Abstract
Clp-HSP100 ATPases are a widespread family of ubiquitous proteins that occur in both prokaryotes and eukaryotes and play important roles in the folding of newly synthesized proteins and refolding of aggregated proteins. They have also been shown to participate in the virulence of several pathogens, including Listeria monocytogenes. Here, we describe a member of the Clp-HSP100 family of L. monocytogenes that harbors all the characteristics of the ClpB subclass, which is absent in the closely related gram-positive model organism, Bacillus subtilis. Transcriptional analysis of clpB revealed a heat shock-inducible σA-type promoter. Potential binding sites for the CtsR regulator of stress response were identified in the promoter region. In vivo and in vitro approaches were used to show that expression of clpB is repressed by CtsR, a finding indicating that clpB is a novel member of the L. monocytogenes CtsR regulon. We showed that ClpB is involved in the pathogenicity of L. monocytogenes since the ΔclpB mutant is significantly affected by virulence in a murine model of infection; we also demonstrate that this effect is apparently not due to a defect in general stress resistance. Indeed, ClpB is not involved in tolerance to heat, salt, detergent, puromycin, or cold stress, even though its synthesis is inducible by heat shock. However, ClpB was shown to play a role in induced thermotolerance, allowing increased resistance of L. monocytogenes to lethal temperatures. This work gives the first example of a clpB gene directly controlled by CtsR and describes the first role for a ClpB protein in induced thermotolerance and virulence in a gram-positive organism.
Listeria monocytogenes is a gram-positive pathogen implicated in food-borne infections and is responsible for meningitis, septicemia, and gastroenteritis—diseases with a high degree of mortality for immunocompromised hosts. During the past few years, this bacterium has been extensively studied, and it has become a model for intracellular growth (51) because of its abilities to escape from the phagosome, grow in the cytosol, and efficiently invade neighboring cells.
Several virulence proteins that are required for the key steps of the infectious process have been identified to date. InlA and InlB are required for entrance of L. monocytogenes into epithelial cells; listeriolysin O is required for escape from the phagosome; ActA is required for actin polymerization, cell-cell mobility, and invasion; and PlcB is required for lysis of the two-membrane vacuole (7). All virulence genes identified so far are under the positive control of a single regulator, PrfA. Due to the secondary structure of its mRNA, this activator, which acts as a thermosensor (28), is present only at the host temperature.
In addition to these major virulence factors are many proteins that are involved in pathogenicity of Listeria. These proteins, known as stress proteins, are important because they allow persistence and rapid adaptation during the infectious process. The accumulating body of data regarding several pathogens indicates that acid or oxidative stress proteins and (more recently) heat shock proteins (HSPs) and chaperones play an important role in virulence. Indeed, synthesis of the two major Staphylococcus aureus chaperones, DnaK and GroESL, was shown to be induced during infection of human epithelial cells (52); in L. monocytogenes, expression of the groESL operon is induced during intracellular infection, while DnaK is required for efficient phagocytosis with macrophages (15, 22). Another class of stress proteins, the Clp family, has also been shown to play a major role in the virulence of several pathogens: ClpP was shown to control expression of the attachment invasion locus (ail) of Yersinia enterocolitica, whereas in Salmonella enterica serovar Typhimurium, inactivation of clpP prevents growth and survival within macrophages (24, 69). Systematic genome-wide approaches such as signature-tagged mutagenesis revealed the roles of several clp genes, including clpE, clpC, and clpL of Streptococcus pneumoniae (23, 33, 48), as well as clpX of S. aureus (38).
Clp proteins are ubiquitous among prokaryotes and eukaryotes, and they function both as proteases and chaperones (19). Bacterial genomes are endowed with different sets of clp paralogs encoding Clp-HSP100 ATPase subunits, belonging to groups A, B, C, D, E, or L, and that are distinguished by their N-terminal domains and the central spacer regions between the two ATP-binding sites. clpP, which encodes the proteolytic subunit of the Clp ATP-dependent protease, is usually present as a single copy, but up to five copies per genome can coexist, as shown in Streptomyces lividans and Streptomyces coelicolor (8, 65, 66). The ClpP proteolytic subunit requires association with an ATPase subunit in order to be active, giving rise to a multimeric complex presenting structural and functional analogies with the eukaryotic proteasome (50). The ATPase subunits can also act in the absence of ClpP, forming a smaller complex with chaperone activity. It is interesting that ClpB of Escherichia coli does not interact with the proteolytic subunit and is exclusively considered a chaperone (68). However, although some ClpB proteins have been characterized for both eukaryotes and bacteria, no phenotypes have been described as yet for low-G+C gram-positive bacteria.
In L. monocytogenes, three clp genes have been shown to play a role in virulence. ClpC is required for intracellular growth and in vivo survival in host tissues by promoting early escape from the phagosomal compartment (54, 55) and is also necessary for cell adhesion and invasion (44). The ClpE ATPase plays a role in L. monocytogenes virulence also (43), and an L. monocytogenes clpP mutant presents a defect in intracellular replication (16).
A ΔclpB mutant of Y. enterocolitica, a major gastrointestinal pathogen, presents a decrease in invasin and flagellin expression, characteristics that are encoded by the two virulence genes inv and fleB (2). For S. enterica serovar Typhimurium, the clpB mutant was discovered during a systematic search for mutants deficient in colonization of the chicken alimentary tract and was shown to be attenuated for virulence in 1-day-old chicks (64). Finally, a ΔclpB mutant of Francisella novicida was isolated during a screen for genes required for in vitro growth in thioglycolate-elicited mouse peritoneal macrophages (21).
Analysis of the complete genome of L. monocytogenes EGDe (18) reveals several uncharacterized genes encoding proteins belonging to the Clp family, two of which are preceded by potential binding sites for the CtsR regulator of stress response (10). Here we have characterized the clpB gene of L. monocytogenes. Using both in vivo and in vitro approaches, regulation of clpB was studied, showing a direct control by CtsR. This repression was demonstrated to be thermosensitive. A deletion mutant was constructed, and functional analysis revealed a role for ClpB in terms of the virulence of L. monocytogenes. We also show that, although ClpB has no obvious role in terms of stress tolerance, it is required for induced thermotolerance of L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and transformation.
Bacterial strains used in this work are listed in Table 1. E. coli K12 strain TG1 [Δ(lac proAB) supE thi hsdΔ5 (F′ traD36 proAB lacIq lacZ ΔM15)] (17) was used for cloning experiments.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| L. monocytogenes | ||
| LO28 | Virulent L. monocytogenes reference strain | 67 |
| LM2000 | ΔclpB | This study |
| LM2001 | ΔctsR::aphA3 | This study |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| QB4991 | trpC2 ΔctsR amyE::(′lacZ aphA3) | 10 |
| QB8059 | trpC2 amyE::(clpB′-bgaB cat) ΔctsR | pDL73/74→QB4991 |
| QB8060 | trpC2 amyE::(clpB′-bgaB cat) ΔctsR thrC::(pxylctsRLmospc) | pxyl59/60→QB8059 |
| Plasmids | ||
| pMAD | pE194 derivative with a thermosensitive origin of replication for deletion replacement of genes in gram-positive bacteria | M. Arnaud and M. Débarbouillé, unpublished |
| pMAD ΔclpB | pMAD derivative, for deletion replacement of the L. monocytogenes clpB gene | This study |
| pMAD ΔctsR | pMAD derivative, for deletion replacement of the L. monocytogenes ctsR gene | This study |
| pDL | Integrative plasmid for constructing transcriptional fusions with the B. stearothermophilus bgaB gene | 70 |
| pDL73/74 | pDL derivative carrying a clpB′-bgaB fusion | This study |
| pXT | Plasmid allowing integration at the thrC locus and expression from the PxylA xylose-inducible promoter | 9 |
| pxyl59/60 | pXT derivative carrying the ctsR coding sequence of L. monocytogenes | 42 |
E. coli was grown in Luria-Bertani (LB) medium. Electroporation procedures were used for transformation with selection on LB plates supplemented with ampicillin (100 μg/ml), erythromycin (200 μg/ml), or kanamycin (25 μg/ml). L. monocytogenes LO28 was routinely grown in brain heart infusion (BHI) complex medium. Constructs were introduced into LO28 strains by electroporation. The following antibiotics were used at the indicated concentrations: erythromycin (8 μg/ml), kanamycin (50 μg/ml), and spectinomycin (60 μg/ml). Bacillus subtilis was grown in LB medium and transformed as previously described by using plasmid DNA (40). Transformants were selected on SP plates supplemented with chloramphenicol (5 μg/ml) or spectinomycin (100 μg/ml).
β-Galactosidase activity was estimated on plates by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) hydrolysis. β-Galactosidase-specific activities were determined as previously described (39-41) and were expressed as Miller units per milligram of protein.
Basal stress resistance experiments were performed as follows. Overnight cultures were diluted 100-fold in BHI medium and grown at 37°C with vigorous shaking until the optical density at 600 nm (OD600) reached 0.3. Exponentially growing cultures were then divided into two parts, and one of which was subjected to one of the following stress conditions: 2% NaCl (wt/vol); 0.01% sodium dodecyl sulfate (SDS) (wt/vol); 15, 30, or 60 μg of puromycin per ml (values indicate final concentrations); or growth at 42, 44, 48, or 55°C. Growth was then monitored for an additional 3 h. For induced thermoresistance, overnight cultures were diluted 100-fold in BHI medium and placed at 37°C with vigorous shaking until the OD600 reached 0.3. Prior to heat treatment at 60°C, cultures were divided into two parts, one of which was maintained at 37°C, while the other half was preincubated at the nonlethal temperature of 48°C for 20 min. Both cultures were then incubated at 60°C, and aliquots were quickly transferred to ice, diluted in ice-cold BHI, and immediately plated on BHI; CFU were then counted. The induced thermoresistance experiment was repeated four times, yielding the same results.
DNA manipulations.
Chromosomal DNA preparation, plasmid isolation, restriction enzyme analysis, and amplification by PCR were performed according to standard protocols (57). DNA sequences were determined by the dideoxy chain termination method (59) using modified T7 DNA polymerase (63) (Pharmacia). DNA concentrations were calculated by measuring UV spectroscopy at 260 nm.
Mutant and plasmid constructions.
All oligonucleotide positions are given relative to the translation initiation codon. The plasmids used in this study are listed in Table 1. Plasmid pDL (71) was used for constructing transcriptional fusions with the Bacillus stearothermophilus bgaB gene, which encodes a thermostable β-galactosidase (25), with subsequent integration at the B. subtilis amyE locus. A clpB′-bgaB transcriptional fusion was constructed using a 189-bp EcoRI/BamHI DNA fragment corresponding to the clpB upstream region, which was generated by PCR by using oligonucleotides ID73 (−193) (5′-GAAGAATTCATGTTCTTACTCCGCC-3′) and ID74 (−5) (5′-GGAGGATCCTTATAAAAGATAAGTC-3′). This fragment was cloned between the EcoRI and BamHI sites of plasmid pDL to give plasmid pDL73/74. Linearization of this plasmid at the unique PstI site and transformation of the B. subtilis QB4991 strain with selection for chloramphenicol resistance yielded strain QB8059, in which the clpB′-bgaB fusion was integrated as a single copy at the amyE locus. The linearized pxyl59/60 plasmid (42) was then introduced in these strains by transformation and selection for spectinomycin to give strain QB8060, in which the L. monocytogenes ctsR gene is placed under the control of the PxylA xylose-inducible promoter and integrated as a single copy at the thrC locus.
A markerless ΔclpB deletion mutant of L. monocytogenes was constructed using plasmid pMADΔclpB. The mutant was constructed by first using PCR to generate two DNA fragments of 829 and 761 bp, using oligonucleotide pairs AC189 (5′-AATGGATCCCACATCCGAGCGAGTAAACAC-3′) and AC190 (5′-TAAGTCGACTCATTCGTCCTCCTTATAAAA-3′) and AC192 (5′-CACGTCGACTGAAAGGGAAAACTTTGGTTG-3′) and AC193 (5′-TATCCATGGAATATTTATTTACTGGTTTTA-3′), corresponding, respectively, to the chromosomal DNA regions that are directly upstream and downstream from the clpB gene. These fragments were cloned in pMAD, a pRN5101 derivative carrying a thermosensitive origin of replication (M. Arnaud and M. Débarbouillé, unpublished data), and the resulting pMADΔclpB plasmid was electroporated in the LO28 strain with selection for erythromycin. Integration and excision of pMADΔclpB was performed as previously described (4) but with a nonpermissive temperature growth of 40°C, thus yielding strain LM2000 (ΔclpB), in which the entire clpB coding sequence was removed. PCR amplifications were performed in order to confirm the gene deletion.
The ctsR deletion mutant was obtained by transforming the LO28 strain by plasmid pMADΔctsR. For this purpose, DNA fragments of 1,044 and 1,031 base pairs corresponding to the upstream and downstream chromosomal DNA regions from ctsR were amplified by using oligonucleotide pairs AC212 (5′-GGCGGATCCCTCCTAAAGAGTAACGGAGGC-3′) and AC213 (5′-ACTGAATTCCAATACTTGTTTCAAATAAGC-3′) and AC214 (5′-TGAGAATTCGGATTTTAGAGGCGATGTTAG-3′) and AC215 (5′-TATCCATGGTCTTTATCAAAAGCATAAC-3′), respectively. The aphA3 kanamycin resistance gene, deprived of its transcription initiation and termination signals, was then cloned at the EcoRI site between the two fragments just described. The resulting pMADΔctsR plasmid was introduced into strain L028, and the integration-excision procedure was performed as was described for the clpB deletion, thus yielding strain LM2001.
Mouse virulence assay.
Six- to 8-week-old pathogen-free Swiss female mice (Janvier, Le Genset St. Isle, France), were used in this study. Groups of five mice were injected intravenously with doses of L. monocytogenes LO28 and ΔclpB mutant ranging between 5 × 105 and 5 × 108 bacteria. Mortality was observed over a 14-day period. The 50% lethal dose was determined by the Probit method. Mice were killed by cervical dislocation in accordance with the policies of the Animal Welfare Committee of the Faculté Necker (Paris, France).
Gel mobility shift DNA-binding assays.
A 189-base-pair EcoRI/BamHI DNA fragment corresponding to the promoter region of clpB was generated by PCR using oligonucleotides ID73 and ID74. Radiolabeling, DNA-binding, and gel shift experiments were performed as previously described (10).
DNase I footprinting.
A 229-base-pair DNA fragment used for DNase I footprinting was prepared by PCR using Pfu DNA polymerase (Stratagene, La Jolla, Calif.) and oligonucleotides ID75 (−147) (5′-AAATTCAGAAGATCTGCCAACC-3′) and ID76 (+82) (5′-CTTATGTTCTGATGCAATAGC-3′). Labeling and DNase I treatment were performed as previously described (10).
RNA extraction and primer extension.
L. monocytogenes strains were grown in BHI medium at 37°C with aeration until the OD600 reached 0.5; half of the culture was then shifted to 42°C, and incubation was undertaken for another 10 min. Cells were pelleted and frozen immediately, and RNA extraction and primer extension were then performed as previously described (5) using radiolabeled oligonucleotide AC209 (+22) (5′-GTGTAAATTTTTGTAAATCCATTC-3′). Radioactive gels were exposed to storage phosphor screens and scanned with a Molecular Dynamics Storm 860 optical scanner. Quantitation of primer extension products was performed using the ImageQuant 5.1 software package (Molecular Dynamics).
RESULTS
Genome sequence analysis reveals ClpB ATPase in L. monocytogenes.
Analysis of the complete genome of L. monocytogenes EGDe reveals a protein with a deduced 63% amino acid sequence identity with ClpB of Lactococcus lactis and 52% with that of E. coli (Fig. 1). The ATG initiation codon of clpB is preceded by a classical ribosome binding site (RBS), tAAGGAGG, at a suitable distance, and this sequence encodes a predicted protein of 866 amino acid residues with a calculated molecular mass of 97.5 kDa. We note that a GTG codon is located 151 codons downstream from the ATG codon and is also preceded by a typical RBS, (AgAGGAGG), at an appropriate distance of 7 bp. This potential internal translation initiation site suggests the existence, as shown for E. coli, of a smaller form of ClpB, with 716 amino acid residues and a theoretical molecular mass of 80.6 kDa (47, 62).
FIG. 1.
Alignment of the ClpB amino acid sequence of L. monocytogenes with those of E. coli and L. lactis. Numbers indicate positions in the amino acid sequence. Identical residues are shaded. The conserved nucleotide-binding regions are boxed. Conserved Walker motifs (A box and B box) and predicted ClpN and coiled-coil motifs are overlined.
Analysis of the amino acid sequence of the protein revealed two typical Clp signature motifs (60, 61). Indeed, two ATP-binding sites are present, one with a single Walker A and two Walker B motifs and the other presenting only one of each Walker motif, a finding that is characteristic of HSP100 proteins (Fig. 1). There are also two repeated Clp amino-terminal domain motifs (ClpN), which are typical of ClpA and ClpB proteins (3) but are also present in most ClpC proteins.
This Clp ATPase also presents a long central domain separating the two ATP-binding sites, approximately 130 amino acids in length, which is characteristic of ClpB proteins. This domain contains a predicted coiled-coil motif (37; http://smart.embl-heidelberg.de), which may be involved in multimerization. Analysis of the carboxy-terminal domain, between the second ATP-binding site and the PDZ-like sensor and substrate discrimination domain, revealed the absence of the IGF loop required for interaction with the ClpP proteolytic subunit (30). This suggests that, as in E. coli, ClpB function in L. monocytogenes is restricted to chaperone activity without any interaction with ClpP.
clpB is a novel member of the L. monocytogenes CtsR regulon.
Three clp genes of L. monocytogenes have been shown to be controlled by the CtsR repressor of stress response genes (42), and many clpB genes are known to be heat shock-induced genes. To investigate a potential mechanism of transcriptional regulation of clpB, we analyzed the sequence of the promoter region, thereby revealing the presence of a potential binding site for CtsR (GGTCAAA AAA GGTCAgA) (see Fig. 3B), suggesting that ClpB may be a novel member of the L. monocytogenes CtsR regulon.
FIG. 3.
(A) Primer extension analysis of clpB expression at 37°C (lanes 1 and 2) or following a 10-min heat shock at 42°C (lanes 3 and 4). Total RNA (20 μg) extracted from L. monocytogenes L028 (lanes 1 and 3) and LM2001 (ΔctsR) (lanes 2 and 4), was used as a template for reverse transcriptase. The corresponding DNA sequence is shown on the left. (B) Nucleotide sequence of the L. monocytogenes L028 clpB promoter region. Potential −35 and −10 promoter sequences are overlined; the transcriptional start site is indicated by +1; the CtsR direct-repeat operator sequence is indicated by arrows; the potential RBS sequence is underlined; the translational start site is boxed, and the deduced amino acid sequence is indicated below the nucleotide sequence.
We used B. subtilis as a heterologous host to test whether CtsR plays a role in controlling clpB expression. For this purpose, a transcriptional fusion was constructed between the L. monocytogenes clpB promoter region and the bgaB gene of B. stearothermophilus, which encodes a thermostable β-galactosidase (25) (see Materials and Methods). The fusion was integrated as a single copy at the amyE locus of B. subtilis strain QB4991, in which the endogenous ctsR gene is deleted (10). The L. monocytogenes ctsR gene was then integrated as a single copy at the thrC locus, under the control of the PxylA xylose-inducible promoter, by using plasmid pxyl59/60 (42), thus leading to strain QB8060.
Strain QB8060 was grown at 37°C in the presence or absence of xylose, and β-galactosidase activities were assayed. As shown in Fig. 2, clpB′-bgaB was expressed up to approximately 550 Miller units · mg−1 of protein at 37°C in the absence of the repressor (without xylose). This expression was repressed 15-fold (35 Miller units · mg−1 of protein) in the presence of CtsR of L. monocytogenes (with xylose). However, when the culture was shifted to high-temperature conditions (48°C) instead of 37°C, expression was induced up to 47-fold (1,650 Miller units · mg−1 of protein) in the presence of CtsR (with xylose; data not shown). These results demonstrate that clpB of L. monocytogenes is under negative regulation by CtsR and that this repression is thermosensitive.
FIG. 2.
clpB is negatively regulated by CtsR in the heterologous host B. subtilis. Levels of expression of clpB′-bgaB (strain QB8060, clpB′-bgaB ΔctsR pxylctsRLmo) in LB medium at 37°C in the presence (□) or absence (▪) of xylose are shown. Symbols indicate β-galactosidase activities expressed as Miller units/mg of protein as a function of time.
clpB is expressed from a σA-dependent heat-inducible promoter.
In order to demonstrate the thermoinducibility of clpB and a role for CtsR in its regulation, an analysis of clpB transcription in L. monocytogenes was performed by using primer extension experiments. First, the transcription initiation site was determined by using RNA from L. monocytogenes cells grown in BHI at 37°C and harvested in mid-exponential phase (see Materials and Methods). This procedure revealed a single transcriptional start site 45 bp upstream from the ATG start codon of clpB (Fig. 3A and B). Consensus −10 and −35 sequences recognized by the EσA RNA polymerase holoenzyme were identified upstream from the transcriptional start site, suggesting a σA-dependent promoter (Fig. 3B). A comparative transcriptional analysis of RNA expression at 37 and 42°C was performed by primer extension. As shown in Fig. 3A, clpB was expressed at a low basal level during growth in BHI at 37°C (Fig. 3A, lane 1), and transcription was increased fourfold when the culture was shifted to 42°C (Fig. 3A, lane 3), a finding which is consistent with a thermosensitive transcriptional regulation.
Repression of clpB by CtsR was examined in vivo in L. monocytogenes. The LM2001 ΔctsR mutant strain was constructed by deleting the entire ctsR coding sequence and replacing it with the aphA3 kanamycin resistance gene. This resistance cassette was deprived of its transcription initiation and termination signals in order to rule out any polar effects on expression of the downstream genes. Expression of clpB at 37°C in the wild-type (L028) (Fig. 3A, lane 1) and ΔctsR (LM2001) (Fig. 3A, lane 2) strains was followed by primer extension analysis, which revealed increased transcription of clpB (6.5-fold) in the absence of CtsR. It is interesting that clpB derepression at 42°C is only partial since expression levels are higher in the ctsR deletion mutant at 37°C (Fig. 3A, lane 2) than in the wild-type strain at 42°C (Fig. 3A, lane 3), thus suggesting a limited inactivation of CtsR at this temperature. In conclusion, the in vivo evidence indicates that clpB expression is repressed by CtsR and is heat shock inducible.
CtsR binds specifically to the clpB promoter region.
An in vitro approach was used to demonstrate a direct interaction between CtsR and the clpB promoter region. Histidine-tagged CtsR of L. monocytogenes, presenting a carboxy-terminal extension of six histidine residues, was overproduced and purified by using a Ni-nitrilotriacetic acid agarose column (42). This recombinant protein was used in gel mobility shift DNA-binding assays with a 189-bp radiolabeled PCR-generated DNA fragment corresponding to the clpB promoter region. This DNA fragment, extending from positions −193 to −5 relative to the translation initiation codon, was incubated with increasing amounts of purified CtsR in the presence of nonspecific competitor DNA [poly-(dI-dC)]. As shown in Fig. 4A, CtsR bound specifically to the radiolabeled fragment, leading to progressive displacement of the probe to the single higher-molecular-weight protein/DNA complex. Although an incomplete displacement was observed even at the highest CtsR concentrations, the single DNA/protein complex suggests the presence of only one CtsR-binding site in this promoter. These results demonstrate that CtsR of L. monocytogenes represses clpB expression by binding directly to the promoter region.
FIG. 4.
(A) CtsR binds specifically to the clpB promoter region. DNA-binding reactions were performed with radiolabeled DNA fragments (10,000 cpm) corresponding to the clpB promoter region. Lane 1, no protein; lane 2, 7 ng; lane 3, 70 ng; lane 4, 700 ng. (B and C) DNase I footprinting analysis of CtsR binding to the clpB promoter region. Each lane contains 50,000 cpm of radiolabeled DNA fragment corresponding to the nontemplate strand (panel B) or the template strand (panel C) of the L. monocytogenes clpB promoter region. Fragments were incubated with increasing amounts of purified CtsR. Lane 1, no protein; lane 2, 35 ng; lane 3, 350 ng; lane 4, 3,500 ng; lane 5, Maxam and Gilbert reactions of the corresponding DNA fragment. Brackets indicate regions protected by CtsR. (D) Nucleotide sequence of the clpB promoter region. The DNase I protected area is boxed, and arrows indicate the CtsR direct-repeat recognition sequence. Positions are numbered relative to the translational initiation codon.
DNase I footprinting assays were performed for L. monocytogenes DNA fragments corresponding to the clpB promoter region in order to precisely determine the location of the CtsR-binding site. When the nontemplate strand of clpB DNA was end labeled, CtsR protected a region extending from positions −42 to −20 (Fig. 4B and D). When the template strand was end labeled, the protected region extended from positions −46 to −24 (Fig. 4C and D). All positions given are relative to the respective translational start site.
A single region within the clpB promoter is protected from DNase I cleavage, a finding which is in agreement with the single protein/DNA complex observed in the gel mobility shift DNA-binding assay (Fig. 4A). This protected region overlaps the transcriptional start site of clpB and contains the predicted CtsR direct-repeat recognition sequence (GGTCAAA AAA GGTCAGA) (Fig. 4D). These results indicate that CtsR negatively regulates clpB expression by directly binding to its operator sequence in the promoter region.
In conclusion, using both in vitro and in vivo approaches, we have shown that L. monocytogenes clpB is a heat shock gene that is under the negative regulation of CtsR, extending the L. monocytogenes CtsR regulon.
ClpB is involved in virulence of L. monocytogenes.
Since ClpP, ClpC, and ClpE of L. monocytogenes have been shown to play a role in virulence (16, 43, 54), we therefore examined the virulence of an L. monocytogenes ΔclpB mutant in a murine model.
For this purpose, we constructed the LM2000 mutant strain of L. monocytogenes, in which the entire coding sequence of clpB was deleted (see Materials and Methods). Virulence of the ΔclpB strain was assayed by intravenous inoculation as described in Materials and Methods and was compared to that of the wild-type L028 strain. The 50% lethal dose of the ΔclpB mutant was 5.4 × 106.3 bacteria, whereas that of L028 was 5.4 × 104.2 bacteria. The ΔclpB mutant thus displays a significant decrease in virulence (100-fold).
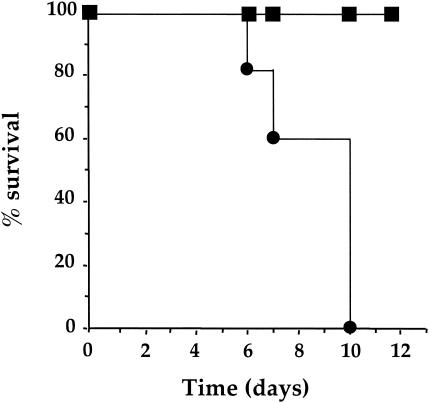
We monitored the survival of mice for 12 days after an inoculation of 5.4 × 105 bacteria. Mice infected with the wild-type strain began to die after 5 days, and all were dead after the 10th day, whereas all animals infected by strain LM2000 (ΔclpB) were still alive after 12 days (Fig. 5). These results clearly show that ClpB plays a significant role in the pathogenicity of L. monocytogenes.
FIG. 5.
ClpB is involved in virulence of L. monocytogenes. Survival curves for Swiss mice after intravenous inoculation with 5 × 105 bacteria of the wild-type L028 (•) or the mutant strain ΔclpB (▪) are shown.
In order to determine whether ClpB of Listeria monocytogenes affects expression of virulence genes, primer extension experiments were carried out for the wild-type and for the ΔclpB and ΔctsR mutants in order to compare expression of the L. monocytogenes hly gene, which encodes listeriolysin O, a major virulence determinant. Expression was identical for all three strains (data not shown), a finding which indicated that the major virulence PrfA regulon is not controlled by ClpB or CtsR and that the role for ClpB in virulence is most likely due to its chaperone activity rather than to a regulatory role in virulence gene expression.
ClpB is not required for general stress response but is necessary for heat shock-induced thermotolerance.
Since several Clp proteins are involved in virulence and because many are also essential for resistance to various stress conditions, one might argue that Clp protein effects on pathogenicity may be indirect consequences of generally lowered cell fitness, thus leading to increased sensitivity to stress when invading the host.
A functional analysis of ClpB was undertaken, during which survival of the ΔclpB mutant was examined under different stress conditions. The LM2000 (ΔclpB) mutant strain had no obvious phenotype, since the mutant cells showed no morphological defects and because the growth curve in BHI at 37°C was identical to that of the LO28 reference strain (data not shown).
The stress resistance of the ΔclpB strain was evaluated under various conditions, such as heat stress, treatment with puromycin, the presence of salt, and SDS-induced stress, all of which are known to require the activity of other Clp proteins. Wild-type and mutant strains were grown in BHI medium until an OD600 of 0.3 was reached, cultures were divided into two parts, and one part was subjected to stress conditions (see Materials and Methods). The results presented in Fig. 6 summarize data obtained for typical growth curves for each stress condition. As shown in Fig. 6, growth of the wild-type and mutant strains was affected when the temperature was equal to or greater than 42°C, when the concentration of puromycin was greater than 30 μg/ml, or in the presence of 0.01% SDS. However, no difference was observed between the ΔclpB and the L028 reference strain, since the two strains grew equally well under all conditions tested. In conclusion, L. monocytogenes ClpB is not required for general stress adaptation, a finding that is contrary to the situation for gram-negative bacteria such as E. coli (29), Brucella suis (12), or Helicobacter pylori (1).
FIG. 6.
ClpB is not required for stress resistance. The L028 wild-type (WT) strain and the ΔclpB mutant strain were grown exponentially at 37°C with aeration in BHI medium until the OD600 reached 0.3. The culture was divided into two parts, one of which was subjected to various stresses. Stresses assayed were temperature shifts to 42, 44, 48, or 55°C; addition of puromycin to a final concentration of 15 (pmc15), 30 (pmc30), or 60 (pmc60) μg/ml; 0.01% SDS; or 2% NaCl. Values represent the percentage of cell growth with respect to the control culture performed in the absence of stress (grown in BHI at 37°C) 2 h after the stress was applied. In all cases, there was no significant difference in the growth curves between the wild-type and the mutant strains.
A recent study reports the induction of L. monocytogenes clpB during growth at low temperature (36), a condition which seems to induce the activity of most of the general stress proteins; for the cyanobacterium Synechococcus sp., ClpB was shown to be involved in cold adaptation (49). Growth at low temperature is an important part of the L. monocytogenes life cycle and is one which favors food contamination and outbreaks of food-borne disease. The role of ClpB in adaptation of L. monocytogenes to cold stress was tested. An overnight culture grown at room temperature was diluted and placed at 5°C for 4 days or was first grown to the mid-exponential phase at 37°C before shifting the culture to a temperature of 5°C. In both cases, the growth rate of the ΔclpB mutant at 5°C was the same as that of the parental strain (data not shown), suggesting that ClpB of L. monocytogenes is not involved with adaptation to cold stress.
It was previously shown that L. monocytogenes has a higher survival rate to lethal temperatures following previous exposure to a sublethal temperature (46, 56). This phenomenon is known as induced thermotolerance, a characteristic which stands in contrast to basal thermotolerance and has been described as occurring in many bacteria. ClpB was shown to be required for induced thermotolerance in the cyanobacterium Synechococcus sp. (13) and in the eukaryote Saccharomyces cerevisiae (58). In order to test the involvement of ClpB in induced thermotolerance of L. monocytogenes, we incubated both wild-type L028 and ΔclpB mutant strains in liquid BHI medium at 37°C until an OD600 of 0.3 was reached. The cultures were divided into two parts; one half was maintained at 37°C, and the other was preincubated at 48°C for 20 min. Both cultures were then subjected to heat treatment at 60°C. As shown in Fig. 7, preincubated wild-type cells presented an increased resistance to lethal heat shock, since after 5 min of incubation at 60°C, the survival rate was approximately 100-fold higher than that for untreated cells. In contrast, no induced thermotolerance could be observed for the ΔclpB strain, which remained as sensitive to lethal temperatures as were the untreated cells. Consequently, contrary to the situation for wild-type L028, a preincubation at 48°C did not protect ΔclpB cells, thus revealing a role for ClpB in induced thermotolerance.
FIG. 7.
ClpB is required for induced thermotolerance. Cultures of wild-type L028 and ΔclpB mutant strains were grown exponentially until the OD600 reached 0.3. Half of the culture was preincubated for 20 min at the nonlethal temperature of 48°C, while the other half was maintained at 37°C. After preincubation, both cultures were incubated at 60°C, and cell survival was evaluated by plating diluted aliquots. White bars indicate CFU values before incubation at 60°C; black bars indicate CFU values after 5 min of stress.
DISCUSSION
Clp-HSP100 proteins make up a ubiquitous family of ATPases that act both as chaperones and as ATPase subunits for the Clp ATP-dependent protease. Most of them are induced by stress and are implicated in stress tolerance. Moreover, Clp proteins are involved in crucial steps of the infectious process for many gram-positive and gram-negative bacteria as well as in lower eukaryotes. In L. monocytogenes, ClpC, ClpP, and ClpE are involved in virulence and are required for thermotolerance and resistance to salt stress (16, 43, 44, 54). Their expression is thermoinducible and is under the negative control of CtsR, a repressor that binds to a heptad operator sequence in the promoter region (42).
Analysis of the complete sequence of L. monocytogenes EGDe (18) revealed several new clp genes. Just as for its closest relative, B. subtilis, we noted the presence of genes encoding orthologs to ATPase subunits ClpY (65.5% identity) and ClpX (81% identity) and to the proteolytic subunit ClpQ (78.5% identity). Surprisingly, and contrary to the situation for B. subtilis, there are two additional clp genes, both of which are preceded by potential operator sites for the CtsR repressor in their promoter regions. One of the encoded proteins shared 40% amino acid sequence identity with the ClpP proteolytic subunit of L. monocytogenes and is now referred to as ClpP2.
The second new clp gene revealed during our analysis has no ortholog in the low-G+C gram-positive model bacterium B. subtilis but shares strong similarities with clpB of L. lactis. We performed a systematic search for ClpB homologs to determine the extent of its distribution among low-G+C gram-positive bacterial genomes. Unlike the situation for B. subtilis, ClpB orthologs are found in all staphylococci, clostridia, and enterococci, as well as in L. lactis, Listeria innocua, and most bacilli. Streptococci seem to be the only group without this homolog, despite the presence in S. mutans of a ClpB-like protein presenting the characteristically long spacer region between both ATP-binding sites (34). However, the very divergent sequence places this paralog far from all the other known eubacterial clpB genes, suggesting a recent acquisition by way of horizontal transfer. Consequently, ClpB ATPases are well represented among low-G+C gram-positive bacteria.
Examination of the clpB promoter sequence revealed a typical σA promoter and a potential CtsR-binding site. In this work, we showed that CtsR represses the expression of clpB in L. monocytogenes. We showed in vitro that CtsR binds directly to the clpB promoter region. The gel mobility shift experiments reveal a single protein/DNA complex. This was confirmed by DNase I footprints in which only one protected region was observed, a finding which corresponded to the predicted CtsR box and overlapping the transcriptional start site.
A detailed DNA motif analysis of the complete genome of L. monocytogenes allowed us to determine that there appear to be only five members of the CtsR regulon: the ctsR-clpC operon, clpP, clpE, clpB, and potentially clpP2. Interestingly, although the dnaK operon is also preceded by a canonical CtsR operator, it does not seem to be controlled by this regulator, although the repressor can bind to this sequence in vitro (4).
Stress induction of clp genes is generally correlated with a role in stress resistance. Data from E. coli suggest that, unlike the other Clp ATPases, ClpB does not associate with the ClpP proteolytic subunit and has no effect on protease activity (68); consequently, ClpB acts exclusively as a chaperone.
In agreement with this activity, ClpB has been shown to have three closely related functions in several gram-negative bacteria and in eukaryotes: (i) resistance to high-temperature stress in H. pylori, B. suis, and E. coli (1, 12, 29, 62); (ii) cold acclimatization in Synechococcus sp.; and (iii) induced thermotolerance to lethal stress in the cyanobacterium Synechococcus sp. (13) and in the eukaryote S. cerevisiae (35, 58). HSP101, a member of the Clp/HSP100 family that is present in plants, has also been shown to be implicated in induced thermotolerance in both Arabidopsis thaliana (53) and maize (45). However, until now, only two clpB genes have been described for gram-positive bacteria, those of Streptomyces albus G (20) and L. lactis (27), and no obvious phenotype was associated with the respective mutants. Indeed, the ΔclpB mutant of L. lactis was still resistant to temperature, salt, and puromycin stresses (27), and no thermosensitivity was observed for the S. albus mutant (C. Grandvalet, personal communication), even though both genes were shown to be thermoinducible. Here, we demonstrate that ClpB is required for induced thermotolerance of L. monocytogenes, which allows for better survival of lethal conditions when cells have been exposed to a nonlethal stress.
ClpB is required for induced thermotolerance of L. monocytogenes; this fact may contribute to the persistence of this bacterium and the health hazard it constitutes. Indeed, this bacterium has the ability to grow in a wide range of temperatures, even during the refrigeration process or after high-temperature short-time pasteurization and is considered one of the most thermotolerant bacteria among non-spore-forming food-borne pathogens (11, 14). It is clear that the ability of L. monocytogenes to grow at high temperatures is an important problem for food processing, and our results suggest that ClpB may be partially responsible for this adaptation faculty.
A deletion of clpB is associated with a reduction in virulence of several eukaryotes and gram-negative bacteria, such as Leishmania major (26), Leishmania donovani (6, 31, 32), S. enterica serovar Typhimurium (64), Y. enterocolitica (2), and F. novicida (21). However, the exact function of ClpB—and more generally, that of Clp homologs—in virulence is still unclear because their targets have not yet been discovered. Clp proteins, because of their central role in protein folding, are important factors for efficient growth and cell fitness. ClpP proteolytic subunits, for example, have pleiotropic roles, and their deletion, even in optimum conditions, greatly affects growth (5, 40). In most cases, Clp proteins involved in virulence are also required for stress survival, and since infection is one of the most stressful conditions encountered by bacteria, one can argue that effects observed in a clp deletion mutant are due to a deficiency in cell fitness. We have shown here that ClpB is not required for general stress survival of L. monocytogenes, with the exception of induced thermotolerance at 60°C. Thus, it is tempting to speculate that the significant reduction in L. monocytogenes virulence of the ΔclpB mutant might not be due to a reduction in survival ability or in adaptation to stressful conditions but rather to a specific alteration in a key process for pathogenic development, where ClpB probably acts as a chaperone. This speculation is supported by the fact that expression of the hly gene, a major virulence determinant belonging to the PrfA regulon, is not modified in the ΔclpB or ΔctsR mutants.
In conclusion, our results demonstrate a role for ClpB in induced thermotolerance and present the first evidence for a role for ClpB in virulence of L. monocytogenes; our work constitutes the first description of phenotypes for a clpB gene in gram-positive bacteria.
Acknowledgments
We are grateful to G. Rapoport for critical reading of the manuscript, and we thank P. Berche, in whose laboratory part of this work was carried out.
This work was supported by research funds from the Institut Pasteur, Centre National de Recherche Scientifique, Université Paris 7, European Commission (grant number QLG2-CT-1999-01455), Ministère de la Défense (Délégation Générale pour l'Armement, grant number 0034069004707501), and the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires of the Ministère de la Recherche. I.D. and A.C. were the recipients of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie, and A.C. was the recipient of a fellowship from the Fondation pour la Recherche Médicale (FRM) and the CANAM (Caisse Nationale d'Assurance Maladie et Maternité des Travailleurs Non Salariés des Professions Non Agricoles).
REFERENCES
- 1.Allan, E., P. Mullany, and S. Tabaqchali. 1998. Construction and characterization of a Helicobacter pylori clpB mutant and role of the gene in the stress response. J. Bacteriol. 180:426-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J. L., B. M. Young, A. J. Darwin, and V. L. Miller. 2000. Yersinia enterocolitica ClpB affects levels of invasin and motility. J. Bacteriol. 182:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. E., A. Zolkiewska, and M. Zolkiewski. 2000. Structure and activity of ClpB from Escherichia coli. Role of the amino- and carboxyl-terminal domains. J. Biol. Chem. 275:37565-37571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 5.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clos, J., L. Klaholz, M. Kroemer, S. Krobitsch, and S. Lindquist. 2001. Heat shock protein 100 and the amastigote stage-specific A2 proteins of Leishmania donovani. Med. Microbiol. Immunol. 190:47-50. [DOI] [PubMed] [Google Scholar]
- 7.Cossart, P., and M. Lecuit. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 17:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Crecy-Lagard, V., P. Servant-Moisson, J. Viala, C. Grandvalet, and P. Mazodier. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32:505-517. [DOI] [PubMed] [Google Scholar]
- 9.Derré, I., G. Rapoport, and T. Msadek. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37°C. Mol. Microbiol. 38:335-347. [DOI] [PubMed] [Google Scholar]
- 10.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-132. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, M. P., K. A. Glass, J. T. Beery, G. A. Garcia, D. J. Pollard, and R. D. Schultz. 1987. Survival of Listeria monocytogenes in milk during high-temperature, short-time pasteurization. Appl. Environ. Microbiol. 53:1433-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekaza, E., J. Teyssier, S. Ouahrani-Bettache, J. P. Liautard, and S. Kohler. 2001. Characterization of Brucella suis clpB and clpAB mutants and participation of the genes in stress responses. J. Bacteriol. 183:2677-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, M. J., and A. K. Clarke. 1996. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 178:4839-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming, D. W., S. L. Cochi, K. L. MacDonald, J. Brondum, P. S. Hayes, B. D. Plikaytis, M. B. Holmes, A. Audurier, C. V. Broome, and A. L. Reingold. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312:404-407. [DOI] [PubMed] [Google Scholar]
- 15.Gahan, C. G., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph. D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 18.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 20.Grandvalet, C., P. Servant, and P. Mazodier. 1997. Disruption of hspR, the repressor gene of the dnaK operon in Streptomyces albus G. Mol. Microbiol. 23:77-84. [DOI] [PubMed] [Google Scholar]
- 21.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215:53-56. [DOI] [PubMed] [Google Scholar]
- 22.Hanawa, T., M. Fukuda, H. Kawakami, H. Hirano, S. Kamiya, and T. Yamamoto. 1999. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones 4:118-128. [PMC free article] [PubMed] [Google Scholar]
- 23.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 24.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 25.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubel, A., S. Krobitsch, A. Horauf, and J. Clos. 1997. Leishmania major Hsp100 is required chiefly in the mammalian stage of the parasite. Mol. Cell. Biol. 17:5987-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingmer, H., F. K. Vogensen, K. Hammer, and M. Kilstrup. 1999. Disruption and analysis of the clpB, clpC, and clpE genes in Lactococcus lactis: ClpE, a new Clp family in gram-positive bacteria. J. Bacteriol. 181:2075-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 29.Kim, K. I., K. M. Woo, I. S. Seong, Z. W. Lee, S. H. Baek, and C. H. Chung. 1998. Mutational analysis of the two ATP-binding sites in ClpB, a heat shock protein with protein-activated ATPase activity in Escherichia coli. Biochem. J. 333:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, Y. I., I. Levchenko, K. Fraczkowska, R. V. Woodruff, R. T. Sauer, and T. A. Baker. 2001. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8:230-233. [DOI] [PubMed] [Google Scholar]
- 31.Krobitsch, S., S. Brandau, C. Hoyer, C. Schmetz, A. Hubel, and J. Clos. 1998. Leishmania donovani heat shock protein 100. Characterization and function in amastigote stage differentiation. J. Biol. Chem. 273:6488-6494. [DOI] [PubMed] [Google Scholar]
- 32.Krobitsch, S., and J. Clos. 1999. A novel role for 100 kD heat shock proteins in the parasite Leishmania donovani. Cell Stress Chaperones 4:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 34.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindquist, S., and G. Kim. 1996. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl. Acad. Sci. USA 93:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse, II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 38.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Msadek, T., V. Dartois, F. Kunst, M.-L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 41.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172:824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 43.Nair, S., C. Frehel, L. Nguyen, V. Escuyer, and P. Berche. 1999. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 31:185-196. [DOI] [PubMed] [Google Scholar]
- 44.Nair, S., E. Milohanic, and P. Berche. 2000. ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68:7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieto-Sotelo, J., L. M. Martinez, G. Ponce, G. I. Cassab, A. Alagon, R. B. Meeley, J. M. Ribaut, and R. Yang. 2002. Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell 14:1621-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagán, R., S. Condón, and F. J. Sala. 1997. Effects of several factors on the heat-shock-induced thermotolerance of Listeria monocytogenes. Appl. Environ. Microbiol. 63:3225-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, S. K., K. I. Kim, K. M. Woo, J. H. Seol, K. Tanaka, A. Ichihara, D. B. Ha, and C. H. Chung. 1993. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J. Biol. Chem. 268:20170-20174. [PubMed] [Google Scholar]
- 48.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porankiewicz, J., and A. K. Clarke. 1997. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 51.Portnoy, D. A., V. Auerbuch, and I. J. Glomski. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qoronfleh, M. W., C. A. Bortner, P. Schwartzberg, and B. J. Wilkinson. 1998. Enhanced levels of Staphylococcus aureus stress protein GroEL and DnaK homologs early in infection of human epithelial cells. Infect. Immun. 66:3024-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Queitsch, C., S. W. Hong, E. Vierling, and S. Lindquist. 2000. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12:479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rouquette, C., C. de Chastellier, S. Nair, and P. Berche. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235-1246. [DOI] [PubMed] [Google Scholar]
- 55.Rouquette, C., M.-T. Ripio, E. Pellegrini, J.-M. Bolla, R. I. Tascon, J.-A. Vázquez-Boland, and P. Berche. 1996. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21:977-987. [DOI] [PubMed] [Google Scholar]
- 56.Rowan, N. J., and J. G. Anderson. 1998. Effects of above-optimum growth temperature and cell morphology on thermotolerance of Listeria monocytogenes cells suspended in bovine milk. Appl. Environ. Microbiol. 64:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Sanchez, Y., and S. L. Lindquist. 1990. HSP104 required for induced thermotolerance. Science 248:1112-1115. [DOI] [PubMed] [Google Scholar]
- 59.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289-296. [PubMed] [Google Scholar]
- 61.Squires, C., and C. L. Squires. 1992. The Clp proteins: proteolysis regulators or molecular chaperones? J. Bacteriol. 174:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Squires, C. L., S. Pedersen, B. M. Ross, and C. Squires. 1991. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 173:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabor, S., and C. C. Richardson. 1987. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 84:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner, A. K., M. A. Lovell, S. D. Hulme, L. Zhang-Barber, and P. A. Barrow. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viala, J., and P. Mazodier. 2002. ClpP-dependent degradation of PopR allows tightly regulated expression of the clpP3 clpP4 operon in Streptomyces lividans. Mol. Microbiol. 44:633-643. [DOI] [PubMed] [Google Scholar]
- 66.Viala, J., G. Rapoport, and P. Mazodier. 2000. The clpP multigenic family in Streptomyces lividans: conditional expression of the clpP3 clpP4 operon is controlled by PopR, a novel transcriptional activator. Mol. Microbiol. 38:602-612. [DOI] [PubMed] [Google Scholar]
- 67.Vicente, M. F., F. Baquero, and J. C. Perez-Diaz. 1985. Cloning and expression of the Listeria monocytogenes haemolysin in Escherichia coli. FEMS Microbiol. Lett. 30:77-79. [Google Scholar]
- 68.Woo, K. M., K. I. Kim, A. L. Goldberg, D. B. Ha, and C. H. Chung. 1992. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J. Biol. Chem. 267:20429-20434. [PubMed] [Google Scholar]
- 69.Yamamoto, T., H. Sashinami, A. Takaya, T. Tomoyasu, H. Matsui, Y. Kikuchi, T. Hanawa, S. Kamiya, and A. Nakane. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan, G., and S. L. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177:6462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan, G., and S. L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the σA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]