Abstract
A high-throughput system to rapidly assess the intracellular replication of Staphylococcus aureus has been developed utilizing S. aureus transformed with a dual gfp-luxABCDE reporter operon under the control of a growth-dependent promoter. Replication of tagged bacteria internalized into bovine mammary epithelial cells (MAC-T) could be measured by monitoring fluorescence and bioluminescence from the reporter operon following removal of extracellular bacteria from the plates. Bacterial replication inside cells was confirmed by a novel ex vivo time-lapse confocal microscopic method. This assay of bacterial replication was used to evaluate the efficacy of antibiotics which are commonly used to treat staphylococcal infections. Not all antibiotics tested were able to prevent intracellular replication of S. aureus and some were ineffective at preventing replication of intracellular bacteria at concentrations above the MIC determined for bacteria in broth culture. Comparison of the fluorescence and bioluminescence signals from the bacteria enabled effects on protein synthesis and metabolism to be discriminated and gave information on the entry of compounds into the eukaryotic cell, even if bacterial replication was not prevented. Elevated resistance of S. aureus to antibiotics inside host cells increases the likelihood of selecting S. aureus strains which are resistant to commonly used antimicrobial agents within the intracellular niche. The approach presented directly assesses intracellular efficacy of antibiotics and provides an evidence-based approach to antibiotic selection for prescribing physicians and medical microbiologists.
Staphylococcus aureus is a versatile pathogen with a broad host range whose infections are causing considerable alarm within the medical community due to the increasing emergence of antibiotic-resistant strains. Infections associated with this organism range from minor wound infections to more serious diseases, including endocarditis, osteomyelitis, and septic shock. These infections are often life-threatening, so there is a pressing need to increase our understanding of staphylococcal disease and develop new antistaphylococcal agents. As S. aureus is also known for its ability to induce long-lasting persistent infections, it is important to ascertain the mechanisms the organism uses to evade host immune responses.
S. aureus has not traditionally been considered an intracellular pathogen; however, it is now well documented that S. aureus can internalize and survive within a wide variety of mammalian cells (2, 4, 19). It has been suggested that the ability of S. aureus to reside in an intracellular niche enables long-term colonization of the host and the maintenance of a chronic infective state (4, 19, 49). Bacterial uptake is triggered through specific interactions between microbial surface components recognizing adhesive matrix molecules (12, 36), which include fibronectin binding proteins in the bacterial cell wall, and the host cell that ultimately leads to the active internalization of bacteria into an endosome. S. aureus is able to escape from this endosome, where it is free to replicate in the host cell cytoplasm (4, 40, 51).
The expression of many S. aureus microbial surface components recognizing adhesive matrix molecules is downregulated upon induction of the of the accessory gene regulator locus (agr) (30, 39, 42). The agr locus is a quorum sensing-regulated system activated by autoinducing peptide pheromone (21, 26). The two divergent transcriptional units of the agr locus, RNAII and RNAIII, are under the control of the P2 and P3 promoters, respectively (reviewed in reference 33). RNAII is a polycistronic mRNA coding for four peptides: AgrB and AgrD, required for the synthesis and export of autoinducing peptide, and AgrA and AgrC, which constitute a classical two-component signal transduction system responsible for sensing and responding to the autoinducing peptide.
RNAIII is the effector molecule in the agr regulon, acting primarily at the level of gene transcription. It is the transcription of RNAIII that is responsible for upregulation of secreted virulence factors as well as the downregulation of surface proteins (29, 34). Different S. aureus strains produce autoinducing peptides with distinct structures. Strains can be grouped on this basis because they will activate the agr response of strains within the same group and inhibit the agr response of strains from different groups by competitive inhibition (21, 27, 33). This inhibitory action of autoinducing peptides on quorum sensing has identified them as potential novel therapeutic and anti-infective agents for S. aureus; indeed a synthetic analogue of autoinducing peptide, (Ala5)AIP-1, has been shown to be a potent inhibitor of agr in all S. aureus agr groups (27).
The intracellular location of S. aureus has important implications for its antibiotic susceptibility. In order to treat intracellular infections, the antibiotic needs to be capable of penetrating the eukaryotic cell to a sufficiently high concentration to be effective against the intracellular bacteria (46). So, penetration and subcellular localization of antibiotics within phagocytic cells as well as the possibility of their intracellular inactivation must be considered when assessing antibiotic efficacy (15). Little is known about the intracellular pharmacodynamics of available antibiotics and how their accumulation within the cell relates to their activity. The intracellular milieu can influence both the metabolism of the intracellular bacterium and the activity of the antibiotic; for example, intracellular enzymes or pH may inactivate certain antibiotics.
One way in which bacterial localization, growth, and gene expression can be studied is through the use of reporter and marker genes. A number of reporters are available for such investigations, for example, green fluorescent protein (GFP) and bacterial luciferase (Lux), which have their own particular advantages and disadvantages. The structural genes that encode the proteins necessary for bacterial bioluminescence are encoded by a polycistronic operon consisting of five genes, luxAB (luciferase) and luxCDE (fatty acid reductase) (17, 28). The bioluminescence process requires energy, in the form of FMNH2 obtained from the bacterium's metabolism, a long-chain aldehyde synthesized and recycled by LuxCDE and molecular oxygen. As the production of bioluminescence from recombinant bacteria containing the lux genes depends on cells' being biochemically active, it can be assumed that any compound that impairs the biochemistry and thus compromises cellular viability will lead to a rapid reduction in luminescence. The effect of antimicrobial compounds on lux-containing recombinant bacteria can therefore be assessed rapidly (43, 44, 48).
Previously bioluminescence emitted by recombinant S. aureus expressing a gfp-luxABCDE reporter operon under the control of a growth-dependent promoter was used to track bacterial replication within mammalian cells in a 24-well microplate format. The use of the GFP-Lux dual reporter system provided GFP fluorescence as an internal control for the luciferase signal and also facilitated analysis of fixed specimens by fluorescence microscopy during the infection process (40).
We have further developed this reporter-based approach to measure intracellular replication of S. aureus and utilized time-lapse confocal microscopy to confirm that the reporter signal is derived from intracellular staphylococci. The assay has been successfully applied to study the efficacy of commonly used therapeutic agents against intracellular S. aureus.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used throughout the study were S. aureus 8325-4 (30), S. aureus RN6390 (32), and S. aureus RN6911 (22). The plasmid pSB2030 (PxylA::gfp-luxABCDE) confers chloramphenicol resistance and provides growth-dependent bioluminescence and fluorescence when used to transform S. aureus (40).
Preparation of the bacterial inoculum for invasion assays.
S. aureus were grown overnight at 30°C statically in HEPES-buffered Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% RPMI (Sigma), 5 μg ml−1 chloramphenicol, and 1.5 nM (Ala5)AIP-1. Overnight cultures were washed twice in an equal volume of fresh medium to remove (Ala5)AIP-1 and then concentrated 10-fold. This cell suspension was used to inoculate fresh HEPES-buffered DMEM (Gibco) supplemented with chloramphenicol (5 μg ml−1) to an OD600 of approximately 0.08.
MIC determination.
S. aureus was grown overnight at 30°C statically in HEPES-buffered DMEM (Gibco) supplemented with 10% RPMI (Sigma) and 5 μg ml−1 chloramphenicol. Cultures were centrifuged at 3,000 × g for 5 min and resuspended in HEPES-buffered DMEM supplemented with 10% RPMI and 5 μg ml−1 chloramphenicol to an OD600 of ≈0.08; 100 μl of this culture was used to inoculate the wells of a 96-well plate containing 100 μl of HEPES-buffered DMEM supplemented with 10% RPMI in which the antibiotics under test had been serially twofold diluted. The plate was then incubated at 37°C, and absorbance, luminescence, and fluorescence were measured in a Tecan Genios Pro multilabel reader at 30-min intervals for 20 h.
MAC-T cell culture and cell invasion assays.
The bovine mammary epithelial cell line MAC-T was routinely cultured in assay medium (DMEM with 10% fetal bovine serum, 2 mM l-glutamine, 5 μg ml−1 insulin, 1 μg ml−1 hydrocortisone) supplemented with 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Sigma) as previously described (20). Antibiotics were omitted from assay medium during internalization studies. MAC-T cells were seeded into 24-well (Costar) or 96-well clear-bottomed (Porvair) microtiter plates in assay medium and grown to confluence as described previously (40), then infected with 1 ml (24-well plate) or 200 μl (96-well plate) of the bacterial inoculum. Microtiter plates were centrifuged for 20 min at 1,000 × g to facilitate interaction of the bacteria with the eukaryotic cell surface. Plates were subsequently incubated for 1 h at 37°C.
Following this, cell monolayers in 96-well plates were washed three times with fresh DMEM in an automated plate washer (Cellwash, Thermo Labsystems); 24-well plates were washed manually. Monolayers were then incubated with lysostaphin (10 μg ml−1; Sigma) in HEPES-buffered DMEM (Gibco) for 20 min at 37°C. Plates were then washed again, and 1.5 ml (24-well plates) or 200 μl (96-well plates) of fresh HEPES-buffered DMEM (Gibco) was put into all wells. The 24-well plates were incubated at 37°C in a Victor 1420 multilabel counter (Perkin-Elmer Instruments), and the 96-well plates were incubated at 37°C in a Lucy 1 luminometer (Anthos), where bioluminescence alone was monitored, or a Tecan Genios Pro multifunctional detector, to concomitantly measure luminescence and fluorescence.
Time-lapse confocal microscopy of intracellular S. aureus growth.
Eukaryotic cells were seeded into 35-mm glass bottom microwell dishes (MatTek) in assay medium and grown to 80% confluence overnight in 5% CO2 at 37°C (40). The monolayers were then washed with DMEM prior to staining of cell membranes with 24 mM FM4-64 (Molecular Probes), a membrane-selective fluorescent dye, for 30 s at room temperature. Cell monolayers were then washed three times in DMEM to remove excess dye. Stained cells were then infected with 1 ml of the bacterial inoculum (OD600 of 0.02); plates were centrifuged at 1,000 × g for 20 min and then incubated for 30 min to allow infection of the monolayers. Extracellular bacteria were then removed from the plates by lysostaphin treatment (40). Specimens were placed on the heated stage of a CO2 incubator on a Zeiss LSM 510 confocal microscope. Image stacks with 1-μm slices were taken sequentially at 10-min intervals over an 8-h time course by a multitrack protocol with a 488-nm laser with a band pass emission filter of 505 to 550 nm for GFP and a 543-nm laser with a long-pass emission filter of 560 nm for FM4-64 visualization.
RESULTS
High-throughput microplate assay of S. aureus intracellular replication.
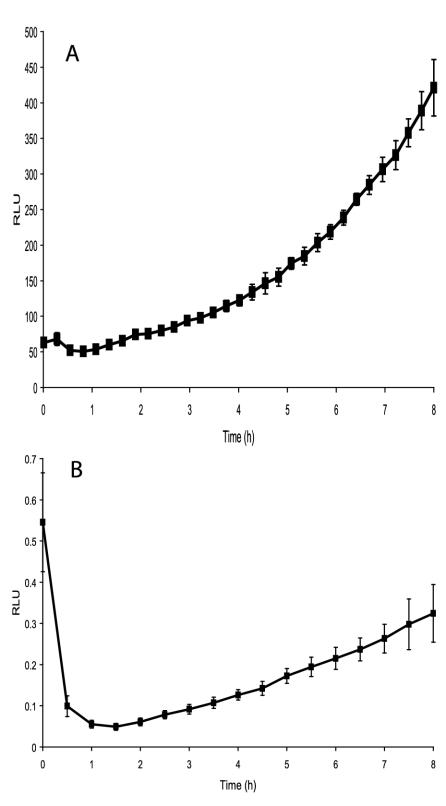
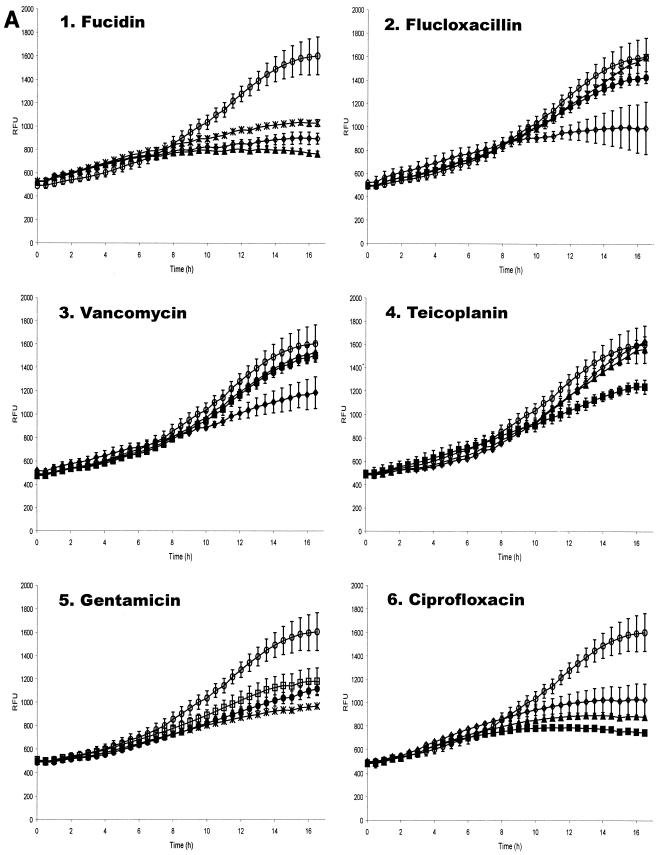
The assay developed previously (40) was sensitive to fluctuations due to differential expression of the agr regulon and variation in the rates of cell adhesion. To overcome this, the infectious process has now been standardized by pretreatment of the bacterial inoculum with (Ala5)AIP-1 to repress agr expression and centrifugation of the inoculum onto the cell monolayer. After a 1-h incubation period, monolayers were washed and treated with lysostaphin, as this has been shown to effectively remove all extracellular and adherent bacteria (40). The bioluminescence of the remaining intracellular replicating bacteria in 24-well plates was monitored in a Victor 1420 multilabel counter (Perkin-Elmer Instruments). For assays in 96-well plates, bioluminescence was measured with an Anthos Lucy 1 luminometer. As the reporter plasmid in the staphylococci (pSB2030) confers growth-dependent bioluminescence, an increase in bioluminescence can be used as a monitor of bacterial replication. Nonreplicating bacteria do not produce light. Figure 1 shows comparative data obtained from the 24-well and 96-well formats from MAC-T cells infected with S. aureus 8325-4(pSB2030).
FIG. 1.
Bioluminescence from S. aureus 8325-4(pSB2030) growing in MAC-T cells in a 24-well (panel A) and 96-well assay (panel B). Light, expressed as relative light units (RLU), was monitored in a Victor 1420 multilabel counter (Perkin-Elmer Instruments) and in an Anthos Lucy 1 luminometer, respectively.
These data show that results obtained from both assay formats and by both instruments are comparable. In each case replication appears to begin approximately 3 h after inoculation (i.e., ≈1 h after the samples are incubated in the instruments). It can be seen (Fig. 1) that at the beginning of the measurement period the bioluminescence from each well of the 96-well plate starts at a high level and then decreases rapidly. This initial high level of bioluminescence can be attributed to preformed Lux proteins in the intracellular bacteria, as the residual volume of wash buffer left in the wells following the postinfection washes is essentially free of bacteria. However, bioluminescence decreases rapidly within the first hour due to the short half-life of luciferase, indicating that the bioluminescence observed after this time point originates from de novo synthesis of the reporter proteins by replicating intracellular organisms.
In the 24-well plate format, bioluminescence levels are seen to increase steadily throughout the experiment after the 1-h time period. This increase in bioluminescence is due to the replication and overall increase in numbers of intracellular S. aureus. In the 96-well plates after 1 h, bioluminescence increases steadily throughout the experiment, again demonstrating the ability of S. aureus to replicate in MAC-T cells. The general patterns in the infection processes and subsequent replication of intracellular bacteria in both plate formats are similar. However, direct quantitative comparisons cannot be made, as different instruments were used to perform the bioluminescent measurements for the different plate types.
Studying S. aureus intracellular replication by time-lapse confocal microscopy.
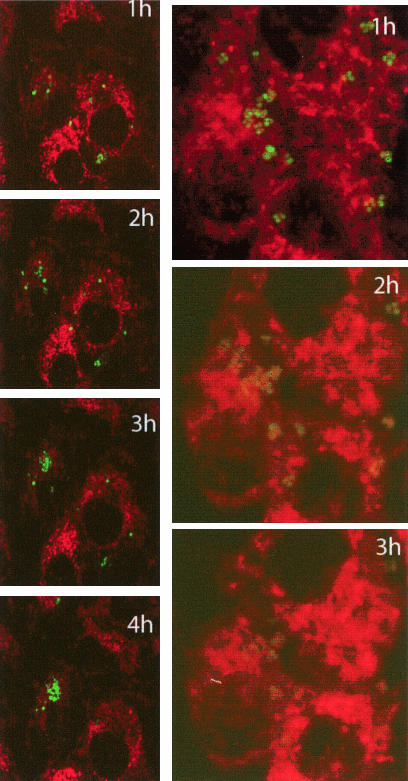
In order to demonstrate that the increase in bioluminescence observed in the microtiter plate assay was due to intracellular replication, S. aureus harboring the PxylA-gfp-luxABCDE reporter were used to infect cell monolayers previously treated with the dye FM4-64 to allow visualization of eukaryotic cell membranes (16). Following infection of the monolayer, specimens were transferred to a Zeiss LSM 510 confocal microscope with a heated stage and CO2 incubator at 37°C. Image stacks were taken as described in Materials and Methods, and images were processed with a Zeiss LSM Image browser.
As can be seen in Fig. 2, intracellular replication of S. aureus RN6390(pSB2030) is readily observable, with countable numbers and fluorescence output of GFP-tagged bacteria increasing over time. Similar time-lapse imaging by the agr mutant S. aureus RN6911(pSB2030) shows that this strain is efficiently internalized due to high-level expression of cell surface adhesins. However, as expected, this strain does not replicate over the time course of this experiment and the GFP signal is seen to degrade.
FIG. 2.
Projections from time-lapse confocal microscopy of S. aureus RN6390(pSB2030) (green; left panel) over 4 h following a 1-h internalization period in eukaryotic cells stained with FM4-64 (red). Similar images of agr S. aureus RN6911(pSB2030) is shown in the right panel over 3 h, after which time the GFP signal is too weak to display.
Efficacy of antibiotics on intracellularly replicating S. aureus.
To investigate the effect of antibiotics on intracellularly replicating S. aureus, a selection of compounds were chosen that are used clinically for the treatment of staphylococcal infections. These different classes of antibiotics are all known to act through different mechanisms on the bacterial cell.
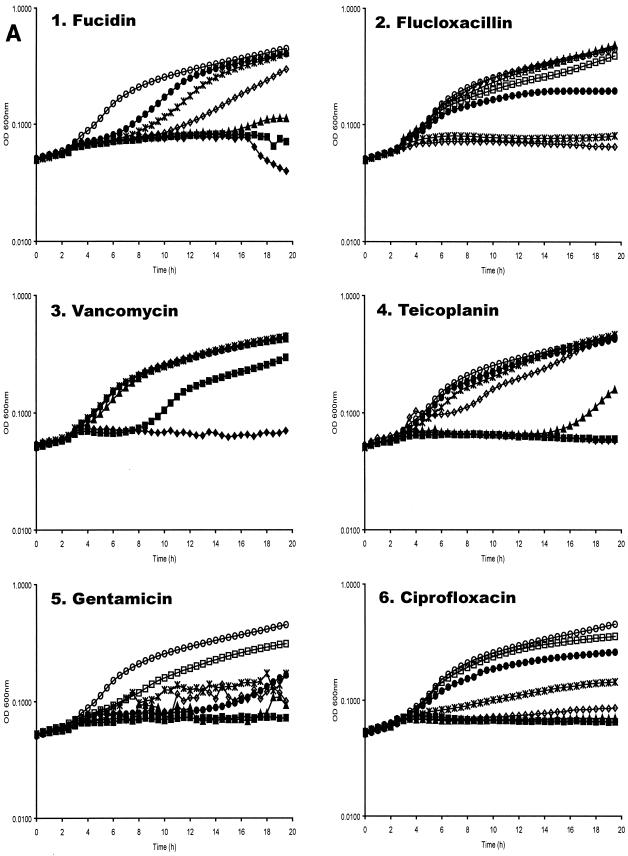
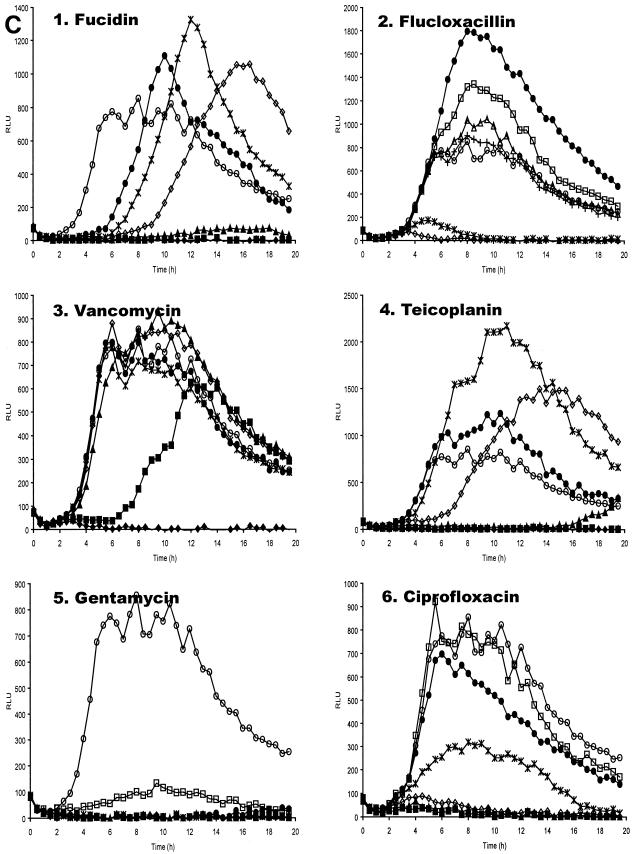
Antibiotic concentrations effective at preventing replication of planktonic S. aureus RN6390(pSB2030) were determined by microtiter broth dilution in tissue culture medium at 37°C. Replication in these experiments was measured by absorbance, bioluminescence, and fluorescence as measured in a Genios Pro multifunctional detector. It has previously been shown for this reporter construct that expression of the reporter operon occurs during bacterial replication (40). It can be seen in Fig. 3 that the signals obtained from both Lux and GFP generally correlate well with the growth of the reporter organism. Inhibition of GFP accumulation in antibiotic-treated samples was seen to correlate with inhibition of replication in a dose-dependent manner (Fig. 3A and 3B). With most antibiotics tested, a reduction in bioluminescence (Lux) signal was seen to correlate with inhibition of replication (Fig. 3A and 3C); however, with some antibiotics an increase in Lux signal was observed at subinhibitory concentrations (Fig. 3C, panels 1 to 4). Addition of fucidin to the bacterial cultures significantly extended the lag phase of growth in a dose-dependent manner.
FIG. 3.
Effect of antibiotics on planktonic S. aureus RN6390(pSB2030). Absorbance (A), fluorescence (B) and bioluminescence (C) from microtiter plate dilution of antibiotics inoculated with S. aureus RN6390(pSB2030). Antibiotics used were fucidin (panel 1), flucloxacillin (panel 2), vancomycin (panel 3), teicoplanin (panel 4), gentamicin (panel 5), and ciprofloxacin (panel 6). Antibiotics were used at the following concentrations: ♦, 4 μg ml−1; ▪, 2 μg ml−1; ▴, 1 μg ml−1; ⋄, 0.5 μg ml−1; *, 0.25 μg ml−1; •, 0.125 μg ml−1; □, 0.063 μg ml−1; ▵, 0.03 μg ml−1; +, 0.015 μg ml−1; ○, control (DMEM + 10% RPMI). Absorbance, fluorescence, and luminescence were read concomitantly on a Genios Pro (Tecan).
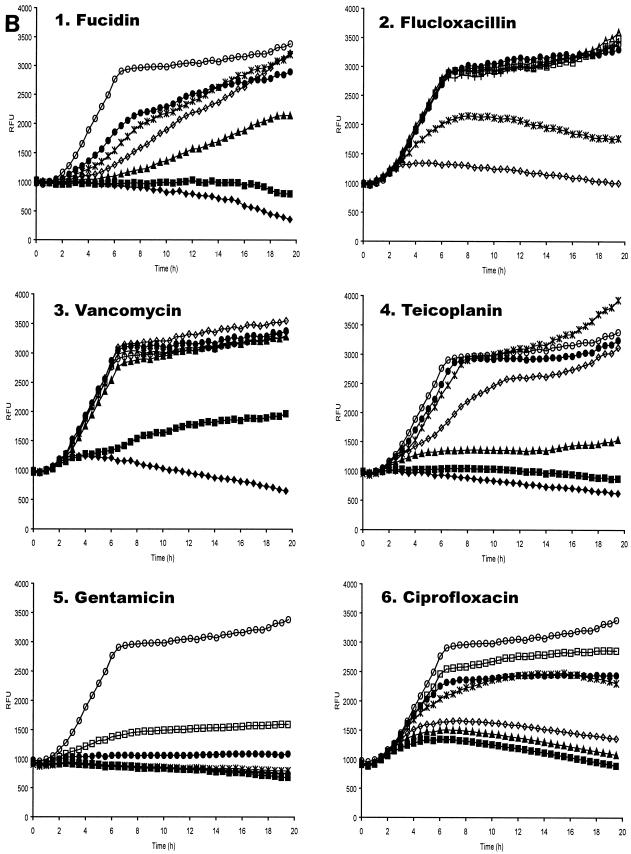
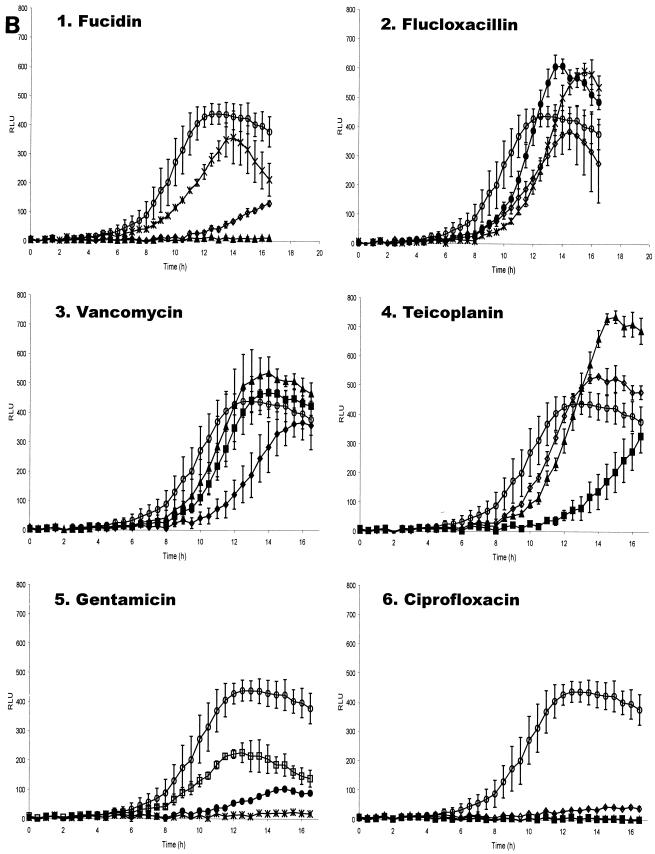
To elucidate the effectiveness of antibiotics at arresting intracellular bacterial growth, 96-well invasion assays were performed as described above. Plates were incubated at 37°C for 3 h after the lysostaphin treatment and subsequent washes in DMEM. After this period, when intracellular S. aureus were anticipated to begin replicating, medium was aspirated from the wells and HEPES-buffered DMEM containing the test antibiotics was added to the wells. Following addition of the antibiotics, the plate was incubated at 37°C in the Genios Pro, where bioluminescence and fluorescence were monitored every 30 min. As illustrated in Fig. 4, the data indicate that the antibiotics enter the cells with different efficiencies, since the changes in patterns of bioluminescence and fluorescence are different for each antibiotic, but in each case the ensuing effect on replication is very marked compared to control wells incubated in medium alone.
FIG. 4.
Bioluminescence from S. aureus RN6390(pSB2030) growing in MAC-T cells following addition of antibiotics. Fluorescence (A) and bioluminescence (B) from S. aureus RN6390(pSB2030) growing in MAC-T cells following the addition of fucidin (panel 1), flucloxacillin (panel 2), vancomycin (panel 3), teicoplanin (panel 4), gentamicin (panel 5), and ciprofloxacin (panel 6). Antibiotics were added at the following concentrations: ♦, 4 μg ml−1; ▪, 2 μg ml−1; ▴, 1 μg ml−1; ⋄, 0.5 μg ml−1; *, 0.25 μg ml−1; •, 0.125 μg ml−1; □, 0.063 μg ml−1. ○, Control (DMEM) with no added antibiotics.
It appears that fucidin is freely able to enter the cell, as bacterial replication is arrested upon addition of the antibiotic, indicated by a dose-dependent reduction of fluorescence and bioluminescence (Fig. 4A and B, panel 1). This is unlike the control, where replication continues throughout the experiment. The effective concentration range of fucidin on intracellular S. aureus appeared similar to that on planktonic bacteria.
Upon addition of the flucloxacillin (Fig. 4A and B, panel 2) intracellular bacteria are still observed to replicate at the concentrations tested, as indicated by an increase in fluorescence and bioluminescence. At 0.5 μg ml−1 flucloxacillin appears to slow replication, as indicated by a significant drop in fluorescence. At concentrations below this, the fluorescence signal is slightly reduced; however, the Lux signal is significantly enhanced, as was observed with sublethal concentrations of flucloxacillin on planktonic S. aureus.
Vancomycin treatment of S. aureus RN6390(pSB2030) gave data similar to that seen with flucloxacillin (Fig. 4A and B, panel 3). Bacterial replication continued in the presence of antibiotic, though at a lower rate than the control sample, as indicated by the GFP signal. The level of bioluminescence was seen to exceed that expressed by the control sample at sublethal concentrations of vancomycin. A vancomycin concentration of 4 μg ml−1, which was sufficient to prevent replication of planktonic bacteria, was unable to prevent replication of the intracellular S. aureus.
The third cell wall synthesis inhibitor tested, teicoplanin, has effects on intracellular replicating S. aureus very similar to those observed with vancomycin (Fig. 4A and B, panel 4). Reduction of growth rate was apparent from the GFP signal, while sublethal concentrations of the antibiotic provoked enhanced bioluminescence from the test samples. A teicoplanin concentration of 2 μg ml−1 was not able to fully arrest intracellular S. aureus replication but was sufficient to prevent growth of planktonic bacteria.
Gentamicin (Fig. 4A and B, panel 5) was clearly able to enter the MAC-T cells, as shown by the reduction in intracellular growth rate measured by both Lux and GFP. The inhibitory levels measured via the reporters were similar to those observed on planktonic S. aureus, suggesting that this antibiotic is able to pass into the eukaryotic cells with ease.
Ciprofloxacin is also seen to influence intracellular bacterial replication in a dose-responsive manner (Fig. 4A and B, panel 6), with higher concentrations inhibiting replication to the greatest degree. Concentrations effective at arresting replication of intracellular S. aureus were similar to those affecting replication of planktonic bacteria.
DISCUSSION
Previous studies by GFP-Lux reporters to elucidate agr expression and subsequent intracellular replication of S. aureus (40) used a 24-well plate assay, which monitored the bioluminescence of internalized staphylococci after addition of bacteria to a eukaryotic monolayer. The variability of the data achieved meant that many replicate samples were required in this assay format and this did not allow numerous specimens to be tested simultaneously. In this work the format has now been transferred to 96-well microplates to allow greater numbers of concurrent samples to be examined.
One of the main laboratory strains of S. aureus studies, S. aureus RN6390, has been shown to carry a deletion in rsbU which is proposed to affect virulence (5, 13, 14, 23). The internalization capabilities of S. aureus rsbU+ and rsbU strains were compared in the 96-well assay and revealed that intracellular replication of both strains was similar, indicating that a deletion in rsbU does not appear to affect internalization and subsequent replication of S. aureus by host cells (41). Although bacterial replication could be measured, signals were often quite low and variable between replicate experiments in this assay format. This was attributed to the fact that bacteria had internalized nonsynchronously during the infection period.
Here we further developed the 96-well microtiter plate assays by growing the initial inoculum with (Ala5)AIP-1 to prevent the induction of agr (27). This ensures that expression of microbial surface components recognizing adhesive matrix molecules, essential for cellular adhesion and uptake, is maximal (12, 36) and that bacteria are in a state optimal for host cell binding. Preliminary experiments showed the equivalence of infection by cycled cells previously used in these kind of experiments (40) and (Ala5)AIP-1-treated cultures. Infection will still not be synchronous if bacteria encounter host cells at different times, so to aid synchronicity, bacteria were centrifuged onto the epithelial cell monolayers seeded into 24- and 96-well plates. This ensured intimate contact between bacteria and host cells, encouraging internalization to occur simultaneously and at higher frequency.
Following a 1-h incubation to allow internalization of S. aureus, external bacteria were removed by washing and lysostaphin treatment. The S. aureus used in these assays contain plasmid pSB2030, which confers growth-dependent bioluminescence (Lux) on the bacteria without the addition of exogenous substrate (40). This enables bacterial growth to be monitored directly, without the need for sampling in luminometers with incubating abilities, providing an assay with great sensitivity and high reproducibility. Our data (Fig. 1) show that bacterial growth can be readily assessed in both 96- and 24-microwell formats.
As the reporter gene operon in pSB2030 also confers a fluorescent phenotype (GFP), we were able to use this, in combination with the vital membrane stain FM4-64, to visualize eukaryotic membranes to develop a novel time-lapse confocal microscopy protocol to image intracellular replication of S. aureus within living host cells. These data confirmed that S. aureus RN6390 was indeed replicating inside the eukaryotic cells (Fig. 2A) and provide temporal data comparable to that obtained from the in vitro assay in 96-well microplates. It was also noted that the GFP signal from the replicating bacteria appears to increase over time, commensurate with its expression from a growth-dependent promoter.
The agr mutant S. aureus RN6911 is able to enter the eukaryotic cell; however, it is not able to escape the endosome and so cannot replicate. Wesson et al. (51) suggested that the increased production of cell surface proteins by the agr mutant could promote the initial binding to the cell membrane and thus lead to higher numbers of internalized bacteria. However, they also demonstrated that while higher numbers of S. aureus RN6911 were able to enter the cell, they were not capable of intracellular growth. Data from confocal time-lapse experiments illustrated in the present study (Fig. 2B) also demonstrate this lack of intracellular replication by the agr mutant. S. aureus RN6911 are seen to be present inside the cells in relatively high numbers, but no replication is seen over the time course of the experiment. It is also noticeable that the level of GFP in individual bacteria diminishes over the period of the experiment. This is likely due to photobleaching of the GFP protein in the bacteria. As agr mutant S. aureus cells are unable to replicate intracellularly, they cannot replenish the damaged GFP, as the promoter driving GFP expression is replication dependent (40). What is also noticeable in these microscopy experiments is the fact that endosomal membranes appear to remain intact around S. aureus RN6911 cells. This further supports the hypothesis that agr expression is necessary for endosomal escape and subsequent replication within MAC-T cells (40).
Many methods are available to evaluate the efficacy of antibacterial compounds in vitro. In these studies bacteria are generally extracellular and are exposed to a constant antibiotic concentration for the test period (approximately 18 h), after which the MIC is determined. Two such standard test methods are the British Society for Antimicrobial Chemotherapy agar dilution (50) and the microtiter broth dilution method (3). While these experiments provide information on inhibition or the bactericidal activity of the compound, they do not provide any kinetic data on the killing rate during the incubation period (6, 8). Our modification of the latter method in which bacterial growth is measured by absorbance, luminescence, and fluorescence provides additional kinetic data on antibiotic efficacy. It is known that antibiotic concentrations in vivo fluctuate according to their pharmacokinetic properties (7); therefore, the response of the antibiotic on bacteria located in an intracellular niche may differ from those results obtained from in vitro experiments. Bacterial growth cannot be assessed intracellularly by absorbance readings; however, the use of Lux/GFP+ bacteria allows the kinetics of intracellular growth to be determined.
Most conventional assays that study the effect of antimicrobial agents on intracellular S. aureus use professional phagocytic cell lines. Generally macrophages are infected with S. aureus and following an infection period all extracellular bacteria are eliminated by incubation in gentamicin (1, 46) or lysostaphin (45). Following this, the intracellular growth is evaluated in the presence and absence of the compound being tested. After various incubation times, the sum total of intracellular associated bacteria is enumerated by plate count assays.
The use of the bioluminescence/fluorescence assay of S. aureus replication is readily adapted to screen mutants of S. aureus which cannot replicate intracellularly or to measure antibacterial efficacy without the need for painstaking bacterial count assays. To demonstrate the utility of this assay method, a number of antibiotics were tested having been chosen on the basis that they are widely used clinically as therapeutic agents against staphylococcal infections. As it is believed that the intracellular location of S. aureus may be responsible for long-term chronic infections (4, 19, 49), it is crucial to elucidate the intrinsic activity of the drug on bacterial replication in an intracellular environment.
Prior to assessment of the action of antibiotics against intracellular S. aureus, the effects of the antibiotics on the growth and GFP/Lux reporter activity of planktonic S. aureus(pSB2030) were examined by a modification of the microtiter broth dilution method (3). S. aureus(pSB2030) was incubated at 37°C in tissue culture medium with doubling dilutions of the test compounds in a 96-well plate. The absorbance, fluorescence, and luminescence intensity of these cultures were measured at 30-min intervals to assess bacterial growth and GFP and Lux expression. Depending upon the mode of action of the antibiotics assayed, very different effects were seen upon Lux/GFP expression of the test organism when compared with the growth data.
Fucidin, an inhibitor of peptide translocation, at lower concentrations (0.125 to 1 μg ml−1) gives a marked dose-dependent increase in the lag phase of the culture (Fig. 3A, panel 1). At the MIC (2 μg ml−1) no growth is seen. These growth data derived from absorbance are closely mirrored by the GFP and Lux signals (Fig. 3B and C, panel 1). The Lux signal from bacteria grown with lower concentrations of fucidin (0.125 to 0.5 μg ml−1), however, is seen to exceed that of the control sample without antibiotic. As the Lux signal is influenced by both protein synthesis and the metabolic state of the cell, the high bioluminescence can be attributed to uncoupling of respiratory metabolism from other cellular processes. This effect is commonly seen when challenging Lux+ bacteria with sublethal concentrations of xenobiotics (47). In contrast the GFP signal, which is not influenced by metabolic activity, never exceeds control values. This observation stresses the advantage of using a GFP-Lux dual reporter operon, not only by providing an internal control for possible reporter-dependent data, but also by giving more insights into the physiological effects of compounds on bacteria.
The cell wall-synthesis inhibitors flucloxacillin, vancomycin, and teicoplanin all provide similar data, with little effect seen upon the growth of the bacteria until the MIC is reached (Fig. 3A, panels 2 to 4). The growth data are closely comparable to that obtained from the GFP signal (Fig. 3B, panels 2 to 4). Once again, an enhancement of the Lux signal over the control, indicative of respiratory uncoupling, is observed at lower concentrations of these antibiotics.
The effect of gentamicin on GFP and Lux expression by S. aureus(pSB2030) shows good dose dependency (Fig. 3B and C, panel 5), with no indication of respiratory uncoupling at concentrations below the MIC (1 μg ml−1) from the Lux data. The growth rate at intermediate concentrations of gentamicin (0.25 to 0.5 μg ml−1) appears to be variable. This may be attributed to error-prone protein synthesis due to mRNA misreading (9) affecting the fitness of the cells. Misreading of the GFP-Lux mRNA would result in nonfunctional reporter proteins, so little or no reporter signal is seen at these concentrations.
A dose-dependent inhibition of replication by ciprofloxacin, a DNA gyrase inhibitor, can be closely correlated with both the GFP and Lux signals (Fig. 3A, B, and C, panel 6). Little or no reporter signal is measured with ciprofloxacin concentrations above the MIC of 0.5 μg ml−1. Reporter signals at lower concentrations are lower than the antibiotic-free control indicative of sublethal injury of the bacteria.
The addition of fucidin to S. aureus(pSB2030) infected MAC-T cells shows a dose-dependent reduction in fluorescence and bioluminescence (Fig. 4A and B, panel 1) indicative of entry of the antibiotic into the cell where it can repress replication of the bacteria. A full reduction of both GFP and Lux reporter signals at 1 μg ml−1, a concentration below the MIC of 2 μg ml−1, suggests that S. aureus may be under stress in the intracellular milieu which makes them more susceptible to this agent. Alternatively, the signal generated by the low number of intracellular bacteria relative to that found in broth-grown cultures may be below the sensitivity of the assay.
Flucloxacillin, vancomycin, and teicoplanin all provide similar data when used to challenge intracellular staphylococci (Fig. 4A and B, panels 2 to 4). A dose-dependent reduction is seen in the accumulation of GFP with all of the compounds. On internalized bacteria with concentrations of vancomycin of 1 to 4 μg ml−1 (MIC = 4 μg ml−1), teicoplanin of 0.5 to 2 μg ml−1 (MIC = 2 μg ml−1) and flucloxacillin of 0.125 to 0.5 μg ml−1 (MIC = 0.25 μg ml−1), GFP and Lux signals are readily measurable indicating the bacteria can still replicate. This may be due an inability of these antibiotics to penetrate the eukaryotic cells. However, an increase in Lux signal above the control (Fig. 4 B, panels 2 to 4), indicative of uncoupling for each of these antibiotics, suggests the agents are capable of penetrating the cells to some extent. The activity of both teicoplanin and vancomycin against S. aureus residing in human neutophils has previously been investigated (37). In this study it was demonstrated that at a concentration of 10 μg ml−1 each of these antibiotics had an inhibitory effect on bacterial replication; however, a total arrest of bacterial replication was not observed.
As the cell wall's function is to preserve cell integrity by withstanding the internal osmotic pressure, it may be that osmotic protection afforded the bacteria by the eukaryotic cytosol may help S. aureus resist cell wall synthesis inhibitors and continue replicating. The growing prevalence of methicillin-resistant S. aureus has led to an increased reliance on the glycopeptides vancomycin and teicoplanin, to which methicillin-resistant S. aureus strains have been uniformly susceptible. However, these data suggest that while these drugs are prescribed to overcome staphylococcal infections they may only show a limited efficiency in inhibiting intracellular S. aureus replication.
If during staphylococcal infections bacteria are residing in an intracellular environment, the fact these antibiotics appear to have lower efficacy inside cells could cause increased problems long term. As bacteria are exposed to sublethal concentrations of antibiotics, it provides a selective environment for the emergence of antibiotic resistant staphylococcal strains against the prescribed drug.
The effect of ciprofloxacin on intracellular S. aureus (Fig. 4A and B, panel 6) shows a clear dose-dependent reduction in GFP and Lux signals. The effective range of concentration is equivalent to that seen in planktonic cells indicating that ciprofloxacin is readily able to enter the MAC-T cells and retains full activity in the cell's cytosol. This correlates well with data obtained of the intracellular killing of S. aureus in human neutophils, where a concentration of 1 μg ml−1 was found to kill 90% of phagocytosed bacteria in comparison to untreated neutrophils (31, 38).
Challenge of intracellular S. aureus with gentamicin at concentration of 0.06 to 0.25 μg ml−1 reveals a dose-dependent reduction of both the GFP and Lux signals (Fig. 4A and B, panel 5), indicative of arrest of replication and showing that this antibiotic can readily enter the MAC-T cells.
The data obtained studying the effects of gentamicin have proved to be very interesting. Gentamicin is an aminoglycoside that interferes with bacterial protein synthesis by binding to the ribosomal 30S subunit. Until recently, it has been assumed that aminoglycosides are poorly effective against treating intracellular pathogens due to their poor ability to penetrate the eukaryotic cell membrane (25). Indeed in conventional assays to enumerate numbers of intracellular S. aureus, gentamicin is used to remove extracellular bacteria from samples. Our data have clearly demonstrated that gentamicin is able to penetrate the cell and inhibit replication of intracellular S. aureus. Indeed, when concentrations of gentamicin of 100 μg ml−1 or 50 μg ml−1, which are routinely used in traditional intracellular replication assays, were added to monolayers infected with Lux+ S. aureus, no bioluminescence was observed, indicating that bacterial replication had been completely abolished (data not shown). This observation concurs with recent reports that have demonstrated that gentamicin is capable of entering macrophages and killing intracellular bacteria (10, 11, 15, 35). Hamrick et al. (15) demonstrated that even at concentrations of 5 μg ml−1 gentamicin was able to affect intracellular bacterial viability. We have demonstrated that concentrations as low as 0.06 μg ml−1 affected replication of intracellular S. aureus tagged with a gfp-lux dual reporter. These results suggest that a reevaluation of the use of gentamicin in the experimental study of persistence and replication of bacterial pathogens within host cells is well overdue.
The ability of intracellular bacteria to be protected against the killing actions of antibiotics that have a poor ability to penetrate the eukaryotic cell as well as intracellular inactivation of the compound could be responsible for the therapy failure and persistent recurrent infections that occur within the host (15, 18, 24). Our data suggest that for efficient antistaphylococcal therapy, it would be pertinent to include antibiotics that can be demonstrated to be effective against intracellular S. aureus.
.
Acknowledgments
We thank the United Kingdom Medical Research Council for funding (G9219778), Vyv Salisbury for providing antibiotics, and Cath Rees for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Ahren, I. L., E. Karlsson, A. Forsgren, and K. Riesbeck. 2002. Comparison of the antibacterial activities of ampicillin, ciprofloxacin, clarithromycin, telithromycin and quinupristin/dalfopristin against intracellular non-typeable Haemophilus influenzae. J. Antimicrob. Chemother. 50:903-906. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, R. A., K. R. Matthews, E. Cifrian, A. J. Guidry, and S. P. Oliver. 1996. Staphylococcus aureus invasion of bovine mammary epithelial cells. J. Dairy Sci. 79:1021-1026. [DOI] [PubMed] [Google Scholar]
- 3.Amsterdam, D. 1996. Susceptibility testing of antimicrobials in liquid media, p. 52-111. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Wilkins and Wilkins, Baltimore, Md.
- 4.Bayles, K. W., C. A. Wesson, L. E. Liou, L. K. Fox, G. A. Bohach, and W. R. Trumble. 1998. Intracellular Staphylococcus aureus escapes the endosome and induces apoptosis in epithelial cells. Infect. Immun. 66:336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarR and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig, W. A., and S. C. Ebert. 1991. Killing and regrowth of bacteria in vitro. Scandinavian J. Infectious Dis. (Suppl.) 74:63-70. [PubMed] [Google Scholar]
- 7.Dalhoff, A. 1999. Pharmocodynamics of fluoroquinolones. J. Antimicrob. Chemother. (Suppl. B) 43:51-59. [DOI] [PubMed] [Google Scholar]
- 8.Dalhoff, A., and U. Ullmann. 1990. Correlation between pharmocodynamics and efficiency of antibacterial agents in animal models. Eur. J. Clin. Microbiol. Infect. Dis. 9:479-487. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J., and B. D. Davies. 1968. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. J. Biol. Chem. 243:3312-3316. [PubMed] [Google Scholar]
- 10.Drevets, D. A., B. P. Canono, P. J. Leenen, and P. A. Campbell. 1994. Gentamicin kills intracellular Listeria monocytogenes. Infect. Immun. 62:2222-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eze, M. O., L. Yuan, R. M. Crawford, C. M. Paranavitana, T. L. Hadfield, A. K. Bhattacharjee, R. L. Warren, and D. L. Hoover. 2000. Effects of opsonisation and gamma interferon on growth of Brucella melitensis 16M in mouse peritoneal macrophages in vitro. Infect. Immun. 68:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, T. J., and M. Hook. 1998. Surface protein adhesions of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 13.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 14.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamrick, T. S., A. H. Diaz, E. A. Havell, J. R. Horton, and P. E. Orndorff. 2003. Influence of extracellular bactericidal agents on bacteria within macrophages. Infect. Immun. 71:1016-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickey, P. C., D. Jacobson, N. D. Read, and L. Glass. 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37:109-119. [DOI] [PubMed] [Google Scholar]
- 17.Hill, P. J., C. E. D. Rees, M. K. Winson, and G. S. A. B. Stewart. 1993. The application of lux genes. Biotechnol. Appl. Biochem. 17:3-14. [PubMed] [Google Scholar]
- 18.Holmes, B., P. G. Quie, D. B. Windhorst, B. Pollara, and R. Good. 1966. Protection of phagocytosed bacteria from the killing action of antibiotics. Nature 210:1131-1134. [DOI] [PubMed] [Google Scholar]
- 19.Hudson, M. C., W. K. Ramp, N. C. Nicholson, A. S. Williams, and M. T. Nousiainen. 1995. Internalisation of S. aureus by cultured osteoblasts. Microb. Pathog. 19:409-419. [DOI] [PubMed] [Google Scholar]
- 20.Hyunh, H. T., G. Robitaille, and J. D. Turner. 1991. Establishment of bovine mammary epithelial cells (MAC-T): an in vivo model for bovine lactation. Exp. Cell Res. 197:191-199. [DOI] [PubMed] [Google Scholar]
- 21.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 22.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 23.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackaness, G. B. 1962. Cellular resistance to infection. Nature 181:381-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurin, M., and D. Raoult. 2001. Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob. Chem. 45:2977-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDowell, P., Z. Affas, C. Reynolds, M. T. G. Holden, S. J. Wood, S. Saint, A. Cockayne, P. J. Hill, C. E. Dodd, B. W. Bycroft, W. C. Chan, and P. Williams. 2001. Structure, activity and evolution of the group 1 thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 41:503-512. [DOI] [PubMed] [Google Scholar]
- 28.Meighen, E. 1993. Bacterial bioluminescence: organisation, regulation, and application of lux genes. FASEB J. 7:1016-1022. [DOI] [PubMed] [Google Scholar]
- 29.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 30.Morfeldt, E., L. Janzon, S. Arvidson, and S. Lofdahl. 1988. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol. Gen. Genet. 211:435-440. [DOI] [PubMed] [Google Scholar]
- 31.Neilsen, S. L., N. Obel, M. Storgaard, and P. L. Anderson. 1997. The effect of quinolones on the intracellular killing of Staphylococcus aureus in neutrophil granulocytes. J. Antimicrob. Chemother. 39:617-622. [DOI] [PubMed] [Google Scholar]
- 32.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 33.Novick, R. P., and T. W. Muir. 1999. Virulence gene regulation by peptides in staphylococci and other Gram positive bacteria. Curr. Opin. Microbiol. 2:40-45. [DOI] [PubMed] [Google Scholar]
- 34.Novick, R. P., H. F. Ross, S. J., Projan, J. Kornblum, B. Kreiswirth. and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohya, S., H. Xiong, Y. Tanabe, M. Arakawa, and M. Mitsuyama. 1998. Killing mechanism of Listeria monocytogenes in activated macrophages as determined by an improved assay system. J. Med. Microbiol. 47:211-215. [DOI] [PubMed] [Google Scholar]
- 36.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. microbial surface components recognizing adhesive matrix molecules-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 37.Pedrera, M. I., C. Barriga, and A. B. Rodriguez. 1995. Intracellular activity of both teicoplanin and vancomycin against Staphylococcus aureus in human neutrophils. Comp. Immunol. Microbiol. Infect. Dis. 18:213-228. [DOI] [PubMed] [Google Scholar]
- 38.Peman, J., E. Canton, M. T. Hernandez, and M. Gobernardo. 1994. Intraphagocytic killing of gram-positive bacteria by ciprofloxacin. Antimicrob. Chemother. 34:965-974. [DOI] [PubMed] [Google Scholar]
- 39.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterisation and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 179:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qazi, S. N. A., E, Counil, J. Morrissey, C. E. D. Rees, A. Cockayne, K. Wizner, W. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qazi, S. N. A., C. E. D. Ress, P. Williams, and P. J. Hill. 2002. A novel dual reporter for studying intracellular bacterial pathogens, p. 365-368. In P. E. Stanley, and L. J. Kricka (ed.), Bioluminescence and chemiluminescence: progress and current applications. World Scientific Publishing Co., Singapore.
- 42.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. Novick. 1986. Regulation of exoprotein gene expression by agr. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 43.Rocchetta, H. L., C. J. Boylan, J. W. Foley, P. W. Iversen, D. L. Letourneau, C. L. McMillian, P. R. Contag, D. E. Jenkins, and T. R. Parr. 2001. Validation of a non-invasive, real time imaging technology using bioluminescent Escherichia coli in the neutropenic mouse thigh model of infection. Antimicrob. Agents Chemother. 45:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salisbury, V., A. Pfoestl, H. Wiesinger-Mayr, R. Lewis, K. E. Bowker, and A. P. MacGowan. 1999. Use of a clinical Escherichia coli isolate expressing lux genes to study the antimicrobial pharmacodynamics of moxifloxacin. J. Antimicrob. Chemother. 43:829-832. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez, M. S., C. W. Ford, and R. J. Yancey, Jr. 1988. Evaluation of antibiotic effectiveness against Staphylococcus aureus surviving within the bovine mammary gland macrophage. J. Antimicrob. Chemother. 21:773-786. [DOI] [PubMed] [Google Scholar]
- 46.Seral, C., S. Carryn, P. M. Tulkens, and F. Van Bembeke. 2003. Influence of P-glycoprotein and MRP efflux pump inhibitors on the intracellular activity of azithromycin and ciprofloxacin in macrophages infected by Listeria monocytogenes or Staphylococcus aureus. J. Antimicrob. Chemother. 51:1167-1173. [DOI] [PubMed] [Google Scholar]
- 47.Shaw, L. J., Y. Beaton, L. A. Glover, K. Killham, and A. A. Meharg. 1999. Development and characterization of a lux-modified 2, 4-dichlorophenol-degrading Burkholderia sp. RASC. Environ. Microbiol. 1:393-399. [DOI] [PubMed] [Google Scholar]
- 48.Tenhami, M., K. Hakkila, and M. Karp. 2001. Measurement of effects of antibiotics in bioluminescent Staphylococcus aureus RN4220. Antimicrob. Agents Chemother. 45:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vann, J. M., and R. A. Proctor. 1987. Ingestion of Staphylococcus aureus by bovine endothelial cells results in time- and inoculum-dependent damage to endothelial cell monolayers. Infect. Immun. 55:2155-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Washington, J. A., and V. L. Sutter. 1980. Dilution susceptibility test: agar and macro-broth dilution procedures, p. 453-458. In E. H. Lennette, A Balows, W. J. Hausler, and J. P. Truant (ed.), Manual of clinical microbiology, 3rd ed. American Society for Microbiology, Washington, D.C.
- 51.Wesson, C. A., L. E. Liou, K. M. Todd, G. A. Bohach, W. R. Trumble, and K. W. Bayles. 1988. Staphylococcus aureus agr and sar global regulators influence internalization and induction of apoptosis. Infect. Immun. 66:5238-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]