Abstract
The Bacillus subtilis genome comprises two paralogous single-stranded DNA binding protein (SSB) genes, ssb and ywpH, which show distinct expression patterns. The main ssb gene is strongly expressed during exponential growth and is coregulated with genes encoding the ribosomal proteins S6 and S18. The gene organization rpsF-ssb-rpsR as observed in B. subtilis is found in many gram-positive as well as some gram-negative bacteria, but not in Escherichia coli. The ssb gene is essential for cell viability, and like other SSBs its expression is elevated during SOS response. In contrast, the paralogous ywpH gene is transcribed from its own promoter at the onset of stationary phase in minimal medium only. Its expression is ComK dependent and its gene product is required for optimal natural transformation.
Single-stranded DNA binding proteins (SSBs) in bacteria play crucial roles in DNA replication, repair, and recombination processes. The function of SSB in these processes, its biochemical properties, and its interaction with other proteins in the cell have been studied extensively in Escherichia coli and several bacteriophages (17, 26). Relatively little is known about the regulation of SSB expression.
In Escherichia coli the ssb gene is preceded by three promoters, one of them being inducible by DNA damage (3, 4). The DNA damage inducibility is due to the presence of a LexA binding site in the upstreammost promoter. Interestingly, this SOS-box is shared with the divergently transcribed uvrA gene, coding for the A subunit of the exonuclease ABC, which is involved in DNA repair (3). The same organization was found for the uvrA and ssb genes in Sinorhizobium meliloti (24). Although this gene organization is also identical in Proteus mirabilis and Serratia marcescens, the ssb genes of these bacteria are not inducible by DNA damage (7). It has been suggested that E. coli SSB negatively autoregulates its own translation, because it is capable of binding to its own mRNA, in this way inhibiting translation (21).
Two paralogous genes coding for SSB were found in the Bacillus subtilis genome, ssb and ywpH. The deduced amino acid sequences of SSB and YwpH show 80% similarity and 63% identity. Notably, YwpH is lacking 66 amino acid residues of the C terminus of SSB. Although the amino acid sequences of bacterial SSBs are highly conserved within the first two thirds of the protein containing the DNA-binding domains, they diverge substantially in the C-terminal third region (7). The C-terminal region of E. coli SSB is not required for DNA binding in vitro, but is essential for its in vivo function (6, 29). In contrast to E. coli, neither of the B. subtilis ssb genes is organized adjacent to uvrA as the ssb gene is in E. coli. The first one, ssb, maps at 358.6o of the B. subtilis genome and is flanked by the rpsF and rpsR genes, coding for the ribosomal proteins S6 and S18, respectively (Fig. 1A). A rho-independent transcriptional terminator is situated downstream of the rpsR gene, and possibly rpsF, ssb, and rpsR belong to one operon. The second ssb-like gene, ywpH, maps at 319.4o and is flanked by a gene of unknown function (ywpG) and the glcR gene, coding for a regulator involved in carbon catabolite repression (23) (Fig. 1B). Between these genes, no obvious terminator structure could be identified.
FIG. 1.
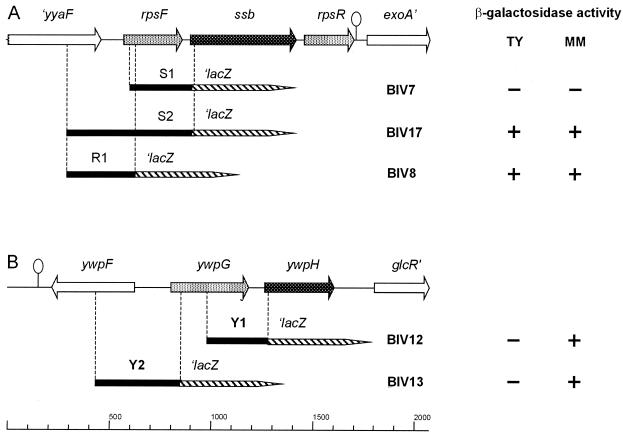
Gene organization of the ssb operon (A) and the ywpH gene (B) in B. subtilis 168, with schematic representation of the constructed lacZ fusions. Black lines in bold type represent the PCR-amplified DNA fragments fused to the promoterless lacZ gene (lacZ gene not drawn to scale). The presence or absence of β-galactosidase activity in a certain construct after growth on rich (TY) or minimal (MM) medium is indicated on the right site by + and −, respectively.
In this paper we studied the transcriptional regulation of the two ssb-like genes in B. subtilis and address the question of why there are two SSBs in this organism.
MATERIALS AND METHODS
Bacterial strains, medium, and growth conditions.
Table 1 lists the bacterial strains and plasmids used in this study. Bacteria were grown at 37°C under vigorous agitation in rich medium (TY [1% tryptone, 0.5% yeast extract, 1.0% NaCl] or BFA [16] when appropriate) or minimal medium (22). For the selection of transformants, appropriate antibiotics were added to the growth media at the following concentrations: for B. subtilis, 5 μg of chloramphenicol per ml, 10 μg of kanamycin per ml, and 100 μg of spectinomycin per ml; for E. coli, 100 μg of ampicillin per ml and 150 μg of spectinomycin per ml. To visualize α-amylase activity, TY plates were supplemented with 1% starch, and to visualize LacZ activity plates were supplemented with 0.004% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference | ||
|---|---|---|---|---|
| Strains | ||||
| 168 | trpC2 | 1 | ||
| BIV7 | trpC2 amyE::(ssb′-lacZ kan) Kmr | This study | ||
| BIV8 | trpC2 amyE::(rpsF′-lacZ kan) Kmr | This study | ||
| BIV12 | trpC2 amyE::(ywpH′-lacZ kan) Kmr | This study | ||
| BIV13 | trpC2 amyE::(ywpG′-lacZ kan) Kmr | This study | ||
| BIV17 | trpC2 amyE::(rpsF ssb′-lacZ kan) Kmr | This study | ||
| BIV24 | trpC2 amyE::(ywpH-lacZ kan) comK::spec Kmr Spr | This study | ||
| BIV27 | trpC2 ywpH::cat Cmr | This study | ||
| BV2004 | trpC2 his met tyr-1 ade nic ura comK::spec Spr | 12 | ||
| Plasmids | ||||
| pBTK2 | E. coli Apr; B. subtilis Kmr, amyE back and front, promoterless lacZ gene for reporter gene expression studies | 15 | ||
| pIC408 | Spr, ori pUC, multiple cloning site of pBluescript II SK(+), suicide plasmid for B. subtilis to construct “knockout” mutants | M. Steinmetz, unpublished | ||
| pBluescript SK- | Apr, T4 and T7 promoter/RNA polymerase site for synthesizing RNA probes | Stratagene | ||
| pUC7c | pUC21 derivative bearing chloramphenicol resistance cassette | 11 |
Strain constructions and transformation.
Cloning and transformation were performed according to established techniques (5), (20) and suppliers' manuals. The nucleotide sequences of the primers used for PCR are listed in Table 2. Enzymes were from Roche Molecular Biochemicals (Mannheim, Germany).
TABLE 2.
Primers used in this study
| Primer | Description | Sequence (5′ to 3′)a |
|---|---|---|
| rpsF-3 | Forward primer to amplify the rpsF (ssb) promoter region without RBS | GGAATTCCTGCAGGTGACTTTGAGCGGGGCTTC |
| rpsF-4 | Reverse primer to amplify the rpsF (ssb) promoter region without RBS | GGCCTCGAGGGCCCATAATGGGCAAGGAGC |
| rpsF-5 | Reverse primer to amplify the rpsF (ssb) promoter region with RBS | CGTACTTTCTCATATGTTTGCACC |
| ssb-1 | Forward primer to amplify the ssb promoter region | GCGAAGCTTCCAAACATTGACGAAGAGTCT |
| ssb-2 | Reverse primer to amplify the ssb promoter region | GCTGGATCCTCGGTTAAGCATAAGAAAGACC |
| rps-1 | Forward primer to amplify the rpsF promoter region | GCGAAGCTTGTGACTTTGAGCGGGGCTT |
| rps-2 | Reverse primer to amplify the rpsF promoter region | GCTGGATCCATCTTCGTCAATGTTTGGGCG |
| yyaE-1 | Forward primer to amplify the yyaF promoter region | GCGAAGCTTGAAGCATGAAGGGCAAGGTG |
| yyaE-2 | Reverse primer to amplify the yyaF promoter region | GCTGGATCCTCCGACGTTCGGCAAACCAA |
| ywpG-1 | Forward primer to amplify the ywpG promoter region | GCGAAGCTTGGCTTCAGATTGGCTGTTTTG |
| ywpG-2 | Reverse primer to amplify the ywpG promoter region | GCTGGATCCTCAGAAGGAACGCCGTCAATA |
| ywpH-1 | Forward primer to amplify the ywpH promoter region | CCCAAGCTTTCAAGCTGTCAATGCCG |
| ywpH-2 | Reverse primer to amplify the ywpH promoter region | CGCGGATCCGATTGAACATGCGATTCC |
| X-ywpH-3 | Forward primer to amplify the ywpGH-glcR region | GCCGCTCGAGGACTATGGATTACGGAGAGATGTGG |
| H-ywpH-4 | Reverse primer to amplify the ywpGH-glcR region | GCCCAAGCTTGCTCCTTTTCCAGCTTGCCTCCC |
The hexameric restriction sites are underlined.
Upstream regions of the ssb and ywpH genes were amplified by PCR with Pwo DNA polymerase and chromosomal DNA of B. subtilis 168 as the template. The following fragments were amplified: fragment S1 (primers ssb-1 and ssb-2; from 295 bp upstream to 11 bp downstream of the start codon of ssb), S2 (primers rps-1, ssb-2; from 602 bp upstream to 11 bp downstream of the start codon of ssb), R1 (primers rps-1, rps-2; from 274 bp upstream to 50 bp downstream of the start codon of rpsF), Y1 (primers ywpH-1, ywpH-2; from 282 bp upstream to 10 bp downstream of the start codon of ywpH), and Y2 (primers ywpG-1, ywpG-2; from 369 bp upstream to 50 bp downstream of the start codon of ywpG). These PCR fragments were cloned into the SmaI-digested promoter-screening vector pBTK2 (15). The resulting plasmids carrying the insert in the correct orientation were linearized and transformed into B. subtilis 168, selecting for kanamycin-resistant transformants. The transformants were screened for an amylase-deficient phenotype to confirm that the construct had integrated in the amyE locus.
The ywpH deletion mutant was constructed as follows. A 1.493-bp DNA fragment containing the ywpH gene flanked by 486 bp of upstream sequence and 665 bp of downstream sequence was amplified by PCR with primers X-ywpH-3 and H-ywpH-4 and chromosomal DNA of B. subtilis 168 as a template. The amplicon was cloned into PvuII-digested plasmid pIC408, a pUC derivative carrying a spectinomycin resistance gene (M. Steinmetz, unpublished data). Subsequently, a 150-bp internal fragment of ywpH was deleted in the resulting plasmid by PvuII and SmaI digestion and replaced with the 1.34-kb PvuII fragment of pUC7c (11) containing the chloramphenicol resistance marker. The resulting plasmid was transformed into B. subtilis 168 and BIV12. Transformants were selected for chloramphenicol resistance and subsequently screened for spectinomycin sensitivity, indicating the successful disruption of ywpH due to a double-crossover event.
The comK disruption mutant was constructed by transforming B. subtilis BIV12 carrying the Y1-lacZ fusion with chromosomal DNA of strain BV2004 carrying a spectinomycin cassette integrated into the comK gene (12). The resulting spectinomycin- and chloramphenicol-resistant strain was designated BIV24 and used for transcription studies.
β-Galactosidase activity assay.
To assay β-galactosidase activities, overnight cultures were diluted in fresh medium and samples were taken at different intervals for optical density readings at 600 nm (OD600) and β-galactosidase activity determinations. The β-galactosidase assay and the calculation of β-galactosidase units (Miller units) were performed as described by Miller (18).
Northern blot analysis.
Total RNA of B. subtilis was isolated from cultures grown in TY, BFA, or MM. Cells were harvested at hourly intervals, from 3 h before until 3 h after transition point (T), and RNA was extracted as previously described (12); 1 μg of total RNA was separated by formaldehyde-agarose gel electrophoresis and blotted onto a nylon membrane. This membrane was hybridized with digoxigenin-labeled probes detecting transcripts containing yyaF, rpsF, ssb, or ywpH, respectively. Probes were constructed by inserting internal fragments of the coding regions of yyaF, rpsF, ssb, or ywpH in the multiple cloning site of the pBluescriptSK(−) plasmid. Subsequently digoxigenin-labeled antisense RNA probes were transcribed in vitro with the T7 or T3 RNA polymerase site present in this plasmid. In vitro RNA labeling, hybridization, and signal detection were carried out according to the manufacturer's instructions (DIG Northern starter kit; Roche Diagnostics, Mannheim, Germany).
Induction of DNA damage.
Cells of the strains carrying either the S1- or the R1-lacZ fusion were grown in TY and cells carrying the Y1-lacZ fusion were grown in MM. After they reached an OD600 of about 0.1, the cultures were divided and mitomycin C was added to one portion of the culture at a final concentration of 100 ng/ml and the expression of ssb and ywpH was monitored.
Assessment of competence.
To test the involvement of YwpH in natural competence a deletion was introduced into the ywpH gene of B. subtilis and transformability assays were performed as described previously (5).
RESULTS AND DISCUSSION
Transcription analysis of the ssb and ywpH genes.
To study the expression of the ssb and ywpH genes, transcriptional fusions of the potential promoter-containing fragments with a promoterless lacZ were constructed and integrated into the B. subtilis chromosome at the site of amyE.
Strains BIV7 (S1), BIV17 (S2), BIV8 (R1), BIV12 (Y1), and BIV13 (Y2) containing the different lacZ fusions, schematically depicted in Fig. 1, were screened on rich (TY) and minimal medium (MM) agar plates containing X-Gal for blue or white phenotypes. This revealed promoter activity only for the constructs in strains BIV17, BIV8, BIV12, and BIV13, but not in BIV7. In BIV12 and BIV13, promoter activity was detected only in MM, indicating a medium-dependent expression of the genes ywpG and ywpH (Fig. 1).
No promoter activity could be detected when the 295 bp immediately upstream of the ssb start codon (S1) were used to drive lacZ expression (Fig. 1). However, strong promoter activity was detected with the S2 fragment containing the complete rpsF gene and 274 bp upstream of the rpsF start codon. Apparently, ssb and rpsF are part of one operon, which presumably also includes rpsR. This is confirmed by the fact that promoter activity was also found from the smaller R1 fragment comprising the 274 bp upstream of the rpsF start codon only. In contrast, ywpH was found to be transcribed from a promoter directly upstream of the gene.
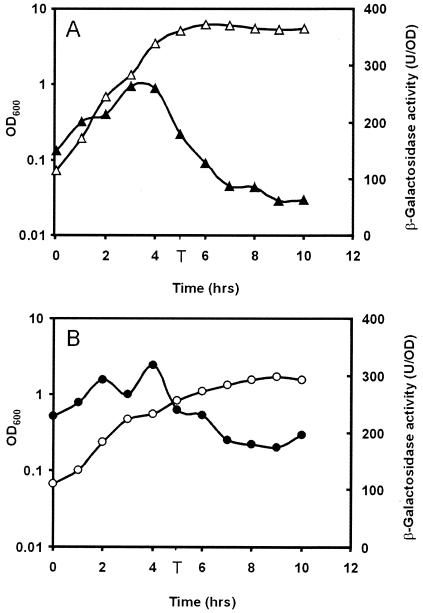
In order to study the expression pattern of ssb and ywpH in more detail, strains containing the S1-, R1-, and Y1-lacZ fusions (BIV7, BIV8, and BIV12, respectively) were grown in TY and MM and β-galactosidase activity was examined. No expression could be detected from the S1 fragment under the conditions employed (data not shown), but the ssb operon appeared to be strongly transcribed from the rpsF promoter in both rich and minimal media. The highest values (between 200 and 300 Miller units per OD) were reached during exponential growth (Fig. 2A and B). After the transition point (T) between logarithmic growth and stationary growth, transcription from the rpsF promoter decreased. There is still transcription in the stationary phase at a two- to fourfold lower level. These results are in agreement with the higher need for SSB protein for DNA replication in fast-growing and thus frequently dividing cells. In contrast, no expression of the ywpH gene could be detected in cells of B. subtilis BIV12 grown in TY and only low expression was observed in exponentially growing cells in MM. However, expression of ywpH was strongly induced when cells entered the stationary phase and reached its highest level after the transition point (Fig. 3).
FIG. 2.
Expression of the ssb operon in B. subtilis. Growth (open symbols) in rich (triangles) or minimal (circles) medium and expression of the transcriptional lacZ fusions (solid symbols) with the rpsF gene, the first gene of the ssb operon, reflected as β-galactosidase activity per OD. T indicates the time point at which transition from logarithmic to stationary growth takes place.
FIG. 3.
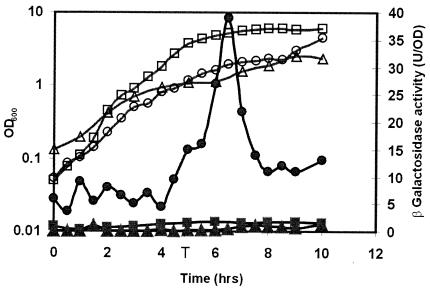
Expression of the ywpH gene in B. subtilis and effect of a comK mutation. Growth (open symbols) in rich (squares) or minimal (circles) medium and expression of the transcriptional lacZ fusions (solid symbols) of the ywpH gene in strain BIV12 reflected as β-galactosidase activity per OD. The effect of a comK mutation on ywpH expression is also shown. Growth in minimal medium (open triangles) and expression of the transcriptional ywpH-lacZ fusion reflected as β-galactosidase activity per OD (solid triangles) in the comK mutant BIV24. T indicates the time point at which transition from logarithmic to stationary growth takes place.
Northern blot analysis.
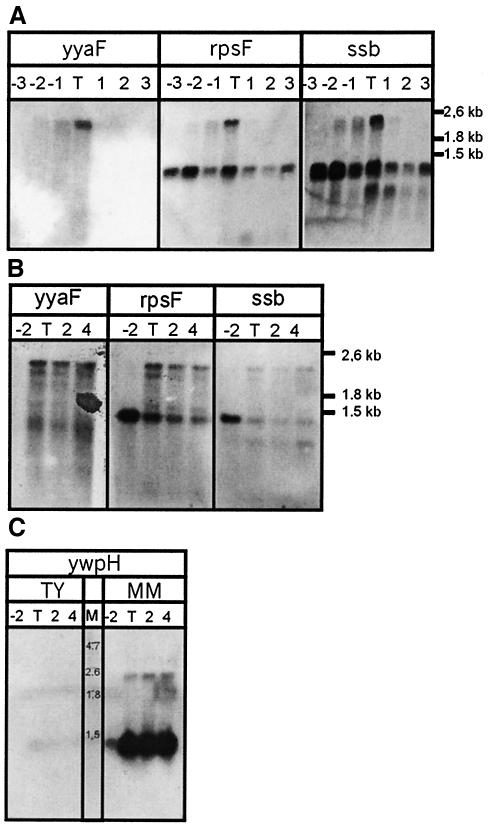
For a more detailed picture of the transcription of ssb and ywpH, Northern blots with probes for yyaF (upstream of rpsF), rpsF, ssb, and ywpH were performed. Upstream of ssb two promoters are present, a promoter upstream of rpsF and a promoter upstream of yyaF. Downstream of rpsR a terminator structure is present. Transcription from these two promoters would result in two mRNA fragments. A transcript containing only rpsF-ssb-rpsR of 1.2 kb and a transcript also containing yyaF of 2.4 kb (Fig. 1A). Figure 4A shows fragment sizes of 2.4 and 1.3 kb with the rpsF probe, which correspond to the expected transcripts. With the ssb probe, a smaller fragment of about 0.9 kb is also visible. Since no promoter activity was detected directly upstream of ssb, this fragment is likely to be caused by selective cleavage or degradation of the mRNA leading to the removal of rpsF from the ssb-rpsR fragment. The size of such a fragment would be 0.85 kb, which corresponds to the size observed.
FIG. 4.
Transcription of the ssb operon and ywpH in B. subtilis. (A) RNA samples were taken from wild-type cells growing in rich medium (BFA) at hourly intervals, from 3 h before until 3 h after transition point (T). The blots were hybridized with RNA probes detecting transcripts containing yyaF, ssb, or rpsF. (B) RNA samples were taken from wild-type cells growing in MM at −2 h, transition point (T), +2 h, and +4 h. The blots were hybridized with RNA probes detecting transcripts containing yyaF, ssb, or rpsF. (C) RNA samples from wild-type cells growing in TY or MM were taken at −2 h, transition point, +2 h, and +4 h. The blots were hybridized with RNA probes detecting transcripts containing ywpH.
In MM ssb and rpsF are transcribed highly during logarithmic growth from their own promoter, and after the transition point they are also transcribed from the yyaF promoter. The results from both the β-galactosidase assay and the Northern blot analyses suggest that ssb is cotranscribed and coregulated with genes coding for ribosomal proteins, thereby coupling the regulation of protein synthesis to DNA metabolism.
In rich medium yyaF is transcribed during log phase and its transcription stops at the onset of stationary phase (Fig. 4A). However, in minimal medium yyaF is highly transcribed after the transition point (Fig. 4B). The RNA fragment that hybridizes with the yyaF probe corresponds to the upper band visible in both the rpsF and ssb blots. This indicates that transcription from the yyaF promoter continues into the rpsF-ssb-rpsR operon, causing additional transcription of this operon from the yyaF promoter. In MM the transcription of rpsF, ssb, and rpsR is therefore boosted from the yyaF promoter, which itself is shown to be ComK dependent by means of DNA array analysis (2, 12, 19). Two of these studies also showed the transcription of rpsF and ssb to be ComK dependent. Our results show that it is likely that this observation is caused by readthrough from the yyaF promoter rather than direct regulation of the rpsF promoter by ComK. The apparent transcription of yyaF in rich medium during late logarithmic phase indicates multiple modes of transcription regulation of this gene.
The transcription of ywpH in TY is virtually absent, but two clear mRNA fragments are present when the cells were grown in MM (Fig. 4C). The fragments observed have sizes of 2.4 and 1.4 kb. The 1.4-kb band corresponds to a transcript containing ywpH and glcR. The longer fragment observed at time points 2 and 4 of 2.4 kb is likely to be a result of readtrough into the downstream gene ywpJ due to an enhanced transcription rate. During logarithmic growth a small amount of ywpH containing mRNA is detected, and when the cells reach stationary growth the transcription of ywpH increases.
In conclusion, the two ssb-like genes obviously show opposite expression patterns, one being expressed to the highest level during exponential growth and the other one being expressed during the stationary phase in MM, indicating a distinct function of their gene products. These results should be interpreted with care, since no direct protein concentration measurements were carried out.
ssb gene is controlled by the SOS response.
It has been reported that the ssb gene of E. coli is induced by DNA damage (3). However, its DNA damage inducibility is still a matter of discussion (17). E. coli SSB is supposed to be involved in the induction of the SOS response by promoting RecA dependent cleavage of LexA (17). Likely SSB serves a similar role in B. subtilis. Since B. subtilis has two paralogues of SSB that are expressed under different growth conditions, we wondered whether one or both of these SSBs could be induced by DNA-damaging agents too.
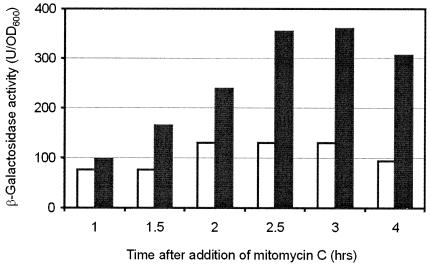
When DNA damage was induced by the addition of mitomycin C to the growth medium, an increase of expression of the ssb gene from the rpsF promoter was observed, starting about 1.5 h after the addition of mitomycin C. A maximum of about threefold elevated expression was reached at 2.5 to 3 h after addition of mitomycin C (Fig. 5). The observed increase of expression is similar to the induction level observed in E. coli, which is very slow compared to that of other genes in the recA-lexA SOS regulon. Even in the presence of mitomycin C there was no promoter activity detectable from the fragment directly upstream of ssb (strain BIV7), confirming the absence of an additional ssb promoter (data not shown). These results suggest the involvement of the ribosomal proteins S6 and perhaps also S18 in the SOS response of B. subtilis, because their expression is subject to the same control, which mediates the SOS response. Several genes under the control of DinR, the B. subtilis LexA homologue, have been identified, and a consensus sequence for its binding to DNA was proposed (30). However, no such target sequence could be found in the regulatory region of the rpsF gene. Therefore, its SOS-dependent induction might be indirect. From the promoter of ywpH no significant mitomycin C-dependent induction could be detected (data not shown).
FIG. 5.
Effect of DNA damage on expression of the ssb operon. Expression of the transcriptional rpsF-lacZ fusion of strain BIV8 in the presence (black bars) or absence (hatched bars) of mitomycin C (100 ng/ml) measured as β-galactosidase activity per OD. The average of two independent experiments is shown.
ywpH gene is regulated by ComK and required for optimal competence.
In the wild-type strain induction of ywpH expression in MM was observed when exponential growth ceased (Fig. 3). ywpH expression reached its maximum 2 h after the onset of stationary phase. This pattern for the regulation of ywpH is similar to that of the late competence genes in B. subtilis. The development of natural genetic competence is a typical postexponential feature of B. subtilis and occurs in response to certain growth conditions such as amino acid limitation in the presence of high glucose concentrations and at high cell density (for reviews see references 8 and 10). The expression of late competence genes is activated by ComK (28). An important step in transformation is recombination, and stable single-stranded DNA is required in this process.
To test the possibility that ywpH is regulated by ComK, we introduced a comK disruption in B. subtilis BIV12, carrying the Y1-lacZ fusion. In this comK null mutant expression of ywpH was totally abolished (Fig. 3), which indicates that ywpH belongs to the late competence genes controlled by ComK. In the course of this study we and others also showed by means of transcription analysis of several B. subtilis comK deletion strains that the transcription of ywpH is indeed ComK dependent (2, 12, 19).
We tested the involvement of YwpH in competence. When ywpH was disrupted the transformation efficiency dropped approximately 50-fold, although not to zero. In the course of this study Ogura et al. and Berka et al. observed the effect of a ywpH disruption on competence within a similar order of magnitude. Competence is not completely annulled in a ywpH mutant. This might be due to the presence of the ssb gene product which might be able to substitute for YwpH, although only partially.
This result demonstrates an important role of the ywpH gene product in competence. In vitro experiments with mutant proteins of the E. coli SSB revealed that the C terminus is not required for DNA binding (29). Moreover, the truncated SSB is functional in promoting RecA protein-dependent homologous pairing and strand exchange in vitro (9). Although YwpH is lacking the C terminus present in SSB, it is likely to be able to bind single-stranded DNA and to allow DNA recombination. However, YwpH is probably not able to replace SSB in DNA replication and/or DNA repair, since the C terminus is required for in vivo function (6). In agreement with that, the ssb gene was found to be essential for viability in B. subtilis (13), whereas the ywpH gene could be knocked out without consequences for bacterial growth under the employed conditions (data not shown). Likely YwpH is involved in homologous DNA recombination processes, which are necessary for acquiring foreign DNA in competent cells of B. subtilis. Presumably SSB also can participate in competence-related recombination, since although transformability is greatly reduced, an ywpH knockout strain can still be transformed.
Organization of ssb genes in other bacteria.
The ssb gene organization in B. subtilis differs from the organization observed in E. coli. We therefore addressed the question of how common this gene organization is in other bacterial species. For that purpose, genomes of bacteria were screened for SSB homologues and their gene organization with the NCBI database (http://www.ncbi.nlm.nih.gov). At this moment 87 complete sequences of bacterial genomes are available, including 69 different species. In all genomes, one or more genes coding for an SSB homologue were found. While only 15 species show the same gene order as in E. coli with an ssb gene divergently situated to the uvrA gene, 35 species show the ssb gene flanked by the rpsF and rpsR genes as it is observed for B. subtilis. These 35 species include all gram-positive bacteria sequenced until now, as well as representatives of the Thermotogales and Thermus/Deinococcus group, and gram-negative species from the phyla Spirochaetales, Aquificales, Thermotogales, and Chlorobi and the epsilon subdivision of the proteobacteria. All other proteobacteria and representatives of the Chlamydiales, Cyanobacteria, and Fusobacteria do not possess this gene organization. On the basis of the ssb gene and number of ssb paralogues per species organization, the sequenced bacteria can be classified in four different groups. An overview is given in Table 3.
TABLE 3.
Classification based on ssb gene organisation
| Group | rpsF-ssb-rpsR | uvrA-ssb | Multiple ssb genes | Gram stain | No. of species in group |
|---|---|---|---|---|---|
| I | + | − | + | +/− | 22 |
| II | + | − | − | +/− | 13 |
| III | − | + | − | − | 15 |
| IV | − | − | +/− | − | 19 |
Group I contains bacteria with the same ssb gene organization as B. subtilis, rpsF-ssb-rpsR. Most bacteria within this group are Gram positive. All bacteria in this group contain multiple SSB paralogues. Group II contains bacteria with the same organization as B. subtilis, rpsF-ssb-rpsR, but they do not possess multiple ssb paralogues. In addition to some gram-positive bacteria, this group also contains bacteria from the epsilon subdivision of the proteobacteria (Helicobacter pylori and Campylobacter jejuni), Ureaplasma urealyticum, and Borrelia burgdorferi. Group III contains bacteria with the same gene organizations as E. coli; ssb is divergently located to uvrA. Ralstonia solanacearum, Salmonella enterica serovar Typhimurium, Shewanella oneidensis, Pseudomonas aeruginosa, and Pseudomonas putida are also classified within this group. They have one gene located between uvrA and ssb. All bacteria in this group contain only one ssb gene in their chromosome. Furthermore, all these bacteria belong to the alpha or gamma subdivision of the proteobacteria (gram negative). Group IV contains bacteria with ssb neither placed between rpsF and rpsR nor divergently located to uvrA. Most of these bacteria contain one ssb gene, but Nostoc sp. strain PCC 7120 and Xylella fastidiosa contain multiple ssb genes.
Since we propose a functional division of the gene products from both SSB paralogues, one being the main SSB and the other being competence related, we checked whether the occurrence of multiple SSB homologues in a bacterial genome could be related to natural competence. Most bacteria known to be naturally transformable (14) are classified in group I, which contains species with multiple ssb homologues. In group IV only Nostoc sp. strain PCC 7120 is known to be naturally transformable, and this species also contains multiple SSBs. The only bacteria that are known to be naturally competent but do not contain multiple ssb genes are H. pylori and C. jenuni (14). These bacteria are both members of the epsilon subdivision of the proteobacteria.
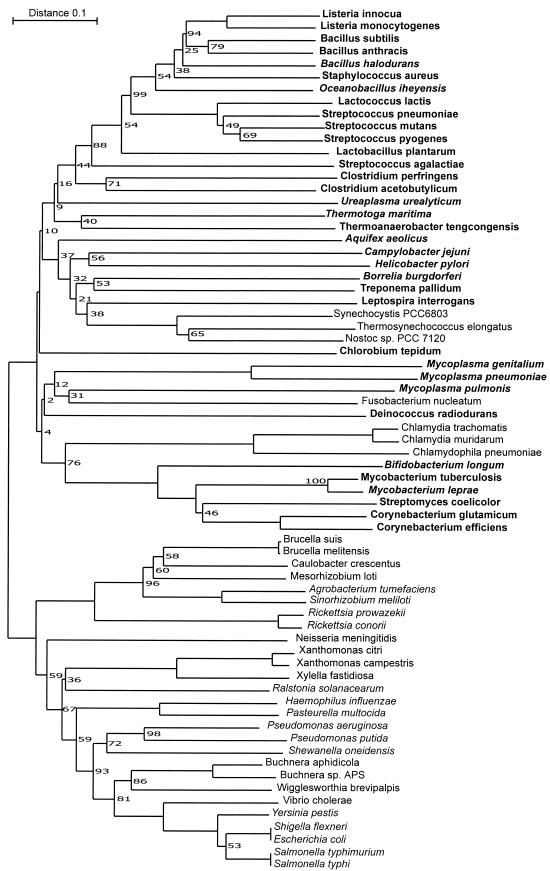
To further investigate if the identified groups can also be divided on the basis of their evolutionary descent, we used all 69 SSB protein sequences to calculate a phylogenetic tree (Fig. 6). The tree shows a clear grouping of groups I and II compared to group III. All the proteobacteria in group IV cluster with the other proteobacteria (group III), but the cyanobacteria, Chlamydiales, and Fusobacterium nucleatum cluster with the gram-positive bacteria from groups I and II. Xanthomonas citri (proteobacteria, gamma subdivision) also clusters outside of the proteobacteria group. Likely this is caused by horizontal gene transfer. In this tree the species from groups I and II are not separated based on their SSB homologies. These results were expected because it is unlikely that the development of the extra SSBs is directly correlated to the sequence of a bacterium's main SSB.
FIG.6.
Unrooted phylogenetic tree of bacterial SSBs. Bacterial SSBs from 69 bacteria were used. When bacteria contained multiple ssb genes, the ssb situated between rpsF and rpsR was used in this tree. When bacteria have multiple ssb genes but do not posses an ssb in an operon structure with rpsF and rpsR, the ssb most homologous to the B. subtilis ssb was used. Alignments were made with ClustalW 1.74 (25) with a gap opening penalty of 30 and a gap extension penalty of 0.5. Dendrogram construction was done with TreeCon 1.3b (27) with the neighbor-joining method and no correction for distance estimation. Bootstrap values (percent) are indicated at the branching points. If no percentage is indicated, the value is 100. The bar indicates 10% difference at the amino acid level. The groups indicated in Table 3 are also indicated in this figure. Bacteria from group I are printed in bold, bacteria from group II are printed in bold italic, bacteria from group III are printed in italic, and bacteria from group IV are printed in normal type.
Next to the well-studied ssb gene organization observed in E. coli and some other gram-negative bacteria, the gene organization rpsF-ssb-rpsR observed in B. subtilis is a suitable model for the study of ssb, since it is found in all gram-positive species sequenced until now and several gram-negative bacteria. Moreover, additional ssb genes are found associated with this configuration, frequently in correlation with natural competence.
Acknowledgments
We thank Wiep Klaas Smits for the gift of strain BV2004 and Sacha van Hijum for excellent technical assistance in the construction of the phylogenetic tree. Piet Nuijten and Johan van den Bosch are acknowledged for valuable discussions.
This work was supported by Intervet International B.V. (Boxmeer, The Netherlands).
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 3.Brandsma, J. A., D. Bosch, C. Backendorf, and P. van de Putte. 1983. A common regulatory region showd by divergently transcribed genes of the Escherichia coli SOS system. Nature 305:243-245. [DOI] [PubMed] [Google Scholar]
- 4.Brandsma, J. A., D. Bosch, M. de Ruyter, and P. van de Putte. 1985. Analysis of the regulatory region of the ssb gene of Escherichia coli. Nucleic Acids Res. 13:5095-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, S., and G. Venema. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat. Res. 15:1-10. [DOI] [PubMed] [Google Scholar]
- 6.Curth, U., J. Genschel, C. Urbanke, and J. Greipel. 1996. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 24:2706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vries, J., J. Genschel, C. Urbanke, H. Thole, and W. Wackernagel. 1994. The single-stranded-DNA-binding proteins (SSB) of Proteus mirabilis and Serratia marcescens. Eur. J. Biochem. 224:613-622. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau, D. 1993. Genetic exchange and homologous recombination, p. 555-584. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 9.Egner, C., E. Azhderian, S. S. Tsang, C. M. Radding, and J. W. Chase. 1987. Effects of various single-stranded-DNA-binding proteins on reactions promoted by RecA protein. J. Bacteriol. 169:3422-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 11.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 12.Hamoen, L. W., W. K. Smits, A. A. Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis with a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meijer, W. J., L. D. van der, G. Venema, and S. Bron. 1995. Effects of the generation of single-stranded DNA on the maintenance of plasmid pMV158 and derivatives in different Bacillus subtilis strains. Plasmid 33:79-89. [DOI] [PubMed] [Google Scholar]
- 16.Mellado, R. 2000. Useful methods for transcriptional analysis, p. 37-40. In W. Schumann, S. D. Ehrlich, and N. Ogasawara (ed.), Functional analysis of bacterial genes: a practical manual. John Wiley & Sons, Ltd., Sussex, England.
- 17.Meyer, R. R., and P. S. Laine. 1990. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 54:342-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1982. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Cold Spring Harbor, N.Y.
- 21.Shimamoto, N., N. Ikushima, H. Utiyama, H. Tachibana, and K. Horie. 1987. Specific and cooperative binding of E. coli single-stranded DNA binding protein to mRNA. Nucleic Acids Res. 15:5241-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spizizen, J. 1985. Conditions for competence in the Bacillus subtilis transformation system. Proc. Natl. Acad. Sci. 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stulke, J., I. Martin-Verstraete, P. Glaser, and G. Rapoport. 2001. Characterization of glucose-repression-resistant mutants of Bacillus subtilis: identification of the glcR gene. Arch. Microbiol. 175:441-449. [DOI] [PubMed] [Google Scholar]
- 24.Tapias, A., and J. Barbe. 1999. Regulation of divergent transcription from the uvrA-ssb promoters in Sinorhizobium meliloti. Mol. Gen. Genet. 262:121-130. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W — improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umezu, K., N. Sugawara, C. Chen, J. E. Haber, and R. D. Kolodner. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148:989-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Peer, Y., and R. de Wachter. 1994. Treecon for Windows — a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Applic. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 28.Van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 29.Williams, K. R., E. K. Spicer, M. B. LoPresti, R. A. Guggenheimer, and J. W. Chase. 1983. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Biol. Chem. 258:3346-3355. [PubMed] [Google Scholar]
- 30.Winterling, K. W., D. Chafin, J. J. Hayes, J. Sun, A. S. Levine, R. E. Yasbin, and R. Woodgate. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]