Abstract
Spores formed by wild-type Bacillus subtilis are encased in a multilayered protein structure (called the coat) formed by the ordered assembly of over 30 polypeptides. One polypeptide (CotB) is a surface-exposed coat component that has been used as a vehicle for the display of heterologous antigens at the spore surface. The cotB gene was initially identified by reverse genetics as encoding an abundant coat component. cotB is predicted to code for a 43-kDa polypeptide, but the form that prevails in the spore coat has a molecular mass of about 66 kDa (herein designated CotB-66). Here we show that in good agreement with its predicted size, expression of cotB in Escherichia coli results in the accumulation of a 46-kDa protein (CotB-46). Expression of cotB in sporulating cells of B. subtilis also results in a 46-kDa polypeptide which appears to be rapidly converted into CotB-66. These results suggest that soon after synthesis, CotB undergoes a posttranslational modification. Assembly of CotB-66 has been shown to depend on expression of both the cotH and cotG loci. We found that CotB-46 is the predominant form found in extracts prepared from sporulating cells or in spore coat preparations of cotH or cotG mutants. Therefore, both cotH and cotG are required for the efficient conversion of CotB-46 into CotB-66 but are dispensable for the association of CotB-46 with the spore coat. We also show that CotG does not accumulate in sporulating cells of a cotH mutant, suggesting that CotH (or a CotH-controlled factor) stabilizes the otherwise unstable CotG. Thus, the need for CotH for formation of CotB-66 results in part from its role in the stabilization of CotG. We also found that CotB-46 is present in complexes with CotG at the time when formation of CotB-66 is detected. Moreover, using a yeast two-hybrid system, we found evidence that CotB directly interacts with CotG and that both CotB and CotG self-interact. We suggest that an interaction between CotG and CotB is required for the formation of CotB-66, which may represent a multimeric form of CotB.
During the process of sporulation in the gram-positive soil bacterium Bacillus subtilis the developing spore is encased in a complex protein structure called the coat, which confers resistance to several physicochemical agents and contributes to the response of spores to the presence of germinants (7, 8, 15). The coat is formed by over 30 polypeptides, ranging in size from about 6 to about 70 kDa, which are assembled into a lamellar inner coat and a thick electron-dense outer coat (7, 8, 15). With only one possible exception (38), synthesis of the coat structural components is restricted to the mother cell chamber of the sporulating cell and is temporally governed by a cascade of transcription factors in the order σE, SpoIIID, σK, and GerE (7, 8, 15, 24, 35, 40). σE and SpoIIID drive synthesis of a class of morphogenetic proteins that (irrespective of their association with the final coat structure) appear to guide the assembly of several structural components into the spore coat (reviewed in references 7, 8, and 15). For instance, spores produced by a cotE mutant fail to assemble the electron-dense outer coat and the remaining coat structure appears to lack, in addition to CotE, several other abundant components (47). The results of a recent study indicate that specific regions in CotE are required for the assembly of different proteins and suggest that CotE might control the assembly of several outer-coat components by direct protein-protein interactions (2, 28). Most of the coat structural components are synthesized (under the control of σK and GerE) at a later stage in coat assembly, and it is only after σK is activated that assembly of the coat is unequivocally recognized by electron microscopy of sporulating cells (7, 8, 15). Activation of σK results in the expression of several genes coding for spore coat proteins and also results in transcription of the gerE gene (4), which encodes an ambivalent transcriptional regulator of coat gene expression. GerE acts together with σK to activate a late class of cot genes, but it also represses transcription of other cot genes (18, 19, 45, 46). These regulatory circuits suggest that the time and level of expression of the genes coding for coat structural components are important for the correct assembly of the coat structure (7, 8, 15). Proper assembly of the coat further relies on mechanisms such as translational control (34) and posttranslational modifications, including proteolytical processing of larger precursors, protein secretion, and protein cross-linking (reviewed in references 7, 8, and 15). These modifications may provide an additional level of control over the timing of assembly of specific components. For example, SafA is a morphogenetic protein of about 45 kDa produced under σE control from hour 2 of sporulation onwards but the main form of SafA detected in the coats is a smaller (approximately 30 kDa) species corresponding to the C-terminal region of the protein (32, 33). This smaller species is produced by internal translation initiation (34). In addition, the full-length and 30-kDa forms of SafA are processed by the YabG protease, which is produced under the control of σK (42, 43). The exact contribution of these mechanisms to the ordered assembly of the various coat components is poorly understood, and determination of their nature and contribution will ultimately rely on the functional and structural characterization of selected components (11, 29). To learn more about the mechanisms involved in the morphogenesis of the coat structure, we analyzed the assembly of the outer-coat protein CotB. The cotB gene was initially found (by reverse genetics) to encode an abundant spore coat component (6), later shown to be in the outer coat (47), which appears to be surface exposed (20). CotB has been utilized as a vehicle for the presentation of heterologous antigens at the spore surface, suggesting its potential use in vaccine development (9, 20). Thus, the study of the assembly of CotB may allow a more precise manipulation of CotB as a fusion partner for heterologous antigen presentation. Also, it will expand our knowledge of the protein-protein interactions underlying assembly of a complex multiprotein structure and may provide us with tools for nanoengineering applications involving the B. subtilis spore. The cotB gene forms a cluster with two cot genes, cotH and cotG (6, 30, 36). Expression of cotH is under the control of σK, whereas both cotG and cotB are expressed later under the dual control of σK and GerE (18, 30, 36, 46, 45). Assembly of CotB-66 was shown to require expression of both cotG and cotH (30, 36).
We now show that cotB encodes a 46-kDa polypeptide (CotB-46) which is posttranslationally converted into a form of about 66 kDa (herein called CotB-66). This form of CotB (CotB-66) is equivalent to the 59-kDa protein previously reported by Donovan et al. (6). We show that formation of CotB-66 requires both cotG and cotH and that CotG does not accumulate in a cotH mutant. This suggests that the requirement for CotH or a CotH-dependent protein for CotB-66 formation results in part from its stabilization of CotG. We also found that CotB is present in complexes with CotG at the time when formation of CotB-66 is detected. Moreover, CotB was found to interact with itself and with CotG in a Saccharomyces cerevisiae two-hybrid assay. We suggest that formation of CotB-66 requires a direct interaction with CotG.
MATERIALS AND METHODS
Bacterial strains, media, and general techniques.
The B. subtilis strains used in this study are listed in Table 1. Sporulation was induced by nutrient exhaustion in Difco sporulation medium (31). Pfu polymerase (Stratagene) was used in all PCRs, and the cloned products were sequenced to ensure that no mutations were introduced. All other general methods were as described previously (5, 16, 17, 31, 38).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype/phenotype | Reference or source |
|---|---|---|
| MB24 | trpC2 metC3/wild type/Spo+ | Laboratory stock |
| DL067a | cotB::catb/Cmr | 6 |
| AH140 | trpC2 metC3 cotA::catb/Cmr | 29 |
| AH141 | trpC2 metC3 cotB::catb/Cmr | This work |
| AH1103 | trpC2 metC3 cotH::catb/Cmr | 48 |
| AH1497 | trpC2 metC3 cotGΩpMS43c/Cmr | 17 |
| AH1816 | BL21 (DE3) (pLysS) (pMS8) | This work |
| AH1834 | BL21 (DE3) (pLysS) (pMS16) | This work |
| AH2036 | trpC2 metC3 cotBΩpRZ24c/Nmr | This work |
| AH2055 | trpC2 metC3 cotBΩpRZ29c/Nmr, Spr | This work |
| AH2088 | trpC2 metC3 ΔamyE::6xHis-cotB/Nmr | This work |
| AH2089 | trpC2 metC3 ΔamyE::PcotEP1-cotB/Nmr | This work |
| AH2119 | trpC2 metC3 ywrJb/Cmr | This work |
Strain DL067 was obtained from the Bacillus Genetic Stock Center as strain 1S102.
The cotB, cotA, cotH, and ywrJ alleles are the result of double-crossover events that replaced part of the chromosomal sequence with a chloramphenicol resistance determinant.
The omega symbol (Ω) denotes that the plasmid was integrated via a single reciprocal crossover in the corresponding region of homology in the B. subtilis chromosome. Integration of pMS43 and pRZ29 resulted in the disruption of the cotG and cotB genes, respectively. Integration of pRZ24 creates an intact copy of the cotB gene under the control of its own promoter and fused at its 3′ end to a six-His-encoding sequence.
Integrational vectors.
A fragment encompassing a chloramphenicol resistance (Cmr) gene was generated by PCR using primers cat-958D and cat-2040R (Table 2) and plasmid pHV33 as the template (41). The 1,080-bp PCR fragment was digested with EcoRI and PstI and inserted between the same sites of pLITMUS 28 and pLITMUS 38 (New England Biolabs), creating the integrational vectors pMS38 and pMS39, respectively.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)/restriction site or six-His taga |
|---|---|
| cat-958D | GGGTAACTAGCCTGCAGGCAATAGTTACCC |
| cat-2040R | GGAGAAGTCGAATTCAGAAAAAGAAGGATATGG |
| cotB-226D | GGAATTTCATATGAGCAAGAGG/NdeI |
| cotB-1345R | CCATTATCTACTCGAGTTTACGTTTCCAGTGATAG/XhoI |
| cotB-235D | ATGAGCAAGAGGAGAATGAAATATC |
| cotB-990R | AATGAACGAATTCGCTGTCCTTATCAT/EcoRI |
| cotB-13D | GCGAGTATATTAAAAAGCTTTCACAATACC/HindIII |
| cotB-1437R | CCTCAACATCTGTGTTAAGGAATTCATTCAAAC/EcoRI |
| cotB-184Dhis | GGAGGAATTTGAATGCATCATCATCATCATCATAGCAAGAGGAGAATG/six-His tag |
| cotB-213Rhis | CATTCTCCTCTTGCTATGATGATGATGATGATGCATTCAAATTCCTCC/six-His tag |
| cotB-150D | GCAGTATGTTTATCATCTAGATATAAGTGACTAGG/XbaI |
| cotB-1373R | CTGAAAAAGGGAATTCACTTTATCTGATGCTCCTCC/EcoRI |
| cotB-7311D | GGAGGAATTTGAATGAGCAAGAGGAG |
| cotB-7706R | GTTTAACGCAGCGTAATCATCTCCCAGC |
| ywrJ-125D | GGAAACGTGAATCTAATAGATAATGGAGGAGC/EcoRI |
| ywrJ-913R | GCCTTCTGCTTTTCTCGAGGGCAGTTTTGTATACACTAGGG/XhoI |
| B/5/Nco | GAGCCATGGGAATGAGCAAGAGGAGA/NcoI |
| B/OMO/3 | GCCTAGGATCCGGGCATCACTTTATC/BamHI |
| G/5/Nco | GAGCCATGGAATTGGGCCACTATTCC/NcoI |
| G/Bam/3 | TACCTCCGCCGGGATCCTATTGAAAC/BamHI |
| H/5/Nde | CTAAGGAGGACATATGATGAAGAATC/NdeI |
| 3/H/Bam | CCGTGGATCCACCCATTTTCACGCAT/BamHI |
| H/5/Bam | GTAAGGAGGGATCCGGATGAAGAAT/BamHI |
| H/3/Xho | CGCGAATACCCTCGAGCACGCATTCA/XhoI |
Restriction sites or sequences encoding the six-His tag are underlined.
cotB, cotG, ywrJ, and cotH null mutants.
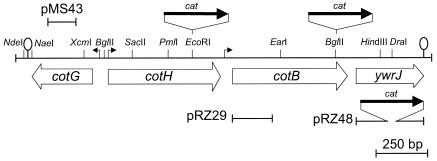
DNA from strain DL067 (obtained from the Bacillus Genetic Stock Center) (6) was used to construct AH141 (cotB::cat; Table 1). The cotH (AH1103) and cotG (AH1497) mutants (Table 1) have been described before (17, 48). To create an insertion within the 5′ end of the cotB gene, a 395-bp PCR fragment (generated with primers cotB-7311D and cotB-7706R) (Table 2) was cloned into pCR 2.1-TOPO (Invitrogen) to create pRZ28. Next, the insert was released with EcoRI and SphI and cloned between the same sites of pUS19 (3), yielding pRZ29. The single-reciprocal-crossover event (Campbell-type recombination) of pRZ29 at the cotB locus of strain MB24 (verified by PCR) produced the spectinomycin-resistant (Spr) strain AH2055. To disrupt the ywrJ gene, a 780-bp PCR product obtained with primers ywrJ-125D and ywrJ-913R (Table 2) was digested with EcoRI and XhoI and inserted into pET30a(+) (Novagen) to yield pRZ47. A Cmr cassette was then released from pMS38 with HindIII and SnaBI and introduced between the HindIII and DraI sites of pRZ47 to produce pRZ48 (Fig. 1). pRZ48 was cut with NdeI and XbaI and used to transform strain MB24 to Cmr, producing AH2119 (Table 1).
FIG. 1.
Diagram of the cotB region of the chromosome. The horizontal line depicts a partial restriction map of the region (only relevant sites are shown for reference), and the arrows below the restriction map indicate the orientation and extent of open reading frames in the region. The positions of previously constructed insertional mutations in cotG, cotH, and cotB are shown above the restriction map. Disruption of cotH, cotB, or ywrJ resulted from double-crossover (marker replacement) events, whereas cotG was disrupted by the single-reciprocal (Campbell-type) integration of pMS43 (36). cotB was also inactivated by the Campbell-type integration of pRZ29. Right-angle arrows represent promoters. The ywrJ gene is presumably transcribed from the cotB promoter. Putative transcriptional terminators are represented by the stem-loop structures.
Construction of strains carrying N- or C-terminal fusions of cotB to the six-His tag at the nonessential amyE locus.
A 1,424-bp PCR product encompassing the cotB promoter and coding region was generated with primers cotB-13D and cotB-1437R (Table 2) and cloned into pCR 2.1 TOPO to yield pRZ33. Plasmid pRZ33 served as a template to insert a six-His tag just downstream of the cotB initiation codon through the use of primers cotB-184Dhis and cotB-213RHis (QuickChange system; Stratagene) (Table 2). The modified cotB gene was released from the resulting plasmid (pRZ36) with EcoRI and HindIII and inserted between the same sites of the amyE integrational vector pMLK83 (21), thereby creating pRZ38. A neomycin-resistant (Nmr) transformant of strain MB24 with PstI- and ScaI-linearized pRZ38 that was AmyE− (indicating a marker replacement event at amyE) was identified and named AH2088 (Table 1). To create a 3′-end fusion of cotB to the six-His tag, a Cmr cassette was released from pMS39 with SpeI and NheI and introduced into XbaI-digested pMS16 (see below). This produced pRZ24, whose Campbell-type integration into the cotB locus of strain MB24 generated AH2036 (Table 1).
Construction of a strain expressing cotB from the cotE P1 promoter.
The cotB coding region and translation initiation signals were PCR amplified with primers cotB-150D and cotB-1373R (Table 2). The resulting 1,233-bp fragment was cleaved with EcoRI and XbaI and cloned downstream of the cotE P1 promoter in an amyE integrational vector (Costa and Henriques, unpublished results) derived from pMLK83 (21). This produced pRZ37, which (following digestion with ScaI) was used to transform strain MB24 with selection for Nmr. A AmyE− transformant was identified and named AH2089 (Table 1).
Overproduction and partial purification of CotB.
Primers cotB-226D and cotB-1345R (Table 2) were used to PCR amplify the 1,120-bp cotB coding region, and the product was cleaved with NdeI and XhoI and inserted between the NdeI and SalI sites of pET-30a(+) (Novagen) to yield pMS16. Primers cotB-235D and cotB-990R (Table 2) were used to PCR amplify a 756-bp fragment coding for the first 252 residues of CotB, which was cleaved with EcoRI and inserted between the EcoRI and EcoRV sites of pET-30a(+), creating pMS8. pMS16 or pMS8 was introduced into BL21(DE3) (pLysS) cells (Novagen), generating AH1834 or AH1816 (Table 1), respectively, in which the full-length CotB (CotB-FL) or its N-terminal half (CotBn) could be synthesized as C-terminal fusions to the six-His tag under the control of the T7lac promoter. Overproduction and partial purification of the His-tagged proteins was essentially as described previously (38). Both CotB-FL- and CotBn-6×His were used for the production of a rabbit polyclonal antiserum (Eurogentec, Herstal, Belgium). However, the antibody raised against CotB-FL was not very specific and the anti-CotBn antibody was used in all experiments herein reported.
Generation of an anti-CotG antibody.
A peptide (CDDYKRHDDYDSKKE) corresponding to residues 166 to 180 of the CotG primary structure (36) was synthesized and conjugated to keyhole limpet hemocyanin, and the conjugate was used to raise rabbit polyclonal antibodies against CotG (Eurogentec).
Preparation of B. subtilis whole-cell extracts and immunoblotting.
B. subtilis whole-cell lysates were prepared, and immunoblotting experiments were conducted as described previously (39) except that gels for sodium dodecyl sulfate-12.5% or -15% polyacrylamide gel electrophoresis (SDS-12.5% or -15% PAGE) were used (as indicated in the figure legends). Antibodies were used at the following concentrations: anti-CotBn, 1:2,000; anti-CotG, 1:20,000; and anti-His (Novagen) (monoclonal), 1:50,000. Secondary anti-rabbit or anti-mouse antibodies (Sigma) were used at concentrations of 1:10,000 or 1:5,000, respectively.
Spore purification and extraction of spore coat proteins.
Spores were harvested 24 h after the onset of sporulation and purified by density gradient centrifugation as described previously (16, 17). Proteins were extracted from purified spores and fractionated on SDS-12.5% or -15% PAGE gels. The gels were stained with Coomassie blue and then transferred to nitrocellulose for immunoblotting or to polyvinylidene difluoride membranes for N-terminal sequence analysis (16, 17).
Ni2+-NTA affinity chromatography purification of complexes containing CotB-6×His.
Samples (100 ml) of Difco sporulation medium cultures of strain MB24 or AH2088 were collected at hour 8 of sporulation, and the cells were collected by centrifugation and resuspended in 1 ml of buffer A (1 mM NaH2PO4, 1 mM Na2HPO4, 50 mM 0.5% [wt/vol] NaCl, 2 mM phenylmethanesulfonyl fluoride). Lysates were prepared by passage (at 19,000 lb/in2) through a French press, and cell debris was removed by centrifugation (7,000 × g for 10 min at 4°C). The clarified lysate was applied to a Ni2+-NTA (nitrilotriacetic acid) agarose column (Qiagen) preequilibrated with 8 volumes of buffer A. The column was washed with 3 volumes of buffer A, and proteins were eluted with increasing concentrations of imidazole (10, 25, 50, 50, 100, 250, and 500 mM) in buffer A. Proteins in the different elution volumes were resolved on SDS-15% PAGE gels and subjected to immunoblot analysis with anti-CotBn or anti-CotG antibodies.
Yeast two-hybrid analysis.
The coding regions of cotB and cotG were PCR amplified with primers B/5/Nco and B/OMO/3 and primers G/5/Nco and G/Bam/3 (Table 2). The cotB and cotG PCR products were digested with NcoI and BamHI and inserted between the same sites of both pAS2-1 and pACT2 (Clontech). The resulting plasmids are indicated in Table 3. The cotH coding sequence was obtained by PCR using two different sets of primers, primers H/5/Nde and 3/H/Bam and primers H/5/Bam and H/3/Xho (Table 2). The cotH PCR products were digested with NdeI and BamHI and inserted between the same sites of pAS2-1 or with BamHI and XhoI and fused to the GAL4 activation domain (AD) in pACT2 (see Table 3). Mating of S. cerevisiae strains and colony lift assays for detection of β-galactosidase activity were performed as described previously (33).
TABLE 3.
Detection of lacZ transcription by colony lift assays of yeast strains expressing fusions of CotH, CotG, or CotB to the GAL4 activation and binding domains
| AD fusiona | DNA-BD fusionb
|
|||
|---|---|---|---|---|
| pAS2-1 (BD) | pAS-B (CotB-BD) | pAS-G (CotG-BD) | pAS-H (CotH-BD) | |
| pACT-2 (AD) | −c | − | − | − |
| pAC-B/(CotB-AD) | − | ++ | + | − |
| pAC-G/(CotG-AD) | − | + | + | − |
| pAC-H/(CotH-AD) | − | − | − | − |
Constructs based on pACT-2 which carry in-frame fusions of CotH, CotG, or CotB to the GAL4 AD. The plasmids were transformed into yeast strain Y190 (Clontech) before mating.
Constructs based on pAS2-1 which carry fusions of CotH, CotG, or CotB to the GAL4 BD. The plasmids were transformed into yeast strain Y187 (Clontech) before mating.
The time of development of blue color on a colony lift assay for the detection of lacZ expression is indicated as follows: ++, development of color in 1 h; +, development of color in up to 5 h. The symbol − indicates no color development in incubations of up to 24 h. Six independent colonies were tested for each combination.
RESULTS
cotB encodes a 46-kDa protein.
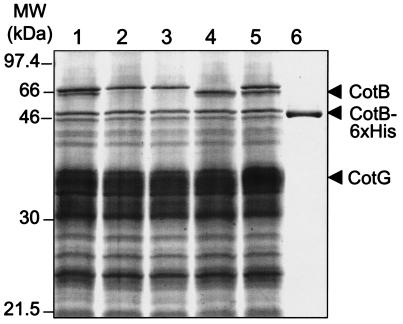
The cotB gene was previously identified (by reverse genetics) as encoding a spore coat component of about 59 kDa (6). Spores from a cotB::cat insertional mutant (strain DL067) (Fig. 1) exhibited the wild-type pattern of Coomassie-stained coat polypeptides on SDS-PAGE except for the absence of a 59-kDa polypeptide (6). Under our electrophoresis conditions, a band of about 66 kDa was absent from the coat of a mutant (AH141) bearing the same cotB::cat allele used by Donovan et al. (6) (Fig. 2, lane 2) or from spores of AH2055 (Fig. 2, lane 3, and Table 1), which bears an insertion of pRZ29 into the 5′ end of cotB (Fig. 1). In addition, the N-terminal sequence of the corresponding band isolated from wild-type spores (Fig. 2, lane 1) was that deduced for the cotB product (data not shown). In two recent studies in which mass spectrometry techniques were used to identify spore- or coat-associated polypeptides, the cotB-encoded polypeptide was estimated to be 66 kDa (26) or 62 kDa (27). We infer that the 66-kDa protein (which we refer to as CotB-66) is equivalent to the protein found by Donovan et al. (6). Under our conditions, the closely migrating CotA protein (6) runs slightly slower than CotB, as shown by its absence from spores of a cotA::cat insertional mutant (AH140) (6, 29) (Fig. 2, lane 4).
FIG. 2.
cotB encodes a protein of 46 kDa in E. coli but accumulates as a 66-kDa form in the spore coat. Material extracted from purified spores of different B. subtilis strains (lanes 1 to 5; see Table 1) was resolved on an SDS-15% PAGE gel and stained with Coomassie blue. Lane 1, wild type (strain MB24); lane 2, cotB::cat (strain AH141); lane 3, cotB::sp (strain AH2055); lane 4, cotA (strain AH140); lane 5, CotB-6×His (strain AH2036). Lane 6 contains about 10 μg of CotB-6×His protein purified from an E. coli strain expressing a CotB-6×His fusion under the control of a T7lac promoter (strain AH1834; Table 1). The arrows mark the position of the CotB-46 or CotB-66 forms. The positions of molecular mass markers (MW) (in kilodaltons) are represented on the left side of the panel.
Inspection of the B. subtilis genome sequence reveals, however, that cotB is capable of encoding a polypeptide of 380 residues and about 43 kDa (25). The disparity between the size of the cotB-dependent polypeptide associated with the coat and that calculated from the gene's sequence (43 kDa) could be due to abnormal electrophoretic mobility. Alternatively, CotB could be modified during its assembly into the spore coat. We reasoned that if migration of CotB in the 66-kDa region of the gel were due to its primary structure, CotB should still migrate at this position when produced in E. coli. Conversely, if the migration of CotB at 66 kDa were due to a modification related to the process of coat assembly, then this alteration of CotB could be restricted to B. subtilis cells. To investigate this, we first cloned the cotB gene with a six-His tag at its 3′ end under the control of the T7lac promoter and introduced the resulting plasmid (pMS16) into cells of the E. coli host BL21(DE3) (Novagen) to produce strain AH1834 (Table 1). During construction of the CotB-6×His fusion, a leucine codon and a glutamate codon were introduced at the 3′ end of cotB (before the six-His-encoding sequence), representing 1.082 kDa. Thus, the CotB-6×His fusion protein has a predicted molecular mass of about 44 kDa. Induction of AH1834 with IPTG (isopropyl-β-d-thiogalactopyranoside) resulted in the production of polypeptides of about 46 kDa that accumulated as inclusion bodies. The 46-kDa polypeptide was solubilized in a urea-containing buffer and was partially purified by affinity chromatography on Ni2+ columns (Fig. 2, lane 6). The N-terminal sequence (MSKRRMKY) confirmed that it derived from the cotB gene. When expressed in E. coli, therefore, cotB encodes a protein whose apparent molecular mass (46 kDa) is in good agreement with the size (43 kDa) predicted from the sequence of the gene (25). To directly compare the size of CotB-6×His made in E. coli to the size of the cotB-derived polypeptide that associates with the B. subtilis spore coat, we made use of strain AH2036 (Table 1), which resulted from the Campbell-type integration of pRZ24 and expressed a CotB-6×His fusion under the control of the cotB promoter. Addition of the six-His tag at the 3′ end of CotB did not prevent its assembly into the coat structure (Fig. 2, lane 5) and did not significantly change the size of the protein associated with the coat relative to that of the wild-type protein (Fig. 2; compare lanes 1 and 5). We also note that an anti-six-His tag antibody reacted with a 66-kDa band in AH2036 spore coat extracts but not in extracts prepared from the wild-type strain MB24 (data not shown), suggesting that the C-terminal region of CotB is represented in the CotB-66 form.
The difference in size between the protein produced in E. coli and the polypeptide that accumulates in the B. subtilis spore coat prompts us to suggest that the form of CotB that prevails in the spore coat (CotB-66) bears a modification that increases its mass by about 20 kDa.
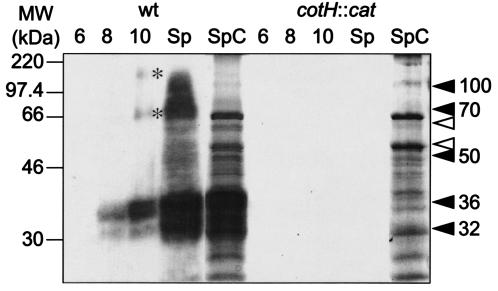
CotB is synthesized as a 46-kDa species (CotB-46) during sporulation and is converted to a 66-kDa form (CotB-66).
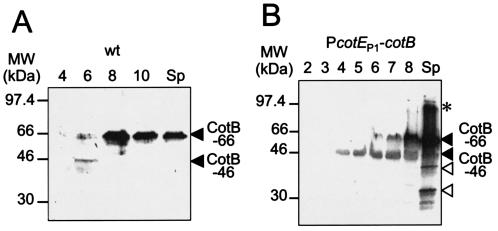
To investigate whether CotB could also accumulate in B. subtilis as a polypeptide of about 43 to 46 kDa before being converted to the CotB-66 form, we used a polyclonal antiserum to follow its accumulation during sporulation. In whole-cell extracts prepared from strain MB24 cells harvested at hour 6 of sporulation (but not in those from the hour-4 sample), the antibody detected a species of approximately 46 kDa (Fig. 3A). This species has about the same size as the CotB-6×His protein produced in E. coli (in good agreement with the predicted size of CotB). In the experiment documented in Fig. 3, this form of CotB (hereinafter designated CotB-46) was not detected in extracts prepared at hour 8 or 10 of sporulation or in spore coat extracts (Fig. 3A). In addition, the antibody detected a 66-kDa species in hour-6 extracts, presumably CotB-66 (see above) (Fig. 3A). In contrast to the results seen with CotB-46, the cellular level of CotB-66 increased from hour 6 to 8 and was the main form of CotB detected at hour 10 of sporulation or in purified coat material (Fig. 3A). The exact temporal profiles with which both CotB-46 and CotB-66 could be detected differed to some extent. In the experiment whose results are presented in Fig. 4A, for example, CotB-46 could still be detected at later times during sporulation whereas CotB-66 was only detected from hour 8 onwards. In any case, CotB-66 tended to predominate at later time points and was the major form of CotB detected in purified coat material. None of the CotB forms were detected in whole-cell extracts or in spore coat extracts prepared from sporulating cells or spores, respectively, of the AH141 (cotB::cat) or AH2055 (cotB::sp) mutants (data not shown).
FIG. 3.
Expression of cotB in sporulating cells. The synthesis of CotB was monitored in extracts from sporulating cells at the indicated times (in hours) after the onset of sporulation or in material extracted from purified spores (Sp) of the following strains (see Table 1): MB24 (wild type) (A) and AH2089 (which expresses cotB from the σE-dependent cotE P1 promoter) (B). CotB was detected by immunoblot analysis with antibodies raised against partially purified CotB. The arrows mark the positions of the CotB-46 and CotB-66 forms. The open arrowheads in panel B indicate the positions of putative CotB degradation products. The asterisk indicates the position of a form of CotB higher than 66 kDa. The positions of molecular mass markers (MW) (in kilodaltons) are shown on the left side of each panel.
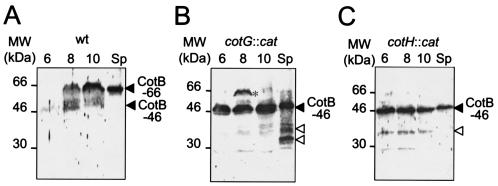
FIG. 4.
Expression of both cotG and cotH is required for the efficient formation of CotB-66. The figure illustrates the accumulation of CotB at the indicated times (in hours) after the onset of sporulation and in spore coat extracts (Sp) in the following strains (Table 1): MB24 (wild type) (A), AH1497 (cotG::cat) (B), and AH1103 (cotH::cat) (C). The accumulation of CotB was monitored with a polyclonal antibody raised against the partially purified protein produced in E. coli cells. The arrowheads on the right sides of the panels indicate the positions of CotB-46 and CotB-66. The open arrowheads in panels B and C indicate the position of putative CotB degradation products. The asterisk in panel B indicates the position of a band of about 66 kDa that accumulates transiently around hour 8 of sporulation in the cotG mutant. The positions of protein molecular mass markers (MW) (in kilodaltons) are indicated to the left of each panel.
The temporal pattern of accumulation of both the 46- and 66-kDa polypeptides is consistent with the time of expression of cotB during spore development which is induced around hour 6 under the joint control of σK and GerE (18, 46). Recent results indicate that cotB expression may also be governed by the early mother cell-specific regulator σE (10). However, this early expression of CotB was not detected under our conditions. Our results suggest that CotB is synthesized around hour 6 of sporulation as a 46-kDa polypeptide (CotB-46) which rapidly undergoes a posttranslational modification to yield a 66-kDa form (CotB-66) which accumulates in the spore coat.
Synthesis and modification of CotB can be uncoupled in sporulating cells.
To independently test whether CotB could be synthesized and accumulate as a 46-kDa form in B. subtilis, we placed the cotB coding region under the control of the strong cotE P1 promoter (PcotE P1), which is utilized by the σE-containing form of RNA polymerase (46). We reasoned that the putative modification of CotB could depend on other late-expressed genes and that the expression of cotB at sufficient levels prior to the onset of σK activity could confirm the size of unmodified CotB in B. subtilis. The PcotE P1-cotB fusion was inserted at the amyE locus in a cotB null mutant, and the accumulation of CotB was monitored in AH2089 by immunoblot analysis throughout sporulation and in purified coat material. Control experiments have shown that the insertion of cotB at the amyE locus resulted in the production and assembly of CotB in a manner indistinguishable from that seen with the wild-type strain MB24 (data not shown). When cotB was expressed from the cotE P1 promoter, CotB was detected as a 46-kDa species from hour 4 of sporulation onwards (Fig. 3B).
Thus, as seen with E. coli cells, cotB seems to encode a protein of about 46 kDa. Even though cotB was expressed at least 2 h earlier than normally, interestingly, CotB-66 was detected only from hour 6 of sporulation onwards, as seen with the wild-type strain MB24 (Fig. 3A). We also found in the coats of AH2089 at least two relatively abundant bands of about 40 and 34 kDa, which we interpret as stable degradation forms of CotB (Fig. 3B and 4B). Donovan et al. (6) also described a coat-associated polypeptide of 34 kDa whose N-terminal sequence matched that of CotB. Inspection of the B. subtilis genome sequence indicates that this polypeptide cannot be the product of a separate gene (25). Since the original insertion into cotB (close to its 3′ end) (Fig. 1) did not prevent its accumulation, the 34-kDa form must derive from the N-terminal portion of the CotB protein as initially hypothesized (6). Presumably this form of CotB represents a stable degradation product of higher-molecular-mass forms of CotB. Spores of AH2089 (PcotE P1-cotB) show wild-type resistance to heat and lysozyme, and except for the differences mentioned above (which were detected by immunoblot analysis), the collection of extractable coat polypeptides does not differ from that of wild-type spores as judged by Coomassie staining (data not shown). However, while spores of the cotB::sp mutant AH2055 are not impaired in their ability to germinate in response to l-alanine (as also reported for a strain bearing the cotB::cat allele) (6), AH2089 spores are slow to germinate upon exposure to l-alanine (data not shown). This suggests that the timing of the expression of cotB is important for the spore's response to the germinant l-alanine.
Transcription of cotB normally requires both σK and GerE (18, 46). That expression of cotB is not sufficient for formation of CotB-66 suggested that at least one additional late gene is needed for the formation of CotB-66. We cannot exclude that formation of CotB-66 involves a factor synthesized early in the mother cell line of gene expression (under the control of σE), but in any case CotB-66 is only formed late, coincidently with the onset of σK-directed gene transcription. Thus, irrespective of the involvement of early gene products, formation of CotB-66 appears to be directly or indirectly dependent on the products of late, σK-dependent genes or morphogenetic events. We also note that in the PcotE P1-cotB strain, CotB-46 accumulates at later times during sporulation and undergoes assembly into the spore coat (Fig. 3B). It appears that in the PcotE P1-cotB strain, CotB-46 cannot be completely converted into the 66-kDa form. Alternatively, the protein at 46 kDa that accumulates from hour 6 onwards may result from degradation of CotB-66. In any event, we infer that formation of CotB-66 is not a strict requirement for assembly of CotB (see also below).
Formation of CotB-66 requires expression of both cotG and cotH.
Previous work has shown that assembly of CotB (CotB-66) into the spore coat requires expression of both the cotG and cotH loci (30, 36). The cotG gene is found just upstream and divergently oriented with respect to cotH, whereas cotB immediately follows cotH (25, 30, 36) (Fig. 1). Both the cotG and cotH genes are under the control of σK, but the expression of cotG further depends on gerE (30, 36). To determine whether cotH or cotG was required for the modification of CotB, we used our polyclonal antibody to monitor the synthesis of CotB in cotH and cotG null mutants (AH1103 and AH1497, respectively) (Table 1). In contrast to the results seen with the wild type (Fig. 4A), we found CotB-46 to be the predominant form of CotB in whole-cell extracts of a cotG mutant prepared 6, 8, or 10 h after the onset of sporulation (Fig. 4B). We found CotB-66 (or a species of approximately the same size) to accumulate transiently at hour 8 in the cotG mutant AH1497, but even in this sample CotB-46 was more abundant (Fig. 4B). At present we do not know whether this species is the same as the CotB-66 form seen in the wild type (see also Discussion). Moreover, CotB-46 was the predominant form of CotB found in coat extracts prepared from cotG spores (Fig. 4B). Several degradation products of CotB were found to accumulate in cells or spores of the cotG mutant (Fig. 4B). We then investigated the accumulation of CotB in sporulating cells and in the coats of the cotH mutant AH1103 (Table 1). The results presented in Fig. 4C show that (together with what appear to be degradation products) only CotB-46 is detected in whole-cell extracts of sporulating cells or in material purified from the coats of cotH mutant spores. Therefore, expression of cotH is also a requirement for the formation of CotB-66. However, CotB-66 was never detected in the cotH mutant, suggesting that cotH and cotG might have different roles in the formation of CotB-66. In any case, the results implicate both cotH and cotG in the formation of CotB-66. We cannot ascertain whether cotH and cotG are also involved in the assembly of CotB-66, but we note that the expression of neither loci is required for the association of CotB-46 with the coat structure.
The cotB gene is followed by ywrJ (25) (Fig. 1), and the two genes appear to be cotranscribed during sporulation (10). To test whether ywrJ had any role in spore coat assembly, we created a ywrJ deletion mutant (AH2119) (Table 1) by a marker replacement event involving plasmid pRZ48 (Fig. 1). We found that disruption of ywrJ did not cause any detectable effect on the coat polypeptide composition, heat or lysozyme resistance of the resulting spores, or accumulation of CotG or CotB-66 (data not shown). Thus, the function of the ywrJ gene remains unknown.
CotH is required for the accumulation of CotG.
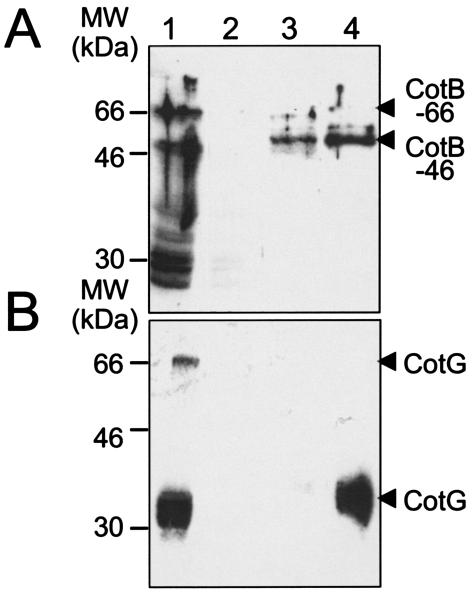
The need for both cotH and cotG for the formation of CotB-66 could indicate that the two loci act together or could reflect a hierarchical control of one locus over the other. To begin to investigate these questions, we first tried to overproduce CotG in E. coli for antibody production. In spite of many attempts to overproduce CotG as N- or C-terminal fusions to the six-His tag or to partners such as maltose binding protein or glutathione S-transferase, we never observed accumulation of the fusion proteins in E. coli (data not shown). We therefore decided to raise a rabbit polyclonal antibody against a peptide derived from residues 166 to 180 of the CotG primary structure (see reference 36 and Materials and Methods). The cotG gene codes for a protein with 195 amino acid residues with a predicted molecular mass of 24 kDa (36). However, CotG is seen mainly as a wide, diffuse band of about 36 kDa in spore coat gels (Fig. 1; see also below) (17, 30, 36). The discrepancy between the predicted size and the observed size of CotG has been attributed to its unusual primary structure, which bears a central region of 117 amino acids organized in nine repeats of a 13-amino-acid sequence motif (36). Using the anti-peptide antibody, we detected CotG as diffuse bands of about 32 and 36 kDa in whole-cell extracts from hour 8 of sporulation onwards (Fig. 5, lane 8 [wild-type strain results]). Migration of the 36-kDa species appears to coincide with that of the 36-kDa cotG-dependent band seen in Coomassie-stained coat gels (Fig. 5, lane SpC [wild-type strain results]) (17, 30, 36). The 32-kDa band, which is the band that is the closest in size to the expected size of the cotG-encoded protein (24 kDa), also coincides with a band seen in the Coomassie-stained gel of the same coat protein extract (Fig. 5, lanes Sp and SpC [wild-type strain results]). Additional higher-molecular-mass forms of CotG (of about 70 and over 100 kDa) were found in hour-10 whole-cell extracts (Fig. 5). In purified coat material, five main forms (around 32, 36, 50, and 70 and over 100 kDa) of CotG were detected (spore coat extract from wild-type spores) (Fig. 5). Presumably, some of these species are the same as those detected in hour-10 extracts. The antibody also revealed several less-distinct CotG forms between the 100-and 32-kDa species found in spore coat extracts (Fig. 5), which could have been caused by extensive cross-linking of CotG or proteolysis or a combination of the two processes. None of the bands seen in immunoblotting analysis of wild-type cell lysates or coat material were detected in extracts of a cotG insertional mutant, demonstrating the specificity of the antibody (data not shown). We then examined the accumulation of CotG in a cotH mutant. Unexpectedly, we did not detect any forms of CotG in whole-cell lysates prepared from sporulating cultures or in material extracted from purified spores of the cotH mutant AH1103 (Fig. 5, lanes 6 to 10 and Sp [cotH::cat mutant results]). Analysis (by SDS-PAGE and Coomassie staining) of the profiles of proteins extracted from AH1103 spores (Fig. 5, lane SpC [cotH::cat mutant results]) revealed the pattern expected for the cotH mutant (30). Since the cotH::cat mutation does not affect transcription of the cotG gene (30), these observations strongly suggest that CotG is unstable and that CotH or a protein controlled by CotH is required for the accumulation of CotG. In any case, the need for cotH expression for the formation of CotB-66 may derive in part from its requirement for the accumulation of CotG. The possibility that CotH has, in addition, a more direct role in formation of CotB-66 is not excluded.
FIG. 5.
Accumulation of CotG in sporulating B. subtilis. The figure depicts the immunoblot analysis of the accumulation of CotG at the indicated times (in hours) after the onset of sporulation in spore coat extracts (Sp) in the wild-type strain (strain MB24) (Table 1) or in a cotH::cat mutant (AH1103). CotG was detected with a polyclonal antibody raised against a peptide derived from residues 166 to 180 of the CotG primary sequence (see Materials and Methods). For comparison, the lanes marked SpC (for strain MB24 or AH1103) show a Coomassie-stained gel of the same sample of extracted coat proteins used for the immunoblot analysis (lanes marked with Sp). Black arrowheads on the right side of the panel indicate the positions of forms of CotG of about 100, 70, 50, 36, and 32 kDa. Other, less-abundant forms of CotG are indicated by open arrowheads. Asterisks indicate a form of CotG of over 100 kDa that was predominantly found in whole-cell extracts at hour 10 of sporulation and a form of about 70 kDa that was found both in the hour 10 sample and in purified coat material (Sp). The positions of protein molecular mass markers (MW) (in kilodaltons) are indicated on the left side of the panel.
CotB and CotG form complexes in vivo.
The observation that cotH and cotG promote formation of CotB-66 prompted us to investigate possible associations in vivo. We used a B. subtilis strain expressing a CotB-6×His fusion protein at the amyE locus (strain AH2088) to purify (by affinity chromatography) complexes containing CotB. We prepared whole-cell extracts from AH2088 cells at hour 8 of sporulation and applied the extract to a Ni2+-NTA agarose column (Fig. 6). The column was first washed and was then eluted with increasing concentrations (from 10 to 500 mM) of imidazole (see Materials and Methods). The proteins were electrophoretically resolved, and the presence of CotB or CotG in the various fractions was assessed by immunoblot analysis. We do not presently have a good antibody against CotH and could not test whether this protein also copurified with CotB. We found that most of the CotB present at hour 8 of sporulation did not bind to the Ni2+-agarose column and that it was detected in the column flowthrough (Fig. 6A, lane 1). Part of this CotB is likely to result from expression of the wild-type cotB gene in AH2088 (Table 1), suggesting that untagged CotB is not retained by the column. To verify this, we prepared a whole-cell lysate from a sporulating culture of strain MB24 at hour 8 of sporulation. The lysate was applied to a Ni2+-NTA column, and the column was eluted with a step gradient of imidazole at concentrations up to 500 mM. No CotB was found in the elution fractions by immunoblot analysis. We conclude that under these conditions, untagged CotB is not retained by a Ni2+ column (data not shown). In strain AH2088 (CotB-6×His), some CotB did bind to the column and was eluted at imidazole concentrations of 50 mM (lane 3) and 100 mM (lane 4). Most of the fusion protein retained by the affinity column was the 46-kDa form (Fig. 6A, lanes 3 and 4). Very little CotB-66 was found in the 50 or 100 mM imidazole fractions, but this form of the protein appeared to be slightly more abundant in the 50 mM fraction (Fig. 6A, lanes 3 and 4). No CotB was detected upon elution of the column with buffers containing higher imidazole concentrations (up to 500 mM; data not shown).
FIG. 6.
CotB and CotG are present in complexes in vivo. Whole-cell extracts were prepared from the wild-type (MB24) or AH2088 (CotB-6×His) strain at hour 8 of sporulation. The extracts were loaded on a Ni2+-NTA-agarose column, and the presence of CotB (A) or CotG (B) in the column flow through or eluate fractions was monitored by immunoblot analysis with specific antibodies. After loading, the column flow through was collected and the column was washed and eluted with increasing concentrations of imidazole (see Materials and Methods). The figure represents the results of analysis of the following samples: the column flowthrough (lane 1) and the 50 mM (lane 3) and 100 mM (lane 4) imidazole eluate fractions after application of the AH2088 extract and the 100 mM imidazole fraction resulting from a similar experiment using an extract prepared from the wild-type strain MB24 (untagged cotB) (lane 2) (Table 1). Molecular mass markers (in kilodaltons) are indicated on the left side of the panels, and the positions of CotB-46, CotB-66, and CotG are indicated on the right side.
We then subjected samples of the same fractions to immunoblot analysis using an anti-CotG antibody (Fig. 6B). We found that a fraction of CotG present in whole-cell extracts prepared at hour 8 of sporulation (and that included at least two forms [about 36 and 70 kDa] of the protein) was not retained by the column (Fig. 6B, lane 1). However, CotG (mainly a form of about 36 kDa) also coeluted with CotB-46 at an imidazole concentration of 100 mM (Fig. 6B, lane 4). No other column fractions (at imidazole concentrations up to 500 mM) contained CotG. Control experiments using the wild-type strain MB24 (Table 1) demonstrated that CotG was not retained by the column when CotB did not carry a six-His tag (Fig. 6B, lane 2, and data not shown). We do not presently know why CotB elutes at two different imidazole concentrations or why CotG only coelutes with CotB at the higher (100 mM) imidazole concentration. Presumably, CotB exists in several forms that interact differently with the column. In any case these results indicate that CotB copurifies with CotG, and we infer that CotB (perhaps mostly as CotB-46) and CotG are present in complexes in vivo.
CotG and CotB directly interact.
Since CotG and CotB appeared to form complexes in sporulating cells, we wanted to investigate whether the two proteins could interact directly. In addition, we wanted to test whether CotH could interact with either CotG or CotB. Interactions among CotH, CotG, and CotB were tested in vivo using a yeast two-hybrid system (12, 13, 14). We fused the entire coding sequence of cotB, cotH, or cotG to either the AD in pACT-2 or the DNA-binding domain (DNA-BD) in pAS2-1 of the yeast transcriptional activator GAL4, and combinations of the fusion plasmids and/or vectors were introduced into appropriate yeast reporter strains (33) (Table 3). Interaction of the fusion proteins in the yeast strains resulted in the expression of a lacZ reporter gene and was detected by a colony lift assay (see Materials and Methods). As shown in Table 3, no β-galactosidase activity was detected when individual fusions were expressed with the corresponding control vector. In addition, background levels (in yeast cells carrying both vectors) were found to be negligible (Table 3). Using this system, we detected interactions between CotB and itself and between CotG and itself. Additionally, we detected an interaction between CotB and CotG (Table 3). All of the interactions were found when the involved proteins were fused to either the GAL4 AD or the GAL4 BD (Table 3). In contrast, we did not find evidence for an interaction between CotH and either CotG, CotB, or itself. These results suggest that CotB is capable of associating with itself and also with CotG and that CotG is also capable of self-association.
DISCUSSION
We found that cotB encodes a polypeptide of about 46 kDa (CotB-46) that (soon after synthesis around hour 6 of sporulation) undergoes a posttranslational modification that converts it into a 66-kDa form (CotB-66). The CotB-66 polypeptide is the form of CotB previously found in the spore coat by Donovan et al. (6). Under our conditions, CotB-66 is the main form of CotB present in the spore coat. That cotB codes for a 46-kDa protein is supported by two lines of evidence. First, expression of cotB in E. coli results in the accumulation of a 46-kDa polypeptide. Second, under genetic conditions in which synthesis of CotB in B. subtilis was artificially induced from the early σE-dependent cotE P1 promoter (46) a 46-kDa polypeptide accumulated from hour 4 of sporulation onwards. Conversion of CotB-46 into CotB-66 in the PcotE P1-cotB strain only occurred around hour 6 of sporulation, as seen with the wild type, indicating the involvement of other, late-expressed genes in the modification of CotB-46. Indeed, our results indicate that the expression of both cotH (which is driven by σK) (30) and cotG (which requires σK and GerE) (36) is needed for the formation of CotB-66. The data also indicate that the involvement of cotH and cotG in CotB-66 formation results in part from a hierarchical sequence of events. In fact, CotG does not accumulate in the absence of CotH, suggesting that the latter is an unstable protein and that CotH or a CotH-dependent factor is somehow needed to stabilize CotG. Even though we did not find evidence for a direct interaction between the two proteins in a yeast two-hybrid system, CotH might act as a chaperone (binding directly to CotG) to promote its stabilization. We note that CotH is found only in minute amounts in the final coat structure (48), whereas CotG accounts for a significant proportion of the protein extractable from the coat of wild-type spores (7, 15). Chaperones play essential roles in the macromolecular assembly of other biological structures (see, for example, references 1 and 37) but have never been proposed to play such roles in the context of the assembly of the bacterial endospore coat. An alternative explanation for the role of CotH is that CotH or a CotH-dependent protein acts to inhibit a protease that uses CotG as a substrate.
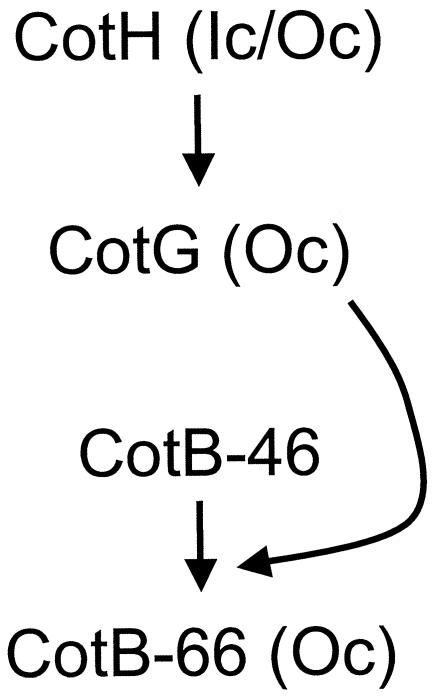
Previous work has shown that in addition to CotH, CotG and CotB are absent from cotH spores (30). In contrast, in addition to CotG, only CotB (CotB-66) is missing from cotG spores (36). The cotH-dependent stabilization of CotG helps to explain its requirement for the assembly of CotG, whereas the cotG-dependent assembly of CotB is in part explained by the role of CotG in CotB-66 formation (Fig. 7). We do not know whether CotG is directly recruited by CotH to the coat structure or whether the assembly of CotB-66 is directly controlled by CotH or CotG (Fig. 7). However, the observation that CotB-46 accumulates in the coats of cotH or cotG mutants indicates that the assembly of at least this form of the protein is independent of the presence of CotH and CotG. Lastly, we note that CotB-66 is detected transiently in the cotG mutant but not in cotH cells, suggesting that other than its role in the stabilization of CotG, CotH might serve yet another role in the formation of CotB-66. However, it is also possible that this band is a cross-reactive species that only accumulates in a cotG mutant.
FIG. 7.
Assembly of CotB. Previous work has shown that CotH is needed for the assembly of CotG and CotB (30, 36). The results of the present study indicate that cotB encodes a protein of about 46 kDa which is rapidly converted into a form of about 66 kDa. The results also indicate that CotH is required directly or indirectly for the stabilization of CotG, which in turn promotes the conversion of CotB-46 into the CotB-66 form, possibly via a direct interaction with CotB-46. It is not known whether CotH directly controls the assembly of CotG or whether CotG is directly involved in the recruitment of CotB-66. However, assembly of CotB-46 can occur in the absence of CotH or CotG. Where known, the location of the indicated proteins within the coa t stucture is indicated (Oc, outer coat; Ic/Oc, inner coat-outer coat interface).
CotG is first detected around hour 8 of sporulation as two main antigenic forms of about 32 and 36 kDa (Fig. 5). The 32-kDa form could represent the unmodified product of the cotG gene (24 kDa) whose abnormal migration may be attributed to its unusual primary structure (36). CotG appears to be converted into forms of about 36 and 70 kDa in sporulating cells and then to be subjected to extensive cross-linking as it is assembled into the spore coat. That CotG is able to form cross-linked multimers has been previously suggested on the basis of the results of analysis of the coat structure in sodA mutant cells (17) and is supported by the results of the yeast two-hybrid assay (this work). A more detailed characterization of the synthesis and assembly of CotG is currently under way.
Our finding that CotG copurifies with CotB strongly suggests that the two proteins are present in complexes at the time in coat assembly during which formation of CotB-66 is detected. In that respect it is noteworthy that CotG (a 36-kDa form of the protein) appears to copurify mainly with CotB-46. On the basis of the results of the yeast two-hybrid experiments, it is tempting to suggest that CotG may directly interact with CotB-46 during assembly of the spore coat to promote formation of CotB-66 (Fig. 7). It is unknown whether CotH also copurifies or interacts with CotB, just as it is unknown whether the presence of CotG and CotH is sufficient to promote the modification of CotB. Spores produced by cotB and cotG mutants are not affected in their resistance to lysozyme or germination properties (6, 36). Therefore, the significance of the interaction between CotB and CotG is presently unclear and may only be apparent under conditions that have not yet been identified.
The nature of the posttranslational modification of CotB remains unclear. On the basis of the size difference from the unmodified form and of the observation that the CotB-66 form presents only one N-terminal sequence (6; this work), the easiest explanation is that CotB-66 is a cross-linked homodimer with irregular mobility (the expected size would be of 92 kDa [in contrast to the observed size of 66 kDa]). It is also possible that CotB-66 is a homodimer cleaved near the C terminus of the protein, but, if so, our observation that a C-terminal six-His sequence is present in the 66-kDa form indicates that at least one of its components is the full-length protein. Our finding that CotB is capable of self-interaction in a yeast two-hybrid system supports the suggestion that CotB undergoes multimerization. Alternatively, CotB-66 is a heterodimer, perhaps containing CotG (given that the two proteins appear to interact); in that case, however, either the cross-linking involves the N terminus of the second component or its N-terminal sequence is blocked and refractory to analysis. Several types of covalent cross-links have been proposed or detected in the coat. These include disulfide bridges (note, however, that cotB does not code for any cysteine), γ-glutamyl-lysil isopeptide bonds mediated by a coat-associated transglutaminase (see, for example, references 16, 22, 23, and 44), or o,o-dityrosine cross-links (reference 17 and references therein). Work in progress aims at determining the nature of the posttranslational modification of CotB and its structural significance as well as the protein components involved and their roles.
Acknowledgments
We thank T. Barbosa for critically reading the manuscript, J. Pohl (Emory Microchemical Facility) for N-terminal sequence determinations, and A. J. Ozin for help with the yeast two-hybrid experiments.
This work was supported by European Union grant QLK5-CT-2001-01729 to E.R. and A.O.H., by MIUR (Cofin 2002 and FIRB 2002) grants to E.R., and by grants GM54395 to C.P.M. and PRAXIS/PCNA/C/BIA/129/96 to R.Z. M.S. holds a Ph.D. fellowship (PRAXIS XXI/BD 18 251/98) from the F.C.T.
REFERENCES
- 1.Auvray, F., J. Thomas, G. M. Fraser, and C. Hughes. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, T., S. Little, A. G. Stöver, and A. Driks. 1999. Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 181:7043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of σB levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 5.Cutting, S. M., and P. B. V. Horn. 1990. Genetics analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 6.Donovan, W., L. B. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driks, A. 2002. Proteins of the spore core and coat, p. 527-535. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 9.Duc, L. H., H. A. Huynh, N. Fairweather, E. Ricca, and S. M. Cutting. 2003. Bacterial spores as vaccine vehicles. Infect. Immun. 71:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J.-E. González-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 11.Enguita, F. J., L. O. Martins, A. O. Henriques, and M. A. Carrondo. 2003. Crystal structure of a bacterial endospore coat component: a laccase with enhanced thermostability properties. J. Biol. Chem. 278:19416-19425. [DOI] [PubMed] [Google Scholar]
- 12.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 13.Fields, S., and R. Sternglanz. 1994. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 10:286-292. [DOI] [PubMed] [Google Scholar]
- 14.Guarente, L. 1993. Strategies for the identification of interacting proteins. Proc. Natl. Acad. Sci. USA 90:1639-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 16.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the α-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 19.Ichikawa, H., R. Halberg, and L. Kroos. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322-8327. [DOI] [PubMed] [Google Scholar]
- 20.Isticato, R., G. Cangiano, H.-T. Tran, A. Ciabattini, D. Medaglini, M. R. Oggioni, M. de Felice, G. Pozzi, and E. Ricca. 2001. Surface display of recombinant proteins on Bacillus subtilis spores. J. Bacteriol. 183:6294-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 163:69-74. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, K., K. Hashiguchi, K. Yokozeki, and S. Yamanaka. 1998. Molecular cloning of the transglutaminase gene from Bacillus subtilis and its expression in Escherichia coli. Biosci. Biotechnol. Biochem. 62:1109-1114. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, K., Y. Kumazawa, K. Miwa, and S. Yamanaka. 1996. ɛ-(γ-glutamyl)lysine cross-links of spore coat proteins and transglutaminase activity in Bacillus subtilis. FEMS Microbiol. Lett. 144:157-160. [Google Scholar]
- 24.Kroos, L., and Y. T. Yu. 2000. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3:553-560. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971-3982. [DOI] [PubMed] [Google Scholar]
- 27.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vasquez, J. A. Vasquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 42:1107-1120. [DOI] [PubMed] [Google Scholar]
- 29.Martins, L. M., C. M. Soares, M. Pereira, M. Teixeira, G. J. Jones, and A. O. Henriques. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase associated with the Bacillus subtilis endospore coat. J. Biol. Chem. 277:18849-18859. [DOI] [PubMed] [Google Scholar]
- 30.Naclerio, G., L. Baccigalupi, R. Zilhão, M. de Felice, and E. Ricca. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 178:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom.
- 32.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182:1828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozin, A. J., C. S. Samford, A. O. Henriques, and C. P. Moran, Jr. 2001. SpoVID guides SafA targeting to the spore coat of Bacillus subtilis. J. Bacteriol. 183:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozin, A. J., T. V. Costa, A. O. Henriques, and C. P. Moran, Jr. 2001. Alternative translation initiation produces a short form of a spore coat protein in Bacillus subtilis. J. Bacteriol. 183:2032-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 36.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer, F. G., J. S. Pinker, G. Waksman, and S. J. Hultgren. 2002. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111:543-551. [DOI] [PubMed] [Google Scholar]
- 38.Serrano, M., R. Zilhão, A. J. Ozin, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seyler, R., A. O. Henriques, A. Ozin, and C. P. Moran, Jr. 1997. Interactions and assembly of cotJ-encoded products, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25:955-966. [DOI] [PubMed] [Google Scholar]
- 40.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan, M. A., R. E. Yasbin, and F. E. Young. 1984. New shuttle vectors for Bacillus subtilis and E. coli which allow rapid detection of inserted fragments. Gene 29:21-26. [DOI] [PubMed] [Google Scholar]
- 42.Takamatsu, H., A. Imamura, T. Kodama, K. Asai, N. Ogasawara, and K. Watabe. 2000. The yabG gene of Bacillus subtilis encodes a sporulation specific protease which is involved in the processing of several coat proteins. FEMS Microbiol. Lett. 192:33-38. [DOI] [PubMed] [Google Scholar]
- 43.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181:4986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 175:3757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226:1037-1050. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]
- 48.Zilhão, R., G. Naclerio, A. O. Henriques, L. Baccigalupi, C. P. Moran, Jr., and E. Ricca. 1999. Assembly requirements and role of CotH in spore coat formation in Bacillus subtilis. J. Bacteriol. 181:2631-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]