Abstract
Ectopically expressed Cdc6 is translocated from the nucleus during S phase in a cyclin A-Cdk2-dependent process, suggesting that reinitiation of DNA replication is prevented by removal of phosphorylated Cdc6 from chromatin after origin firing. However, whether endogenous Cdc6 translocates during S phase remains controversial. To resolve the questions regarding regulation of endogenous Cdc6, we cloned the cDNA encoding the Chinese hamster Cdc6 homolog and specifically focused on analyzing the localizations and chromatin affinities of endogenous and exogenous proteins during S phase and following overexpression of cyclin A. In agreement with other reports, ectopically expressed Cdc6 translocates from the nucleus during S phase and in response to overexpressed cyclin A. In contrast, using a combination of biochemical and immunohistochemical assays, we show convincingly that endogenous Cdc6 remains nuclear and chromatin bound throughout the entire S period, while Mcm5 loses chromatin affinity during S phase. Overexpression of cyclin A is unable to alter the nuclear localization of Cdc6. Furthermore, using a phosphospecific antibody we show that phosphoserine-54 Cdc6 maintains a high affinity for chromatin during the S period. Considering recent in vitro studies, these data are consistent with a proposed model in which Cdc6 is serine-54 phosphorylated during S phase and functions as a chromatin-bound signal that prevents reformation of prereplication complexes.
Initiation of DNA replication in eukaryotic chromosomes is governed by multiprotein complexes that assemble at origins of replication in a temporally specified manner (2). In Saccharomyces cerevisiae, cis-regulatory replicator elements at plasmid and chromosomal origins are bound throughout the cell cycle by an aggregate of six proteins termed the origin recognition complex (2, 3). During late mitosis or early G1, the origin recognition complex serves as a loading site for the binding of Cdc6 and Cdt1, followed by recruitment of the minichromosome maintenance (MCM) complex (2, 7, 8, 13, 37-40, 45, 47). The resulting aggregate is known as the prereplication complex (pre-RC).
Regulation of Cdc6 abundance and subcellular localization is thought to be important during the cell cycle for licensing origins and preventing further rounds of initiation in the same cell cycle (5, 10, 11, 27-29, 31, 33). Cdc6 is required for initiation of replication in yeast (7, 21, 31, 45), Xenopus laevis (8), and likely mammals (33, 46, 48). Once replication begins, Cdc6 is degraded in yeast (5, 7, 30, 31, 45), whereas for mammals it has been suggested that Cdc6 is translocated out of the nucleus during S phase in a cyclin A-Cdk2- and phosphorylation-dependent manner (10, 11, 27-29, 33) and then subject to degradation by the anaphase-promoting complex (10, 23, 29). However, the majority of data supporting the translocation model have been obtained using ectopically expressed or microinjected Cdc6 cDNAs (11, 18, 28, 33) or with recombinant Cdc6 protein added to in vitro replication systems (10, 27). In addition, many of these studies employed tumor cell lines to obtain data describing normal Cdc6 regulation (10, 11, 15, 16, 18, 28, 33), and synchronization regimens utilizing hydroxyurea, aphidicolin, or thymidine blocks were often performed (10, 11, 15, 16, 18, 28, 33), which could easily have created stress or checkpoint response conditions within the cells (1, 4). For these reasons, it is difficult to ascertain whether endogenous Cdc6 is likewise regulated by translocation during a normal S phase.
In apparent contradiction of the translocation model are a few reports that have shown that some endogenous Cdc6 protein is detergent resistant during S phase (10, 15, 16, 23, 26, 36), while one of these groups has shown by immunofluorescence that some endogenous Cdc6 is possibly nuclear at some unknown point in S phase (15, 16), although the latter studies utilized hydroxyurea-synchronized tumor cells and failed to show that cells displaying Cdc6 in the nucleus were simultaneously in S phase (15). However, as with most of the ectopic studies described above, the majority of these endogenous approaches also utilized tumor cell lines (10, 15, 16, 23, 36) and/or employed hydroxyurea, aphidicolin, or thymidine-blocking methods to analyze endogenous Cdc6 regulation during S phase (10, 15, 16, 26, 36), thus requiring caution in the interpretation of these studies with regard to Cdc6 regulation in normal cells. Even more important, advocates of the translocation model can point out that the experiments utilizing detergent-resistant approaches (10, 15, 16, 23, 26, 36) do not conclusively show that a recovered protein is indeed nuclear prior to cell fractionation. Indeed, if endogenous Cdc6 is translocating from nuclei along the cytoskeletal components, such as microtubules, then it is plausible that any such Cdc6 molecules might also be resistant to detergent extraction and be recovered in the so-called “nuclear” fraction, resulting in potential overinterpretation of any such chromatin-binding assays. For these reasons, it becomes necessary that biochemical fractionation procedures be complemented by immunofluorescence experiments on fixed cells to support or refute models for endogenous Cdc6 regulation by translocation. It is also important that any such studies utilize non-tumor-derived cells, as well as experimental approaches that do not involve any drug treatments that may complicate interpretation of the results. To date, no such clear and comprehensive analysis focusing specifically on regulation of endogenous Cdc6 subcellular localization during S phase has been performed.
To investigate more thoroughly the regulation of endogenous Cdc6 during S phase, we have cloned the cDNA encoding the Chinese hamster (Cricetulus griseus) Cdc6 homolog and obtained several effective antibodies that allowed us to focus specifically on the issue of whether endogenous and exogenous Cdc6 proteins behave the same way in tumor and non-tumor-derived cells, both with and without synchronization. Using biochemical fractionation together, for the first time, with thorough immunohistochemical techniques, we compared the subcellular distribution of endogenous and ectopically expressed Cdc6 during G1 and S phases. We demonstrate that, in contrast to ectopically expressed Cdc6, a significant fraction of the endogenous Cdc6 protein not only remains nuclear throughout the entire S period but also displays a high affinity for chromatin at all stages of S phase. We also demonstrate for the first time that endogenous Cdc6 phosphorylated specifically on serine-54 appears during S phase but does not lose its affinity for chromatin despite its apparent Cdk2-cyclin A-mediated modification. In agreement with this result, we additionally show that overexpressed cyclin A fails to yield any changes in the nuclear localization of endogenous Cdc6, whereas ectopically expressed Cdc6 is clearly translocated from the nucleus under similar conditions.
Our analysis more thoroughly addresses the weaknesses of other reports and strengthens the idea that endogenous Cdc6 does not appreciably translocate from the nucleus during S phase or following ectopic cyclin A expression, instead becoming phosphorylated on serine-54 in a chromatin-bound state. Taking these data together with recent in vitro data published by Laskey and coworkers (9), we propose a model in which endogenous Cdc6 is phosphorylated when its associated pre-RC is activated to initiate but remains chromatin bound, perhaps functioning as a block to further pre-RC formation or as a signal to later mitotic events. In this manner, chromatin-bound and phosphorylated Cdc6 may represent another mechanism that prevents premature reinitiation of DNA replication by inhibiting new pre-RC formation.
MATERIALS AND METHODS
Cell culture and synchronization.
Chinese hamster ovary (CHO) or HeLa cells were maintained in minimal essential medium (Invitrogen) supplemented with 10% Fetal Clone II (HyClone). CHO cells were synchronized in G0 by starving them for isoleucine for 36 h followed by release into complete minimal essential medium. Bromodeoxyuridine (BrdU; Sigma) labeling (15 μM, 30 min) was used to verify synchrony. Transfections were performed in 35-mm-diameter plates with FuGENE 6 according to the protocol of the manufacturer (Roche). Transfections lasted 24 h, after which cells were analyzed as described below.
Antibodies.
Monoclonal antihemagglutinin (anti-HA), nonconjugated or conjugated with fluorescein isothiocyanate (FITC) (1:1,000; HA.11; Covance), and rabbit polyclonal anti-Cdc6 (sc-8341, antibody 1) and anti-Cdc6-phosphoserine-54 (sc-12920; 1:500) and blocking peptide (sc-12920p) were from Santa Cruz Biotechnology; rabbit polyclonal anti-Mcm5 (1:50; provided by Rolf Knippers, University of Konstanz, Konstanz, Germany), monoclonal anti-BrdU (1:20; Roche), monoclonal anti-Cdc6 37314 (against amino acids 366 to 560 of human Cdc6; antibody 2; 1:1,000), and monoclonal anti-Cdc6 16724 (against amino acids 1 to 224 of human Cdc6; antibody 3; 1:1,000 on immunoblots and 1:500 in immunohistochemistry) were provided by Nicholas Heintz (University of Vermont); monoclonal anti-BrdU-Alexa-594 (1:20) and monoclonal anti-Cdc6 (antibody 4; immunogenic peptide shown in Fig. 1; 1:20) were from Molecular Probes; monoclonal anti-lamin A/C (1:500) and monoclonal antitubulin (1:1,000) were from Calbiochem; rabbit polyclonal anti-cyclin A (1 μg/ml) was from Upstate Biotechnology Inc.
FIG. 1.
Alignment of the amino acid sequences of Chinese hamster Cdc6, human Cdc6, and murine Cdc6. The alignment was performed using Clustal W algorithms, followed by shading with the program Boxshade, both available at the Internet location http://restools.sdsc.edu. Residues identical between two or among three species are indicated by dark shading, and similar residues are indicated by light shading. The immunogenic peptide to which the Molecular Probes anti-Cdc6 monoclonal antibody (antibody 4) was made is indicated above the amino acid sequence, as are the Walker A and B boxes (43). Conserved serines that are putative substrates of cyclin-dependent kinases in vitro and in vivo are indicated below the amino acid sequence (see text for references). GenBank accession numbers are as follows: human Cdc6, NM_001254; murine Cdc6, NM_011799; Chinese hamster Cdc6, AY491989.
Cloning and subcloning.
Human and murine Cdc6 sequences (GenBank accession numbers NM_001254 and NM_011799, respectively) were aligned, and identical stretches of DNA sequences were used to design primers for use in 5′ rapid amplification of cDNA ends (5′-RACE) and reverse transcription-PCRs (RT-PCRs) on Chinese hamster cDNA. 5′-RACE was performed according to the instructions of the manufacturer (Invitrogen) to obtain a hamster Cdc6 5′ fragment. PCR conditions and primer sequences are available upon request. The 5′-RACE product was sequenced to determine the hamster 5′ coding region. These sequence data were used to design a new primer (Cdc6-5′Met), with an in-frame BamHI site. PCR using the Cdc6-5′Met primer and the Cdc6-3′B primer (based on identical murine and human sequence in the 3′ tail of Cdc6) was performed on hamster cDNA with High Fidelity Platinum Taq polymerase (Invitrogen). PCR conditions and primer sequences are available upon request. The 1.9-kb product was subcloned, and four independent clones were sequenced two to four times on each strand to determine the wild-type Chinese hamster Cdc6 coding sequence.
A Homo sapiens cyclin A (HsCyclin A) plasmid was provided by Joseph Nevins (Duke University). HsCyclin E (E1 isoform 2; see GenBank entry NP_476530) was obtained from human cDNA by RT-PCR. PCR using Pfu-Turbo (Stratagene) was performed on the HsCyclin A plasmid or on human cDNA. Primers contained an in-frame BamHI or EcoRI site at the 5′ end of the gene and a NotI site at the 3′ end (primers and methods are available upon request). PCR products were verified by sequencing.
The C. griseus Cdc6 (CgCdc6), HsCyclin A, and HsCyclin E cDNAs were transferred to pcDNA3 (Invitrogen) and to pcDNA3-3×HA (containing three HA tags) by using BamHI and NotI (for CgCdc6 and HsCyclin A genes) or EcoRI and NotI (for HsCyclin E). This produced the expression vectors pc2HA-CgCdc6, pc2HA-HsCyclin A, pc3HA-HsCyclin E, pcDNA3-CgCdc6, pcDNA3-HsCyclin A, and pcDNA3-HsCyclin E. CgCdc6 was transferred to the vectors pGEX-4T1 (Amersham Pharmacia) and pET28a (Novagen) for bacterial expression with an in-frame glutathione S-transferase (GST) tag or six-histidine tag. CgCdc6 was also subcloned into the vector pRcLac (49), allowing expression with an in-frame LacI tag.
Immunoblotting.
CHO, 3T3, or HeLa cells were washed with cold phosphate-buffered saline (150 mM NaCl, 5 mM Na2HPO4, 1.7 mM KH2PO4, pH 7.4). Cells were lysed, and total cell extract (TCE) was analyzed by immunoblotting. For antibody blocking, a fivefold excess of blocking peptide (by weight) was incubated with antibody on ice for 2 h in phosphate-buffered saline. Membranes were probed with antisera in the presence of 3% dried milk in TBS-T (20 mM Tris base, 137 mM NaCl [pH 8], 0.1% Tween 20), followed by incubation with a 1:10,000 dilution of secondary antibody conjugated with horseradish peroxidase (anti-mouse immunoglobulin G [IgG], Amersham Pharmacia; anti-rabbit IgG, Jackson Immunoresearch; anti-mouse IgM, Pierce). Membranes were subjected to enhanced chemiluminescence (Amersham Pharmacia).
To collect detergent-resistant chromatin and detergent-soluble (cytosolic and nucleosolic) fractions, a published protocol (23) was followed with some modifications. Monolayer CHO cells were collected at each time point following synchronization. Samples were normalized to cell number prior to fractionation (cell numbers never varied by more than 5%). CHO cells were separated into S1 and P3 fractions as described previously (23). The S2 samples in the previous report are referred to here as S1. In the end, all S1 and P3 sample pairs had equal volumes of CHO cell equivalent extract. TCE samples were collected at each time point without fractionation. Equal amounts of TCE, S1, and P3 were analyzed by immunoblotting.
Bacterial expression.
GST-CgCdc6 was purified from bacteria according to the suggestions of the manufacturer (Amersham Pharmacia). Histidine-tagged CgCdc6 (His6-CgCdc6) was purified using BD Talon Resin (BD Biosciences). For phospho-Cdc6 antibody analysis, approximately 0.75 μg of full-length GST-CgCdc6 was used. For analysis of the monoclonal anti-Cdc6 antibody (antibody 3), approximately 1 μg of purified His6-CgCdc6 protein was used.
Immunohistochemistry.
CHO or HeLa cells on coverslips were fixed with 2% formaldehyde. In some experiments, cells were incubated in a Triton X-100 buffer (0.1% Triton X-100, 10 mM HEPES-KOH [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol) prior to formaldehyde fixation to remove detergent-sensitive proteins. Immunohistochemistry was performed as described previously (49). BrdU incorporation was detected after treatment with 1.5 N HCl for 30 min. Secondary antibodies consisted of the following: anti-mouse-Texas Red, anti-mouse-FITC, and anti-rabbit-Texas Red (Jackson Immunoresearch). Coverslips were mounted with Prolong Antifade reagent (Molecular Probes) and examined with a Nikon Microphot-SA fluorescence microscope equipped with a Spot digital camera and software.
For double labeling experiments with anti-BrdU or anti-HA and anti-Cdc6, signals from the monoclonal antibodies were visualized using sequential probings. Primary, nonlabeled antibodies to Cdc6 were incubated with fixed cells and then bound by secondary antibody. The cells and antibodies were fixed again with formaldehyde, and anti-HA conjugated with FITC and anti-BrdU-Alexa-594 were used as probes.
Nucleotide sequence accession number.
The wild-type Chinese hamster Cdc6 coding cDNA sequence is deposited in GenBank under accession number AY491989.
RESULTS
Cloning of CgCdc6 cDNA and characterization of Cdc6 antibodies.
Using a combination of 5′-RACE and RT-PCR techniques, we have cloned the 1.9-kb sequence corresponding to the Chinese hamster Cdc6 cDNA (CgCdc6; Fig. 1). Primers were designed using the alignments of human and murine Cdc6 sequences. Clustal W alignments showed that hamster and human coding regions are 82% identical and 13% similar, while murine and hamster regions show 83% identity and 12% similarity. This suggests that Cdc6 performs the same function in all mammals, presumably regulating replication origin activity.
The CgCdc6 gene encodes a 561-amino-acid polypeptide with a predicted size of ∼62 kDa (33, 46). To test the integrity of the cloned CgCdc6 cDNA, it was inserted into an expression vector carrying two in-frame HA tags and the fusion protein was transiently expressed in CHO cells. Immunoblotting with anti-HA detected a 65-kDa polypeptide, which corresponds to the expected 62-kDa Cdc6 polypeptide when corrected for the size of the tags (data not shown).
Several antibodies proved effective in the various applications employed here. Polyclonal anti-Cdc6 (antibody 1) and monoclonal anti-Cdc6 (antibody 2) recognized a ∼62-kDa polypeptide in immunoblots of TCEs from three mammalian species, as well as CgCdc6 fused to the LacI DNA-binding domain (Fig. 2A) (49). Both antibodies detect a second band (and sometimes a third) at a slightly higher molecular weight in hamster cells (Fig. 2A and C; see also Fig. 3 below). Others have also detected a species of similar size in human cells (10). Since antibodies 1 and 2 did not efficiently recognize Cdc6 in formaldehyde-fixed cells, we used two additional monoclonal antibodies (antibodies 3 and 4) that were effective in cytological assays (see below). On immunoblots, antibody 3 recognized endogenous mammalian Cdc6, as well as bacterially expressed histidine-tagged CgCdc6 (Fig. 2B), and could be blocked with purified Cdc6 (data not shown). Antibody 4 did not efficiently recognize Cdc6 on immunoblots following cell lysis and protein denaturation.
FIG. 2.
Characterization of antibodies. (A) (Left) Western analysis of TCEs from asynchronous HeLa, 3T3, and CHO cells or from CHO cells transfected with pRcLac-CgCdc6, probed with polyclonal anti-Cdc6 (antibody 1). Arrowheads indicate Cdc6 bands. (Right) Western analysis of the same samples probed with monoclonal anti-Cdc6 (antibody 2). (B) (Left) TCEs from asynchronous HeLa, 3T3, and CHO cells analyzed by immunoblotting using mouse monoclonal anti-Cdc6 (antibody 3). (Right) Histidine-tagged CgCdc6 purified from Escherichia coli was analyzed by immunoblotting using monoclonal anti-Cdc6 (antibody 3). Note the degradation product below the predominant full-length Cdc6 band. (C) Asynchronous HeLa and CHO TCEs, HeLa and CHO nuclear fractions (Nucl; isolated as described for log-phase P3 pellet in Fig. 3), bacterially expressed GST-tagged CgCdc6, and log-phase CHO cells transfected with pRcLac-CgCdc6 were used to analyze the specificity of the anti-Cdc6-phosphoserine-54 polyclonal antibody. (Top) Cells probed with phosphoserine-54-specific anti-Cdc6 antibody. (Bottom) Cells probed with nonphosphospecific anti-Cdc6 polyclonal antibody 1. Arrowheads in the lower panel indicate two Cdc6-specific bands in this experiment, while a third is visible upon longer exposure (data not shown). Numbers at left of panels indicate molecular masses in kilodaltons.
FIG. 3.
Chromatin association and detergent sensitivity of Cdc6 and Cdc654P during G0, G1, and S phases. CHO cells were synchronized in G0 by isoleucine deprivation, followed by release into G1 and S phases. TCEs, cytosolic-nucleosolic S1 fractions, and detergent-resistant P3 chromatin fractions were isolated at the times indicated above. Asynchronous CHO cells (Log) were also collected as a reference sample. Western blotting with anti-Cdc6 polyclonal antibody 1 (Cdc6), anti-Cdc654P polyclonal antibody (Cdc6-P), and anti-cyclin A polyclonal antiserum was performed. Probing with anti-lamin A/C and antitubulin monoclonal antibodies confirmed that the fractionation procedure was effective. BrdU analysis using immunohistochemistry was performed to verify that cells had been adequately synchronized (Fig. 6).
We also obtained a rabbit polyclonal antibody directed against Cdc6 phosphorylated on serine-54 (Cdc654P). This antiserum efficiently recognized endogenous human Cdc654P and, less efficiently, the endogenous hamster counterpart (Fig. 2C, top panel). Preincubation with an excess of the immunogenic Cdc654P peptide blocked reactivity (data not shown). Transfected LacI-CgCdc6 protein was efficiently recognized by this antibody, suggesting that hamster Cdc654P is recognized but that there are low levels of endogenous Cdc654P in extracts prepared from unsynchronized cultures. As shown in Fig. 2C (upper panel), an excess of bacterially expressed, unphosphorylated, GST-tagged CgCdc6 is not recognized by this phosphospecific antibody whereas antibody 1 to Cdc6 gives a very strong signal (Fig. 2C, lower panel), demonstrating the specificity of this preparation.
Endogenous Cdc6 and Cdc654P are associated with chromatin throughout S phase.
We first asked how endogenous Cdc6 levels in CHO cells fluctuate during the G1-to-S-phase transition following release from a G0 block. Immunoblotting of TCE at the times after release indicated in Fig. 3 showed that Cdc6 levels are slightly reduced during starvation itself (Iso−) relative to log-phase samples but rise somewhat following release from the G0 block (compare to the lamin A/C and tubulin loading controls). These data are consistent with results published by other groups (28, 35, 36, 46). The polyclonal anti-Cdc6 used here (antibody 1) detects three distinct Cdc6 species in this experiment, the middle one of which is absent in G0 but reappears ∼6 h after release from the block, around the time at which the population begins to enter the S period (see below). A corresponding increase in signal strength with the Cdc654P antibody suggests that this band represents a phosphorylated isoform of Cdc6 (Fig. 3, TCE samples). Assignments of approximate cell cycle positions were made by monitoring the appearance of cyclin A (Fig. 3) and by pulse-labeling parallel cultures with BrdU to determine the percentage of cells in S phase at each time point (see Fig. 6).
FIG. 6.
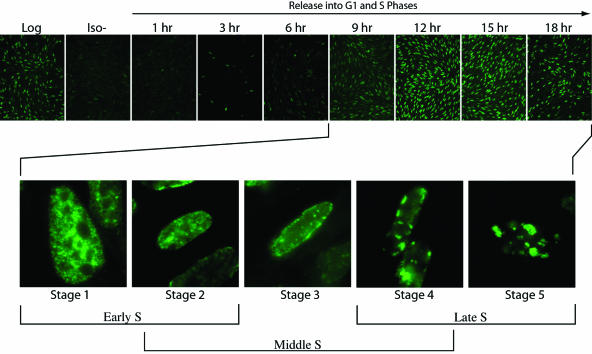
BrdU incorporation in synchronized CHO cells and staging of BrdU patterns. CHO cells were synchronized in G0 by isoleucine deprivation (Iso− sample) and were released into G1 and S phase in complete medium for the times indicated above (see Materials and Methods). Cells were pulsed with BrdU for 30 min prior to fixing. Asynchronous cells were also labeled with BrdU (log-phase sample). BrdU labeling was visualized using anti-BrdU monoclonal antibody followed by secondary anti-mouse antibody conjugated with FITC. (Top) Multiple fields were observed for each time point, and representative fields are shown at ×20 magnification. Log-phase cells generally showed 30 to 50% BrdU incorporation. At the peak periods (12 and 15 h), 90 to 95% of cells were labeled. (Bottom) Magnifications of ×100 showing details of nuclei at different stages of S phase. The 9-h point showed primarily stage 1 BrdU patterns. The 12-h point showed a few stage 1, many stage 2, and some stage 3 BrdU patterns. The 15-h point showed primarily stage 3 and 4 BrdU patterns. The 18-h point showed mostly stage 4 and 5 BrdU patterns. At least 100 nuclei were analyzed at each time point, and representative photographs are shown.
To determine the affinity of endogenous Cdc6 for chromatin during G1 and S phases, parallel cultures were subjected to a modified chromatin binding assay published previously (23) in which Triton X-100 is used to separate nucleosolic and cytosolic proteins (fraction S1) from chromatin-bound, detergent-resistant material (fraction P3). Immunoblotting with anti-lamin A/C and antitubulin verified that the fractionation was effective (Fig. 3), and more importantly, it suggested that if any cytoplasmic proteins were tightly associated with microtubules then they were unlikely to be present in the chromatin pellets in our experiments, since tubulin itself was detergent sensitive and not associated with the pellets. Note, however, that a minor portion of the total tubulin appears to become more resistant to detergent ∼18 h after release from G0, a time when some cells have finished S phase and are entering G2 and mitosis.
Interestingly, the endogenous Cdc6 isoforms partition into different fractions (Fig. 3, top rows) during the cell cycle. Notably, the middle Cdc6 (presumably phosphorylated) band that appears in late G1 and S phase fractionates with the P3 chromatin fraction, while the two other isoforms are largely (but not completely) solubilized by Triton X-100. Probing with the Cdc654P antibody suggests that the middle band corresponds to phospho-Cdc6 (Fig. 3, Cdc6-P). A small portion of Cdc654P begins to appear in the soluble S3 fraction only in the later stages of S phase. Thus, phosphorylation of Cdc6 may lead to its increased affinity for chromatin.
Subcellular distributions of endogenous and ectopically expressed Cdc6 in unsynchronized cell populations.
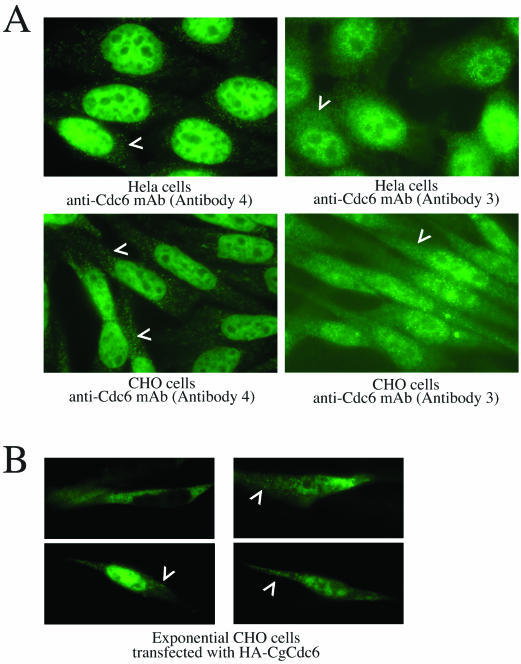
We also analyzed the subcellular distribution of endogenous Cdc6 in cells by immunohistochemical methods. In unsynchronized CHO and HeLa cells, both monoclonal antibodies to Cdc6 (antibodies 3 and 4) display a predominantly nuclear pattern (Fig. 4A). However, both reagents also detect a less pronounced extranuclear signal distributed throughout the cytoplasm in many discrete foci (arrowheads in Fig. 4A).
FIG. 4.
Immunohistochemical analysis of endogenous Cdc6 in CHO and HeLa cells and transfected CgCdc6 in CHO cells. (A) Asynchronous HeLa or CHO cells were analyzed by immunohistochemistry with two independently derived monoclonal antibodies to mammalian Cdc6 (antibodies 3 and 4). The secondary antibody was anti-mouse antibody conjugated with FITC. Arrowheads indicate discrete cytoplasmic focal staining with either antibody. At least 300 nuclei were observed; representative photographs are shown. (B) Asynchronous CHO cells were transfected with the pc2HA-CgCdc6 vector and fixed and analyzed by immunohistochemistry with anti-HA monoclonal antibody followed by anti-mouse antibody conjugated with FITC. Arrowheads indicate diffuse and focal cytoplasmic staining for exogenous HA-CgCdc6. More than 50 transfected cells were observed, and representative cells were photographed.
HA-tagged CgCdc6 also displays a predominantly nuclear staining pattern when expressed in unsynchronized CHO cells, again with a less-pronounced focal cytoplasmic signal (Fig. 4B, arrowheads). However, in 20 to 30% of transfected cells examined, the HA-CgCdc6 protein was completely cytoplasmic, with little staining detectable in the nucleus (Fig. 4B, upper left panel). Neither of the monoclonal antibodies detected this “nuclear ghost” pattern with endogenous Cdc6, in either CHO or HeLa cells. Thus, these immunohistochemical studies suggest that endogenous Cdc6 is primarily nuclear, while exogenous Cdc6 is localized to the cytoplasm in some cells. Since the biochemical fractionation studies presented in Fig. 3 show that a significant portion of endogenous Cdc6 is detergent soluble at all time points, much of the Cdc6 protein detected here by immunohistochemical methods may actually be nucleoplasmic rather than tightly bound to chromatin.
Transfected Cdc6 is cytoplasmic in S-phase cells.
The differences in behavior between endogenous and ectopically expressed Cdc6 in the experiments presented above suggested that cells may regulate the intracellular localization of the two species differently. Several studies have suggested that ectopically expressed Cdc6 is lost from the nucleus during S phase (10, 18, 27, 28, 33). Therefore, the possibility arose that the lack of nuclear staining in some of our transfected cells was due to their being in S phase at the time of transfection, resulting in the appearance of exogenous Cdc6 in the cytoplasm.
To determine whether there is a strict correlation between cytoplasmic staining of HA-CgCdc6 and residence in S phase, we performed dual-labeling assays on transfected CHO cells with antibodies directed toward HA-CgCdc6 and BrdU (the HeLa cell line used here was resistant to lipid-based transfection methods and was not analyzed in this experiment). As shown in Fig. 5A, transfected CHO cells that displayed a clear nuclear staining pattern with anti-Cdc6 did not stain with anti-BrdU, indicating that these cells were not in S phase during the transfection procedure. In contrast, cells that displayed only the extranuclear staining pattern with anti-HA-CgCdc6 also exhibited distinct BrdU labeling, indicating that the cells were in S phase, or had entered S phase, during the transfection process (Fig. 5B).
FIG. 5.
Transfected CgCdc6 is cytoplasmic in S-phase cells. Asynchronous CHO cells were transfected with pc2HA-CgCdc6 and analyzed for exogenous protein expression and BrdU incorporation by immunohistochemistry. BrdU was added to cells for 30 min prior to fixation. HA-CgCdc6 was detected with anti-HA monoclonal antibody conjugated with FITC (green), and BrdU was detected with anti-BrdU monoclonal antibody conjugated with Alexa-594 fluorophore (red). More than 100 transfected cells were analyzed, and representative photographs are shown. (A) Cells expressing Cdc6 in the nucleus do not simultaneously show BrdU labeling and are therefore non-S-phase cells. Arrowheads indicate nuclei with a clear reciprocal relationship between anti-BrdU and anti-HA (Cdc6p) staining. (B) Cells in S phase are labeled with BrdU and consistently show ectopic Cdc6 expression in the cytoplasm.
Endogenous Cdc6 is nuclear and chromatin bound throughout S phase in mammalian cells.
It appears that when HA-CgCdc6 is expressed ectopically during S phase, it is translocated to the cytoplasm (our data and references 10, 18, 27, 28, and 33). However, the immunohistochemical studies presented in Fig. 4 showed that endogenous Cdc6 is detected primarily in the nucleus in the majority of asynchronous CHO and HeLa cells, many of which are likely to be in S phase. Furthermore, in fractionation studies from our own and other laboratories (Fig. 3) (10, 23, 26, 35, 36), a significant portion of endogenous Cdc6 is nuclear and bound to chromatin in S phase. These data suggested a potentially significant difference in behavior between endogenous and ectopically delivered Cdc6.
Therefore, we determined the subcellular location of endogenous Cdc6 at various stages of S phase under circumstances in which the cells had not been synchronized. To accomplish this, we first characterized the early-, middle-, and late-S-phase BrdU staining patterns in synchronized cultures of CHO cells that were arrested in G0 and were then released from this block and pulse-labeled with BrdU at the time points shown in Fig. 6. After fixation and staining with anti-BrdU, microscopic examination showed that most cells entered S phase 6 to 9 h after release from the G0 block and that S phase was complete in the majority of cells between 15 and 18 h (top panels). High-magnification views showed distinctly different patterns of BrdU staining at various stages of S phase (lower panels), in agreement with earlier reports (20, 25), and are classified in Fig. 6 according to the five stages identified in those studies.
When unsynchronized CHO and HeLa cells were pulsed with BrdU, fixed, and probed simultaneously with anti-Cdc6 and anti-BrdU, the resulting patterns show clearly that endogenous Cdc6 localizes primarily to the nucleus in all stages of S phase (Fig. 7A). Identical results were obtained with both monoclonal anti-Cdc6 antibodies 3 and 4 (compare left two panels with right two in each case). We also tested whether the affinity of Cdc6 for the nucleus (and likely chromatin) was changed in S-phase versus non-S-phase nuclei. After labeling with BrdU, cells were incubated in Triton X-100 buffer prior to fixation with formaldehyde. Although the level of fluorescence was reduced relative to that in non-detergent-treated samples, endogenous Cdc6 was again observed to localize to the nucleus and to resist extraction with Triton X-100 at all stages of S phase in both CHO and HeLa cells (Fig. 7B). These findings are consistent with those of earlier studies in which Cdc6 was suggested to be nuclear at some unknown point in S phase (15, 16). (Note that all five S-phase stages were detected in both CHO and HeLa preparations with each of the two monoclonal antibodies to Cdc6, but only selected examples are shown.)
FIG. 7.
Endogenous Cdc6 remains nuclear and chromatin bound throughout the entire S phase. Asynchronous HeLa and CHO cells were labeled for 30 min with BrdU, fixed immediately (A) or following Triton X-100 treatment (B), and analyzed by immunohistochemistry for BrdU incorporation and endogenous Cdc6 localization. Cdc6-labeled nuclei and BrdU-labeled nuclei are shown in horizontal pairs. Cdc6 was visualized with anti-Cdc6 monoclonal antibodies 3 and 4, and both Cdc6 monoclonal antibodies were detected with anti-mouse secondary antibody conjugated with FITC (green). Following Cdc6 probing, cells and Cdc6 primary and secondary antibodies were fixed again, and then BrdU was detected with anti-BrdU monoclonal antibody conjugated with Alexa-594 (red). Labeled arrowheads point to S-phase nuclei staged according to the results of Fig. 6. Unlabeled arrowheads indicate non-S-phase cells. For each pair of pictures, at least 100 nuclei were analyzed, and representative pictures are shown.
As a control for the effectiveness of the Triton X-100 pretreatment, HeLa cells were either fixed immediately with formaldehyde or treated with Triton X-100 prior to fixation, and the nuclear localization and affinity of Mcm5 were determined. In agreement with previously published results for other MCM family members (14, 19, 22, 32, 41), when fixed immediately, Mcm5 clearly localized to the nucleus in non-S-phase cells and during all stages of S phase (Fig. 8A). However, when cells were pretreated with Triton X-100 prior to fixation, Mcm5 became sensitive to extraction in early S phase and was virtually undetectable in late-S-phase nuclei (Fig. 8B). We conclude from these immunohistochemical data that a subpopulation of endogenous Cdc6 not only is nuclear but remains tightly bound to some structure in the nucleus during the entire S phase. These results are in general agreement with the biochemical fractionation data presented above. (Note that the fluor used to detect BrdU in Fig. 8 is FITC [green], while in Fig. 5 and 7 it is Alexa-594 [red].)
FIG. 8.
Pretreatment with Triton X-100 effectively removes Mcm5 from S-phase nuclei. Asynchronous HeLa cells were labeled for 30 min with BrdU, fixed immediately (A) or following Triton X-100 treatment (B), and analyzed by immunohistochemistry for BrdU incorporation and Mcm5 localization. Mcm5 was detected with polyclonal anti-Mcm5 followed by secondary anti-rabbit antibody conjugated with Texas Red (red). BrdU was detected with monoclonal anti-BrdU followed by secondary anti-mouse antibody conjugated with FITC (green). Arrowheads point to non-S-phase cells or S-phase cells staged according to the results in Fig. 6. At least 100 nuclei were examined, and representative photographs are shown.
Overexpression of cyclin A causes ectopically expressed, but not endogenous, Cdc6 to translocate to the cytoplasm.
It has been shown that when Cdc6-expressing plasmids are comicroinjected into unsynchronized cell populations with a cyclin A expression vector (but not with a cyclin E vector), the ectopic Cdc6 localizes to the cytoplasm (28). However, we showed above that, while overexpressed HA-CgCdc6 is efficiently translocated to the cytoplasm during S phase, endogenous Cdc6 is retained in the nucleus throughout S phase. Thus, the question arose whether the localization of endogenous Cdc6 is regulated by cyclin A.
As shown in Fig. 9A, ectopically expressed HA-CgCdc6 does, indeed, localize exclusively to the cytoplasm when unsynchronized cells are cotransfected with a cyclin A expression plasmid, but not with a vector encoding cyclin E. In contrast, neither HA-cyclin A nor HA-cyclin E overexpression in unsynchronized CHO cell populations provoked a relocalization of endogenous Cdc6 to the cytoplasm: the endogenous Cdc6 signal remained nuclear in all cells examined (Fig. 9B and C). Identical results were obtained with both monoclonal anti-Cdc6 preparations. We were unable to test whether endogenous Cdc6 became detergent sensitive when either cyclin A or cyclin E was overexpressed in log-phase cells, since the exogenous cyclin proteins were themselves removed from nuclei by the Triton X-100 treatment, precluding the identification of cyclin-overexpressing cells (data not shown). However, we can conclude that the overexpression of either cyclin A or cyclin E does not provoke the relocalization of endogenous Cdc6 from the nucleus to the cytoplasm.
FIG. 9.
Overexpression of cyclin A causes exogenous, transfected CgCdc6 to translocate to the cytoplasm but has no effect on endogenous CgCdc6. (A) Asynchronous CHO cells were transfected with the HA-tagged CgCdc6 expression vector and a fivefold molar excess of either untagged pcDNA3-HsCyclin A (left panels) or untagged pcDNA3-HsCyclin E (right panels). Cells expressing HA-CgCdc6 were detected with monoclonal anti-HA followed by secondary anti-mouse antibody conjugated with FITC. (B and C) Asynchronous CHO cells were transfected with pc2HA-HsCyclin A (B) or pc3HA-HsCyclin E (C) and analyzed for endogenous Cdc6 in HA-cyclin-expressing cells. Cdc6 was detected with monoclonal anti-Cdc6 antibody 4 (panel pairs on left) or monoclonal anti-Cdc6 antibody 3 (panel pairs on right), followed by secondary anti-mouse antibody conjugated with Texas Red (red images). Cells and Cdc6 primary and secondary antibodies were fixed again, and HA-cyclin A (B) or HA-cyclin E (C) was detected with monoclonal anti-HA conjugated with FITC (green images). In all experiments, ∼50 HA-expressing cells were analyzed, and representative photographs are shown.
DISCUSSION
There are conflicting reports in the literature regarding the subcellular distribution of mammalian Cdc6 during G1-to-S-phase progression. Several studies have shown that ectopically expressed Cdc6 translocates from the nucleus to the cytoplasm in an S-phase- and cyclin A-Cdk2-dependent manner (10, 11, 27, 28, 33). This has led to a model in which the subcellular distribution of mammalian Cdc6 is regulated by cyclin A during S phase as a means of preventing reinitiation of DNA replication. A few studies of the behavior of endogenous Cdc6 have indicated that, while some of the protein exits the nucleus upon entry into S phase, a subpopulation remains chromatin bound (10, 15, 16, 23, 26, 35, 36). It could be that the differences in behaviors of endogenous versus ectopically expressed Cdc6 result from differences in the cell lines used (i.e., tumor derived versus non-tumor derived) or from the application of inhibitors or double-thymidine blocks to synchronize cells in different stages of the cycle (10, 15, 16, 23, 26, 28, 33). To attempt to resolve these issues, we have utilized both immunohistochemical and biochemical fractionation methods to determine how endogenous and ectopically expressed Cdc6 proteins behave in synchronized and unsynchronized cultures of tumor-derived HeLa cells and non-tumor-derived CHO cells.
The biochemical fractionation method of Mendez and Stillman (23) was modified here to allow efficient separation of chromatin-bound proteins from cytosolic and nucleosolic proteins in synchronized monolayer cultures of CHO cells. Our studies show that a significant portion of endogenous Cdc6 can be solubilized with detergent during G1 and S phase, while an isoform of Cdc6 appears in late G1 and throughout S phase that is reduced in mobility and remains detergent resistant throughout S phase (Fig. 3). Based on the observation that Cdc6 is phosphorylated during S phase in mammalian cells (10, 18, 27-29), we suspected that this isoform of Cdc6 was phosphorylated. Support for this conclusion came from immunoblotting (Fig. 3) with a phosphoserine-54-specific anti-Cdc6 antibody, which showed that Cdc654P is indeed present in late-G1- and S-phase cells and is detergent resistant (and likely chromatin bound) throughout S phase. The latter result is also supported by data published by others (10). Overall, the biochemical fractionation data in the present report are consistent with work from other laboratories showing that a subpopulation of endogenous Cdc6 remains bound to chromatin during S phase (10, 15, 16, 23, 26, 35, 36).
Biochemical fractionation approaches are useful for determining the relative detergent sensitivities of proteins. However, proteins solubilized by detergent may actually have resided in the nucleus, either loosely affixed to chromatin or in the nucleosolic phase, prior to fractionation. In addition, proteins that are detergent resistant, and therefore operationally defined as chromatin bound, may actually be cytoplasmic but tightly associated with structures such as microtubules that may themselves interact with detergent-resistant nuclear proteins (e.g., lamins).
We therefore employed a thorough set of immunohistochemical assays to independently assess the intracellular distribution of endogenous Cdc6. The results from these studies showed that endogenous Cdc6 is primarily nuclear in HeLa and CHO cells, with some evidence of cytoplasmic staining. Simultaneous staining of endogenous Cdc6 and BrdU showed that the protein is present in the nucleus at all stages of S phase. Pretreatment of cells with Triton X-100 prior to fixation indicated that, although some of the Cdc6 was lost from the nucleus, a significant fraction remained tightly associated at all stages of S phase and is likely to be bound to chromatin (Fig. 7). In combination with the fractionation data, these studies suggest that the majority of the detergent-sensitive Cdc6 detected on immunoblots (Fig. 3, S1 samples) was probably nuclear prior to the fractionation procedure but was either nucleoplasmic or loosely affixed to chromatin.
The overexpression of either cyclin A or cyclin E clearly did not affect the nuclear localization of endogenous Cdc6. These results contrast sharply with those obtained by us and others (28) with transfected or microinjected HA-CgCdc6, respectively, which cycles from the nucleus to the cytoplasm during S phase but which is exclusively cytoplasmic when coexpressed with cyclin A. Our results are consistent with other studies demonstrating that mammalian cells phosphorylate both endogenous and ectopic Cdc6 in S phase on serine-54 (11, 18, 27, 28) in a cyclin A-dependent manner (10, 28); however, our data clearly show that the cells partition these phospho-Cdc6 proteins into different compartments.
The immunohistochemical data presented here differ somewhat from those of Tsurumi and colleagues (15, 16), in that we did not detect a significant translocation of endogenous Cdc6 from the nucleus to the cytoplasm during any stage of S phase (Fig. 7A and B). However, Triton X-100 treatment prior to fixation yielded results nearly identical to theirs: during S phase, endogenous Cdc6 was clearly nuclear and the Cdc6 staining patterns appeared more punctate and reduced in the S-phase nuclei (Fig. 7B). Our findings extend these earlier studies by showing that this is true throughout the entire S period. It seems unlikely that epitope masking could explain our inability to detect an increase in cytoplasmic endogenous Cdc6 in S-phase cells, since two independently derived monoclonal antibodies gave the same results. It is conceivable that hydroxyurea, which was used to prepare synchronized cultures in the studies by Fujita et al. (15, 16), could have affected the intracellular location of Cdc6 in some way, perhaps by eliciting a cellular damage-sensing response (1, 4). In addition, the studies by Fujita et al. did not directly demonstrate that the cells they examined were actually in S phase, which complicates interpretation of their data. Here we used BrdU to definitively correlate Cdc6 localization with S-phase state in unsynchronized populations.
Helin and colleagues also have used immunohistochemical methods to monitor the distributions of endogenous and exogenously delivered Cdc6 under various conditions (28). Like Tsurumi and colleagues (15, 16), they concluded in one experiment that endogenous Cdc6, like its ectopically expressed counterpart, translocates out of the nucleus into the cytoplasm during S phase, due primarily to a detectable increase in cytoplasmic staining. However, the authors also noted that a significant fraction of endogenous Cdc6 could still be detected in the nucleus when the majority of the cell population was in S phase. Importantly, the most prominent cytoplasmic Cdc6 signal was observed 24 h after release from a G0 block, by which time many of the cells in the population likely would have completed S phase.
Taking into account the results from our own and other laboratories, we suggest the following general model for the regulation of Cdc6. The endogenous protein is normally localized to the nucleus, where it participates in the formation of pre-RCs (reviewed in reference 2). Once organized, the pre-RCs recruit other replication proteins such as DNA polymerases (12, 24, 44), and the complex is triggered to initiate DNA synthesis by cyclin-dependent kinases, including cyclin A-Cdk2 (9, 10, 17, 18, 28). Endogenous Cdc6 is one substrate for this kinase (10, 18, 27, 28), and after phosphorylation on one or more serines (18, 28), the phospho-Cdc6 could serve as a distinguishing chromatin-bound mark for pre-RCs that have already fired, possibly preventing rebinding of MCMs and thus disallowing reinitiation. Indeed, Laskey and coworkers have recently presented in vitro evidence supporting such a mechanism (9). In their studies it was shown that, in contrast to what was seen with untreated Cdc6 protein, cyclin A-Cdk2-phosphorylated Cdc6 protein added to mid-G1 nuclei failed to allow these nuclei to initiate replication when incubated with S-phase cytosolic extracts. More importantly, this lack of initiation was coincident with a failure to recruit Mcm2 to chromatin (9). Alternatively, and not mutually exclusive with the reinitiation mechanism, chromatin-bound phospho-Cdc6 might function as a negative signal to prevent mitosis until S phase is complete or as a positive signal that S phase is complete and mitosis can occur. Such a role for Cdc6 has recently been proposed by others (6, 42).
We also suggest that some endogenous Cdc6 can be phosphorylated by cyclin A-Cdk2, even though it has not been incorporated into pre-RCs (i.e., is nucleosolic) or resides in pre-RCs that have not functioned in an initiation reaction (the latter have been shown to exist in yeast cells [34]). The nucleosolic or nonutilized Cdc6 then could either be translocated to the cytoplasm (10, 11, 16, 28, 33) or have its affinity for chromatin reduced but still remain in the nucleus (as our immunohistochemical and biochemical data would suggest); this would prevent inappropriate pre-RC formation and reinitiation of DNA replication. It is likely that the majority of ectopically expressed Cdc6 would fall into the nucleosolic category, since it might not be able to integrate into preformed pre-RCs because of the timing of transfection with respect to the cell cycle and endogenous pre-RC assembly. Our studies raise the possibility that it may not be possible to design valid studies using transient, ectopically delivered Cdc6 as a marker for endogenous Cdc6 behavior, since the exogenous proteins may not actually be incorporated into pre-RCs.
Altogether, in this report we have thoroughly addressed the weaknesses of other reports and strengthened the idea that endogenous Cdc6 does not appreciably translocate from the nucleus to the cytoplasm during any stage of S phase, or following ectopic cyclin A or E expression. Endogenous Cdc6 instead becomes phosphorylated on serine-54 and remains nuclear and in a chromatin-bound state. We propose that chromatin-bound and phosphorylated Cdc6 may represent another mechanism that prevents premature reinitiation of DNA replication by inhibiting new pre-RC formation.
Acknowledgments
We thank Nicholas Heintz (University of Vermont) for the anti-Cdc6 monoclonal antibodies, Rolf Knippers (University of Konstanz) for anti-Mcm5 antibody, Joseph Nevins (Duke University) for the cyclin A plasmid, and Rong Li (University of Virginia) for plasmid pRcLac. We also thank Carlton White for technical assistance and the other members of the Hamlin laboratory for valuable discussions.
This work was supported by National Institutes of Health grant RO1 GM 26108 (to J.L.H.) and National Institutes of Health postdoctoral fellowship 5F32 GM19304-02 (to M.G.A.).
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 4.Brush, G. S., and T. J. Kelly. 2000. Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res. 28:3725-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bueno, A., and P. Russell. 1992. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 11:2167-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clay-Farrace, L., C. Pelizon, D. Santamaria, J. Pines, and R. A. Laskey. 2003. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J. 22:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocker, J. H., S. Piatti, C. Santocanale, K. Nasmyth, and J. F. X. Diffley. 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379:180-182. [DOI] [PubMed] [Google Scholar]
- 8.Coleman, T. R., P. B. Carpenter, and W. G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87:53-63. [DOI] [PubMed] [Google Scholar]
- 9.Coverley, D., H. Laman, and R. A. Laskey. 2002. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 4:523-528. [DOI] [PubMed] [Google Scholar]
- 10.Coverley, D., C. Pelizon, S. Trewick, and R. A. Laskey. 2000. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113:1929-1938. [DOI] [PubMed] [Google Scholar]
- 11.Delmolino, L. M., P. Saha, and A. Dutta. 2001. Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 276:26947-26954. [DOI] [PubMed] [Google Scholar]
- 12.Desdouets, C., C. Santocanale, L. S. Drury, G. Perkins, M. Foiani, P. Plevani, and J. F. X. Diffley. 1998. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J. 17:4139-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimitrova, D. S., T. A. Prokhorova, J. J. Blow, I. T. Todorov, and D. M. Gilbert. 2002. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 115:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, M., Y. Ishimi, H. Nakamura, T. Kiyono, and T. Tsurumi. 2002. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 277:10354-10361. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, M., C. Yamada, H. Goto, N. Yokoyama, K. Kuzushima, M. Inagaki, and T. Tsurumi. 1999. Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human mcm complex, and CDC2 kinase-mediated hyperphosphorylation. J. Biol. Chem. 274:25927-25932. [DOI] [PubMed] [Google Scholar]
- 17.Hua, X. H., and J. Newport. 1998. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and Cdc6, but dependent on Cdk2. J. Cell Biol. 140:271-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, W., N. J. Wells, and T. Hunter. 1999. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 96:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krude, T., C. Musahl, R. A. Laskey, and R. Knippers. 1996. Human replication proteins hCdc21, hCdc46, and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J. Cell Sci. 109:309-318. [DOI] [PubMed] [Google Scholar]
- 20.Li, G., G. Sudlow, and A. S. Belmont. 1998. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140:975-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, C., M. Weinreich, and B. Stillman. 1995. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81:667-676. [DOI] [PubMed] [Google Scholar]
- 22.Madine, M. A., C.-Y. Khoo, A. D. Mills, C. Musahl, and R. A. Laskey. 1995. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol. 5:1270-1279. [DOI] [PubMed] [Google Scholar]
- 23.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mimura, S., and H. Takisawa. 1998. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 17:5699-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Keefe, R. T., S. C. Henderson, and D. L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116:1095-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelizon, C., M. A. Madine, P. Romanowski, and R. A. Laskey. 2000. Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 14:2526-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen, B. O., J. Lukas, C. S. Sorensen, J. Bartek, and K. Helin. 1999. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18:396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. L. Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatti, S., T. Bohm, J. H. Cocker, J. F. X. Diffley, and K. Nasmyth. 1996. Activation of S-phase-promoting CDKs in late G1 defines a ‘point of no return' after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 10:1516-1531. [DOI] [PubMed] [Google Scholar]
- 31.Piatti, S., C. Lengauer, and K. Nasmyth. 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14:3788-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romanowski, P., M. A. Madine, and R. A. Laskey. 1996. XMcm7, a novel member of the Xenopus MCM family, interacts with XMcm3 and colocalizes with it throughout replication. Proc. Natl. Acad. Sci. USA 93:10189-10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha, P., J. Chen, K. C. Thome, S. J. Lawlis, Z.-H. Hou, M. Hendricks, J. D. Parvin, and A. Dutta. 1998. Human CDC6/Cdc18 associates with Orc1 and cyclin-Cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 18:2758-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santocanale, C., and J. F. X. Diffley. 1996. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 15:6671-6679. [PMC free article] [PubMed] [Google Scholar]
- 35.Stoeber, K., A. D. Mills, Y. Kubota, T. Krude, P. Romanowski, K. Marheineke, R. A. Laskey, and G. H. Williams. 1998. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 17:7219-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoeber, K., T. D. Tlsty, L. Happerfield, G. A. Thomas, S. Romanov, L. Bobrow, E. D. Williams, and G. H. Williams. 2001. DNA replication licensing and human cell proliferation. J. Cell Sci. 114:2027-2041. [DOI] [PubMed] [Google Scholar]
- 37.Tada, S., J. P. Chong, H. M. Mahbubani, and J. J. Blow. 1999. The RLF-B component of the replication licensing system is distinct from Cdc6 and functions after Cdc6 binds to chromatin. Curr. Biol. 9:211-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka, S., and J. F. Diffley. 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 4:198-207. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 41.Todorov, I. T., A. Attaran, and S. E. Kearsey. 1995. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J. Cell Biol. 129:1433-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D. S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11:997-1008. [DOI] [PubMed] [Google Scholar]
- 43.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 45.Weinreich, M., C. Liang, and B. Stillman. 1999. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, R. S., R. V. Shohet, and B. Stillman. 1997. A human protein related to yeast Cdc6p. Proc. Natl. Acad. Sci. USA 94:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]
- 48.Yan, Z., J. DeGregori, R. Shohet, G. Leone, B. Stillman, J. R. Nevins, and R. S. Williams. 1998. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 95:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye, Q., Y.-F. Hu, H. Zhong, A. C. Nye, A. S. Belmont, and R. Li. 2001. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 155:911-921. [DOI] [PMC free article] [PubMed] [Google Scholar]