Abstract
Saccharomyces cerevisiae snR30 is an essential box H/ACA small nucleolar RNA (snoRNA) required for the processing of 18S rRNA. Here, we show that the previously characterized human, reptilian, amphibian, and fish U17 snoRNAs represent the vertebrate homologues of yeast snR30. We also demonstrate that U17/snR30 is present in the fission yeast Schizosaccharomyces pombe and the unicellular ciliated protozoan Tetrahymena thermophila. Evolutionary comparison revealed that the 3′-terminal hairpins of U17/snR30 snoRNAs contain two highly conserved sequence motifs, the m1 (AUAUUCCUA) and m2 (AAACCAU) elements. Mutation analysis of yeast snR30 demonstrated that the m1 and m2 elements are essential for early cleavages of the 35S pre-rRNA and, consequently, for the production of mature 18S rRNA. The m1 and m2 motifs occupy the opposite strands of an internal loop structure, and they are located invariantly 7 nucleotides upstream from the ACA box of U17/snR30 snoRNAs. U17/snR30 is the first identified box H/ACA snoRNA that possesses an evolutionarily conserved role in the nucleolytic processing of eukaryotic pre-rRNA.
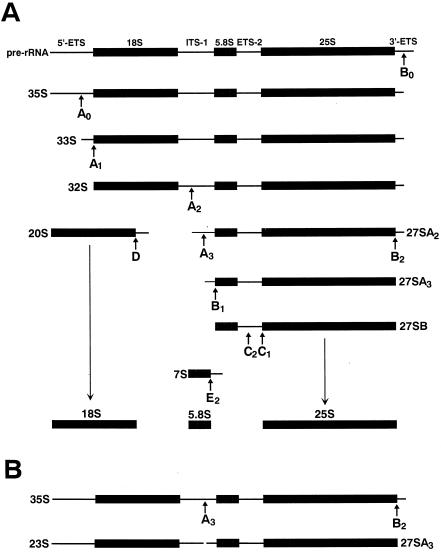
In eukaryotes, biogenesis of cytoplasmic ribosomes is a complex process that takes place mainly in the nucleolus (12, 27). The RNA polymerase I synthesizes the 18S, 5.8S, and 25/28S rRNAs within a large precursor rRNA (pre-rRNA) that also contains long externally (5′ ETS and 3′ ETS) and internally (ITS-1 and ITS-2) transcribed spacers (Fig. 1A). To release mature-size 18S, 5.8S, and 25/28S rRNAs, the noncoding spacer regions are removed by an ordered series of endo- and exonucleolytic cleavages. During processing, the maturing rRNAs associate with 80 ribosomal proteins to form the 40S and 60S ribosomal subunits. Maturation of rRNAs and their assembly into functional ribosomes require about 150 trans-acting protein and about the same number of small nucleolar RNA (snoRNA) factors (12, 27, 52).
FIG. 1.
Processing of pre-rRNA in yeast S. cerevisiae. (A) The major processing pathway of pre-rRNA. The primary rRNA transcript (pre-rRNA) contains externally (ETS) and internally (ITS) transcribed spacers. Arrows, sites of endo- and exonucleolytic cleavage (12, 27, 52). (B) Processing of 35S pre-rRNA in snR30-depleted yeast cells.
The snoRNAs, with the sole exception of 7-2/MRP, fall into two distinct families which contain either the evolutionarily conserved C (UGAUGA) and D (CUGA) or H (ANANNA) and ACA box motifs, which are essential for the expression, nucleolar accumulation, and function of snoRNAs (13, 24, 47, 49, 54). The two classes of snoRNAs are associated with two distinct sets of proteins to form small nucleolar ribonucleoproteins (snoRNPs). The box C/D snoRNAs are associated with the Snu13p (15.5-kDa), Nop56p, Nop58p, and Nop1p/fibrillarin snoRNP proteins; the box H/ACA snoRNAs are complexed with Gar1p, Nhp2p, Nop10p, and Cbf5p/dyskerin. The box C/D snoRNPs are responsible for the site-specific synthesis of the numerous 2′-O-methylated nucleotides in rRNAs, while the box H/ACA snoRNPs synthesize ribosomal pseudouridines (13, 24, 25, 47). The snoRNAs specify the substrate nucleotides through forming transient base pairing interactions with the appropriate rRNA sequences. The 2′-O-methyltransferase (Nop1p/fibrillarin) and pseudouridine synthase (Cbf5p/dyskerin) enzymes are found among the snoRNP proteins.
A few box C/D and H/ACA snoRNPs, however, function in the nucleolytic processing of rRNAs. These snoRNPs, in contrast to the modification guide snoRNPs, are usually essential for cell viability (34, 49). The most extensively studied processing snoRNP, the U3 box C/D snoRNP, functions in the early cleavages of pre-rRNA at the A0, A1, and A2 sites (Fig. 1A). In the absence of U3, accumulation of mature 18S rRNA is inhibited both in Saccharomyces cerevisiae cells and Xenopus oocytes (21, 23, 37, 43). Likewise, depletion of yeast and Xenopus U14 (29, 30) or Xenopus U22 (51) box C/D snoRNAs abolishes 18S rRNA production. Two other snoRNAs, the Xenopus U8 box C/D (39) and the yeast 7-2/MRP (33) snoRNAs, were demonstrated to function in the processing of 5.8S and 25S rRNAs. With the exception of 7-2/MRP, which is an RNase P-related endoribonuclease (34, 49), none of the processing snoRNPs have been demonstrated to possess ribonucleolytic activities. The function that box C/D snoRNAs have in rRNA processing is underlined by formation of transient base pairing interactions with pre-rRNA sequences. These snoRNAs likely function as chaperones. They prevent premature or incorrect folding events and/or facilitate the formation of pre-rRNA structures competent for nucleolytic processing (3, 4, 20, 31, 38, 45).
Less is known about the function that box H/ACA snoRNPs have in rRNA processing. Thus far, it has only been unambiguously demonstrated that two box H/ACA snoRNAs, yeast snR10 and snR30, participate in rRNA processing. While yeast cells lacking snR10 are only slightly impaired in cell growth and in processing of 35S pre-rRNA, snR30 is essential for cell viability (2, 48). In the absence of snR30, cleavages of 35S pre-rRNA at the A0, A1, and A2 processing sites are inhibited, and, therefore, accumulation of mature 18S rRNA and its immediate precursor, the 20S pre-rRNA, is abolished (Fig. 1A) (36). Instead, the 35S pre-RNA is cleaved into 23S and 27SA3 products (Fig. 1B). While the 27SA3 RNA is processed into mature 5.8S and 25S rRNAs, the aberrant 23S product, encompassing the 18S rRNA, is rapidly degraded (36). In vitro processing studies also implicated the vertebrate U17/E1, U19/E2, and E3 box H/ACA snoRNAs in the early A0 cleavage of pre-rRNA (11, 35). However, these observations have been called into question by the finding that at least two of these snoRNAs, U19/E2 and E3, are genuine pseudouridylation guide snoRNAs that function in 28S modification (6, 14).
The molecular basis of the function of yeast snR30 in rRNA processing is unknown, and, so far, no functional homologues of snR30 have been identified in other organisms. In this study, we undertook a detailed structural and functional analysis of yeast snR30 to determine the sequence and structural elements supporting its function. We show that snR30 is an evolutionarily conserved snoRNA that is present in vertebrates, the fission yeast Schizosaccharomyces pombe, and the ciliate Tetrahymena thermophila. We demonstrate that all snR30 RNAs contain two short, highly conserved sequence motifs that are essential for the production of 18S rRNA.
MATERIALS AND METHODS
General procedures.
Standard manipulations of DNA, RNA, and oligodeoxynucleotides, as well as Escherichia coli and Saccharomyces cerevisiae, were performed as described previously (42, 46). The identities of all constructs were confirmed by sequence analyses. Yeast strain D190 (MATa ura3-52 leu2-3 ade1-100 his4-519 URA3-GAL10::snR30) was kindly provided by D. Tollervey (University of Edinburgh, Edinburgh, United Kingdom). The GAL::snR30 yeast strain (MATa ade2 leu2-3 ilv1 met8 lys2 Trp1Δ his3Δ URA3-GAL10::snR30) was obtained by crossing strains D190 and 578.SC (MATα ade2 ilv1 leu2-3 met8 Trp1Δ his3Δ ura3-53 lys2) (kindly provided by J.-P. Gélugne, Laboratoire de Biologie Moléculaire Eucaryote du Centre National de la Recherche Scientifique, Toulouse, France). Expression plasmids were introduced into yeast cells by the lithium acetate transformation procedure (22). The following oligodeoxynucleotides were used in this study: 1, CAACCTGCAGACATC; 2, TCGAGATGTCTGCAGGTTGGTAC; 3, CTCGGTACCAACCATAGTCTCGTG; 4, CGTCTGCAGTATGGTTTTACC; 5, TGTGTGCATTTCTTGCTATTGGACGAGTTTAACTTAGATT; 6, CGTCCTACCGCAGTATCTTTCGAACACTATGAAATGACCC; 7, GTTTAGGAATATACTGCGGCTTAATCTCTTCCAGCCATTTGCGCTT; 8, AGATCATGCGTCCTACTTAATCTCTTCCAGCCATTTGCGCT; 9, ATAGGTACCATCATTCAATAAACTGA; 10, CCCGTTGACGAGTTTAACTTAGATTAAGCCGCAGTATATTCCTA; 11, CCCGTTGACGAGTTTAACTTCGCTTAAGCCGCGTATATT; 12, TTGCATGCCTGCAGTAGATCGTTACCCAAATGATCATGG; 13, ATAGGTACCATCATGAATCTGAAGTTAC; 14, ATACTCGAGAAATGTCAACTGTATGGTT; 15, AGATGTCTGCAGTATGGTTT; 16, TTGTGTCTGGACCTGGTGAG; 17, ACACCCTCTATGTCTCTTCAC; 18, TTAAGCGCAGGCCCGGCTGG; 19, NNTGTATATTCCTANP.
Plasmid constructs.
Construction of the pFL45/SNR yeast expression vector, containing the promoter and terminator regions of the yeast SNR5 gene, has been described (5). To facilitate the cloning of full-length snR30, oligonucleotides 1 and 2 were annealed and inserted into the KpnI and XhoI sites of pFL45/SNR. The resulting pFL45/SNRP construct carried 12 3′-terminal nucleotides of snR30, including the PstI site at position 601. The coding region of the yeast SNR30 gene from position 1 to 602 was amplified by PCR by using yeast genomic DNA as a template and oligonucleotides 3 and 4 as 5′- and 3′-specific primers, respectively. The resulting fragment was digested with KpnI and PstI and inserted into the same sites of the pBluescript (Stratagene) cloning vector, resulting in pBS-R30. To obtain pFL45/SNR/R30, the KpnI and PstI fragment of pBS-R30 was inserted into the same sites of pFL45/SNRP. Construction of pFL45/SNR/R30-d5′ and pFL45/SNR/R30m1 was performed by a two-step PCR using pBS-R30 as a template. The 3′ half of snR30 was amplified with oligonucleotides 5 (R30-d5′) and 6 (R30m1) as 5′-end-specific primers and oligonucleotide 4 as a common 3′-specific primer. The resulting PCR products were used as megaprimers in the second PCR together with oligonucleotide 3 as a 5′-specific primer. The amplified fragments were digested with KpnI and PstI and inserted into the same sites of pFL45/SNRP. A similar approach was used to construct pFL45/SNR/R5-R30S and pFL45/SNR/R5-R30L except that, in the first PCR, the 5′ half of snR5 RNA was amplified with oligonucleotides 7 (R5-R30S) and 8 (R5-R30L) as 3′-specific primers, with oligonucleotide 9 as a 5′-specific primer and yeast genomic DNA as a template. The PCR products were used as megaprimers in the second amplification reaction, which also contained oligonucleotide 4 as a 3′ primer and pBS-R30 as a template. The amplified fragments were cloned into the KpnI and PstI sites of pFL45/SNRP. To generate pFL45/SNR/R30-dIH and pFL45/SNR/R30-H, fragments of the SNR30 gene were amplified by using oligonucleotide 4 as a common 3′ primer and oligonucleotides 10 and 11 as 5′ primers, respectively. After digestion with HincII and PstI, the resulting PCR fragments were used to replace the HincII-PstI fragment of pFL45/SNR/R30. To construct pFL45/SNR/R30m2, the coding region of snR30 was amplified by using oligonucleotide 3 as a 5′ primer and a mutagenic 3′ primer (oligonucleotide 12). The PCR product was digested with KpnI and PstI and cloned into the same sites of pFL45/SNRP. To generate pFL45/SNR/U17pombe, the coding region of the S. pombe U17 snoRNA gene was amplified by using oligonucleotide 13 as a 5′-specific primer, oligonucleotide 14 as a 3′ primer, and genomic DNA as a template. After digestion with KpnI and XhoI, the resulting PCR fragment was cloned into pFL45/SNRP.
RNA analysis.
Cellular RNA from yeasts S. cerevisiae and S. pombe was isolated by the guanidine thiocyanate-phenol-chloroform extraction method (50). T. thermophila cellular RNA was kindly provided by K. Collins (University of California). For Northern analysis of rRNAs, total cellular RNA corresponding to 1.8 × 107 exponentially growing yeast cells was size fractionated on 1.2% agarose-formaldehyde gel. For analysis of snoRNAs, about 5 μg of total RNA was fractionated on 6% denaturing polyacrylamide gels. RNAs were electroblotted (acrylamide gels) or transferred by capillary blotting (agarose gels) onto a Hybond-N nylon membrane (Amersham Biosciences). The filters were probed with terminally labeled oligonucleotides specific for the snR30 (oligonucleotide 15), 18S (oligonucleotide 16), 25S (oligonucleotide 17), or 20S (oligonucleotide 18) RNA. RNase A/T1 mapping was described previously (17). To generate an antisense RNA probe for mapping snR30, snR30m1, and snR30m2 RNAs, the pBS/R30 recombinant plasmid was linearized by XhoI and used as a template for synthesis of an internally labeled RNA probe by the T3 RNA polymerase.
The 3′-terminal portions of S. pombe and T. thermophila U17 snoRNAs were cloned by the oligonucleotide ligation-PCR amplification procedure (26), except that a degenerate oligonucleotide (19) was used as an upstream primer. Further upstream sequences of the S. pombe and T. thermophila U17 snoRNAs were determined by dideoxy sequencing, using total RNA as a template and 5′-end-labeled oligonucleotides complementary to the 3′-terminal sequences of the S. pombe (oligonucleotide 20) and T. thermophila (oligonucleotide 21) U17 snoRNAs as primers for avian myeloblastosis virus reverse transcriptase.
RESULTS
Secondary structure of yeast snR30.
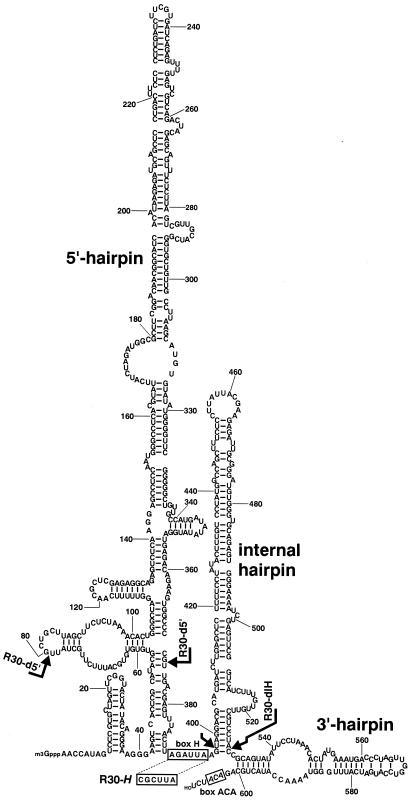
The yeast S. cerevisiae snR30 consists of 608 nucleotides and represents the longest known snoRNA. To facilitate the functional analysis of snR30, we decided to establish its secondary structure. The facts that snR30 contains an ACA triplet 3 nucleotides before its 3′ end (1) and associates with all box H/ACA snoRNPs (16, 19, 28, 53) demonstrate that it belongs to the family of box H/ACA snoRNAs. Therefore, snR30 is expected to fold into the consensus “hairpin-hinge-hairpin-tail” structure of box H/ACA snoRNAs (15). Keeping in mind that the putative box H motif (consensus ANANNA) of snR30 should occur in the single-stranded “hinge” region and should follow the 5′ hairpin of the RNA (5, 15), several alternative structures were developed by a computer folding algorithm (55). Although the sequence of snR30 contains five potential H box motifs, we found that only one located at position 390 (AGAUUA) could fully satisfy the above-described structural requirements (Fig. 2). According to the most probable structural model, snR30 consists of three major hairpin structures, a long 5′ hairpin (A42 to U389), a short 3′ hairpin (G532 to C600), and an additional, medium-size internal hairpin (G397 to C530). In fact, internal hairpins following the hinge region and preceding the 3′-terminal hairpin are frequently found in both yeast and mammalian box H/ACA snoRNAs (15). The 5′ end of snR30 commences with a 41-nucleotide leader that is again a characteristic feature of yeast box H/ACA snoRNAs transcribed from independent genes (1, 15). We propose that, in spite of extended length of yeast snR30, its secondary structure conforms well to the evolutionarily conserved consensus structure of box H/ACA snoRNAs.
FIG. 2.
Predicted two-dimensional structure of yeast snR30. The nucleotide sequence of snR30 was first published by Bally et al. (2) and later modified by Balakin et al. (1). The box ACA and the putative box H motifs are boxed. Arrows, regions deleted in R30-d5′ and R30-dIH RNAs. The nucleotide sequence of the altered box H motif of R30-H is shown.
Elements essential for cell viability are located in the 3′ hairpin of snR30.
Being armed with a secondary structure of snR30, we performed a deletion analysis to define the functionally essential elements of yeast snR30. The coding region of the SNR30 gene was inserted between the promoter and terminator of the SNR5 snoRNA gene within the pFL45/SNR expression vector (Fig. 3A) (5). The resulting expression construct, pFL45/SNR/R30, was transformed into the GAL::snR30 yeast test strain (see Materials and Methods), in which the authentic SNR30 promoter had been replaced by the GAL10 promoter. Since the GAL10 promoter is active only when cells are grown on galactose, the GAL::snR30 cells cannot grow on glucose-containing medium due to the repression of snR30 synthesis (36). However, after transformation with the pFL45/SNR/R30 expression construct, but not the pFL45/SNR vector, the transformed GAL::snR30 cells (R30) could grow on glucose (Fig. 3C). As expected, Northern blot analysis demonstrated that the R30 cells accumulated snR30 (Fig. 3D, lane 1). First, we tested the functional importance of the long 5′ hairpin of snR30. To this end, a large portion of this structural domain of snR30, from G80 to G373 (Fig. 2), was deleted and the resulting pFL45/SNR/R30-d5′ construct was transformed into the GAL::snR30 test cells. As demonstrated by Northern analysis, the truncated R30-d5′ RNA accumulated in the transformed cells (Fig. 3D, lane 4) and supported cell growth on glucose-containing medium (Fig. 3C). The same results were obtained when a shorter fragment of the 5′ hairpin (C171 to C304, C171 to G373, C123 to C170, or C123 to U175) was removed (data not shown). Likewise, removal of the internal hairpin of snR30 (from U398 to A529) had no influence on the accumulation of the resulting R30-dIH RNA (Fig. 3D, lane 5). Moreover, the accumulating R30-dIH RNA supported the growth of the resulting R30-dIH strain on glucose (Fig. 3C). These results demonstrate that the distal part of the 5′ hairpin and the internal hairpin do not contain elements essential for the accumulation and function of yeast snR30.
FIG. 3.
Determination of the functionally essential regions of yeast snR30. (A) Schematic structure of the construct used to express the wild-type and mutant versions of snR30. The coding region of the yeast SNR30 gene (open arrow) was placed between the promoter (SNR5-P) and terminator (SNR5-T) of the yeast SNR5 gene (5). Relevant restriction sites are indicated (H, HindIII; K, KpnI; X, XhoI; B, BamHI). (B) Proposed secondary structure of the snR5-snR30 chimeric RNA (R5-R30S). Nucleotides are numbered according to the wild-type snR5 and snR30 snoRNAs. The box H/ACA elements and the evolutionarily conserved m1 and m2 sequence motifs are boxed. (C) Growth properties of yeast GAL::snR30 cells transformed with the pFL45/SNR, pFL45/SNR/R30, pFL45/SNR/R30-H, pFL45/SNR/R30-dIH, pFL45/SNR/R30-d5′, pFL45/SNR/R5-R30S, or pFL45/SNR/R5-R30L expression plasmid on galactose- (GAL) and glucose-containing (GLU) solid medium. (D) Northern blot analysis of the expression of wild-type and mutant snR30 RNAs. Total RNAs extracted from GAL::snR30 cells transformed with the pFL45/SNR, pFL45/SNR/R30, pFL45/SNR/R30-H, pFL45/SNR/R30-dIH, pFL45/SNR/R30-d5′, pFL45/SNR/R5-R30S, or pFL45/SNR/R5-R30L expression construct were separated on a 6% sequencing gel, electroblotted onto a nylon membrane, and probed with a terminally labeled oligodeoxynucleotide complementary to the 18 3′-terminal nucleotides of snR30. Lane M, size markers (terminally labeled HaeIII- and TaqI-digested pBR322).
For accumulation of box H/ACA snoRNAs, the conserved A1 and A3 residues in the consensus ANANNA sequence of the H box are indispensable (15). To test whether the predicted box H motif of snR30 (390-AGAUUA-305) is essential for RNA accumulation, the A390 and A392 residues were replaced with C residues in the R30-dIH RNA gene. Indeed, the resulting mutant R30-H RNA did not accumulate in the transformed GAL::snR30 strain (Fig. 3D, compare lanes 3 and 5). Consistently, the GAL::snR30 cells carrying the pFL45/SNR/R30-H expression plasmid failed to grow on glucose (Fig. 3C). These results, besides demonstrating that the 390-AGAUUA-305 region represents the authentic box H motif of snR30, strongly support the correctness of the proposed structural model of yeast snR30.
To test whether the 5′ leader or the proximal part of the 5′ hairpin of snR30 contains functionally important elements, a chimeric snR5-snR30 (R5-R30S) RNA was created (Fig. 3B). The R5-R30S RNA contained the 5′-terminal half of the yeast snR5 box H/ACA pseudouridylation guide snoRNA (A1-G85), the hinge region (A390 to G398), and the 3′-terminal hairpin (C530 to U608) of snR30. When placed under the control of the SNR5 promoter in the pFL45/SNR expression vector, the chimeric R5-R30S RNA was efficiently expressed in the GAL::snR30 test strain (Fig. 3D, lane 6) and, more importantly, supported cell growth on glucose (Fig. 3C). However, compared to that of the control R30 strain, expressing wild-type snR30, the growth rate of the R5-R30 strain was reduced by about 40% (Fig. 3C and data not shown). Since yeast small nuclear and nucleolar RNAs are generally much longer than their vertebrate counterparts (18), we hypothesized that the decreased functional efficiency of the R5-R30S RNA might result from its reduced molecular mass rather than the lack of important elements. To test this assumption, the inserted hairpin element of snR30 that had been demonstrated to lack functionally important elements (Fig. 3C, R30-dIH) was reintroduced into the R5-R30S RNA. The resulting R5-R30L chimeric RNA, which contained the 5′-terminal region of snR5 (A1-G84) and the 3′ half of snR30 (A390 to U608), accumulated in GAL::snR30 cells (Fig. 3D, lane 7). When grown on glucose, the GAL::snR30 cells expressing R5-R30L had a growth rate that was highly comparable to that of the control R30 cells (Fig. 3C), indicating that increasing the molecular mass of R5-R30S could improve its functional efficacy. In conclusion, the results of the deletion analysis of snR30, taken together with the finding that R5-R30S chimeric RNA can support cell growth, indicate that all the elements essential for cell viability are contained within the 3′-terminal hairpin of snR30.
snR30 is a ubiquitous RNA.
The notion that the 3′-terminal hairpin of snR30 contains the functionally important elements that are likely conserved during evolution prompted us to undertake a closer inspection of the 3′ hairpins of all known box H/ACA snoRNAs. We noticed that the 3′ hairpin of the human U17 snoRNA contains two short sequence motifs, called m1 (141-AUAUUCCUA-149) and m2 (188-AAACCAU-194) (Fig. 4A), which are also present in yeast snR30 (Fig. 3B). The nucleotide sequences of U17 for a variety of vertebrate species, including four mammal, one bird, eight reptile, one amphibian, and four fish species, have been determined (7-9, 26, 41, 44). We found that perfect copies of the m1 and m2 motifs are present in the 3′-terminal hairpin of all vertebrate U17 snoRNAs. In Fig. 4A, the computer-predicted structures of the 3′-terminal hairpins of Caretta caretta (turtle), Xenopus laevis (frog), and Fugu rubripes (fish) U17 snoRNAs are shown as representative examples.
FIG. 4.
Evolutionary conservation of U17/snR30. (A) Proposed structures of the 3′-terminal hairpins of vertebrate U17 snoRNAs. Sequences (with GenBank accession numbers in parentheses) of the human U17a (26) (L16791), C. caretta U17f (9) (AJ306558), X. laevis U17f (8) (X71081), and F. rubripes U17f (7) (X94942) snoRNAs have been reported. The conserved m1 and m2 sequence motifs are boxed. The ACA boxes are underlined. (B) Structure of the 3′-terminal hairpin of putative U17 snoRNAs obtained from S. pombe (AJ544685) and T. thermophila (AJ544686). (C) Expression of S. pombe and T. thermophila U17 snoRNAs. Total cellular RNAs were separated on a 6% sequencing gel, electroblotted onto nylon membranes, and hybridized with sequence-specific oligodeoxynucleotide probes. Lane M, size markers.
To assess the evolutionary conservation of the U17 snoRNA by using a reverse transcription-PCR approach (see Materials and Methods), we identified the 3′-terminal sequences of potential homologues of U17 in the fission yeast S. pombe and the unicellular protozoan T. thermophila (Fig. 4B). Northern blot analysis demonstrated that the novel putative S. pombe and T. thermophila U17 snoRNAs are efficiently expressed and are composed of about 325 and 240 nucleotides, respectively (Fig. 4C). Since the new sequences carried an ACA motif 3 nucleotides before their 3′ termini and folded into a hairpin structure, we concluded that they represent authentic box H/ACA snoRNAs. More importantly, the putative U17 snoRNAs of S. pombe and T. thermophila contained the conserved m1 and m2 motifs, except that the T. thermophila RNA carried a G rather than an A residue in the last position of its m1 element.
Upon a detailed scrutiny of all known U17/snR30 snoRNAs, including the new S. pombe and T. thermophila RNAs, it became apparent that not only the sequences but also the positions of the m1 and m2 motifs are strictly conserved during evolution. The m1 and m2 motifs occupy the proximal (lower) part of an internal stem-loop-stem structure within the 3′ hairpins of all U17/snR30 snoRNAs (Fig. 4). While the first 2 residues of the m1 motif (AU) and the last 2 nucleotides of the m2 motif (UA) are predicted to base pair to each other, the other conserved nucleotides of the m1 and m2 motifs occupy unpaired positions in the opposite strands of the internal loop. We also noticed that the distance between the conserved m1-m2 structural motif and the ACA box of U17/snR30 snoRNAs is perfectly conserved; they are separated always by 7 nucleotides. Taken together, these observations led us to conclude that U17/snR30 is an evolutionarily conserved snoRNA that is present in many, if not all, eukaryotic organisms.
S. pombe U17 snoRNA can restore depletion of snR30 in yeast S. cerevisiae.
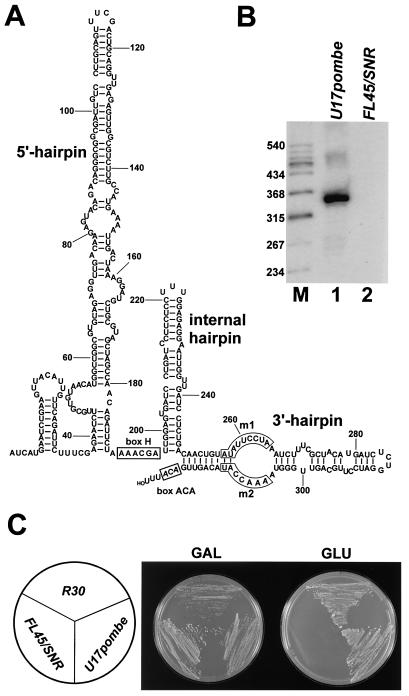
A database search revealed that the nucleotide sequence of the S. pombe U17 gene had been already determined to be part of chromosome 1 (GenBank accession number AL357232). Primer extension analysis showed that the S. pombe U17 snoRNA is composed of 325 nucleotides and, as predicted by computer modeling, it folds into a 5′-hairpin hinge-internal hairpin-3′-hairpin structure highly reminiscent of the architecture of the yeast S. cerevisiae snR30 (Fig. 5A). Inspired by the significant structural conservation of U17/snR30, we tested whether the S. pombe U17 snoRNA could restore the growth of the GAL::snR30 test strain on glucose-containing medium when accumulation of snR30 is inhibited. The coding region of the S. pombe U17 gene was amplified by PCR and inserted into the pFL45/SNR expression vector (Fig. 3A). Upon transformation of the resulting pFL45/SNR/U17pombe expression plasmid into the GAL::snR30 strain, the S. pombe U17 RNA (U17pombe) was faithfully expressed (Fig. 5B). When streaked on glucose-containing medium, GAL::snR30 cells expressing U17pombe or yeast snR30 showed similar growth properties (Fig. 5C), demonstrating that the fission yeast S. pombe U17 snoRNA can fully restore the function of snR30 in S. cerevisiae. In a similar experiment, the human U17a snoRNA, though it accumulated efficiently, failed to restore the growth of GAL::snR30 cells on glucose (data not shown; see also Discussion).
FIG. 5.
Functional characterization of S. pombe U17 snoRNA. (A) Proposed secondary structure of S. pombe U17. The conserved sequence elements are boxed. (B) Northern analysis. Total RNAs extracted from GAL::snR30 cells transformed with either the pFL45/SNR (FL45/SNR) or the pFL45/SNR/U17pombe (U17pombe) expression vector were separated on a 6% sequencing gel, transferred onto a nylon membrane, and probed with an oligonucleotide probe specific for S. pombe U17. Lane M, size markers. (C) Growth properties of yeast FL45/SNR, R30, and U17pombe strains on galactose- (GAL) and glucose-containing (GLU) media.
The m1 and m2 elements of snR30 are required for 18S production.
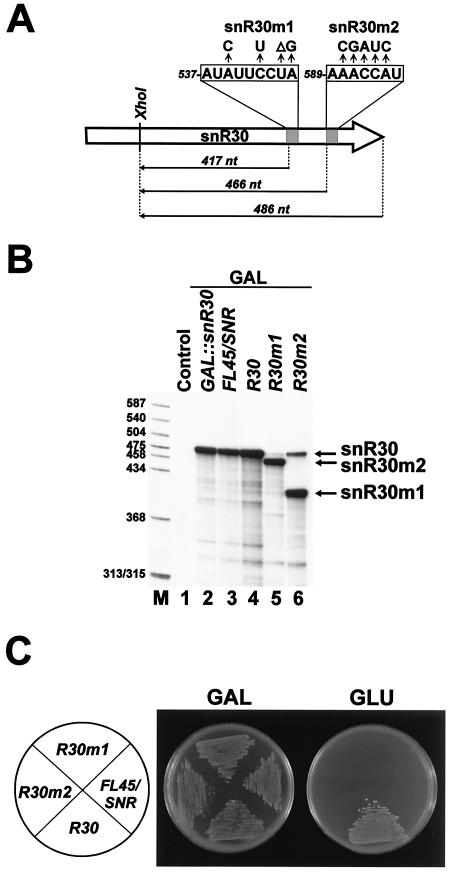
To assess the functional importance of the conserved m1 and m2 elements, two mutant versions of yeast snR30, snR30m1 and snR30m2 (Fig. 6A), carrying altered m1 and m2 motifs, respectively, were expressed in the GAL::snR30 test strain with the help of the pFL45/SNR expression vector. Accumulation of snR30m1 and snR30m2 was demonstrated by RNase A/T1 mapping by using an antisense RNA probe complementary to the wild-type snR30 (Fig. 6A and B, lanes 5 and 6). In contrast to the efficient expression of snR30m1 and snR30m2, neither the R30m1 nor the R30m2 cells could grow on glucose-containing medium (Fig. 6C), demonstrating that the m1 and m2 motifs of snR30, although not required for RNA accumulation, are essential for cell viability.
FIG. 6.
The conserved m1 and m2 elements of snR30 are essential for cell viability. (A) Schematic structure of the mutant snR30m1 and snR30m2 RNAs. The wild-type (boxed) and mutant sequences of the m1 and m2 motifs of snR30 are indicated. The predicted lengths of antisense RNA probes protected by snR30m1 (417 nucleotides [nt]), snR30m2 (466 nt), and snR30 (486 nt) are indicated. (B) RNase A/T1 mapping. RNAs obtained from the GAL::snR30, FL45/SNR, R30, R30m1, and R30m2 strains were annealed with an internally labeled antisense RNA probe complementary to the 3′-terminal part of snR30. Upon incubation with a mixture of RNase A and T1, protected RNAs were separated on a 6% denaturing polyacrylamide gel. The control lane shows a mapping reaction with E. coli RNA. Lane M, size markers. (C) Growth of yeast FL45/SNR, R30, R30m1, and R30m2 strains on galactose- (GAL) and glucose-containing (GLU) media.
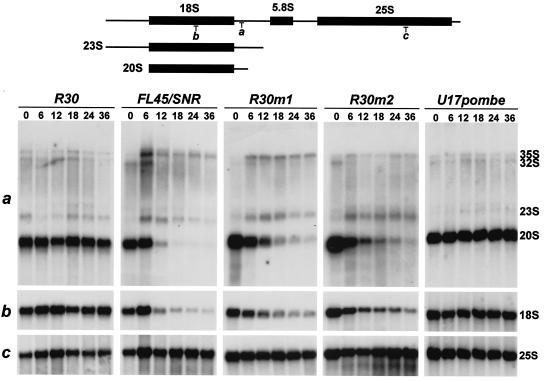
Genetic depletion of yeast snR30 disrupts cleavages of the 35S pre-rRNA at the A0, A1, and A2 processing sites and inhibits accumulation of the 20S pre-rRNA and mature 18S rRNA (36) (Fig. 1A). Instead, in the absence of snR30, processing of the 35S pre-rRNA at the A3 site results in an aberrant 23S processing intermediate that is not utilized as a substrate by the processing machinery (Fig. 1B). To test whether the critical role played by the m1 and m2 motifs of snR30 in cell viability is connected to 18S production, the processing of pre-rRNA in the R30m1 and R30m2 strains was investigated. After 0, 6, 12, 18, 24, and 36 h of growth on glucose, RNA was extracted from both strains and accumulation of the mutant snR30m1 and snR30m2 RNAs and depletion of snR30 were verified by RNase mapping (data not shown). As a negative control, we used the FL45/SNR strain, which does not express snR30 on glucose. The R30 strain that constitutively expresses snR30 was utilized as a positive control. In each cell line, the steady-state levels of the mature 18S and 25S rRNAs and their precursors were assessed by Northern blot analysis with specific oligonucleotides (a, b, and c) depicted in Fig. 7. Hybridization of the rRNA blots with the 18S-specific oligonucleotide b probe demonstrated that depletion of snR30 in the R30m1 and R30m2 strains during growth on glucose inhibited 18S production. Similar results were obtained when 18S accumulation in the FL45/SNR control cells was monitored. But it is noteworthy that in the FL45/SNR cells, probably due to the complete lack of snR30, the level of 18S rRNA fell much faster upon growth on glucose than in the test strains accumulating snR30m1 or snR30m2. Probing the same blots with oligonucleotide a, complementary to the 5′-terminal part of ITS-1, demonstrated that in the R30m1 and R30m2 strains, just like in the control FL45/SNR strain, accumulation of the 20S precursor of 18S was abolished. In contrast, accumulation of mature 25S rRNA was not affected in any of the observed cell lines (oligonucleotide c), supporting the previous conclusion that snR30 is not involved in 25S production (36).
FIG. 7.
Alteration of the m1 and m2 motifs of snR30 disrupts the production of 18S rRNA. The steady-state levels of the 20S and 23S processing intermediates and the mature 18S and 25S rRNAs were determined by Northern analyses. Total RNA isolated from the R30, FL45/SNR, R30m1, R30m2, and U17pombe cells grown for 0, 6, 12, 18, 24, and 36 h on glucose was separated on agarose-formaldehyde gels and transferred onto nylon membranes. The RNA blots were probed with terminally labeled oligonucleotides complementary to 18S (b), 25S (c), or ITS-1 (a) sequences.
We also noticed that, in the R30m1 and R30m2 cells, there were significant and reproducible differences in the levels of accumulation of some rRNA processing intermediates. For example, an elevated 35S accumulation was observed in the R30m1 strain, compared to accumulation in the R30m2 and control FL45/SNR cells. In turn, the 23S aberrant processing product accumulated more efficiently in the R30m2 strain. The observed differences in the pre-rRNA processing pathways of the R30m1 and R30m2 cells might indicate that the m1 and m2 motifs of snR30 possess different functions. Finally, Northern blot analysis of RNAs extracted from GAL::snR30 cells transformed with pFL45/SNR/U17pombe expression plasmid demonstrated that the S. pombe U17 snoRNA can restore the normal processing of rRNAs in S. cerevisiae cells depleted of snR30. In summary, our results demonstrate that U17/snR30 is an ubiquitous box H/ACA snoRNA that carries two evolutionarily highly conserved sequence elements, m1 and m2, which are essential for the nucleolytic processing of 18S rRNA.
DISCUSSION
Maturation of eukaryotic rRNAs is assisted by a large number of snoRNAs that either direct the posttranscriptional 2′-O-methylation and pseudouridylation of rRNAs or, less frequently, function in the nucleolytic processing of pre-rRNA. While the molecular function of modification guide snoRNAs is well understood, much less is known about the role that snoRNAs play in rRNA processing. The yeast snR30 is an essential box H/ACA snoRNA that is required for the nucleolytic processing of 35S pre-rRNA at the A0, A1, and A2 sites (36). In this study, we have demonstrated that snR30 is an evolutionarily conserved snoRNA that is also present in all vertebrates, the fission yeast S. pombe, and the unicellular ciliated protozoan T. thermophila. More recently, we have identified a potential plant homologue of snR30 in Medicago truncatula (our unpublished results), further supporting the notion that U17/snR30 is a ubiquitous snoRNA that is present in most, if not all, eukaryotic organisms.
Secondary structure modeling demonstrated that all U17/snR30 snoRNAs fold into the consensus hairpin-hinge-hairpin-tail structure of box H/ACA snoRNAs, except that they carry an additional internal hairpin located before the 3′-terminal hairpin of the RNA (Fig. 2, 4, and 5 and data not shown). As indicated by deletion analysis of yeast snR30, neither the 5′-terminal nor the internal hairpin of U17/snR30 contains functionally important elements (Fig. 3). Consistent with this notion, both the nucleotide compositions and sizes of these hairpins show a high level of variation during evolution. Expression of an snR5-snR30 chimeric RNA (R5-R30S) demonstrated that the 3′-terminal hairpin of snR30 carries all the elements that are critical for cell viability (Fig. 3). The 3′ hairpin of each U17/snR30 RNA carries the conserved m1 (AUAUUCCUA) and m2 (AAACCAU) sequence elements, which occupy an invariant position in the proximal (lower) part of an internal loop, exactly 7 nucleotides upstream to the ACA box of the RNA (Fig. 4). Mutational analyses demonstrated that the m1 and m2 motifs are crucial for cell viability and 18S production (Fig. 6 and 7). Apart from the essential m1 and m2 elements, the nucleotide sequence of the 3′-terminal hairpin of U17/snR30 RNAs shows no significant conservation, indicating that it contains no additional functionally important sequence elements.
One possibility is that the m1 and m2 motifs might function as specific protein-binding signals. Besides the Cbf5p/dyskerin (65 kDa), Gar1p (25 kDa), Nhp2p (22 kDa), and Nop10p (10 kDa) box H/ACA snoRNP core proteins (1, 16, 19, 53), purified yeast snR30 snoRNP was reported to be associated with three additional proteins of 36, 46, and 48 kDa (32). However, in vitro reconstitution experiments performed with the human U17 snoRNA could detect only four U17-associated proteins, which, based on their apparent molecular masses, likely represent the human Cbf5p/dyskerin (60 kDa), Gar1p (29 kDa), Nhp2p (23 kDa), and Nop10p (14 kDa) box H/ACA core proteins (10). Notably, in vitro reconstitution studies also revealed that the 3′-terminal hairpin of U17 could be packaged into a stable snoRNP particle (10). In marked contrast, full-length snoRNAs were required for the assembly of the human U64 and U19 box H/ACA pseudouridylation guide snoRNPs. This observation strongly suggests that the 3′-terminal hairpin of U17 has specific structural and functional features which distinguish this RNA from the authentic pseudouridylation guide snoRNAs. Apparently, further experiments are required to clarify whether the m1 and m2 motifs of U17/snR30 can bind specific proteins.
The high degree of evolutionary conservation of the m1 and m2 motifs might also indicate that they represent antisense sequence recognition elements. In fact, the overwhelming majority of box H/ACA snoRNAs function as guide RNAs which select ribosomal pseudouridylation sites by forming transient base pairing interactions with complementary rRNA sequences. However, the fact that the m1 and m2 elements occupy the proximal part of the putative pseudouridylation loop of U17/snR30 rather than its distal part, as would have been expected for a genuine pseudouridylation guide RNA (14), strongly argues against a modification guide function for U17/snR30. Moreover, human and yeast rRNAs lack sequences with known pseudouridylation sites that could form a canonical rRNA-guide RNA interaction with U17/snR30. At the moment, however, we cannot unambiguously rule out the formal possibility that U17/snR30 directs pseudouridylation of another, not yet identified RNA that plays a crucial role in pre-rRNA processing. Currently, we are testing whether the mouse U17 snoRNA is capable of directing pseudouridylation of appropriately designed artificial substrate RNAs expressed in the nucleoli of mouse cells.
According to another, more likely scenario, U17/snR30 might function as a molecular chaperone that facilitates the correct folding of pre-rRNAs, as has been proposed for the U3, U14, and U8 box C/D snoRNAs (see the introduction). Previous psoralen photo-cross-linking experiments found the human U17 (E1) and yeast snR30 snoRNAs to be associated with large pre-rRNAs (36, 40). Although vertebrate and yeast pre-rRNAs lack perfect, evolutionarily conserved complementarities to the m1 and m2 motifs of U17/snR30, numerous shorter, either perfect or imperfect base pairing interactions could be formed between the m1 and m2 motifs of U17/snR30 and pre-rRNA sequences (data not shown). This also implies that the m1 and m2 elements might form functionally important hydrogen bonding interactions with distinct regions of the pre-rRNA. Notably, alteration of the m1 or the m2 motif of snR30 gave rise to slightly different pre-rRNA processing pathways (Fig. 7). In the absence of an intact m1 motif, processing of 35S pre-rRNA is delayed, while alteration of the m2 motif results in an increased accumulation of the aberrant 23S processing intermediate. Moreover, we have recently found that the viability of yeast cells expressing snR30m2, but not snR30m1, could be restored by coexpression of the human U17a snoRNA (our unpublished data). This observation, besides demonstrating that the human U17 snoRNA is indeed the functional homologue of yeast snR30, provides further support for the assumption that the m1 and m2 elements might work, at least to some extent, independently from one another. To identify the putative sites of interaction between U17/snR30 and pre-rRNA, we are currently performing a detailed cross-linking analyses of human and yeast pre-rRNAs.
Thus far, most box C/D and H/ACA snoRNAs implicated in rRNA processing have been assumed to be unique to either vertebrates (U8 and U22) or the yeast S. cerevisiae (snR30 and snR10). Only the U3 and U14 box C/D snoRNAs have been found in both yeast and metazoan cells. Demonstration that the U17/snR30 box H/ACA snoRNA is also present in vertebrates, the fission yeast S. pombe, and the protozoan T. thermophila further supports the idea that the snoRNA-assisted processing mechanism of eukaryotic pre-rRNAs has an ancient evolutionary origin. More importantly, determination of the functionally essential elements of U17/snR30 will greatly facilitate the future dissection of the molecular mechanism underlying the function of this snoRNA in pre-rRNA processing.
Acknowledgments
We thank Y. de Préval for synthesis of oligonucleotides. We are grateful to D. Tollervey, J.-P. Gélugne, and K. Collins for providing us with yeast strains D190 and 578.SC and Tetrahymena cellular RNA, respectively. We thank Y. Henry and J.-P. Gélugne for helpful discussions.
V.A. was funded by the Deutscher Akademischer Austauschdienst and Fondation pour la Recherche Médicale. P.F. was funded by CNR-NATO and CNRS. Our work was supported by grants from la Ligue Nationale contre le Cancer, Association pour le Recherche contre le Cancer, and MURST (RBNE015MPB and RBNE01KXC9).
REFERENCES
- 1.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 2.Bally, M., J. Hughes, and G. Cesareni. 1988. SnR30: a new, essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 16:5291-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrame, M., and D. Tollervey. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borovjagin, A. V., and S. A. Gerbi. 2000. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 300:57-74. [DOI] [PubMed] [Google Scholar]
- 5.Bortolin, M. L., P. Ganot, and T. Kiss. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 18:457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bortolin, M. L., and T. Kiss. 1998. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4:445-454. [PMC free article] [PubMed] [Google Scholar]
- 7.Cecconi, F., C. Crosio, P. Mariottini, G. Cesareni, M. Giorgi, S. Brenner, and F. Amaldi. 1996. A functional role for some Fugu introns larger than the typical short ones: the example of the gene coding for ribosomal protein S7 and snoRNA U17. Nucleic Acids Res. 24:3167-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecconi, F., P. Mariottini, F. Loreni, P. Pierandrei-Amaldi, N. Campioni, and F. Amaldi. 1994. U17XS8, a small nucleolar RNA with a 12 nt complementarity to 18S rRNA and coded by a sequence repeated in the six introns of Xenopus laevis ribosomal protein S8 gene. Nucleic Acids Res. 22:732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervelli, M., F. Cecconi, M. Giorgi, F. Annesi, M. Oliverio, and P. Mariottini. 2002. Comparative structure analysis of vertebrate U17 small nucleolar RNA (snoRNA). J. Mol. Evol. 54:166-179. [DOI] [PubMed] [Google Scholar]
- 10.Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, C. A., E. S. Maxwell, G. L. Eliceiri, and B. Sollner-Webb. 1996. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 2:1094-1099. [PMC free article] [PubMed] [Google Scholar]
- 12.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 13.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 14.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 15.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 16.Girard, J. P., H. Lehtonen, M. Caizergues-Ferrer, F. Amalric, D. Tollervey, and B. Lapeyre. 1992. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 11:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodall, G. J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148-161. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie, C., and B. Patterson. 1988. Spliceosomal snRNAs. Annu. Rev. Genet. 22:387-419. [DOI] [PubMed] [Google Scholar]
- 19.Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J. P. Gelugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes, J. M. 1996. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 259:645-654. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, J. M., and M. Ares, Jr. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kass, S., K. Tyc, J. A. Steitz, and B. Sollner-Webb. 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60:897-908. [DOI] [PubMed] [Google Scholar]
- 24.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 26.Kiss, T., and W. Filipowicz. 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 12:2913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafontaine, D. L., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange, T. S., A. Borovjagin, E. S. Maxwell, and S. A. Gerbi. 1998. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 17:3176-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H. D., J. Zagorski, and M. J. Fournier. 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, W. Q., and M. J. Fournier. 1995. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 9:2433-2443. [DOI] [PubMed] [Google Scholar]
- 32.Lubben, B., P. Fabrizio, B. Kastner, and R. Luhrmann. 1995. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem. 270:11549-11554. [DOI] [PubMed] [Google Scholar]
- 33.Lygerou, Z., C. Allmang, D. Tollervey, and B. Seraphin. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268-270. [DOI] [PubMed] [Google Scholar]
- 34.Maxwell, E. S., and M. J. Fournier. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897-934. [DOI] [PubMed] [Google Scholar]
- 35.Mishra, R. K., and G. L. Eliceiri. 1997. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc. Natl. Acad. Sci. USA 94:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 13:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mougey, E. B., L. K. Pape, and B. Sollner-Webb. 1993. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5′ external transcribed spacer of Xenopus precursor rRNA. Mol. Cell. Biol. 13:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peculis, B. A. 1997. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 17:3702-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peculis, B. A., and J. A. Steitz. 1993. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73:1233-1245. [DOI] [PubMed] [Google Scholar]
- 40.Rimoldi, O. J., B. Raghu, M. K. Nag, and G. L. Eliceiri. 1993. Three new small nucleolar RNAs that are psoralen cross-linked in vivo to unique regions of pre-rRNA. Mol. Cell. Biol. 13:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruff, E. A., O. J. Rimoldi, B. Raghu, and G. L. Eliceiri. 1993. Three small nucleolar RNAs of unique nucleotide sequences. Proc. Natl. Acad. Sci. USA 90:635-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Savino, R., and S. A. Gerbi. 1990. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 9:2299-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selvamurugan, N., O. H. Joost, E. S. Haas, J. W. Brown, N. J. Galvin, and G. L. Eliceiri. 1997. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 25:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma, K., and D. Tollervey. 1999. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 19:6012-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-23. [DOI] [PubMed] [Google Scholar]
- 47.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10:17-39. [PMC free article] [PubMed] [Google Scholar]
- 48.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 50.Tollervey, D., and I. W. Mattaj. 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 6:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tycowski, K. T., M. D. Shu, and J. A. Steitz. 1994. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 266:1558-1561. [DOI] [PubMed] [Google Scholar]
- 52.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 53.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, Y. T., E. C. Scharl, S. C. M., and J. A. Steitz. 1999. The growing world of small nuclear ribonucleoproteins, p. 487-524. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.