed effects of Mnt knockdown on tumorigenesis. MEFs were transduced with MSCV-MntRNAi-IRES-GFP virus and pBabe-Ha-RasV12-puro virus (which expresses activated Ha-Ras and the puromycin resistance gene) or with control GFP- and puromycin-only-expressing viruses. Infected cells were selected for GFP and for growth in puromycin medium for 2 days and immediately plated in soft agar or injected into immunocompromised Nu/Nu mice (n = 5). In soft-agar assays, MEFs expressing MntRNAi alone were capable of forming colonies, although these were much smaller and were reduced 10- to 20-fold relative to the numbers of colonies observed in MEFs expressing both MntRNAi and Ha-Ras (Fig. 6A and data not shown). Furthermore, although all mice injected with MEFs expressing MntRNAi or Ras alone failed to develop tumors, all five animals injected with MEFs expressing both MntRNAi plus Ras developed rapidly growing fibrosarcomas (Fig. 6B). Furthermore, BALB/c-3T3 fibroblasts expressing MntRNAi alone were capable of forming tumors (Fig. 6B), indicating that removal of Mnt is sufficient to provoke tumorigenesis in cells that have undergone immortalization.
FIG. 6.
Mnt knockdown provokes transformation in conjunction with activated Ha-Ras. (A) A representative field of soft-agar assays of MEFs expressing GFP, GFP plus Ras, MntRNAi, and MntRNAi plus Ras are shown. MntRNAi-expressing MEFs were capable of growing in an anchorage-independent manner, although colonies were reduced 10- to 20-fold in their numbers and were much smaller than colonies generated by MEFs expressing both Ha-Ras and MntRNAi. (B) MEFs expressing both Ras and MntRNAi are tumorigenic in immunocompromised Nu/Nu mice (n = 5 for each group). BALB/c-3T3 fibroblasts expressing MntRNAi alone were also tumorigenic in this assay (n = 5).
Collectively, these data demonstrate that knockdown of Mnt mimics Myc overexpression in its effects on cell growth, survival, and transformation. Importantly, since these effects occur even in c-myc−/− Rat1a fibroblasts that lack all forms of Myc (27), the phenotypes provoked by Mnt knockdown can be Myc independent.
Mnt loss triggers “Myc” transcriptional programs.
The surprising similarities in the biological responses to knockdown of Mnt versus Myc overexpression, even in fibroblasts lacking c-myc, suggested that gene programs regulated by Myc might be due to relief of Mnt-mediated repression. We therefore assessed pathways triggered by Myc in fibroblasts expressing MntRNAi. Indeed, the ablation of Mnt protein expression was similar to the effects of Myc in inducing E2f1 expression in MEFs (5) and in inducing Odc in all three fibroblast strains (Fig. 7A and B). Previously it has been established that the ability of Myc to provoke accelerated proliferation is E2f1 dependent (5). We therefore transduced E2f1-deficient MEFs with MSCV-MntRNAi-IRES-GFP virus or control GFP-only-expressing virus and sorted GFP-expressing cells by FACS. Strikingly, knockdown of Mnt expression in E2f1−/− MEFs (data not shown) failed to alter their morphology or to provoke accelerated growth (Fig. 3C). Therefore, the effects of Mnt knockdown on cell proliferation are, as in the scenario of Myc overexpression, E2f1 dependent. Finally, another hallmark of the ability of Myc to provoke accelerated cell cycle traverse is the down-regulation of the universal cyclin-dependent kinase inhibitor p27KipI (39), and all three fibroblast strains expressing MntRNAi displayed markedly reduced levels of p27KipI (Fig. 7A).
The induction of apoptosis in MntRNAi-expressing fibroblasts when deprived of serum suggested that knockdown of Mnt may provoke apoptosis via typical mediators of Myc-induced cell death, for example by activating the Arf-p53 tumor suppressor pathway (40) and/or by suppressing the expression of the anti-apoptotic Bcl-XL protein (13). Indeed, in MEFs expressing MntRNAi, Arf and p53 expression were highly elevated, and in all three MntRNAi-expressing fibroblast strains, the expression of Bcl-XL protein was effectively abolished (Fig. 7C). Myc is also capable of stabilizing and activating p53 in the absence of Arf (23) and in c-myc−/− Rat1a fibroblasts and immortal BALB/c-3T3 fibroblasts Mnt RNAi expression was associated with massive increases in p53 protein levels (Fig. 7C). Thus, knockdown of Mnt also mimics Myc in its ability to regulate apoptotic pathways.
DISCUSSION
Through reinterrogating how Myc actually induces Odc transcription, we uncovered the surprising finding that the gene is actually repressed by another member of the Max network, Mnt, and that Myc stimulates Odc by relieving this repression. Odc is considered a direct transcription target induced by c-Myc both in cells and in vivo in transgenic mice (1, 34) and contains two conserved CACGTG elements in intron 1 that are required for Myc to activate the Odc promoter (7). However, Myc activates Odc expression in scenarios where it is overexpressed, which is certainly relevant to cancer but perhaps less so for physiological control of Odc transcription, since the gene is responsive to many signals that regulate cell proliferation (12). Indeed, the ability of other effectors to regulate Odc may explain why the expression of Odc is unaffected by the somatic deletion of c-myc in immortal fibroblasts (11). Nevertheless, Odc expression is reduced in c-myc−/− ES cells, a scenario in which the DNA binding activity of Mnt is enhanced, which we posit may contribute to transcriptional repression.
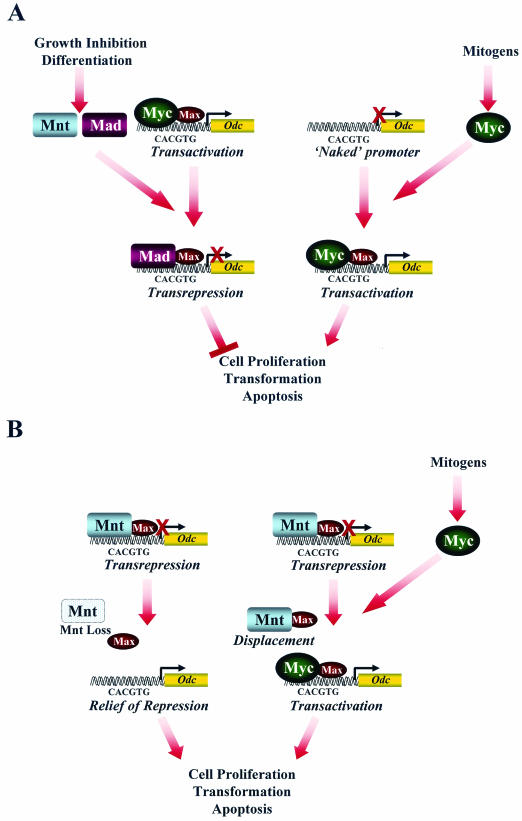
Stable RNA interference targeting Mnt indeed directly supports the notion that Odc is regulated by Mnt-mediated transrepression, and promoter-reporter assays established that Myc can relieve this suppression. EMSA and ChIP paint an interesting picture of how Odc regulation is orchestrated. EMSA established that the abundance of the Mnt-Max complex is not regulated by the proliferative state of fibroblasts or myeloid cells, but ChIP revealed that this complex is displaced by a c-Myc-Max complex. The binding of the c-Myc-Max complex thus may activate Odc, and by inference its other targets, by performing two functions: actively displacing Mnt and relieving active repression and simultaneously recruiting the transcriptional co-activators such as TRRAP and associated histone acetylases (28) (Fig. 8). However, a totally unexpected finding was that Mnt ablation essentially mimicked many of the transcriptional programs provoked by Myc and that this could occur even in cells lacking all forms of Myc. This would then argue that Mnt-mediated repression is the focal point for transcriptional control (Fig. 8) and that Myc oncoproteins really activate transcription indirectly by effectively blocking Mnt functions.
FIG.8.
Myc antagonizes Mnt to regulate cell fate. (A) (Left) Conventional models of transcriptional regulation by the Max network. In these models, Mad and Mnt work as antagonists of Myc. (Right) Alternatively, the Myc-Max complex can activate promoters by direct binding to E-boxes not previously occupied by other complexes. (B) In the revised model, Myc functions as an antagonist of Mnt, since Mnt loss triggers most of the“Myc” response, including Myc's transcription targets (e.g., Odc), apoptosis, accelerated proliferation, and transformation. (Right) In this scenario Myc may operate in two steps to induce its targets. First, it may displace Mnt-Max complexes from response elements, thus relieving repression. Second, when bound the Myc-Max complex may transactivate the gene. (Left) The fact that Mnt loss can activate some “Myc” transcription targets even in cells lacking all forms of Myc indicates that Myc binding may not be required to activate some of its targets, which instead are controlled through active repression.
The parallels between Mnt knockdown and Myc overexpression are striking at many levels. Although interrogating the genomic response is clearly a priority, it is already evident that besides Odc, many other genes that are normally triggered by Myc are also regulated by Mnt loss. The biological responses to Mnt knockdown also recapitulate the effects of Myc overexpression, including accelerated cell proliferation (40), apoptosis following the withdrawal of survival factors (1, 14), and transformation in conjunction with activated Ha-Ras (22). Interestingly, the notion that relieving Mnt-mediated transrepression is sufficient to activate genes involved in transformation is supported by experiments demonstrating that a deletion mutant of Mnt lacking the Sin3 interaction domain is capable of transforming primary rat embryo fibroblasts in conjunction with Ha-Ras (20). This strengthens our view that Myc activates some target genes by merely relieving Mnt-mediated transrepression.
Surprisingly modest phenotypes have been observed in the mouse knockouts of the Mad family members of the Max network. For example, hematopoietic cells from Mad1 knockout mice do exhibit an increased sensitivity to apoptosis following growth factor withdrawal ex vivo, yet Mad1 loss does not alter the hematopoietic compartment in vivo (15). Further, MEFs from Mxi1-deficient mice are more prone to transformation, yet this is manifest only in the presence of both activated Ras and Myc (36). Finally, Mad3-deficient thymocytes and neural progenitor cells exhibit increased sensitivity to DNA damage (35), a phenotype also manifested following Myc overexpression (26). However, none of these knockouts come close to having the profound effects observed following Mnt knockdown, which in many respects appears to phenocopy the effects of Myc overexpression as seen in cancer. Indeed our studies are largely supported by the very recent and initial characterization of the mouse Mnt knockout, since MEFs from these mice display accelerated growth and apoptosis and since loss of Mnt can lead to breast cancer (21). However, there are important differences in the cast of “Myc” targets expressed in MEFs from these mice and those expressed in our Mnt knockdown cells, suggesting that some level of compensation for Mnt loss may be operational during development (as would be expected for a proapoptotic signal).
Most importantly, the results presented here challenge many of the conventional notions of how Myc functions to regulate gene transcription, cell proliferation, apoptosis, and transformation. Heretofore, the Mad and Mnt transcriptional repressors have been suggested to antagonize Myc (Fig. 8A). However, the data herein suggest that it may be Myc that functions as an antagonist, by specifically relieving Mnt-mediated repression (Fig. 8B). These results specifically support a model whereby the function of Myc as an oncogene relies on its ability to block Mnt activity. At face value, these findings would predict that Mnt is a bona fide tumor suppressor and that it should be inactivated in human cancers. Here we establish that this is probably true for every tumor that overexpresses Myc. Alternatively, the current failure to detect direct inactivation of MNT in human tumors (37, 38) could reflect the profound apoptotic response observed following Mnt loss, since, for example, knockdown of Mnt is more lethal than Myc overexpression in primary hematopoietic progenitors (J. A. Nilsson and J. L. Cleveland, unpublished data). Finally, this model also explains why Myc overexpression is such a pervasive event in cancer—increasing the thresholds of Myc would specifically override the transcriptional repression and tumor suppressor functions of Mnt.
Acknowledgments
We thank R. E. Lewis, J. M. Bishop, A. E. Pegg, P. J. Hurlin, and R.N. Eisenman for providing reagents. We thank P. Brindle, E. Parganas, and G. Zambetti for critical review and E. White, S. Norton, C. Yang, A. Bizet, and the cell-sorting core facility at St. Jude Children's Research Hospital for excellent technical assistance.
This work was supported by National Institutes of Health grants DK44158 and CA100603 (to J.L.C.), by Cancer Center CORE grant CA21765, and by the American Lebanese Syrian Associated Charities (ALSAC). J.A.N is a recipient of the George J. Mitchell endowed fellowship from ALSAC. U.K. is a fellow of the Deutsche Forschungssemeinschoff.
REFERENCES
- 1.Askew, D. S., R. A. Ashmun, B. C. Simmons, and J. L. Cleveland. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6:1915-1922. [PubMed] [Google Scholar]
- 2.Ayer, D. E., and R. N. Eisenman. 1993. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 7:2110-2119. [DOI] [PubMed] [Google Scholar]
- 3.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. [DOI] [PubMed] [Google Scholar]
- 4.Baudino, T. A., and J. L. Cleveland. 2001. The Max network gone Mad. Mol. Cell. Biol. 21:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudino, T. A., K. H. Maclean, J. Brennan, E. Parganas, C. Yang, A. Aslanian, J. A. Lees, C. J. Sherr, M. F. Roussel, and J. Cleveland. 2003. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compomised by E2f1 loss. Mol. Cell 11:1-20. [DOI] [PubMed] [Google Scholar]
- 6.Baudino, T. A., C. McKay, H. Pendeville-Samain, J. A. Nilsson, K. H. Maclean, E. L. White, A. C. Davis, J. N. Ihle, and J. L. Cleveland. 2002. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 16:2530-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell, T. K., L. Kretzner, E. M. Blackwood, R. N. Eisenman, and H. Weintraub. 1990. Sequence-specific DNA binding by the c-Myc protein. Science 250:1149-1151. [DOI] [PubMed] [Google Scholar]
- 9.Blackwood, E. M., B. Luscher, and R. N. Eisenman. 1992. Myc and Max associate in vivo. Genes Dev. 6:71-80. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard, C., O. Dittrich, A. Kiermaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush, A., M. Mateyak, K. Dugan, A. Obaya, S. Adachi, J. Sedivy, and M. Cole. 1998. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 12:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, R. H., D. R. Morris, and P. Coffino. 1992. Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol. Rev. 56:280-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eischen, C. M., D. Woo, M. F. Roussel, and J. L. Cleveland. 2001. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol. Cell. Biol. 21:5063-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 15.Foley, K. P., G. A. McArthur, C. Queva, P. J. Hurlin, P. Soriano, and R. N. Eisenman. 1998. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 17:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Roman, N., C. Grandori, R. N. Eisenman, and R. J. White. 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421:290-294. [DOI] [PubMed] [Google Scholar]
- 18.Harrington, E. A., M. R. Bennett, A. Fanidi, and G. I. Evan. 1994. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 13:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemann, M. T., J. S. Fridman, J. T. Zilfou, E. Hernando, P. J. Paddison, C. Cordon-Cardo, G. J. Hannon, and S. W. Lowe. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33:396-400. [DOI] [PubMed] [Google Scholar]
- 20.Hurlin, P. J., C. Queva, and R. N. Eisenman. 1997. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes. Dev. 11:44-58. [DOI] [PubMed] [Google Scholar]
- 21.Hurlin, P. J., Z. Q. Zhou, K. Toyo-oka, S. Ota, H. L. Walker, S. Hirotsune, and A. Wynshaw-Boris. 2003. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J. 22:4584-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Land, H., L.-F. Parada, and R. A. Weinberg. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304:596-602. [DOI] [PubMed] [Google Scholar]
- 23.Lindstrom, M. S., and K. G. Wiman. 2003. Myc and E2F1 induce p53 through p14ARF-independent mechanisms in human fibroblasts. Oncogene 22:4993-5005. [DOI] [PubMed] [Google Scholar]
- 24.Littlewood, T. D., B. Amati, H. Land, and G. I. Evan. 1992. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene 7:1783-1792. [PubMed] [Google Scholar]
- 25.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maclean, K. H., U. Keller, C. Rodriguez-Galindo, J. A. Nilsson, and J. L. Cleveland. 2003. c-Myc augments gamma-irradiation-induced apoptosis by suppressing Bcl-XL. Mol. Cell. Biol. 23:7256-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 28.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 29.Meroni, G., A. Reymond, M. Alcalay, G. Borsani, A. Tanigami, R. Tonlorenzi, C. L. Nigro, S. Messali, M. Zollo, D. H. Ledbetter, R. Brent, A. Ballabio, and R. Carrozzo. 1997. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 16:2892-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momand, J., and G. P. Zambetti. 1996. Analysis of the proportion of p53 bound to mdm-2 in cells with defined growth characteristics. Oncogene 12:2279-2289. [PubMed] [Google Scholar]
- 31.Nilsson, J., S. Koskiniemi, K. Persson, B. Grahn, and I. Holm. 1997. Polyamines regulate both transcription and translation of the gene encoding ornithine decarboxylase antizyme in mouse. Eur. J. Biochem. 250:223-231. [DOI] [PubMed] [Google Scholar]
- 32.Packham, G., and J. L. Cleveland. 1997. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene 15:1219-1232. [DOI] [PubMed] [Google Scholar]
- 33.Pelengaris, S., M. Khan, and G. Evan. 2002. c-myc: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 34.Pelengaris, S., T. Littlewood, M. Khan, G. Elia, and G. Evan. 1999. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell 3:565-577. [DOI] [PubMed] [Google Scholar]
- 35.Queva, C., G. A. McArthur, B. M. Iritani, and R. N. Eisenman. 2001. Targeted deletion of the S-phase-specific Myc antagonist Mad3 sensitizes neuronal and lymphoid cells to radiation-induced apoptosis. Mol. Cell. Biol. 21:703-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber-Agus, N., Y. Meng, T. Hoang, H. Hou, Jr., K. Chen, R. Greenberg, C. Cordon-Cardo, H. W. Lee, and R. A. DePinho. 1998. Role of Mxil in ageing organ systems and the regulation of normal and neoplastic growth. Nature 393:483-487. [DOI] [PubMed] [Google Scholar]
- 37.Sommer, A., A. Waha, J. Tonn, N. Sorensen, P. J. Hurlin, R. N. Eisenman, B. Luscher, and T. Pietsch. 1999. Analysis of the Max-binding protein MNT in human medulloblastomas. Int. J. Cancer 82:810-816. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, T., H. Konishi, K. Kozaki, H. Osada, and S. Saji. 1998. Molecular analysis of a Myc antagonist, ROX/Mnt, at 17p13.3 in human lung cancers. Jpn. J. Cancer Res. 89:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlach, J., S. Hennecke, K. Alevizopoulos, D. Conti, and B. Amati. 1996. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 15:6595-6604. [PMC free article] [PubMed] [Google Scholar]
- 40.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]