Abstract
EDA-A1 and EDA-A2 are members of the tumor necrosis factor family of ligands. The products of alternative splicing of the ectodysplasin (EDA) gene, EDA-A1 and EDA-A2 differ by an insertion of two amino acids and bind to distinct receptors. The longer isoform, EDA-A1, binds to EDAR and plays an important role in sweat gland, hair, and tooth development; mutations in EDA, EDAR, or the downstream adaptor EDARADD cause hypohidrotic ectodermal dysplasia. EDA-A2 engages the receptor XEDAR, but its role in the whole organism is less clear. We have generated XEDAR-deficient mice by gene targeting and transgenic mice expressing secreted forms of EDA-A1 or EDA-A2 downstream of the skeletal muscle-specific myosin light-chain 2 or skin-specific keratin 5 promoter. Mice lacking XEDAR were indistinguishable from their wild-type littermates, but EDA-A2 transgenic mice exhibited multifocal myodegeneration. This phenotype was not observed in the absence of XEDAR. Skeletal muscle in EDA-A1 transgenic mice was unaffected, but their sebaceous glands were hypertrophied and hyperplastic, consistent with a role for EDA-A1 in the development of these structures. These data indicate that XEDAR-transduced signals are dispensable for development of ectoderm-derived organs but might play a role in skeletal muscle homeostasis.
The ectodysplasin (EDA) gene was cloned because of its mutation in families with X-linked hypohidrotic ectodermal dysplasia (HED), a disease affecting hair, sweat gland, and tooth development (11). Mutation of the mouse ectodysplasin gene was subsequently found to be responsible for the HED-like phenotype of the mutant Tabby mouse (4, 19). The predominant splice variants of EDA, EDA-A1 and EDA-A2, are transmembrane proteins belonging to the tumor necrosis factor (TNF) family of ligands, and they bind to the receptors EDAR and XEDAR, respectively (21). Binding of EDA-A1 to EDAR recruits the cytoplasmic adaptor EDARADD and leads to activation of the IκB kinase (IKK) complex necessary for translocation of NF-κB transcription factors into the nucleus. This signaling pathway, rather than that initiated by EDA-A2/XEDAR, seems to be essential for the development of epidermal appendages because mutations in EDAR, EDARADD, and NEMO/IKKγ, a component of the IKK complex, also produce HED in humans and mice (2, 7, 8, 13, 22). Consistent with this notion, recombinant EDA-A1 protein or an EDA-A1 transgene can rescue the mutant Tabby phenotype (6, 18).
EDA-A2 has two fewer amino acids in its extracellular domain than EDA-A1 and specifically binds to the receptor XEDAR. Like EDA-A1, EDA-A2 signals NF-κB activation. Coimmunoprecipitation experiments following transient expression of XEDAR and TRAF6 suggest that TRAF6 may be recruited to ligated XEDAR and contribute to activation of the IKK complex (21). Consistent with this idea, cross-linking of a chimeric receptor composed of the extracellular domain of CD40 fused to the intracellular domain of XEDAR induced NF-κB DNA binding activity in wild-type, but not TRAF6-deficient, mouse embryo fibroblasts (15). TRAF6-deficient mice are reported to have HED (15), but evidence that this phenotype is due to disrupted XEDAR signaling is lacking. Specifically, mutation of the xedar gene in individuals with HED has not been reported. We determined the contribution of the EDA-A2/XEDAR signaling axis to the development of ectoderm-derived organs by using XEDAR-deficient mice generated by gene targeting and transgenic mice that express a secreted form of EDA-A1 or EDA-A2. Mice lacking XEDAR were fertile, appeared healthy, and did not exhibit HED, indicating that XEDAR is dispensable for normal mouse development. However, a potential role for XEDAR in skeletal muscle homeostasis was revealed by EDA-A2 transgenic mice, which developed multifocal myodegeneration that was dependent on expression of XEDAR.
MATERIALS AND METHODS
Generation of xedar mutant mice.
The Institutional Animal Care and Use Committee approved all of protocols used in this study. A genomic xedar clone was isolated from a 129/SvJ library (Incyte Genomics) and used to construct a targeting vector (Fig. 1A) that was electroporated into 129 R1 embryonic stem (ES) cells. A single homologous recombinant (clone 13 B8) was identified by PCR and by Southern blotting with a probe 5′ of the genomic sequence present in the targeting vector. The hemizygous xedar−/Y ES cell clone was microinjected into C57BL/6N blastocysts, and chimeric offspring were backcrossed to C57BL/6N mice. Germ line transmission was confirmed by PCR and Southern blot analysis of tail DNA. Mice analyzed in this study were backcrossed to C57BL/6N mice for two or three generations.
FIG. 1.
Generation of XEDAR-deficient mice. (A) A PGK-neo selection cassette replaced the sequence in exon 4 encoding the transmembrane domain of XEDAR. (B) Southern blot analysis of genomic DNA from wild-type (+/Y) and hemizygous xedar mutant (−/Y) male mice. SpeI-digested DNAs were hybridized to a probe that binds a sequence that is 5′ of the targeting construct. The 7.4- and 3.6-kb fragments correspond to the wild-type and mutant xedar alleles, respectively. (C) Immunoprecipitation and Western blot (IP/WB) analysis of XEDAR in wild-type and XEDAR-deficient embryo fibroblasts. XEDAR protein was immunoprecipitated and blotted with a mouse monoclonal antibody (clone 11D7.1.6) that was raised in xedar mutant mice against the extracellular domain of mouse XEDAR (amino acids 1 to 136). Arrowheads indicate mouse XEDAR, which has a predicted molecular mass of 33 kDa but might appear larger because of glycosylation of its extracellular domain. An asterisk marks the light chain (IgL) of the precipitating antibody. (D) Western blot (WB) analysis of IκBα in wild-type and XEDAR-deficient embryo fibroblasts treated with 500 ng of recombinant human EDA-A2 (amino acids 179 to 389) per ml. Blots were probed with polyclonal antibodies that recognize either a phosphorylated form of IκBα (top) or all forms of IκBα (bottom). (E) Western blot analysis of IκBα in wild-type and XEDAR-deficient embryo fibroblasts treated with 20 ng of recombinant human TNF per ml.
Generation of EDA-A1 or EDA-A2 transgenic mice.
For skeletal muscle-specific expression, cDNA encoding human EDA-A1 (amino acids 179 to 391) or EDA-A2 (amino acids 179 to 389) with an N-terminal Flag epitope tag and signal sequence was cloned downstream of the rat myosin light-chain 2 (MLCH) promoter and an optimized chimeric intron. Downstream of the cDNA was a minigene composed of human growth hormone exons 4 and 5 plus 3′ untranslated and polyadenylation sequences (5). For skin-specific expression, constructs contained the human keratin 5 (K5) promoter, a simian virus 40 intron, and a simian virus 40 polyadenylation site (20). Linearized constructs were injected into the pronuclei of inbred FVB/N mouse zygotes. Transgene-positive mice were identified by PCR, and expression of Flag-tagged EDA-A1 or EDA-A2 was determined by immunoprecipitation with M2 anti-Flag antibodies conjugated to agarose (Sigma), followed by Western blotting with rabbit anti-Flag antibodies (Sigma).
Mouse embryo fibroblasts and human skeletal muscle cells.
Fibroblasts derived from embryos at 13.5 to 14.5 days postcoitus by trypsinization were cultured for up to three passages on gelatin-coated plates in the high-glucose version of Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Normal human skeletal muscle cells (Clonetics) at passage 3 were differentiated into myotubes for 5 days in the high-glucose version of Dulbecco's modified Eagle's medium supplemented with 2% horse serum. Fibroblasts and skeletal muscle cells were treated with 20 ng of recombinant human TNF (Genentech) per ml or 500 ng of recombinant N-terminal Flag-tagged human EDA-A1 (amino acids 179 to 391) or EDA-A2 (amino acids 179 to 389) (Genentech) per ml.
Western blotting.
Cell lysates were prepared in 20 mM Tris-HCl (pH 7.5)-135 mM NaCl-1.5 mM MgCl2-2 mM EGTA-1% Triton X-100-10% glycerol-1 mM Na3VO4-10 mM NaF-1 mM sodium pyrophosphate-0.1 mM β-glycerophosphate supplemented with a complete protease inhibitor cocktail (Roche). Proteins were resolved in 4 to 20% gradient Tris-glycine polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen). Blots were probed with 11D7.1.6 mouse anti-XEDAR (Genentech), AC-15 mouse anti-β-actin (Abcam), rabbit anti-IκBα (Cell Signaling), rabbit anti-phospho-IκBα (Ser32) (Cell Signaling), or rabbit anti-Flag (Sigma) antibodies.
Reverse transcription (RT)-PCR.
First-strand cDNAs prepared from a panel of mouse tissues (Origene) were used as templates in PCRs to amplify XEDAR or, as a control, hypoxanthine phosphoribosyltransferase (HPRT) sequences. The primers used to amplify a 435-bp XEDAR cDNA fragment were 5′-GGAGATGCACACTGCATAGTCTGC and 5′-TTCAGCCTCATATTGCAACGAATC. The primers used to amplify a 249-bp HPRT cDNA fragment were 5′-GCTGGTGAAAAGGACCTCT and 5′-CACAGGACTAGAACACCTGC. To confirm their identities, PCR products were blotted onto a GeneScreen Plus nylon membrane (NEN) and probed with an internal 32P-labeled oligonucleotide (XEDAR, 5′-GCTTAGTGAAGGTAGATGCACACACTGTTC; HPRT, 5′-GGATACAGGCCAGACTTTGT).
In situ hybridization.
33P-labeled murine XEDAR riboprobes were used to evaluate gene expression in murine skeletal muscle. A 928-bp fragment of xedar spanning nucleotides 349 to 1276 of the sequence with accession no. XM_142010 was amplified by PCR with primers that contained 27-nucleotide T7 or T3 RNA polymerase initiation sites for in vitro transcription of sense or antisense probes, respectively (upper, 5′-TAGTCTGCCCTCCCCGAAAGT; lower, 5′-GCTCTGAAGACCCCATTTTTG). Sections were deparaffinized, deproteinized in 4 mg of proteinase K per ml for 30 min at 37°C, and processed for in situ hybridization as previously described (9). Sense and antisense riboprobes were hybridized to sections at 55°C overnight. Unbound probe was removed by incubation in 20 mg of RNase A per ml for 30 min at 37°C, followed by a high-stringency wash at 55°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 h and dehydration through graded ethanols. The slides were dipped in NBT2 nuclear track emulsion (Eastman Kodak), exposed in sealed plastic slide boxes containing desiccant for 4 weeks at 4°C, developed, and counterstained with hematoxylin and eosin.
RESULTS AND DISCUSSION
Targeted disruption of xedar.
XEDAR-deficient mice were generated with a targeting vector designed to remove the sequence in exon 4 encoding the transmembrane domain (Fig. 1A). We obtained one 129 R1 ES cell clone with a disrupted xedar allele, the gene being located on the X chromosome, and this was injected into blastocysts to generate chimeric mice. Chimeras were crossed to C57BL/6N mice, and their heterozygous xedar+/− female offspring also were crossed to C57BL/6N mice to yield hemizygous xedar−/Y male mice at the expected frequency. ES cells and mice were genotyped by PCR (data not shown) and Southern blotting (Fig. 1B). We confirmed that no XEDAR protein was made in xedar−/Y male or xedar−/− female mice with a monoclonal antibody raised against the extracellular domain of mouse XEDAR in immunoprecipitation and Western blot analyses (Fig. 1C and data not shown). Indeed, this monoclonal antibody was generated by immunizing xedar mutant mice with mouse XEDAR (amino acids 1 to 136) fused to the Fc region of human immunoglobulin G1. Two bands approximately 45 kDa in size were detected in xedar+/+ and xedar+/− embryo fibroblasts but were absent from xedar−/Y and xedar−/ cells. Recombinant epitope-tagged mouse XEDAR expressed in 293T cells produced a similar doublet (data not shown), suggesting that the size discrepancy from the predicted molecular mass of mouse XEDAR, which is 33 kDa, might be due to variable glycosylation of the XEDAR extracellular domain. XEDAR lacks an N-terminal signal peptide (21), but it was possible that xedar mutant mice produced a soluble form of the receptor. Sequencing of xedar transcripts from xedar−/Y skeletal muscle revealed two forms of splicing from exon 3 to exon 5. These splicing events produced frameshifts that would lead to either a 13-kDa or a 25-kDa protein containing the first 115 amino acids of XEDAR. However, no proteins of these sizes were detected in xedar mutant embryo fibroblasts with our XEDAR-specific monoclonal antibody, nor could we detect secreted proteins of these sizes in serum from xedar−/Y mice (data not shown). Expression of XEDAR in wild-type mice is not limited to the developing embryo; XEDAR transcripts also were detected in multiple adult tissues (see Fig. 4A). Nevertheless, XEDAR-deficient mice were indistinguishable from their wild-type littermates; they were fertile and appeared healthy, and histological analysis did not reveal abnormalities in any of their major organs (data not shown). These findings indicate that XEDAR, unlike EDAR, is dispensable for normal mouse development.
FIG. 4.
Skeletal muscle cells express XEDAR and activate NF-κB in response to EDA-A2 but not EDA-A1. (A) XEDAR mRNA expression in different mouse tissues was assessed by RT-PCR (top). The pattern of XEDAR expression was compared to that of the housekeeping gene HPRT (bottom). (B) Representative sections of MLCH.EDA-A2 transgenic skeletal muscle displaying myodegeneration by hematoxylin and eosin staining (left side) were probed for XEDAR expression by in situ hybridization (right side). (C) Western blot (WB) analysis of normal human skeletal muscle cells treated with 500 ng of recombinant human EDA-A1 or EDA-A2 per ml. Blots were probed with polyclonal antibodies to phosphorylated IκBα or, as a loading control, with antibodies to β-actin (bottom).
XEDAR is essential for NF-κB signaling by EDA-A2.
Previously, 293E cells treated with Flag-tagged recombinant human EDA-A2 (amino acids 179 to 389) were shown to activate the IKK complex, leading to phosphorylation of IκBα. It was assumed that EDA-A2 had engaged endogenous XEDAR because phosphorylation of IκBα was blocked by a monoclonal antibody to human XEDAR (21). To determine whether XEDAR is essential for EDA-A2-induced IκBα phosphorylation, we performed Western blot analysis of IκBα in wild-type and XEDAR-deficient embryo fibroblasts after EDA-A2 treatment (Fig. 1D). Total IκBα levels in wild-type and XEDAR-deficient cells were comparable during the first 20 min of EDA-A2 exposure but only wild-type cells showed phosphorylation of IκBα. This phosphorylation normally is transient because it targets IκBα for ubiquitination and subsequent degradation by the 26S proteasome (10). The defect in NF-κB signaling by XEDAR-deficient cells was specific to EDA-A2 because XEDAR-deficient embryo fibroblasts exhibited normal phosphorylation and degradation of IκBα in response to recombinant human TNF, which specifically engages mouse TNF receptor 1 (12) (Fig. 1E). These results prove that XEDAR is essential for EDA-A2-induced IκBα phosphorylation. Furthermore, they establish that our failure to detect XEDAR protein in xedar−/Y or xedar−/− cells (Fig. 1C) is coincidental with a loss of XEDAR function.
EDA-A1 or EDA-A2 secretion in transgenic mice.
To gain further insight into the roles of EDA-A1 and EDA-A2, we also generated transgenic mice expressing secreted forms of the ligands in a skin- or skeletal muscle-specific manner. Thus, the extracellular domains of EDA-A1 (amino acids 179 to 391) and EDA-A2 (amino acids 179 to 389) were given an N-terminal signal sequence, followed by a Flag epitope tag, and placed under the control of the K5 or rat MLCH promoter. These secreted forms of EDA-A1 and EDA-A2 were chosen because of their demonstrated biological activity on cells expressing EDAR and XEDAR, respectively (21) (Fig. 1D). In addition, a soluble form of EDA-A1 appears to be of functional importance in vivo because the furin cleavage site required for shedding of EDA-A1 from the cell surface in vitro is mutated in some individuals with HED (1, 3, 16, 17).
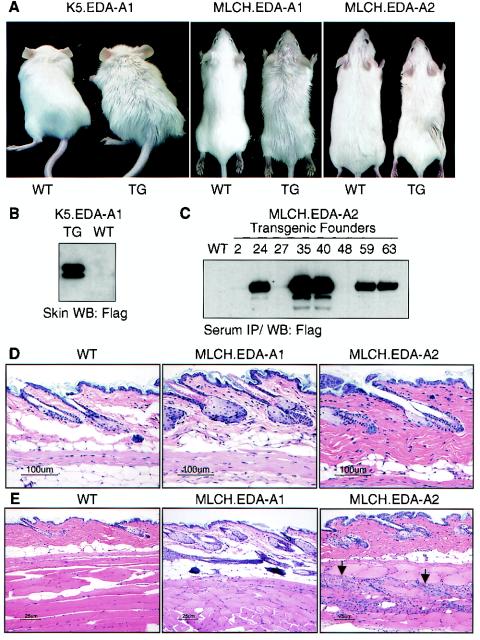
Fifteen K5.EDA-A1 transgenic founder mice on an FVB/N genetic background were obtained that expressed detectable EDA-A1 protein in skin as determined by Western blotting (Fig. 2B). These mice could be readily distinguished from their wild-type littermates by their greasy, unkempt appearance (Fig. 2A). A similar phenotype was observed for four MLCH.EDA-A1 transgenic founder mice secreting EDA.A1 from skeletal muscle cells (Fig. 2A). Histological analysis of skin from these animals revealed prominent sebaceous glands. Sebocytes within each unit were increased in both size and number (Fig. 2D). Hair follicle density in the MLCH.EDA-A1 transgenic mice appeared increased as well, but scanning electron microscopy revealed no qualitative difference in the guard, awl, auchene, and zigzag hair types (data not shown). Transgenic mice expressing full-length mouse EDA-A1 from the K14 promoter were reported to have a similar phenotype (14).
FIG. 2.
Sebaceous glands in EDA-A1 transgenic mice are hypertrophied, and EDA-A2 transgenic mice exhibit multifocal myodegeneration. (A) Mice expressing a secreted form of human EDA-A1 (amino acids 179 to 391) in skin (K5.EDA-A1) or skeletal muscle (MLCH.EDA-A1) have a greasy, unkempt appearance. The coats of mice expressing a secreted form of human EDA-A2 (amino acids 179 to 389) in skeletal muscle are indistinguishable from those of wild-type littermates. TG, transgenic; WT, wild type. (B) Western blot (WB) analysis of Flag-tagged EDA-A1 in the skin of wild-type and K5.EDA-A1 transgenic mice with anti-Flag antibodies. (C) Immunoprecipitation and Western blotting (IP/WB) of Flag-tagged EDA-A2 in the serum of viable MLCH.EDA-A2 transgenic founder mice. Animals 35 and 40 exhibited subclinical multifocal myodegeneration at 6 months of age. (D) Hematoxylin and eosin staining of skin sections from wild-type and MLCH.EDA-A1 and MLCH.EDA-A2 transgenic mice. EDA-A1, but not EDA-A2, transgenic mice had abnormally large sebaceous glands. (E) Hematoxylin and eosin staining of skeletal muscle sections from wild-type and MLCH.EDA-A1 and MLCH.EDA-A2 transgenic mice. Myodegeneration and resulting satellite cell proliferation (indicated by arrowheads) were observed in skeletal muscle from EDA-A2, but not EDA-A1, transgenic mice.
More than 30 MLCH.EDA-A2 and K5.EDA-A2 transgenic founder mice analyzed did not have an abnormal coat appearance, and their sebaceous glands were comparable to those found in wild-type littermates (Fig. 2A and D). However, many founder animals bearing an EDA-A2 transgene became thin and listless and died within 1 month of birth, the severity of the phenotype correlating with EDA-A2 transcript levels in skeletal muscle or skin and the level of EDA-A2 protein detected in the serum (Fig. 2C and data not shown). Previously described K14.EDA-A2 transgenic mice lacked a detectable phenotype, presumably owing to a low level of EDA-A2 expression (14). Histological analysis of affected MLCH.EDA-A2 transgenic mice revealed skeletal muscle degeneration in both weight-bearing and non-weight-bearing muscles. The myodegeneration was multifocal, affected random muscle bundles, and was frequently accompanied by a regenerative response in the form of satellite cell proliferation (Fig. 2E). No myodegeneration was ever detected in MLCH.EDA-A1 transgenic mice (data not shown). To determine whether myodegeneration was a consequence of XEDAR signaling rather than a nonspecific effect of excessive amounts of EDA-A2 protein, crosses were set up between MLCH.EDA-A2 transgenic and XEDAR-deficient mice. The MLCH.EDA-A2 transgenic mice used in these crosses were derived from two viable MLCH.EDA-A2 transgenic founder males that expressed intermediate levels of EDA-A2 protein (35 and 40 in Fig. 2C) and exhibited moderate myodegeneration upon histological examination at 6 months of age (data not shown). The impact of loss of XEDAR on EDA-A2-induced myodegeneration was assessed by comparing 6-month-old EDA-A2 transgenic littermates that had either wild-type or mutant xedar alleles. When sections of gastrocnemius and epaxial muscle were examined histologically, myodegeneration was evident in 11 out of 12 EDA.A2 transgenic mice expressing XEDAR and in 0 out of 11 EDA-A2 transgenic mice lacking XEDAR (Fig. 3). These data strongly suggest that protracted XEDAR signaling is responsible for the myodegeneration in EDA-A2 transgenic mice.
FIG. 3.
XEDAR deficiency prevents myodegeneration in EDA-A2 transgenic mice. Representative skeletal muscle sections from an MLCH.EDA-A2 transgenic mouse (left panel) and an MLCH.EDA-A2 transgenic, XEDAR-deficient mouse (right panel) were stained with hematoxylin and eosin. Arrows indicate areas of myodegeneration and satellite cell proliferation in MLCH.EDA-A2 transgenic muscle expressing XEDAR.
To investigate whether EDA-A2 might target skeletal muscle cells directly, we first examined XEDAR expression in various mouse tissues by RT-PCR (Fig. 4A). XEDAR transcripts were detected in skin, consistent with previous in situ hybridization studies (21), as well as in heart, kidney, small intestine, lung, testis, prostate, breast, uterus, and skeletal muscle tissues. To determine whether particular skeletal muscle fibers might express XEDAR, skeletal muscle from wild-type and MLCH.EDA-A2 transgenic mice was examined by in situ hybridization. Intriguingly, an XEDAR riboprobe gave the greatest signal at sites of muscle damage in EDA-A2 transgenic mice (Fig. 4B). Healthy muscle in wild-type and EDA-A2 transgenic mice was labeled weakly or not at all. Next we tested whether XEDAR mRNA in skeletal muscle tissue corresponded to functional XEDAR on myotubes. Normal human skeletal muscle cells expressing XEDAR transcripts (data not shown) were differentiated into myotubes in culture, treated with recombinant EDA-A2, and analyzed for IκBα phosphorylation by Western blotting (Fig. 4C). Phosphorylated IκBα was detected within 5 min of EDA-A2 treatment and then gradually disappeared at later time points. An equivalent amount of recombinant EDA-A1 did not promote IκBα phosphorylation in these cells. These results suggest that EDA-A2 might act directly on skeletal muscle cells to induce their degeneration. However, the downstream events culminating in myodegeneration are unclear. EDA-A2 had no adverse effect on the short-term viability of human skeletal muscle cells in culture (data not shown). Nor did it affect the skeletal muscle of wild-type mice when a large dose was injected subcutaneously on a daily basis for 5 days such that systemic levels of EDA-A2 were similar to or exceeded those found in EDA-A2 transgenic mice (data not shown). These results suggest that chronic rather than acute stimulation of XEDAR is necessary for myodegeneration. One possibility is that cytokines produced by skeletal muscle in response to EDA-A2 elicit an inflammatory response that leads to muscle damage. However, an absence of infiltrating cells at sites of muscle damage in the EDA-A2 transgenic mice would argue against this scenario. Whatever the mechanism behind EDA-A2-induced myodegeneration, our results point to a potential role for XEDAR in skeletal muscle homeostasis. Future studies might investigate whether XEDAR deficiency has an impact on other models of muscle injury.
Acknowledgments
We thank Luz Orellana, Jessica Kloss, and Meg Fuentes for animal husbandry; Sharon Fong, Ellen Filvaroff, and Ralph Schwall for helpful advice; and Michele Bauer, Merone Roose-Girma, Willis Su, Lucrece Tom, Marjie Van Hoy, Joel Morales, Khiem Tran, Christine Tan, Aparna Draksharapu, and Peter Wong for technical assistance.
REFERENCES
- 1.Chen, Y., S. S. Molloy, L. Thomas, J. Gambee, H. P. Bachinger, B. Ferguson, J. Zonana, G. Thomas, and N. P. Morris. 2001. Mutations within a furin consensus sequence block proteolytic release of ectodysplasin-A and cause X-linked hypohidrotic ectodermal dysplasia. Proc. Natl. Acad. Sci. USA 98:7218-7223. [DOI] [PMC free article] [PubMed]
- 2.Doffinger, R., A. Smahi, C. Bessia, F. Geissmann, J. Feinberg, A. Durandy, C. Bodemer, S. Kenwrick, S. Dupuis-Girod, S. Blanche, P. Wood, S. H. Rabia, D. J. Headon, P. A. Overbeek, F. Le Deist, S. M. Holland, K. Belani, D. S. Kumararatne, A. Fischer, R. Shapiro, M. E. Conley, E. Reimund, H. Kalhoff, M. Abinun, A. Munnich, A. Israel, G. Courtois, and J. L. Casanova. 2001. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat. Genet. 27:277-285. [DOI] [PubMed] [Google Scholar]
- 3.Elomaa, O., K. Pulkkinen, U. Hannelius, M. Mikkola, U. Saarialho-Kere, and J. Kere. 2001. Ectodysplasin is released by proteolytic shedding and binds to the EDAR protein. Hum. Mol. Genet. 10:953-962. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson, B. M., N. Brockdorff, E. Formstone, T. Ngyuen, J. E. Kronmiller, and J. Zonana. 1997. Cloning of Tabby, the murine homolog of the human EDA gene: evidence for a membrane-associated protein with a short collagenous domain. Hum. Mol. Genet. 6:1589-1594. [DOI] [PubMed] [Google Scholar]
- 5.Filvaroff, E. H., S. Guillet, C. Zlot, M. Bao, G. Ingle, H. Steinmetz, J. Hoeffel, S. Bunting, J. Ross, R. A. Carano, L. Powell-Braxton, G. F. Wagner, R. Eckert, M. E. Gerritsen, and D. M. French. 2002. Stanniocalcin 1 alters muscle and bone structure and function in transgenic mice. Endocrinology 143:3681-3690. [DOI] [PubMed] [Google Scholar]
- 6.Gaide, O., and P. Schneider. 2003. Permanent correction of an inherited ectodermal dysplasia with recombinant EDA. Nat. Med. 9:614-618. [DOI] [PubMed] [Google Scholar]
- 7.Headon, D. J., S. A. Emmal, B. M. Ferguson, A. S. Tucker, M. J. Justice, P. T. Sharpe, J. Zonana, and P. A. Overbeek. 2001. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature 414:913-916. [DOI] [PubMed] [Google Scholar]
- 8.Headon, D. J., and P. A. Overbeek. 1999. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat. Genet. 22:370-374. [DOI] [PubMed] [Google Scholar]
- 9.Holcomb, I. N., R. C. Kabakoff, B. Chan, T. W. Baker, A. Gurney, W. Henzel, C. Nelson, H. B. Lowman, B. D. Wright, N. J. Skelton, G. D. Frantz, D. B. Tumas, F. V. Peale, Jr., D. L. Shelton, and C. C. Hebert. 2000. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 19:4046-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin, M., and M. Delhase. 2000. The IκB kinase (IKK) and NF-κB: key elements of proinflammatory signalling. Semin. Immunol. 12:85-98. [DOI] [PubMed] [Google Scholar]
- 11.Kere, J., A. K. Srivastava, O. Montonen, J. Zonana, N. Thomas, B. Ferguson, F. Munoz, D. Morgan, A. Clarke, P. Baybayan, E. Y. Chen, S. Ezer, U. Saarialho-Kere, A. de la Chapelle, and D. Schlessinger. 1996. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat. Genet. 13:409-416. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, M., L. A. Tartaglia, A. Lee, G. L. Bennett, G. C. Rice, G. H. Wong, E. Y. Chen, and D. V. Goeddel. 1991. Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species specific. Proc. Natl. Acad. Sci. USA 88:2830-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monreal, A. W., B. M. Ferguson, D. J. Headon, S. L. Street, P. A. Overbeek, and J. Zonana. 1999. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat. Genet. 22:366-369. [DOI] [PubMed] [Google Scholar]
- 14.Mustonen, T., J. Pispa, M. L. Mikkola, M. Pummila, A. T. Kangas, L. Pakkasjarvi, R. Jaatinen, and I. Thesleff. 2003. Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev. Biol. 259:123-136. [DOI] [PubMed] [Google Scholar]
- 15.Naito, A., H. Yoshida, E. Nishioka, M. Satoh, S. Azuma, T. Yamamoto, S. Nishikawa, and J. Inoue. 2002. TRAF6-deficient mice display hypohidrotic ectodermal dysplasia. Proc. Natl. Acad. Sci. USA 99:8766-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paakkonen, K., S. Cambiaghi, G. Novelli, L. V. Ouzts, M. Penttinen, J. Kere, and A. K. Srivastava. 2001. The mutation spectrum of the EDA gene in X-linked anhidrotic ectodermal dysplasia. Hum. Mutat. 17:349. [DOI] [PubMed] [Google Scholar]
- 17.Schneider, P., S. L. Street, O. Gaide, S. Hertig, A. Tardivel, J. Tschopp, L. Runkel, K. Alevizopoulos, B. M. Ferguson, and J. Zonana. 2001. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J. Biol. Chem. 276:18819-18827. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava, A. K., M. C. Durmowicz, A. J. Hartung, J. Hudson, L. V. Ouzts, D. M. Donovan, C. Y. Cui, and D. Schlessinger. 2001. Ectodysplasin-A1 is sufficient to rescue both hair growth and sweat glands in Tabby mice. Hum. Mol. Genet. 10:2973-2981. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava, A. K., J. Pispa, A. J. Hartung, Y. Du, S. Ezer, T. Jenks, T. Shimada, M. Pekkanen, M. L. Mikkola, M. S. Ko, I. Thesleff, J. Kere, and D. Schlessinger. 1997. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc. Natl. Acad. Sci. USA 94:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie, J., M. Murone, S. M. Luoh, A. Ryan, Q. Gu, C. Zhang, J. M. Bonifas, C. W. Lam, M. Hynes, A. Goddard, A. Rosenthal, E. H. Epstein, Jr., and F. J. de Sauvage. 1998. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature 391:90-92. [DOI] [PubMed] [Google Scholar]
- 21.Yan, M., L. C. Wang, S. G. Hymowitz, S. Schilbach, J. Lee, A. Goddard, A. M. de Vos, W. Q. Gao, and V. M. Dixit. 2000. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science 290:523-527. [DOI] [PubMed] [Google Scholar]
- 22.Yan, M., Z. Zhang, J. R. Brady, S. Schilbach, W. J. Fairbrother, and V. M. Dixit. 2002. Identification of a novel death domain-containing adaptor molecule for ectodysplasin-A receptor that is mutated in crinkled mice. Curr. Biol. 12:409-413. [DOI] [PubMed] [Google Scholar]