Abstract
We have analyzed 75 isolates of Plasmodium falciparum, collected in Venezuela during both the dry (November) and rainy (May–July) seasons, with a range of genetic markers including antigen genes and 14 random amplified polymorphic DNA (RAPD) primers. Thirteen P. falciparum stocks from Kenya and four other Plasmodium species are included in the analysis for comparison. Cross-hybridization shows that the 14 RAPD primers reveal 14 separate regions of the parasite's genome. The P. falciparum isolates are a monophyletic clade, significantly different from the other Plasmodium species. We identify three RAPD characters that could be useful as “tags” for rapid species identification. The Venezuelan genotypes fall into two discrete genetic subdivisions associated with either the dry or the rainy season; the isolates collected in the rainy season exhibit greater genetic diversity. There is significant linkage disequilibrium in each seasonal subsample and in the full sample. In contrast, no linkage disequilibrium is detected in the African sample. These results support the hypothesis that the population structure of P. falciparum in Venezuela, but not in Africa, is predominantly clonal. However, the impact of genetic recombination on Venezuelan P. falciparum seems higher than in parasitic species with long-term clonal evolution like Trypanosoma cruzi, the agent of Chagas' disease. The genetic structure of the Venezuelan samples is similar to that of Escherichia coli, a bacterium that propagates clonally, with occasional genetic recombination.
Plasmodium falciparum, the agent of the most malignant form of malaria, causes 1.5–2.7 million deaths each year, mostly among children in Sub-Saharan Africa. The incidence of malaria in the world is estimated to be from 300 to 500 million infections annually, and more than 2.3 billion people live in areas with malaria risk (1). P. falciparum shows remarkable genetic diversity, which allows the parasite to adapt to drugs and the host's immune response. This genetic diversity has been manifested by isoenzyme and antigenic methods, drug tests, and gene sequencing (2–5).
The human infective state of P. falciparum is haploid, whereas the diploid form occurs in the mosquito vector, where gamete fertilization takes place, rapidly followed by the formation of haploid sporozoites. The sporozoites are inoculated to the human host by the mosquito in association with its blood meal. The mating system and the population structure of P. falciparum have been the subject of debate. Some authors have postulated a panmictic population structure based on the following evidence: (i) sex is an obligatory event for the life cycle of the parasite to be completed, (ii) genetic recombinants have been obtained in experimental models (6), and (iii) epidemiological data derived from several regions of the world that have different endemicities and intensities of transmission (7–9).
The clonality hypothesis is supported by the following evidence: (i) statistically significant linkage disequilibrium in some populations analyzed by isoenzymes, showing significant departures from panmictic expectations (10–12), (ii) low frequency of P. falciparum heterozygotes in the mosquito vector from regions with different transmission intensities such as Tanzania (8) and Papua New Guinea (13), (iii) high rate of self-fertilization in endemic areas (13), (iv) female-biased sex ratio in P. falciparum (14), which could lead to a situation of de facto clonality (offspring genetically identical to the parents), and (v) rates of intragenic recombination and strength of linkage disequilibrium between nucleotide sites that are consistent with a clonal population structure (15).
The results available therefore are not fully consistent, which may suggest that P. falciparum has different population structures in different places, ranging from clonality to panmixia depending on the incidence of transmission. The population structure impacts the propagation and stability in space and time of multilocus genotypes, which has important consequences for strain typing, pathogenicity, vector specificity, and susceptibility to drugs and vaccines. Given the epidemiological significance of the matter, it would seem desirable to ascertain the population structure of this parasite in several endemic areas with different rates of transmission.
We study the genetic diversity and the population structure of P. falciparum in natural isolates from the State of Bolivar, the main endemic area in Venezuela. To our knowledge, this is the first study conducted in an endemic area with low transmission intensity (compared with Africa's) while using a significant number of natural isolates of P. falciparum analyzed by a heterogeneous set of multilocus markers, which increases the robustness of the results.
Materials and Methods
Study Area and Collection of Blood Samples.
Samples were collected in 1995 during the dry and rainy seasons in several communities of the malarious region of the State of Bolivar in a tropical humid forest in southeastern Venezuela. Malaria transmission in Venezuela is seasonal, with two well marked periods, one of high transmission during the rainy season (May–October) followed by low transmission during the dry season (November–April).
The 75 new isolates of P. falciparum were collected from malaria-infected miners and Amerindians, who manifested moderate parasitemia detected by microscopy and had not received any antimalarial drugs during the previous 14 days. The parasites were cultured for 24–36 h according to the protocol of Trager and Jensen (16) to increase the number of parasites, and they were immediately cryopreserved. The collection and short-term culture of samples were performed in the Field Institute “Francisco Vitanza” and in Tumeremo's Hospital, both located in Tumeremo, State of Bolivar, Venezuela.
As a reference group from a high transmission region, 13 isolates of P. falciparum from Kenya were investigated also. In addition, four other Plasmodium species (P. cynomolgi, P. vivax, P. malariae, and P. chabaudi) were included as outgroups. The study protocols were approved by the Biomedical Committee of Malariology, Venezuelan Ministry of Health.
Extraction of Genomic DNA.
Parasite DNA was extracted from infected red blood cells by using proteinase K, followed by extraction with phenol-chloroform and ethanol precipitation.
Amplification by PCR of the Genes Msp-1, Msp-2, Resa, and Csp.
Multiple co-infecting parasite genotypes within an individual patient are frequent (2, 7, 17), which renders tentative the delimitation of multilocus genotypes. To detect (and eliminate) mixed infections in the sample under study, we used a PCR protocol (18). We used genes that have conserved 5′ and 3′ ends but contain an intermediate region with blocks of repeated segments that vary in length and DNA sequence from strain to strain (19).
We assayed four surface antigen genes encoding (i) the precursor of the major merozoite surface protein, MSP-1 (20), (ii) the major surface protein 2, MSP-2 (21, 22), (iii) the ring-infected erythrocyte surface antigen, RESA (23), and (iv) the circumsporozoite surface protein, CSP (24).
Because the primers are similar in size (about 20 bp) and about 50% in G/C content, we used the same PCR conditions for all four genes. The genes were amplified by using 100 ng of genomic DNA for each isolate of P. falciparum under the following conditions: 1 μM of each primer, 200 μM of each dNTP, buffer (50 mM KCL/10 mM Tris, pH 8.4/1.5 mM MgCl2/0.1 mg/ml gelatin), and 2.5 units of Taq polymerase in 100-μl reactions, in the presence of negative control. The genes were amplified in a GeneAmp PCR system 9600 thermocycler (Perkin–Elmer/Cetus) with initial denaturation at 94°C for 2 min followed by 37 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 2 min, and final extension at 72°C for 5 min. The PCR products were analyzed by electrophoresis on a 2% agarose gel and stained with ethidium bromide for UV visualization.
Random Amplified Polymorphic DNA (RAPD).
We have used this method for the genetic fingerprinting of several parasitic protozoan species including P. falciparum (25). It was performed as described (26) with slight modifications. Forty 10-mer primers (Operon Technologies, Alameda, CA) were tested initially, of which 14 primers (Table 1) were retained because they produced clear and highly reproducible amplification profiles. RAPD polymorphism was analyzed on ethidium bromide-stained agarose gels.
Table 1.
Sequences of the 14 RAPD primers
| Primer | Sequence | Primer | Sequence |
|---|---|---|---|
| A2 | 5′-TGCCGAGCTG-3′ | A17 | 5′-GACCGCTTGT-3′ |
| A7 | 5′-GAAACGGGTG-3′ | A18 | 5′-AGGTGACCGT-3′ |
| A8 | 5′-GTGACGTAGG-3′ | A20 | 5′-GTTGCGATCC-3′ |
| A9 | 5′-GGGTAACGCC-3′ | R5 | 5′-GACCTAGTGG-3′ |
| A10 | 5′-GTGATCGCAG-3′ | R6 | 5′-GTCTACGGCA-3′ |
| A11 | 5′-CAATCGCCGT-3′ | U10 | 5′-ACCTCGGCAC-3′ |
| A12 | 5′-TCGGCGATAG-3′ | B17 | 5′-AGGGAACGAG-3′ |
DNA Probe Labeling.
RAPD-amplified products were purified by Wizard PCR preps (Promega). The DNA concentration was measured by spectrophotometer and 32P-labeled by nick translation (Roche Molecular Biochemicals) following the manufacturer's protocol. The labeled DNA was purified by chromatocentrifugation on column Quik Spin Sephadex G-50 (Roche Molecular Biochemicals).
Dot-Blot and Hybridization.
To ascertain the independence of the genetic profiles generated by the different RAPD primers, we performed cross-hybridization experiments by using the dot-blot technique. The amplified product obtained from a given 10-mer primer for a given DNA was applied on the membrane and hybridized with a probe consisting of the whole amplification product derived from another primer.
Dot blot was performed as follows: the RAPD-amplified product (10 ng) was denatured by heating at 95°C for 10 min followed by rapid cooling in ice water for 10 min. After the addition of 1 volume (100 μl) of cold 20× SSC (0.3 M Tri-sodium citrate/3 M NaCl, pH 7.0), samples were applied onto a nylon membrane (Hybond N+, Amersham Pharmacia) by using manifold vacuum filtration (“dot-blot”) apparatus (BRL). The membranes were treated with denaturing solution followed by neutralizing solution and were fixed by alkaline (27). Filters were prehybridized and hybridized as described (27) by using probe-labeled DNA.
Phylogenetic Analysis.
Phylogenetic relationships among the Venezuelan isolates were inferred from RAPD data by computing Jaccard's distance (28), which measures the proportion of band mismatches between pairs of isolates. Jaccard's distance is Dij = 1 − [a/(a + b + c)], where a = the number of bands that are common to the i and j stocks, b = the number of bands present in the first genotype and absent in the second, and c = the number of bands absent in the first genotype and present in the second.
The unweighted pair-group method with arithmetic means (UPGMA; ref. 29) and neighbor-joining (NJ) methods (30) were used to construct dendrograms. These two methods are based on genetic distances. In addition, we carried out a cladistic analysis relying on Wagner's parsimony model (31, 32) with the presence/absence of each RAPD fragment as a character state. The robustness of the branches was assessed by bootstrap (33).
Linkage disequilibrium (nonrandom association between genotypes occurring at different loci) was tested by using the f test (10, 11, 34) based on the null hypothesis that recombination occurs at random in the population under investigation (panmixia). The f test was performed by using Monte Carlo simulations with 104 iterations.
Results
Genetic Polymorphism in the Venezuelan Sample.
Of the 75 P. falciparum isolates from Venezuela, 16 were excluded because the genetic markers (Msp-1, Msp-2, Resa, and Csp) manifested mixed infections or did not show a good amplification signal by RAPD. Among the 59 remaining stocks, 58 different multilocus genotypes were detected (genotypic diversity = 0.98); only two stocks, V19Nov and V23Nov, exhibited the same genotype. Jaccard's genetic distances ranged from 0 to 0.70 (maximum possible = 1.0) with a mean of 0.37 ± 0.14. Fig. 1 shows an NJ dendrogram that includes the other Plasmodium species in addition to the Venezuelan stocks. The sample distribution for both NJ and UPGMA methods was very similar, although these two phylogenetic methods are based on different principles. UPGMA assumes that rates of evolution are constant, whereas NJ accepts variable rates of evolution. In the NJ tree (Fig. 1), the P. falciparum stocks are clearly distinct from the other species. The P. falciparum parasites from Venezuela are subdivided into two major branches that include all stocks but V31Jul. The top major branch is divided into three lesser subdivisions. The two first subdivisions correspond to all parasites collected in November except stock V1Nov. The third subdivision only includes parasites collected in July. The lower major branch is genetically very heterogeneous and shows no clear subdivisions. It only includes stocks collected either in July or May except V1Nov, which was collected in November. The isolate V31Jul falls outside of the two major subdivisions.
Figure 1.
NJ tree (30) depicting the phylogenetic diversity of the P. falciparum stocks from Venezuela and four other Plasmodium species. Stocks of P. falciparum are designated by a ‘V,’ for Venezuela, followed by the number of the sample within the month of collection. The other Plasmodium species are: P. chabaudi, P. malariae, P. vivax, and P. cynomolgi.
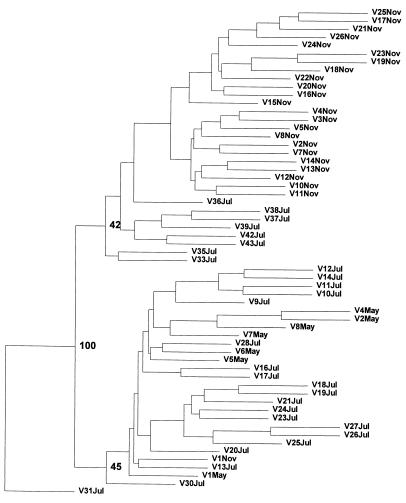
A tree obtained by cladistic analysis with the Venezuelan stocks only, using V31Jul as outgroup (Fig. 2), shows a similar distribution into two main branches, although the bootstrap values supporting these branches are low (42 and 45%, respectively). When including the other species in the cladistic analysis, the P. falciparum group is supported by a bootstrap value of 99%.
Figure 2.
Cladistic tree depicting the genetic diversity of the Venezuelan P. falciparum stocks. Bootstrap values based on 100 replications are shown at the nodes of the three major subdivisions.
Linkage Disequilibrium.
Significant linkage disequilibrium (P < 10−4) exists in the entire sample of the 59 Venezuelan stocks analyzed as well as within each of the two seasonal samples treated separately. For the November sample, we performed the test with and without V1Nov, which is phylogenetically distant from the rest of the November sample. The results remain equally significant. Linkage disequilibrium remains unchanged when the repeated genotype (stocks V19Nov and V23Nov) is counted only once. Last, we performed the analysis on the two main subgroups manifest in Fig. 1. V31Jul, which is outside of the two main subgroups, was not included in this analysis. These results are also significant (P < 10−4).
Genetic Polymorphism in the Stocks from Kenya.
For comparison purposes, we analyzed the genetic polymorphism of 13 Kenyan stocks of P. falciparum by using eight different RAPD primers (A-7, A-9, A-10, A-11, A-12, A-17, A-18, and A-20; Table 1). Thirteen different genotypes (genotypic diversity = 1) appeared. The f test shows no statistically significant linkage disequilibrium for this Kenyan sample.
Discussion
Genetic Diversity of the Venezuelan Sample.
Previous studies conducted in a low-transmission area (French Guiana) have shown that P. falciparum populations have in that region low genetic diversity (35). In contrast, we have found much genetic polymorphism in the natural populations of P. falciparum in Venezuela. We found 58 different genotypes among the 59 parasite isolates examined. The genetic diversity of these isolates is large. The mean value of Jaccard's genetic distance is 0.37 ± 0.14, and the largest value observed is 0.70; the maximum possible is 1.0. These values are comparable to those obtained with RAPD markers in Trypanosoma brucei, the agent of human African trypanosomiasis, and are higher than the values recorded in several species of Leishmania (36).
Phylogenetic Individualization of P. falciparum.
The position of the four other Plasmodium species in the phylogenetic analysis indicates that P. falciparum is monophyletic. Three RAPD synapomorphic characters (“tags”) (37) are identified in the present study (R5, 1-3-6-7; R6, 3-4-5-6; and U10, 2-3-4-6-7-8-9; the numbers listed after the name of the primer refer to the RAPD profile). These RAPD fragments are being sequenced presently to design specific PCR primers for fast and easy identification of P. falciparum in epidemiological surveys. The rate of molecular evolution of the RAPD markers, which conditions their resolving value (36), is suitable for ascertaining the discreteness of the P. falciparum set of isolates in relation to the four other Plasmodium species (Fig. 1). However, this rate of molecular evolution is too fast for reliable discrimination of the phylogenetic relationship among these species. At high levels of phylogenetic divergence, the RAPD markers (as well as enzyme-electrophoresis markers) get “saturated” and generate much homoplasy (38).
Seasonal Distribution of the Genotypes.
The rainy season (May–October) is a period of high malarial transmission in Venezuela, whereas the dry season (November–April) is a period of low transmission. Our phylogenetic analysis shows an obvious association between season of transmission and genetic diversity. All stocks collected in November (dry season) except V1Nov fall into the same genetic lesser subdivision of the top branch (Fig. 1). Moreover, again with the exception of V1Nov, the November sample shows much lower genetic diversity than the May–July sample.
Population Structure.
The most remarkable result of this study is the finding of highly significant linkage disequilibrium in the presence of high levels of genetic variation. Highly significant linkage disequilibrium is the case when all Venezuelan samples are examined as a set as well as when parasites are separated by season. It is not parsimonious to recourse to the Wahlund effect to account for the results for two reasons. First, all samples were collected from a relatively small area (the maximum distance among localities is 200 km), and the vectors (anopheline mosquitoes) and hosts (humans) of P. falciparum move considerably. Second, there is no association between locality and genotype distribution. The localities are distributed randomly on the phylogenetic trees. When the November and May/July samples are treated separately, the level of significance remains the same. The linkage disequilibrium in the whole sample cannot be explained therefore by different genotype distribution between seasons of transmission. The result of the November sample remains the same when the VNov1 isolate, which has a genotype drastically different from the others, is excluded. Moreover, the level of significance remains the same when the repeated genotype (stocks VNov19 and VNov23) is counted only once, a procedure recommended (39) for distinguishing between clonal evolution and epidemic clonality (a basically sexual species that undergoes occasional bouts of clonality). To verify whether linkage could be attributable mainly to the presence of two main subdivisions tending to discreteness, we have analyzed the two main subgroups apparent in Figs. 1 and 2 separately. For the bottom subgroup, we have done the analysis with and without V1Nov. In all cases, linkage disequilibrium remains highly significant (P < 10−4).
This linkage disequilibrium cannot be explained by the hypothesis that different RAPD primers do not explore independent regions of the genome and therefore convey redundant information. We have shown by cross-hybridization experiments that the different RAPD primers have no overlapping specificity, and consequently they explore distinct genome regions.
The strong linkage disequilibrium in the populations surveyed suggests that clonal propagation plays an important role in the population structure of P. falciparum in Venezuela. This inference is consistent with the results obtained in other Venezuelan P. falciparum populations characterized with three single-copy genes (A. Tami, personal communication) and with the model of clonal population structure inferred by Rich et al. (15) from gene sequencing data for a worldwide sample. It is worth noting that mutations of the genes Dhfr and Dhps associated with pirymethamine/sulfadoxine resistance (40) in samples from Venezuela have shown the joint presence of the specific mutations in both genes with high incidence (96%). The high prevalence and fast diffusion of resistant genotypes could be a direct consequence of clonality, which would facilitate the rapid spread of multiresistance to antimalarial drugs.
It is informative to compare our present results with others from Africa. Our Kenyan sample shows no linkage disequilibrium. However, a study conducted on other African populations has shown linkage disequilibrium with multilocus enzyme electrophoresis; this linkage was absent with RAPD data (12). This and the present study were performed by the same team of investigators by using the same molecular techniques and the same statistics. Lack of linkage disequilibrium in the Kenyan populations of P. falciparum has been recorded also by Qari's study (41) of genetic diversity in three antigen genes.
It is thus apparent that the population structure of P. falciparum is notably different between Venezuela and Africa. A reasonable hypothesis to account for such difference is that lower rates of transmission such as occur in Venezuela favor high rates of self-fertilization, which lead to de facto clonal propagation (offspring genotypes identical to the parent's genotypes), whereas high rates of transmission (like in Africa) favor cross-fertilization (42). Recent studies conducted in nine localities worldwide confirm that dramatic differences in P. falciparum population structure can be observed because six of the nine populations studied showed strong linkage disequilibrium (43).
But even in the case of the Venezuelan populations, the population structure of P. falciparum seems quite different from that of other parasitic protozoa such as T. cruzi, the agent of Chagas' disease. Tibayrenc (36) has proposed that the relevant boundary with respect to the population structure of pathogenic microorganisms is between those species that exhibit clear-cut genetic subdivisions (discrete typing units or DTUs; refs. 37 and 44) and those species in which the persistence of such DTUs is thwarted by genetic exchange. Although T. cruzi seems to have undergone exceptional events of hybridization between different clonal lines (44, 45), it is clearly subdivided into two main DTUs (25, 36, 38), of which one subdivided into five lesser DTUs (44). All these subdivisions are supported by high bootstrap values (44), whereas the clusters revealed by phylogenetic analysis in the present sample of P. falciparum stocks are supported by low bootstrap values. It is apparent that the impact of genetic recombination in the Venezuelan population of P. falciparum is higher than in T. cruzi. An instructive comparison can be made with Escherichia coli, a bacterium able of both clonal propagation and genetic recombination. RAPD analysis of a large E. coli sample (46, 47) has confirmed the occurrence of strong linkage disequilibrium, consistent with the results obtained by multilocus enzyme electrophoresis (48). However, the genetic subdivisions revealed within E. coli by our phylogenetic analysis (46, 47) are supported by weak bootstrap values only, whereas these values are very high for the DTUs that subdivide T. cruzi (44). The RAPD analyses of E. coli and of the Venezuelan sample of P. falciparum give similar results in terms of population structure, equally different but in different directions, from the results related in one direction to T. cruzi (clonal evolution with very rare events of hybridization and strong structuration) and in the opposite direction to African populations of P. falciparum (tending to panmixia).
Acknowledgments
We thank S. Qari for providing the P. falciparum isolates from Kenya, O. Mercereau-Puijalon for supplying the P. cynomolgi and P. chabaudi reference stocks, and the teams at the Field Institute “Francisco Vitanza” and Tumeremo Hospital for their help in the collection of samples. This work was supported by a fellowship of the World Bank-Malariologia, Venezuela.
Abbreviations
- RAPD
random amplified polymorphism DNA
- UPGMA
unweighted pair-group method with arithmetic means
- NJ
neighbor joining
- DTU
discrete typing unit
References
- 1.World Health Organization. Wkly Epidemiol Rec. 1997;72:269–292. [Google Scholar]
- 2.Carter R, Voller A. Trans R Soc Trop Med Hyg. 1975;69:371–376. doi: 10.1016/0035-9203(75)90191-1. [DOI] [PubMed] [Google Scholar]
- 3.Creasey A, Fenton B, Walker A, Thaithong S, Oliveira S, Mutambu S, Walliker D. Am J Trop Med Hyg. 1990;42:403–413. doi: 10.4269/ajtmh.1990.42.403. [DOI] [PubMed] [Google Scholar]
- 4.McBride J S, Walliker D, Morgan G. Science. 1982;217:254–257. doi: 10.1126/science.6178159. [DOI] [PubMed] [Google Scholar]
- 5.Kemp D J, Cowman A F, Walliker D. Adv Parasitol. 1990;29:75–149. doi: 10.1016/s0065-308x(08)60105-0. [DOI] [PubMed] [Google Scholar]
- 6.Walliker D, Quakyi I A, Wellems T E, McCutchan T F, Szarfman A, London W T, Corcoran L M, Burkot T R, Carter R. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 7.Babiker H A, Creasey A M, Fenton B, Bayoumi R A L, Arnot D E, Walliker D. Trans R Soc Trop Med Hyg. 1991;85:572–577. doi: 10.1016/0035-9203(91)90347-2. [DOI] [PubMed] [Google Scholar]
- 8.Babiker H A, Ranford-Cartwright L C, Currie D, Charlwood J D, Billingsley P, Teuscher T, Walliker D. Parasitology. 1994;109:413–421. doi: 10.1017/s0031182000080665. [DOI] [PubMed] [Google Scholar]
- 9.Conway D J, McBride J S. Parasitology. 1991;103:7–16. doi: 10.1017/s0031182000059229. [DOI] [PubMed] [Google Scholar]
- 10.Tibayrenc M, Kjellberg F, Ayala F J. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tibayrenc M, Kjellberg F, Ayala F J. Bioscience. 1991;41:767–774. [Google Scholar]
- 12.Ben Abderrazak S, Ouri B, Altaf L, Bosseno M F, Force-Barge P, Dujardin J P, Fandeur T, Molez J F, Kjellberg F, Ayala F, Tibayrenc M. Exp Parasitol. 1999;92:232–238. doi: 10.1006/expr.1999.4424. [DOI] [PubMed] [Google Scholar]
- 13.Paul R E L, Packer M J, Walmsley M, Lagog M, Ranford-Cartwright L C, Paru R, Day K P. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 14.Read A, Anwar M, Shutler D, Nee S. Proc R Soc London. 1995;260:359–363. doi: 10.1098/rspb.1995.0105. [DOI] [PubMed] [Google Scholar]
- 15.Rich S M, Hudson R R, Ayala F J. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trager W, Jensen J B. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 17.Paul R E L, Hackford I, Brockman A, Muller-Graf C, Price R, Luxemburger C, White N J, Nosten F, Day K P. Am J Trop Med Hyg. 1998;58:195–203. doi: 10.4269/ajtmh.1998.58.195. [DOI] [PubMed] [Google Scholar]
- 18.Wooden J, Gould E E, Paull A T, Sibley C H. Exp Parasitol. 1992;75:207–212. doi: 10.1016/0014-4894(92)90180-i. [DOI] [PubMed] [Google Scholar]
- 19.Kemp D J, Coppel R L, Anders R F. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 20.Mackay M, Goman M, Bone N, Hyde J E, Scaife J, Certa U, Stunnenberg H, Bujard H. EMBO J. 1985;4:3823–3829. doi: 10.1002/j.1460-2075.1985.tb04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenton B, Clark J T, Khan C M A, Robinson J V, Walliker D, Ridley R, Scaife J G, McBride J S. Mol Cell Biol. 1991;11:963–971. doi: 10.1128/mcb.11.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smythe J A, Coppel R L, Day K P, Martin R K, Oduola A M, Kemp D J, Anders R F. Proc Natl Acad Sci USA. 1991;88:1751–1755. doi: 10.1073/pnas.88.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favalaro J M, Coppel R L, Corcoran L M, Foote S J, Brown G V, Anders R F, Kemp D J. Nucleic Acids Res. 1986;14:8265–8277. doi: 10.1093/nar/14.21.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dame J B, Williams J L, McCutchan T F, Weber J L, Wirtz R A. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 25.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Sarkeski D, Ayala F J. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouri B, Dutrait N, Bastrenta B, Tibayrenc M. J Parasitol. 1997;83:52–57. [PubMed] [Google Scholar]
- 28.Jaccard P. Bull Soc Vaud Sci Nat. 1908;44:223–270. [Google Scholar]
- 29.Sneath P H A, Sokal R R. Numerical Taxonomy. Vol. 39. San Francisco: Freeman; 1973. pp. 783–791. [Google Scholar]
- 30.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Kluge A G, Farris J S. Syst Zool. 1969;18:1–32. [Google Scholar]
- 32.Farris J S. Syst Zool. 1970;19:83–92. [Google Scholar]
- 33.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 34.Zang Q, Tibayrenc M, Ayala F J. J Protozool. 1988;35:81–85. doi: 10.1111/j.1550-7408.1988.tb04081.x. [DOI] [PubMed] [Google Scholar]
- 35.Ariey F, Chalvet W, Hommel D, Peneau C, Hulin A, Mercereau-Puijalon O, Duchemin J B, Sarthou J L, Reynes J M, Fandeur T. Am J Trop Med Hyg. 1999;61:978–985. doi: 10.4269/ajtmh.1999.61.978. [DOI] [PubMed] [Google Scholar]
- 36.Tibayrenc M. Adv Parasitol. 1995;36:47–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 37.Tibayrenc M. Int J Parasitol. 1998;28:85–104. doi: 10.1016/s0020-7519(97)00180-x. [DOI] [PubMed] [Google Scholar]
- 38.Tibayrenc M. Annu Rev Microbiol. 1996;50:401–429. doi: 10.1146/annurev.micro.50.1.401. [DOI] [PubMed] [Google Scholar]
- 39.Maynard S, Smith N H, O'Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urdaneta L, Plowe C V, Goldman I, Lal A. Am J Trop Med Hyg. 1999;61:457–462. doi: 10.4269/ajtmh.1999.61.457. [DOI] [PubMed] [Google Scholar]
- 41.Qari S H. Ph.D. thesis. France: University of Montpellier; 1998. [Google Scholar]
- 42.Paul R E L, Day K. Parasitol Today. 1998;14:197–202. doi: 10.1016/s0169-4758(98)01226-5. [DOI] [PubMed] [Google Scholar]
- 43.Anderson T J, Haubold B, Williams J T, Estrada-Franco J G, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, et al. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 44.Brisse S, Barnabé C, Tibayrenc M. Int J Parasitol. 2000;30:35–44. doi: 10.1016/s0020-7519(99)00168-x. [DOI] [PubMed] [Google Scholar]
- 45.Carrasco H J, Frame I A, Valente S A, Miles M A. Am J Trop Med Hyg. 1996;54:418–424. doi: 10.4269/ajtmh.1996.54.418. [DOI] [PubMed] [Google Scholar]
- 46.Grandhomme M. Pharm.D. thesis dissertation. France: University of Montpellier; 1999. [Google Scholar]
- 47.Tibayrenc M. Annu Rev Genet. 1999;33:449–477. doi: 10.1146/annurev.genet.33.1.449. [DOI] [PubMed] [Google Scholar]
- 48.Whittam T S, Ochman H, Selander R K. Proc Natl Acad Sci USA. 1983;80:1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]