Abstract
The Hierarchical Technology for Quantitative Structure - Activity Relationships (HiT QSAR) was applied to 95 diverse nitroaromatic compounds (including some widely known explosives) tested for their toxicity (50% inhibition growth concentration, IGC50) against the ciliate Tetrahymena pyriformis. The dataset was divided into subsets according to putative mechanisms of toxicity. Classification and Regression Trees (CART) approach implemented within HiT QSAR has been used for prediction of mechanism of toxicity for new compounds. The resulting models were shown to have ~80% accuracy for external datasets indicating that the mechanistic dataset division was sensible. Then, Partial Least Squares (PLS) statistical approach was used for the development of 2D QSAR models. Validated PLS models were explored to (i) elucidate the effects of different substituents in nitroaromatic compounds on toxicity; (ii) differentiate compounds by probable mechanisms of toxicity based on their structural descriptors; (iii) analyze the role of various physical-chemical factors responsible for compounds’ toxicity. Models were interpreted in terms of molecular fragments promoting or interfering with toxicity. It was also shown that mutual influence of substituents in benzene ring plays the determining role in toxicity variation. Although chemical mechanism based models were statistically significant and externally predictive (R2ext=0.64 for the external set of 63 nitroaromatics identified after all calculations have been completed), they were also shown to have limited coverage (57% for modeling and 76% for external set).
Keywords: Tetrahymena pyriformis, QSAR, chemical toxicants, prediction of toxicity
1. Introduction
Nitroaromatics compounds and their numerous derivatives are of use as explosives and propellants in the military and in industry [1, 2]. Waste from nitro compounds are easily disseminated during manufacturing, storage, transportation, and utilization of munitions, leading to a potential hazard for humans and the environment [3]. A number of studies have shown that nitro compounds, as well as their metabolites of environmental transformation, by-products of synthesis, or incomplete combustion are harmful for the biosphere due to their toxicity [3–6]. For instance, toxic effects in humans after dermal, oral, or respiratory exposures include gastrointestinal, neurological and reproductive disorders, cirrhosis of the liver, hepatitis, cataracts, respiratory and skin irritation, nephrotoxicity, and hematological defects. Moreover, nitroaromatic compounds are widely used in medicine, industry and agriculture. Nitroaromatic pesticides as well as the explosive residues are considered as toxic environmental pollutants. Some of these compounds have mutagenic or carcinogenic activity and may accumulate in the food chain (bioaccumulation). Therefore, the presence of aromatic and nitroaromatic xenobiotics in the environment may present serious public health and environmental problems, and both nature and degree of aromatic substitutions may have profound effects on the chemical toxicity of nitroaromatic compounds [7].
Chemical toxicity can be associated with many hazardous biological effects such as gene damage, carcinogenicity, or the induction of lethal rodent or human diseases. It is important to evaluate the toxicity of all commercial chemicals, especially the High Production Volume compounds. To address this need, standard experimental protocols have been established by chemical industry, pharmaceutical companies, and government agencies to test chemicals for their toxic potential.
Although the experimental protocols for toxicity testing have been developed for many years and the cost of compound testing has decreased significantly, computational chemical toxicology continues to be a viable approach to reduce both the amount of effort and the cost of experimental toxicity assessments. Significant savings could be achieved if the potential toxicity of a new chemical could be predicted before its synthesis and experimental testing. To address this challenge, many Quantitative Structure Activity/Toxicity Relationship (QSAR/QSTR)1 studies have been conducted and reported for different toxicity endpoints, e.g., [8–10].
The toxicity of nitrobenzenes against the aquatic ciliate Tetrahymena pyriformis has been extensively studied by several research groups [8, 9, 11, 12] using 2D and 3D QSAR methodologies. There are multiple mechanisms of nitrobenzene toxic action, with hydrophobicity and electrophilic reactivity being the most important structural factors contributing to the mode of action [12]. Hydrophobicity is considered to be important for compounds’ transport from the environment to the site of action, whereas the electrophilicity is related to an intrinsic reactivity pattern. Reactivity of nitrobenzenes can be due to: (i) reduction of the nitro group and (ii) the tendency to act as an electrophile in SNAr reactions [9, 12]. The reduction of a nitro group can occur by at least two mechanisms: the single-step reduction with an enzyme such as nitroreductase and the so-called redox cycling, during which multiple back-oxidation of the reduced nitro compound can occur.
Agrawal and Khadikar [11] built multiple regression models based solely on topological descriptors. Cronin and co-workers [12] employed 3D descriptors and postulated a separate toxicity mechanism for para-substituted nitrobenzenes, which were detected as statistical outliers. The effect of different chemical narcotics on Tetrahymena pyriformis was investigated by Bearden and Schultz [13]. Various aromatic compounds display distinct types of narcosis: the toxicity of molecules with strong electron-releasing amino and hydroxyl groups was explained by polar narcosis mechanism [14, 15]. However, Vaes et al. [16] showed that the distinction between different types of narcosis (polar and apolar) is only an experimental artifact from using octanol as surrogate for the cell membrane lipids. Estrada and Uriarte [17] applied their original Topological Sub-Structural Molecular Design (TOPS-MODE) approach based on topological descriptors to a data set of 43 substituted nitrobenzenes. Although mechanistic interpretation of the correlation is complex, it can be used for the prediction of molecular toxicity through the summation of toxicity contributions by individual structural groups. The most relevant efforts to develop QSAR models for toxicity of nitroaromatics are listed in the Table 1. All these studies [9, 11, 12, 18, 19] have the same drawbacks: obtained models have only internal cross-validation, they have no AD estimation and no prove of passing Y-randomization test; small sets of compounds has been used for model development. Detailed interpretation of the developed models is reported in each article, however the absence of external validation, DA and Y-scrambling is causing the serious doubts of reliability of this interpretation. Certainly, there were many published studies devoted to QSAR modeling of this endpoint, but they were no focused on nitroaromatic compounds. However we should mention the international collaborative QSAR modeling of Tetrahymena pyriformis [20] which was resulted in, robust, predictive and pretty comprehensive model for this endpoint. However, many of compounds of military interest are outside AD of this model.
Table 1.
The most relevant QSAR models of nitroaromatics toxicity.
| Investigated activity |
Number of compounds (Modeling + External) |
Descriptors | Statistical Methods |
Models | Validation | DA | Y scrambling |
Ref./ Year |
|
|---|---|---|---|---|---|---|---|---|---|
| 1 | Toxicity to Tetrahymena pyriformis |
47 | logP, molar refractivity, logD; Molecular connectivities, electrotopological indices and kappa indices. |
MLR | Q2=0.81– 0.91 |
Internal cross- validation |
no | no | 1995 [62] |
| 2 | Toxicity to Tetrahymena pyriformis |
42 | The logarithm of the 1- octanol/water partition coefficient (log Kow), The lowest unoccupied molecular orbital energy (Elumo), Maximum acceptor superdelocalizability (Amax) |
MLR | Q2=0.74– 0.89 |
Internal cross- validation |
no | no | 1998 [12] |
| 3 | Toxicity to Scenedesmus vacuolatus |
19 | log Kow Whole-molecule descriptors: CLOGP, Ehomo, Elumo, Esomo, Electronegativity |
PCA MLR |
radj2=0.90– 0.93 |
no | no | no | 2000 [63] |
| 4 | Toxicity | 40 | Szeged index, Sz, Padmakar–Ivan index, PI, Balaban index, J, Molecular redundancy index, MRI, Indicator parameters (Ip1, Ip2, Ip3) |
MLR | R2CV=0.75 | Internal cross- validation |
no | no | 2001 [11] |
| 5 | Toxicity to Tetrahymena pyriformis |
97 | CODESSA The energy of the singly occupied molecular orbital ESOMO |
MLR | R2CV=0.79 | Internal cross- validation |
no | no | 2003 [9] |
In spite of earlier effort [9] to develop acceptable QSAR models for given dataset using topological, quantum-chemical and some other chemical parameters generated by CODESSA, many questions pertinent to the toxicity of nitroaromatic compounds remain unanswered. One of them, addressed in this paper in great detail, is the relationship between chemical structure (especially the influence of substituents in the aromatic ring) and toxicity. This analysis could provide useful knowledge regarding the acceptance or rejection of the proposed mechanisms of chemical toxicity for this group of molecules. Another common vulnerability of all investigations mentioned above is the absence of any external validation of reported QSAR models, i.e., all these models are well-fitted, but there is no information as to how predictive they are when applied to external datasets. Therefore, the aim of the present study is to extend recent investigation [9] by applying Hierarchical Technology for Quantitative Structure-Activity Relationships (HiT QSAR) for: (i) generation and external validation of QSAR models describing the influence of the structure of 95 various nitroaromatic compounds (including some widely known explosives) on their toxicity against the ciliate Tetrahymena pyriformis; (ii) elucidation of the effects of different substituents in the benzene ring on toxicity of nitroaromatic compounds; (iii) differentiation of compounds by probable mechanisms of toxicity based on their structural descriptors; (iv) analysis of the role of various physical-chemical factors (e.g., electrostatics, hydrophobicity, hydrogen bonding, atomic identity, etc.) in compounds’ toxicity; and, ultimately, (v) development of a reliable computational tool for accurate environmental risk assessment of novel untested nitroaromatic compounds.
2. Materials and methods
2.1 Dataset
The modeling set for the present investigation was created from the data set compiled from well-characterized previous studies [12, 18, 21, 22], totally 115 records. After removal of 20 duplicates, 95 compounds remained for QSAR analysis. The inverse logarithm of the concentration causing 50% growth inhibition of Tetrahymena pyriformis after 40 hours log (IGC50)−1, mM, was used as a measure of compounds’ toxicity (Table 2). The whole database of 95 nitrobenzenes was divided into two overlapping clusters (60 compounds in total) based on mechanistic considerations outlined in [9, 19, 23]: 41 compounds which caused the appearance of oxidative stress in a living cell due to the redox cycling during nitro group reduction (mechanism A) and 48 species which are predisposed to the nucleophilic attack (mechanism B). 35 remaining compounds that could not be assigned to either of these two groups formed a separate subset. For nitroaromatic compounds that may exert oxidative stress by acting as redox cyclers mode of action, the nitroaromatic radical anion formed by one-electron reduction is oxidized back to the parent compound while forming superoxide (O2-•), which then leads to the generation of hydrogen peroxide (H2O2) and hydroxyl radical (OH•) as highly reactive oxidants. Apparently, redox cycling potency thus competes with the ability of further reduction [19]. Schmitt et al. [19] suggested to use ESOMO (energy of single occupied molecular orbital) window of −0.30 to 0.55 to relate dinitrobenzenes as well as multiply chlorinated nitrobenzenes to redox cyclers. For compounds acting by mechanism B the presence of strong electron-attracting groups can activate the halogen or pseudohalogen toward substitution via SNAr mechanism. Mekenyan et al. [23] suggested that toxic behavior of such chemicals involves covalent binding to protein and the difference in lowest unoccupied molecular orbital energy between the parent compounds and their Meisenheimer complexes together with the maximum acceptor superdelocalizabilities determined over the aromatic reaction sites were found to discriminate correctly such nucleophilic compounds [23]. Then, mentioned above criteria were used by Katritzky et al. [9] to classify investigated nitroaromatics by mode of toxic action. In the given we used the same classification.
Table 2.
Investigated compounds
| No | Compound | Ref. | −lg(IGC50) | Mechanism A | Mechanism B | |||
|---|---|---|---|---|---|---|---|---|
| Obs. | Pred. | Obs. | Pred. | Obs. | Pred. | |||
| 1 | nitrobenzene | [12, 18] | 0.14 | 0.41 | 0 | 1 | 0 | 0 |
| 2 | 2-nitrophenol | [18, 22] | 0.67 | 0.45 | 0 | 0 | 0 | 0 |
| 3 | 2-nitrotoluene | [12, 18] | 0.05 | 0.47 | 0 | 0 | 0 | 0 |
| 4 | 2-chloronitrobenzene | [12, 18] | 0.68 | 0.73 | 1 | 1 | 0 | 0 |
| 5 | 2-bromonitrobenzene | [12, 18] | 0.75 | 0.85 | 1 | 1 | 0 | 0 |
| 6 | 2-nitrobenzyl alcohol | [18] | −0.16 | −0.07 | 0 | 0 | 0 | 0 |
| 7 | 2-nitrobiphenyl | [18] | 1.3 | 1.30 | 0 | 1 | 0 | 0 |
| 8 | 3-nitroaniline | [18] | 0.03 | 0.32 | 0 | 1 | 0 | 0 |
| 9 | 3-nitrophenol | [18, 22] | 0.51 | 0.32 | 0 | 0 | 0 | 0 |
| 10 | 3-nitrotoluene | [12, 18] | 0.05 | 0.52 | 0 | 0 | 0 | 0 |
| 11 | 3-chloronitrobenzene | [12, 18] | 0.73 | 0.69 | 1 | 1 | 0 | 0 |
| 12 | 3-nitrobenzonitrile | [18, 22] | 0.45 | 0.45 | 1 | 1 | 0 | 0 |
| 13 | 3-nitrobenzaldehide | [18] | 0.14 | 0.22 | 1 | 1 | 0 | 0 |
| 14 | 1,3-dinitrobenzene | [12, 18] | 0.89 | 0.98 | 1 | 1 | 1 | 1 |
| 15 | 3-nitroanisole | [18] | 0.67 | 0.60 | 1 | 1 | 0 | 0 |
| 16 | 4-nitrotoluene | [12, 18] | 0.17 | 0.49 | 0 | 0 | 0 | 0 |
| 17 | 4-ethylnitrobenzene | [18] | 0.8 | 0.67 | 0 | 0 | 0 | 0 |
| 18 | 4-nitroanisole | [18] | 0.54 | 0.81 | 1 | 1 | 0 | 0 |
| 19 | 4-chloronitrobenzene | [12, 18] | 0.43 | 0.81 | 1 | 1 | 0 | 0 |
| 20 | 4-bromonitrobenzene | [12, 18] | 0.38 | 0.85 | 1 | 1 | 0 | 0 |
| 21 | 4-nitrobenzyl alcohol | [18] | 0.1 | 0.16 | 0 | 0 | 0 | 0 |
| 22 | 4-nitrobenzamide | [18] | 0.18 | −0.16 | 1 | 1 | 0 | 0 |
| 23 | 4-nitrobenzaldehyde | [18] | 0.2 | 0.27 | 1 | 1 | 1 | 0 |
| 24 | 2-nitrobenzoic acid | [18] | −1.64 | −1.45 | 0 | 0 | 1 | 1 |
| 25 | 3-nitrobenzoic acid | [18] | −1.09 | −0.64 | 0 | 0 | 0 | 0 |
| 26 | 4-nitrobenzoic acid | [18] | −0.86 | −0.59 | 0 | 0 | 1 | 0 |
| 27 | 2-nitrobenzamide | [18] | −0.72 | −0.74 | 1 | 1 | 1 | 1 |
| 28 | 3-nitrobenzyl alcohol | [18] | −0.22 | 0.16 | 0 | 0 | 0 | 0 |
| 29 | 3-nitrobenzamide | [18] | −0.19 | −0.21 | 1 | 1 | 0 | 0 |
| 30 | 2,4,6-trinitrophenol | [22] | −0.16 | 1.05 | 0 | 1 | 0 | 1 |
| 31 | 4-nitrophenylacetonitrile | [18] | 0.13 | 0.46 | 0 | 0 | 0 | 0 |
| 32 | 2-nitrobenzaldehyde | [18] | 0.17 | 0.13 | 1 | 1 | 1 | 1 |
| 33 | 4-fluoronitrobenzene | [18] | 0.25 | 0.66 | 0 | 1 | 0 | 0 |
| 34 | 3,4-dinitrophenol | [22] | 0.27 | 0.80 | 1 | 1 | 1 | 1 |
| 35 | 3-nitroacetophenone | [18] | 0.32 | 0.28 | 1 | 1 | 0 | 0 |
| 36 | 5-hydroxy-2-nitrobenzaldehyde | [18] | 0.33 | 0.15 | 1 | 0 | 1 | 1 |
| 37 | 2,3-dinitrophenol | [22] | 0.46 | 0.75 | 1 | 1 | 1 | 1 |
| 38 | 2-amino-4-nitrophenol | [22] | 0.48 | 0.53 | 0 | 0 | 0 | 0 |
| 39 | 3,5-dinitrobenzyl alcohol | [12] | 0.53 | 0.51 | 0 | 0 | 1 | 1 |
| 40 | 2,6-dinitrophenol | [22] | 0.54 | 0.79 | 1 | 1 | 1 | 1 |
| 41 | 4-nitrobenzonitrile | [18, 21] | 0.57 | 0.46 | 1 | 1 | 1 | 0 |
| 42 | 4-methyl-2-nitrophenol | [22] | 0.57 | 0.81 | 0 | 0 | 0 | 0 |
| 43 | 2,6-dichloro-4-nitrophenolc | [22] | 0.63 | 1.08 | 0 | 0 | 1 | 1 |
| 44 | 2-chloro-6-nitrotoluene | [12] | 0.68 | 0.83 | 0 | 0 | 0 | 0 |
| 45 | ethyl-4-nitrobenzoate | [18] | 0.71 | 0.26 | 1 | 1 | 1 | 0 |
| 46 | 4-methyl-3-nitrophenol | [22] | 0.74 | 0.72 | 0 | 0 | 0 | 0 |
| 47 | 2-chloromethyl-4-nitrophenol | [22] | 0.75 | 0.93 | 0 | 0 | 0 | 0 |
| 48 | 4-chloro-2-nitrotoluene | [12] | 0.82 | 0.84 | 0 | 0 | 0 | 0 |
| 49 | 4-nitrophenetole | [18] | 0.83 | 0.94 | 1 | 1 | 0 | 0 |
| 50 | 2,4,6-trimethylnitrobenzene | [12] | 0.86 | 0.99 | 0 | 0 | 0 | 1 |
| 51 | 6-methyl-1,3-dinitrobenzene | [12] | 0.87 | 1.26 | 0 | 0 | 1 | 1 |
| 52 | 4-amino-2-nitrophenol | [22] | 0.88 | 0.78 | 0 | 0 | 0 | 0 |
| 53 | 2,5-dinitrophenol | [22] | 0.95 | 0.96 | 1 | 1 | 1 | 1 |
| 54 | 2,4-dichloronitrobenzene | [12] | 0.99 | 1.25 | 1 | 1 | 0 | 0 |
| 55 | 3-bromonitrobenzene | [12, 18] | 1.03 | 0.73 | 0 | 1 | 0 | 0 |
| 56 | 2,3-dichloronitrobenzene | [12] | 1.07 | 0.96 | 0 | 1 | 0 | 0 |
| 57 | 2-nitrobenzonitrile | [18, 21] | 1.08 | 0.45 | 1 | 1 | 1 | 0 |
| 58 | 2,4-dinitrophenol | [22] | 1.08 | 0.99 | 1 | 1 | 1 | 1 |
| 59 | 3,4-dinitrobenzyl alcohol | [12] | 1.09 | 0.72 | 0 | 0 | 0 | 1 |
| 60 | 5-fluoro-2-nitrophenol | [22] | 1.13 | 0.68 | 1 | 0 | 0 | 0 |
| 61 | 3-methyl-4-bromonitrobenzene | [12] | 1.16 | 1.06 | 1 | 0 | 0 | 0 |
| 62 | 3,4-dichloronitrobenzene | [12] | 1.16 | 1.29 | 1 | 1 | 1 | 0 |
| 63 | 2-amino-4-chloro-5-nitrophenol | [22] | 1.17 | 0.99 | 0 | 0 | 0 | 1 |
| 64 | 4-nitrobenzyl chloride | [18] | 1.18 | 0.60 | 1 | 0 | 1 | 0 |
| 65 | 2,6-dinitro-4-cresol | [22] | 1.23 | 1.21 | 0 | 0 | 1 | 1 |
| 66 | 1,2-dinitrobenzene | [22] | 1.25 | 1.01 | 1 | 1 | 1 | 1 |
| 67 | 1,4-dinitrobenzene | [12, 18] | 1.3 | 1.11 | 1 | 1 | 1 | 1 |
| 68 | 2,6-dibromo-4-nitrophenol | [18] | 1.35 | 1.42 | 0 | 0 | 1 | 1 |
| 69 | 2,5-dibromonitrobenzene | [12] | 1.37 | 1.42 | 1 | 1 | 1 | 0 |
| 70 | 4-nitrophenol | [22] | 1.42 | 0.62 | 0 | 0 | 0 | 0 |
| 71 | 4-butoxynitrobenzene | [18] | 1.42 | 1.43 | 1 | 1 | 0 | 0 |
| 72 | 2,4,6-trichloronitrobenzene | [12] | 1.43 | 1.71 | 1 | 1 | 1 | 1 |
| 73 | 2,3,4-trichloronitrobenzene | [12] | 1.51 | 1.46 | 1 | 1 | 1 | 1 |
| 74 | 5-methyl-1,2-dinitrobenzene | [12] | 1.52 | 1.28 | 1 | 0 | 1 | 1 |
| 75 | 2,4,5-trichloronitrobenzene | [12] | 1.53 | 1.79 | 1 | 1 | 1 | 1 |
| 76 | 3-nitrobiphenyl | [18] | 1.57 | 1.34 | 0 | 1 | 0 | 0 |
| 77 | 2-chloro-4-nitrophenol | [22] | 1.59 | 0.94 | 0 | 0 | 0 | 0 |
| 78 | 4-chloro-6-nitromcresol | [22] | 1.64 | 1.58 | 0 | 0 | 0 | 1 |
| 79 | 2,6-diiodo-4-nitrophenol | [22] | 1.71 | 1.65 | 0 | 0 | 1 | 1 |
| 80 | 4,6-dinitro-2-cresol | [22] | 1.72 | 1.54 | 0 | 0 | 1 | 1 |
| 81 | 3-methyl-4-nitrophenol | [22] | 1.73 | 1.05 | 0 | 0 | 0 | 0 |
| 82 | 2,4-chloro-6-nitrophenol | [22] | 1.75 | 1.56 | 0 | 0 | 1 | 1 |
| 83 | 2,3,4,5-tetrachloronitrobenzene | [12] | 1.78 | 1.87 | 1 | 1 | 1 | 1 |
| 84 | 2,3,5,6-tetrachloronitrobenzene | [12] | 1.82 | 1.93 | 1 | 1 | 1 | 1 |
| 85 | 4-nitrodiphenylamine | [18] | 1.89 | 1.66 | 0 | 1 | 0 | 0 |
| 86 | 4-chloro-2-nitrophenol | [22] | 2.05 | 1.31 | 0 | 0 | 0 | 0 |
| 87 | 6-iodo-1,3-dinitrobenzene | [12] | 2.12 | 2.03 | 1 | 1 | 1 | 1 |
| 88 | 2,4,6-trichloro-1,3-dinitrobenzene | [12] | 2.19 | 2.36 | 1 | 1 | 1 | 1 |
| 89 | 1,2-dinitro-4,5-dichlorobenzene | [12] | 2.21 | 2.16 | 1 | 1 | 1 | 1 |
| 90 | 6-bromo-1,3-dinitrobenzene | [12] | 2.31 | 1.99 | 1 | 1 | 1 | 1 |
| 91 | 2,4,5-trichloro-1,3-dinitrobenzene | [12] | 2.59 | 2.29 | 1 | 1 | 1 | 1 |
| 92 | 4,6-dichloro-1,2-dinitrobenzene | [12] | 2.42 | 2.09 | 1 | 1 | 1 | 1 |
| 93 | 2,3,5,6-tetrachloro-1,4-dinitrobenzene | [12] | 2.74 | 2.58 | 1 | 1 | 1 | 1 |
| 94 | 1,3-dimethyl-2-nitrobenzene | [12] | 0.3 | 0.62 | 0 | 0 | 0 | 0 |
| 95 | 2,3-dimethylnitrobenzene | [12] | 0.56 | 0.66 | 0 | 0 | 0 | 0 |
| 96 | 3,5-dichloronitrobenzene | [20] | 1.13 | 1.19 | 1 | 0 | ||
| 97 | 4-chloro-3-nitrophenol | [20] | 1.27 | 1.24 | 0 | 0 | ||
| 98 | 4,5-dichloro-2-nitroaniline | [20] | 1.66 | 1.44 | 1 | 1 | ||
| 99 | 3-chloro-4-fluoronitrobenzene | [20] | 0.8 | 1.01 | 1 | 0 | ||
| 100 | 2.5-dichloronitrobenzene | [20] | 1.13 | 1.29 | 1 | 0 | ||
| 101 | 2,4-dinitroaniline | [20] | 0.72 | 1.13 | 1 | 1 | ||
| 102 | 1,2,3-trifluoro-4-nitrobenzene | [20] | 1.89 | 0.99 | 1 | 1 | ||
| 103 | 4-(tert)butyl-2,6-dinitrophenol | [20] | 1.8 | 1.71 | 0 | 1 | ||
| 104 | 2,3,4,6-tetrafluoronitrobenzene | [20] | 1.87 | 1.35 | 1 | 1 | ||
| 105 | 1-chloro-2,4-dinitrobenzene | [20] | 2.16 | 1.61 | 1 | 1 | ||
| 106 | 2,4-dinitro-1-fluorobenzene | [20] | 1.71 | 1.27 | 1 | 1 | ||
| 107 | pentafluoronitrobenzene | [20] | 2.43 | 1.61 | 1 | 1 | ||
| 108 | 1,5-difluoro-2,4-dinitrobenzene | [20] | 2.08 | 1.70 | 1 | 1 | ||
| 109 | 4-chloro-3,5-dinitrobenzonitrile | [20] | 2.66 | 1.51 | 1 | 1 | ||
| 110 | 2-methyl-5-nitrophenol | [20] | 0.66 | 0.71 | 0 | 0 | ||
| 111 | 4-nitrophenyl-phenyl-ether | [20] | 1.58 | 1.29 | 1 | 0 | ||
| 112 | 2-nitroanisole | [20] | −0.07 | 0.76 | 1 | 0 | ||
| 113 | 3-methyl-2-nitrophenol | [20] | 0.61 | 0.81 | 0 | 0 | ||
| 114 | 5-methyl-2-nitrophenol | [20] | 0.59 | 0.81 | 0 | 0 | ||
| 115 | 2,5-difluoronitrobenzene | [20] | 0.33 | 0.89 | 1 | 0 | ||
| 116 | 2,4-dibromo-6-nitroaniline | [20] | 1.62 | 1.58 | 1 | 1 | ||
| 117 | 4-hydroxy-3-nitrobenzaldehyde | [20] | 0.61 | 0.40 | 0 | 0 | ||
| 118 | 2-chloro-6-nitrobenzaldehyde | [20] | 0.16 | 0.42 | 1 | 1 | ||
| 119 | 4-nitro-catechol | [20] | 1.17 | 0.58 | 0 | 0 | ||
| 120 | 1,2-dimethyl-4-nitrobenzene | [20] | 0.59 | 0.69 | 0 | 0 | ||
| 121 | 4-ethoxy-2-nitroaniline | [20] | 0.76 | 0.87 | 1 | 0 | ||
| 122 | 2-nitroaniline | [20] | 0.08 | 0.40 | 1 | 0 | ||
| 123 | 1-fluoro-3-iodo-5-nitrobenzene | [20] | 1.09 | 1.14 | 1 | 0 | ||
| 124 | 2-chloro-4-nitroaniline | [20] | 0.75 | 0.87 | 1 | 0 | ||
| 125 | 2,4-dichloro-6-nitroaniline | [20] | 1.26 | 1.45 | 1 | 1 | ||
| 126 | 2-chloro-5-nitrobenzaldehyde | [20] | 0.53 | 0.60 | 1 | 0 | ||
| 127 | 2,6-dinitroaniline | [20] | 0.84 | 0.80 | 1 | 1 | ||
| 128 | 6-chloro-2,4-dinitroaniline | [20] | 1.12 | 1.32 | 1 | 1 | ||
| 129 | 2-bromo-4,6-dinitroaniline | [20] | 1.24 | 1.54 | 1 | 1 | ||
| 130 | methyl-4-nitrobenzoate | [20] | 0.39 | 0.08 | 1 | 0 | ||
| 131 | dimethylnitroterephthalate | [20] | 0.43 | −0.26 | 1 | 1 | ||
| 132 | 2-nitroresorcinol | [20] | 0.66 | 0.43 | 0 | 0 | ||
| 133 | methyl-4-chloro-2-nitrobenzoate | [20] | 0.82 | 0.49 | 1 | 1 | ||
| 134 | 1-fluoro-2-nitrobenzene | [20] | 0.23 | 0.53 | 1 | 0 | ||
| 135 | 3-hydroxy-4-nitrobenzaldehyde | [20] | 0.27 | 0.39 | 0 | 0 | ||
| 136 | 4,5-difluoro-2-nitroaniline | [20] | 0.75 | 0.87 | 1 | 1 | ||
| 137 | 3-nitro-2-hydroxybenzaldehyde | [20] | 0.87 | 0.12 | 0 | 0 | ||
| 138 | 5-chloro-2-nitrobenzamide | [20] | −0.32 | 0.10 | 1 | 1 | ||
| 139 | 1-fluoro-3-nitrobenzene | [20] | 0.2 | 0.45 | 1 | 0 | ||
| 140 | 4-methyl-2-nitroaniline | [20] | 0.37 | 0.79 | 0 | 0 | ||
| 141 | 2-methyl-4-nitroaniline | [20] | 0.49 | 0.84 | 0 | 0 | ||
| 142 | 4-amino-3,5-dinitrobenzamide | [20] | 0.51 | 0.47 | 1 | 1 | ||
| 143 | 2-chloro-5-nitroaniline | [20] | 0.6 | 0.96 | 1 | 0 | ||
| 144 | 2-methyl-3-nitrophenol | [20] | 0.78 | 0.58 | 0 | 0 | ||
| 145 | 2-bromo-2'-hydroxy-5'-nitroacetanilide | [20] | 0.87 | 0.94 | 0 | 0 | ||
| 146 | 3-fluoro-4-nitrophenol | [20] | 0.93 | 0.68 | 0 | 0 | ||
| 147 | 3,5-dinitroaniline | [20] | 0.94 | 0.94 | 1 | 1 | ||
| 148 | 4-bromo-2-fluoro-6- v nitroanisole | [20] | 0.97 | 1.63 | 1 | 1 | ||
| 149 | 4-chloro-2,6-dinitroaniline | [20] | 1.15 | 1.68 | 1 | 1 | ||
| 150 | 4-chloro-3-nitrobenzaldehyde | [20] | 1.19 | 0.72 | 1 | 0 | ||
| 151 | 2-amino-6-chloro-4-nitrophenol | [20] | 1.2 | 0.76 | 0 | 1 | ||
| 152 | 3,5-dinitrobenzonitrile | [20] | 1.22 | 1.21 | 1 | 1 | ||
| 153 | 1,2,3-trichloro-5-nitrobenzene | [20] | 1.55 | 1.56 | 1 | 1 | ||
| 154 | 4-bromo-2-fluoro-6-nitrophenol | [20] | 1.62 | 1.20 | 0 | 1 | ||
| 155 | 2,4-dinitro-5-fluoroaniline | [20] | 1.69 | 1.45 | 1 | 1 | ||
| 156 | 4-chloro-3-nitrobenzonitrile | [20] | 1.71 | 0.94 | 1 | 0 | ||
| 157 | 5-chloro-2,4-dinitrotoluene | [20] | 2.33 | 2.14 | 0 | 1 | ||
| 158 | 1,3-dichloro-4,6-dinitrobenzene | [20] | 2.72 | 2.13 | 1 | 1 | ||
| 159 | 1,3,5-trinitrobenzene | 1.37 | 1 | 1 | ||||
| 160 | 2,4,6-trinitrotoluene | 1.46 | 0 | 1 | ||||
| 161 | pentachloronitrobenzene | 2.16 | 1 | 1 | ||||
| 162 | 1,2,4-trichloro-6-nitrobenzene | 1.68 | 1 | 1 | ||||
| 163 | 1,2,4-trichloro-3-nitrobenzene | 1.53 | 1 | 1 | ||||
| 164 | 1,2,3,5-tetrachloro-4-nitrobenzene | 1.93 | 1 | 1 | ||||
| 165 | 3-methyl-2,4-dinitrophenol | 1.38 | 0 | 1 | ||||
| 166 | 3,5-dinitro-p-cresol | 1.01 | 0 | 1 | ||||
| 167 | 3-methyl-4,6-dinitrophenol | 1.57 | 0 | 1 | ||||
| 168 | nitroglycerin | 0.65 | 1 | 1 | ||||
| 169 | 2-amino-4,6-dinitrotoluene | 1.13 | 0 | 1 | ||||
| 170 | 4-hydroxylamino-2,6-dinitrotoluene | 1.2 | 0 | 1 | ||||
| 171 | ANTA (3-nitro-1,2,4-triazol-5-amine) | 0.29 | 1 | 1 | ||||
| 172 | CL-14 | 1.29 | 1 | 1 | ||||
| 173 | CL-20 | 1.1 | 1 | 1 | ||||
| 174 | HMX | 0.47 | 1 | 1 | ||||
| 175 | 3-nitro-1,2,4-triazol-5-ol | 0.16 | 0 | 1 | ||||
| 176 | 1-octanol | 0.58 | 0.89 | 0 | 1 | |||
| 177 | pentryl | 0.45 | 1 | 1 | ||||
| 178 | RDX | 0.27 | 1 | 1 | ||||
| 179 | Tetryl | 1.36 | 1 | 1 | ||||
| 180 | diaminoazofurazan | −0.02 | 1 | 1 | ||||
| 181 | K-55 | 0.65 | 1 | 1 | ||||
| 182 | dinitroglycouril | 0.4 | 1 | 1 | ||||
| 183 | 2,4-dinitroimidazole | 0.29 | 1 | 1 | ||||
| 184 | 3-nitro-1,2,4-triazol-5-one | 0.25 | 1 | 1 | ||||
| 185 | 2-nitroimino-5-nitro-hexahydro-1,3,5-triazine | 0.42 | 1 | 1 | ||||
| 186 | nitrosobenzene | 0.09 | 1 | 0 | ||||
| 187 | 2,3-dinitrotoluene | 1.27 | 0 | 1 | ||||
| 188 | 2,5-dinitrotoluene | 1.43 | 0 | 1 | ||||
| 189 | 2,3,4-trinitrotoluene | 1.85 | 0 | 1 | ||||
| 190 | 2,3,6-trinitrotoluene | 1.98 | 0 | 1 | ||||
| 191 | 2,4,5-trinitrotoluene | 2.26 | 0 | 1 | ||||
| 192 | 1,3,5-trinitrosobenzene | 0.41 | 1 | 0 | ||||
| 193 | 1,3-dinitrosobenzene | 0.12 | 1 | 0 | ||||
| 194 | 1,3-dinitro-5-nitroso-1,3,5-triazine | 0.6 | 1 | 1 | ||||
| 195 | 1,3-dinitroso-5-nitro-1,3,5-triazine | 0.45 | 1 | 1 | ||||
| 196 | 1,3,5-trinitroso-1,3,5-triazine | 0.42 | 1 | 1 | ||||
| 197 | 1-hydroxylamino-3,5dinitro-1,3,5-triazine | 0.52 | 1 | 1 | ||||
| 198 | 1-hydroxylamino-3-nitroso-5-nitro-1,3,5-triazine | 0.3 | 0 | 1 | ||||
| 199 | 1-hydroxylamino-3,5-dinitroso-1,3,5-triazine | 0.17 | 0 | 1 | ||||
| 200 | EGDN | 0.41 | 1 | 1 | ||||
| 201 | 1,4-dinitrosobenzene | 0.33 | 1 | 0 | ||||
| 202 | 3-amino-2,6-dinitrotoluene | 1.46 | 0 | 1 | ||||
| 203 | 2-hydroxilamino-4,6-dinitrotoluene | 1 | 0 | 1 | ||||
| 204 | 2,4,6-trinitroanisole | 1.98 | 1 | 1 | ||||
| 205 | FOX-7 | 0.3 | 1 | 1 | ||||
| 206 | HBT | 0.06 | 1 | 1 | ||||
Notes: Obs. – observed value;
Pred. – Predicted values of −lg(IGC50) obtained using consensus model 16;
An additional dataset of nitroaromatic compounds [20] was identified after the completion of the modelling studies reported herein. After identification and removal of 78 duplicates, remaining 63 compounds from this dataset were used for external validation of the developed models. All the structures (including nitro group representation) and related activity values were carefully curated and checked according to procedures described by Fourches et al. [24].
2.2. HiT QSAR
All studies using real values of toxicity were completed with the HiT QSAR software based on Simplex representation of molecular structure (SiRMS) [25]. This method afforded good results in previous studies for solving different "structure-activity" problems [26–30]. 2D Simplex descriptors (number of tetratomic fragments with fixed composition and topology) were used for molecular structure representation. Thus, SiRMS accounts not only for the atom type, but also for other atomic characteristics that may impact biological activity of molecules, e.g., partial charge [31], lipophilicity [32], refraction [33], and atom ability for being a donor/acceptor in hydrogen-bond formation (H-bond). For atom characteristics, which have real values (charge, lipophilicity, refraction) the division of the entire value range into definite discrete groups has been carried out [34]. The number of groups is a tuning parameter and can be varied. In the present study the atoms have been divided into groups corresponding to their (i) partial charge A≤−0.3<B≤−0.1<C≤0<D≤0.03<E≤0.07<F≤0.2<G, (ii) lipophilicity A≤−1.6<B≤−0.35<C≤0.04<D≤0.05<E≤0.3<F≤1.6<G and (iii) refraction A≤1.5<B≤3<C≤8<D. For H-bond characteristics, the atoms have been divided into three groups: A (acceptor of hydrogen in H-bond), D (donor of hydrogen in H-bond), and I (indifferent atom). The usage of sundry variants of differentiation of simplex vertexes (atoms) represents the principal feature of this approach. The main advantages of SiRMS are its ability to analyze molecules with significant apparent structural differences as well as the possibility to reveal individual molecular fragments (simplex combinations) promoting or interfering with the investigated antiviral activity [35].
2.3. Statistical approaches
Because of a large number of simplex descriptors generated in the HiT QSAR approach, Partial Least Squares or Projections to Latent Structures (PLS) method [36, 37] was used for statistical model development. Genetic algorithm [38], trend-vector method [39–41] and automatic variable selection strategy [25] based on interactive [42] and evolutionary [43] variables selection were used for descriptor selection in PLS. Briefly this scheme can be represented in the following way: elimination of non-significant and highly correlated descriptors →TV procedure→AVS ↔ GA → partial or complete enumeration methods → best QSAR model. Selection of the best QSAR model on every stage of this process was carried out according to maximum of Fitness Function (FF) criterion, where FF = R2 + 2Q2 and FF → max, i.e. the best selected QSAR model represents the model with the maximum FF value [44]. After the selection of these best models, they have been validated using corresponding external test set, i.e., selected test set compounds were eliminated from initial work set before QSAR modeling and were used only for validation of the chosen models. R2 and Q2 are parameters of model goodness-of-fit and robustness, correspondingly. They are obligatory but insufficient conditions of model acceptance. Certainly, neither R2 nor Q2 are not the characteristic of model quality (predictivity), which is related to model ability to predict activity for compounds from external test set. General scheme of the PLS models generation and selection applied in HiT QSAR can be found in the literature [25].
Classification models were built with the Classification and Regression Trees (CART) approach [45], which is a nonparametric statistical method of analysis. In the CART approach, the resulting models represent hierarchical sets of rules based on parameters selected for the description of the investigated property. The rule represents an "IF-THEN" logical construction. For example, the simple rule can be "IF lipophilicity > 3 THEN compound is active". In fact, such a model is realized as a set of consecutive nodes, and each of them contains certain sets of compounds which correspond to that node’s rule. The CART method has several advantages: obtaining intuitively understandable models using natural language, quick learning and predicting processes, nonlinearity of models, and the ability to develop models using ranked values of the activity (it allows for the analysis of sets of compounds with heterogeneous experimental activity values).
2.4. Consensus approaches and model comparison
In modern QSAR analysis the most effective predictions are realized as the result of using consensus approaches [46–49], i.e., when several single models are used concurrently. In this study, the prognosis of activity/property was developed by averaging (using different schemes, see Results and Discussion) the predictions generated by an ensemble of QSAR models. The success of the consensus approach depends on the selection of models; it is expected that the use of multiple models leads to the compensation of errors thereby improving the overall predictive power as compared to any contributing single model. Most probably, it is related to the fact that a multidimensional response surface (activity, property) in structural space is locally non-linear with multiple maxima and minima. Hence, any single QSAR model, even a non-linear one, as a rule, is not able to approximate such a complicated response surface. Evidently, the combination of different QSAR models affords a more successful approximation. The power of consensus approach has been validated in the study [20] where consensus model was constructed by averaging all available predicted values taking into account the AD of each individual model. The advantage of this data treatment is that the overall coverage of the prediction is still high because it was rare to have an external compound outside of the ADs of all available models. The authors [20] demonstrated superior performance of the consensus modeling approach. Both the predictive accuracy and coverage of the final consensus QSAR models were superior as compared to these parameters for individual models. Moreover, the coverage of this consensus model was actually 100% for all three data sets. Thus, the consensus models appeared robust in terms of being insensitive to both incorporating individual models with low prediction accuracy and the inclusion or exclusion of the AD [48]. However, consensus modeling is not a panacea and it cannot establish the relationship between the structure and activity in case of its absence.
Models included in the ensemble used for consensus prediction should be different in terms of how they relate compound descriptors to the target property(-ies). Thus, the problem of estimation of similarity/difference between QSAR models becomes important. Each individual model has its own structural space defined by the descriptors involved. The method used for the estimation of models’ similarity/diversity was developed by Todeschini [50, 51]. In this approach similarity/dissimilarity of models has been estimated as Hamming distance between binary vectors, where 0 and 1 reflected the absence or presence of specific descriptor. Such estimation, in our opinion, is too rough, because the degree and direction of influence of selected descriptors on the investigated property are not taken into account. Moreover, this approach is not applicable to models containing the same or completely different descriptors.
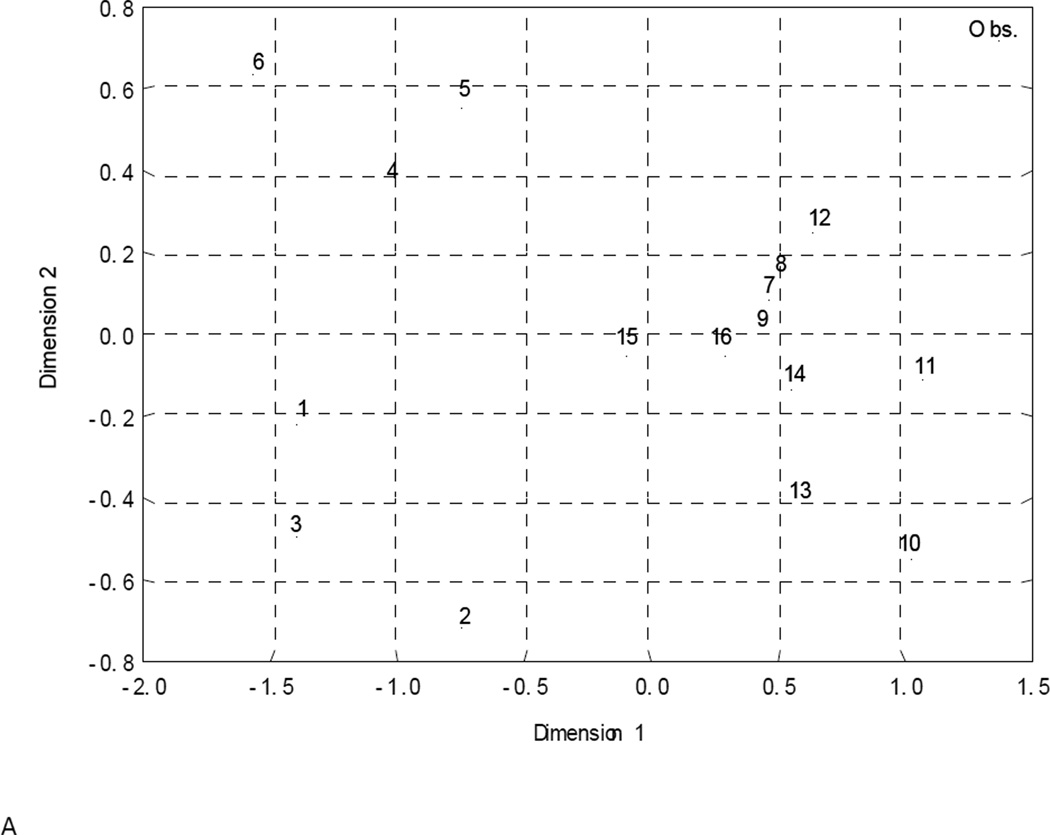
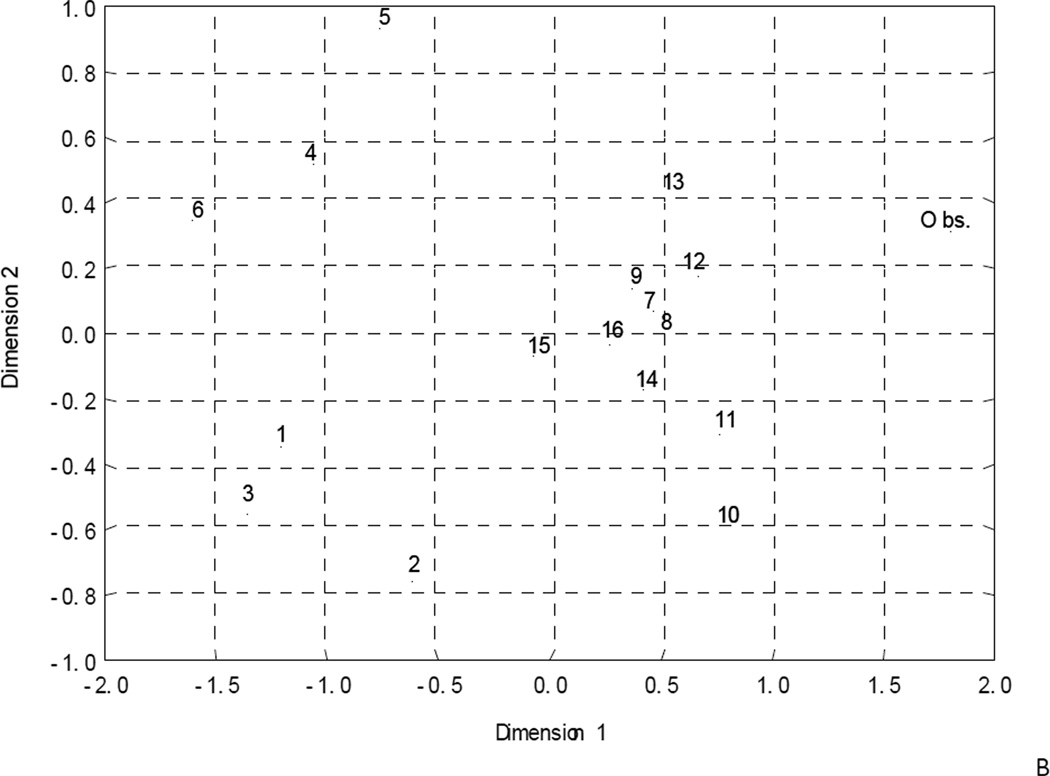
Herein, we have employed an alternative method for estimation of the similarity/dissimilarity for a model, in which the models are compared in the specially defined space. Consider an n×m matrix, where n is the number of models selected including the consensus and "experimental" (observed values of investigated activity) ones and m is the number of molecules. Thus, each model will be presented in m-dimensional molecular space by vectors formed by m molecules used in QSAR modeling. Each of the training or test set molecules specifies the basis vector; its components are calculated from each molecule’s activity values predicted by every selected model. Then, one can compare different models using either a correlation coefficient (normalized characteristic of similarity/difference of QSAR models) or Euclidean distance between vectors obtained. This approach could be applied to both training or test set molecules. Since the observed (experimental) activity values are also considered, it is obvious, that the correlation coefficients or Euclidean distances between the model vectors are the characteristics of the quality of approximation of investigated activity by selected QSAR models. Both approaches (correlation coefficient and Euclidean distance) have been used in this study to estimate the similarity/difference of QSAR models in molecular space. Distribution of the QSTR models obtained was visualized in two-dimensional space (Fig. 1) using multidimensional scaling [52] of these matrices.
Figure 1.
QSAR models similarity/dissimilarity in the structural space estimated by correlation (A, C) and Euclidian distances methods (B, D) for modeling (A, B) and external test (C, D) sets.
2.5. Applicability domain
According to the OECD principles [53] the estimation of applicability domain (AD) for all QSAR models is strictly required. However, correct estimation of AD is still one of the biggest challenges in QSAR analysis [47, 54]. In this study, we have applied a new algorithm for estimation of AD that involves the following steps:
Generation of the distance matrix between training set molecules in the descriptor space of each QSAR model (the coordinates of latent variables in the PLS model were used).
Detection of the shortest distances between molecules using the aforementioned matrix and subsequent building of minimal spanning tree (see [55] for details) for all training set molecules. This tree characterizes clustering of molecules in the structural space.
Finding the average distance (dav) and its root-mean-square deviation (σ) for the spanning tree. This distance could be regarded as a measure of the average density of molecular distribution in the structural space.
Finally, test set molecules are projected onto the space of latent variables. If the distance between test set molecule and the nearest training set point is larger than dav+3σ, this molecule is considered outside of the AD. Respectively, molecules belonging to the AD are situated at the distance smaller than dav+3σ from the training set points.
This scheme of AD estimation is shown in Fig. 2. Training set compounds projected onto two-dimensional space of PLS latent variables T1 T2 are represented as points. Each point is connected with its nearest neighbor in this structural space. Altogether these points create minimal spanning tree. Each point is surrounded by sphere with radius R= dav+3σ. The ensemble of such spheres created local AD. Integral AD for the same model is depicted by big oval. External set compounds within the local AD are depicted by stars and molecules falling outside local AD - by "X". This approach for AD estimation is similar, in some ways, to the methods described in [56]. It defines AD locally as opposed to integral approaches, e.g., [35], that usually define the AD in terms of the convex region (hyper-sphere or polyhedron or ellipsoid) in the multidimensional descriptor space, which could contain vast cavities. As obvious from Fig. 2, local AD is defined by the union of regions surrounding every training set point and contains no ambiguous cavities.
Figure 2.
Local AD approach.
3. Results and discussion
More than 12,000 simplex descriptors were generated for the initial modeling set of 95 molecules (Table 2). Exclusion of non-significant and highly correlated (r ≥ 0.9) descriptors reduced the number of descriptors to ca. 2000 that were used for subsequent PLS modeling. Within the total set, two overlapping subsets of 41 structures known to cause oxidative stress in living cells (mechanism A) and 48 compounds known to participate in nucleophilic attack (mechanism B) were selected [9]. The third set consisted of 35 compounds not included in either subsets A or B. We expected that taking into account the mode of action (mechanism A or B) would allow us to generate robust and predictive QSAR models. Indeed, successful QSAR models 7, 1 and 4 (Table 3) were obtained for both the entire training set and subsets A and B, respectively. The model obtained for the third set (compounds not belonging to either mechanism A or B) was rejected because of the lack of predictivity as described below. 1000 rounds of Y-scrambling test for the whole dataset (95 compounds) resulted in Q2YS ≤ 0.35 indicating the absence of chance correlations.
Table 3.
Statistics of QSAR models
| Model | Modeling set |
R2 | Q2 | R2test (ext folds) |
Sws | Scv | Sts | A | M | D | R2test (other) |
R2test (ext) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Subset A (redox cyclers) |
0.91 | 0.86 | 0.23 | 0.28 | 2 | 48 | 27 | 0.27 | |||
| 2 | 0.88 | 0.81 | 0.74 | 0.28 | 0.35 | 0.33 | 1 | 38 | 96 | 0.39 | ||
| 3 | 0.92 | 0.86 | 0.57 | 0.23 | 0.30 | 0.48 | 2 | 38 | 39 | 0.24 | ||
| 4 | Subset B (nucleophilic attack) |
0.92 | 0.83 | 0.25 | 0.39 | 2 | 41 | 59 | 0.22 | |||
| 5 | 0.95 | 0.79 | 0.61 | 0.22 | 0.45 | 0.49 | 2 | 33 | 95 | 0.22 | ||
| 6 | 0.93 | 0.68 | 0.72 | 0.24 | 0.53 | 0.52 | 2 | 33 | 154 | 0.07 | ||
| 7 | Whole set (all 95 compounds) |
0.84 | 0.77 | 0.32 | 0.38 | 2 | 95 | 31 | ||||
| 8 | 0.84 | 0.75 | 0.86 | 0.32 | 0.40 | 0.30 | 2 | 76 | 58 | |||
| 9 | 0.85 | 0.74 | 0.74 | 0.31 | 0.41 | 0.38 | 2 | 76 | 47 | |||
| 10 | 0.82 | 0.73 | 0.70 | 0.33 | 0.41 | 0.47 | 3 | 76 | 24 | |||
| 11 | 0.90 | 0.84 | 0.67 | 0.25 | 0.31 | 0.49 | 3 | 76 | 28 | |||
| 12 | 0.82 | 0.73 | 0.77 | 0.34 | 0.41 | 0.38 | 2 | 76 | 39 | |||
| 13 | 0.86 | 0.80 | 0.64 | 0.29 | 0.35 | 0.48 | 3 | 76 | 33 | |||
| 14* | mechanism-based consensus model (models 1–6) |
0.91 | 0.76 | 0.23 | 0.38 | 63 | 154 | 0.64 | ||||
| 15 | mechanism-free consensus model (models 7–13) |
0.83 | 0.76 | 0.33 | 0.38 | 95 | 119 | 0.54 | ||||
| 16 | global consensus model (models 1–13) |
0.85 | 0.77 | 0.31 | 0.37 | 95 | 257 | 0.65 |
R2 - determination coefficient for training set;
Q2 – cross-validation determination coefficient for training set;
R2test- determination coefficient for test set formed by corresponding external fold;
R2test(other) - determination coefficient for set of other mechanisms;
Sws - standard error of a prediction for training set;
Scv - standard error of prediction for training set in cross-validation terms;
Sts - standard error of a prediction for test set;
A - number of PLS latent variables;
D - number of descriptors;
M - number of molecules in the training set;
R2test(ext) - determination coefficient for external test set;
Coverage of consensus model 14 for training set and external folds is 57%; for external test set – 76%.
The most critical limitation of many traditional QSAR studies is their low external predictive power, i.e., inability to predict accurately the underlying end point toxicity for compounds that were not used for model development. The low external prediction accuracy of QSAR models in spite of their high accuracy on the training set fitting QSAR models is a well known phenomenon named as Kubinyi paradox [57]. There could be many reasons for the discrepancy between internal and external predictive power of QSAR models. The most common is that training set models are based on data interpolation and, therefore, they inherently have limited applicability in the chemical space, whereas any external prediction implies inherent and, frequently, excessive extrapolation of the training set models. To assess the external predictivity of models, the initial dataset (95 compounds) was divided into training and test sets. Approximately 20% of compounds from different groups of activity were randomly selected into the test set [58]; the remaining compounds were assigned to the training set.
In addition, an external test set was generated by selecting a subset of 10 or 8 compounds (~20% of mechanism-based subsets A and B respectively) most similar to the corresponding training set (see reference [25] for details). Ultimately, the external predictions were obtained for all molecules of the initial dataset, since each of them belonged to one of the test sets. This approach allows one to minimize the dependence of the predictivity estimation on the test set compounds selection; however, subsequent external validation on independent test set is still highly desirable [46, 49, 58, 59]. Good models 2 (Mechanism A, minimal dissimilarity set), 3 (Mechanism A, randomly chosen set), 5 (Mechanism B, minimal dissimilarity set), and 6 (Mechanism B, randomly chosen set) (Table 3) were obtained for every set, except for models generated for compounds without any known mechanism of action (data not shown). The latter models were shown to be well-fitted and robust; however, they did not show any significant predictive power when applied to test sets and, consequently, they were excluded from subsequent studies. Models 1–6 were also applied to another test set – compounds with unknown mechanism without any success (R2test(other) in Table 3).
Our results (Table 3) indicate that the models obtained for compounds acting via a particular mechanism were unable to predict toxicity of compounds of a different structural class with a different presumed mechanism of toxicity (R2testother = 0.07 – 0.39). In other words, models developed for structures acting via one mechanism could predict external compounds acting via the same mechanisms but lacked predictive power when applied to the molecules possessing different mode of action.
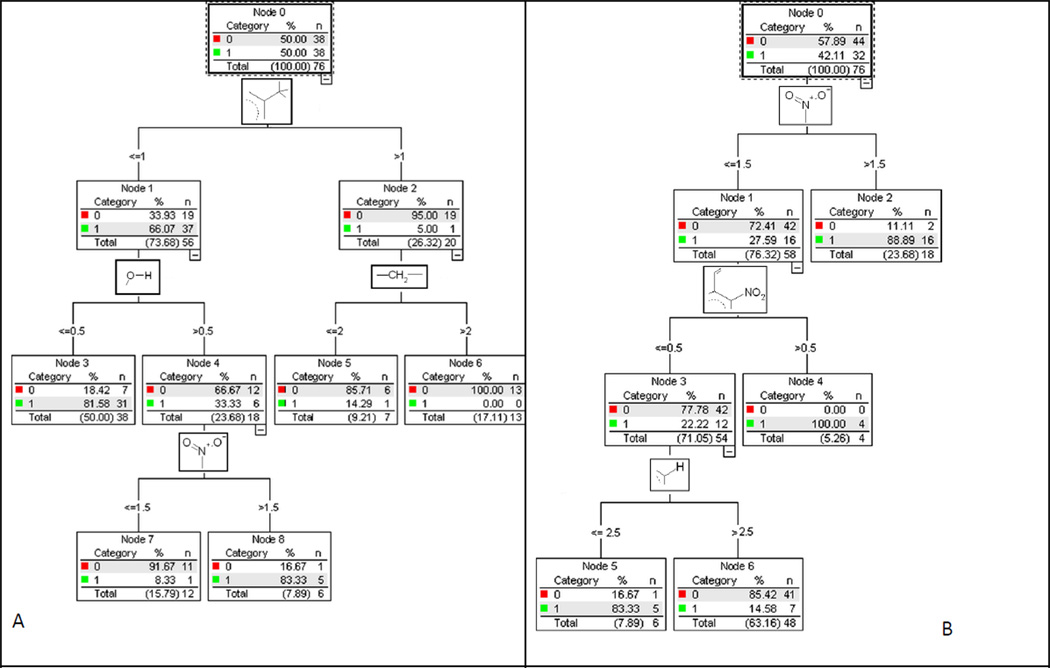
These observations suggest that the putative mechanism of toxic action must be determined to enable the correct prognosis of toxicity using the respective QSAR model. For this purpose, two classification models were obtained using data from [9] (in case of compounds with identified toxicity mechanisms) with the CART approach [45]. The first of them classified nitroaromatic compounds in two classes based on whether the molecule acted via the mechanism A (redox cyclers) or not. Similarly, the second model divides all compounds in two classes, depending on whether the compound acts via mechanism B (nucleophilic attack) or not. Corresponding classification trees (mechanism A vs. not mechanism A and mechanism B vs. not mechanism B) which represent the set of structural rules are shown on the Fig. 3. Compounds belonging to certain mechanism are marked by 1 and others – by 0. Simplex descriptors corresponding to structural rules are graphically represented in the knots of each tree. The final models have only ~15% of misclassification errors. Only 3–4 simplex descriptors have been used to develop each of these models. The models predicted the mechanism of action for test set compounds correctly with 79%–84% accuracy. Using these structural filters (value of depicted simplex descriptors) it is possible to classify any new compound by its mode of toxic action, i.e., mechanism A or B in our case.
Figure 3.
Decision trees for mechanisms A (left) and B (right).
According to selected structural filters (Fig. 3A), compound will act by mechanism A if: (i) it has one or less methyl substituents in the aromatic ring and no hydroxyl groups; (ii) it has one or less methyl substituents in the aromatic ring, one or more hydroxyl groups, and two or more nitro groups. Compound will not act by mechanism A if: (i) it has more than one methyl substituents in the aromatic ring; (ii) it has one or less methyl substituents in the aromatic ring, one or more hydroxyl groups, and less than two nitro groups.
According to selected structural filters (Fig. 3B), compound will act by mechanism B if: (i) it has more than one nitro groups; (ii) it has one nitro group and it has unsaturated hydrocarbon substituent in the aromatic ring next to nitro group; (iii) it has one nitro group, no unsaturated hydrocarbon substituent in the aromatic ring next to nitro group, and four or more substituents in the aromatic ring. Compound will not act by mechanism B if it has one nitro group, no unsaturated hydrocarbon substituent in the aromatic ring next to nitro group, and less than four substituents in the aromatic ring.
In spite of reasonably high accuracy of the mechanism-based models described above they have a natural limitation in terms of chemical diversity of compounds that could be predicted with these models. For instance, as mentioned above, none of the mechanism-based models could accurately predict the toxicity of compounds acting via an alternative mechanism. For this reason, we have considered building global QSAR models for the entire available dataset of diverse 95 chemicals regardless of their mechanism of action. As stated above, we were able to generate a well-fitted QSAR model 7 for the entire dataset (Table 3). Y-scrambling test repeated 1000 times revealed the absence of chance correlations (Q2YS = 0.35). To demonstrate that the global model is externally predictive, a subset of 19 compounds, most similar to the training set (the remaining 76 compounds) was selected for external validation of model 8. In addition, for better evaluation of the predictive power of QSAR models, several other external test sets were used in this analysis following principles of n-fold external cross-validation [60] (n=5 in our case). In this case the entire set of 95 compounds was divided randomly into five non-overlapping subsets and used each subset systematically as an external test set. Robust and predictive models 9–13 (Table 3) were obtained for every external fold.
After our calculations reported above were completed, we have identified toxicity data for 63 additional nitroaromatic compounds 96–158 [20] (Table 2). Three different consensus models 14–16 (Table 3) were used to predict toxicity for this external test set keeping in mind the desire to obtain predictions that are both statistically accurate and, if possible, informative of the underlying mechanism of action. The workflow development of these consensus models is shown at Fig. 4. Models 1–6 were combined in the mechanism-based consensus model 14. In this model, compounds with predicted by CART model (Fig. 3) mechanism A were treated by models 1–3, and compounds with predicted mechanism B – by models 4–6. If both mechanisms were assigned to the same compound, it was predicted by models 1–6. Compounds that were predicted (by CART model) to have unknown mechanism of toxicity (not A and not B) were considered out of AD of mechanism-based consensus model 14. Models 7–13 obtained with all 95 compounds were combined in mechanism-free consensus model 15. All 13 models (1–13) and CART models for mechanisms A (redox cyclers) and B (nucleophilic attack) were used for the development of global consensus model 16. Here, for models 1–6 the similar logic as for mechanism-based consensus model 14 was used (i.e. compounds with predicted by CART model (Fig. 3) mechanism A were treated by models 1–3, 7–12, and compounds with predicted mechanism B – by models 4–6, 7–12). If both mechanisms were assigned to the same compound, it was predicted by all models 1–12, if no mechanisms (not A and not B), compound, it was predicted by only models 7–12. For any consensus model, toxicity of every compound was estimated as an arithmetic average of predicted values from one to thirteen individual QSAR models (i.e., only models for which a compound is found within their AD are used). As can be seen in Table 3, predictivity of global consensus model 16 for the external test set is higher (R2test=0.65) than that of mechanism-free model 15 (R2test=0.54). At the same time mechanism-based consensus model 14 (with similar predictivity, R2test=0.64) covers only 76% of external set. Thus, the results of external validation show that the global consensus model 16, i.e., combination of mechanism-based models with the models obtained using all available compounds gives the best results in both coverage and predictivity.
Figure 4.
Workflow of consensus models development.
Comparative analysis of all sixteen QSAR models has been carried out separately for modeling and external test sets. Estimation of their similarity/difference either by correlation or by Euclidean distance between corresponding activity vectors (see Materials and Methods) shows that, as expected, consensus model 16 has the highest accuracy of prediction based on both similarity metrics. Multidimensional scaling has been used to visualize the results of this analysis (Fig. 1). Although Fig. 1 shows the distribution of pairwise distances between models only approximately, it does reflect the distribution pattern. It is obvious from Fig. 1 that such distribution is substantially dependent not only on the method used to estimate the similarity/dissimilarity (correlation or Euclidean distance) but also on the dataset used. Thus, these distributions are markedly different for the training (Figs. 1A and B) and external sets (Figs. 1C and D). For the modeling set distribution (Fig. 1A and B) models 1–3, 4–6, and 7–15 form separate clusters, but for the external test set (Fig. 1C and D) they are “mixed”.
Since our consensus model 16 was shown to have the highest predictive power and coverage, it was applied to predict toxicity of 48 novel explosives such as RDX, HMX, CL-20, FOX-7, HBT, theirs derivatives, and other compounds of military interest. The results (Table 2) suggest rather moderate levels of toxicity for polychloronitrobenzenes, e.g., pentachloronitrobenzene or 1,2,3,5-tetrachloro-4-nitrobenzene, but higher levels for the newest explosives such as FOX-7 and HBT.
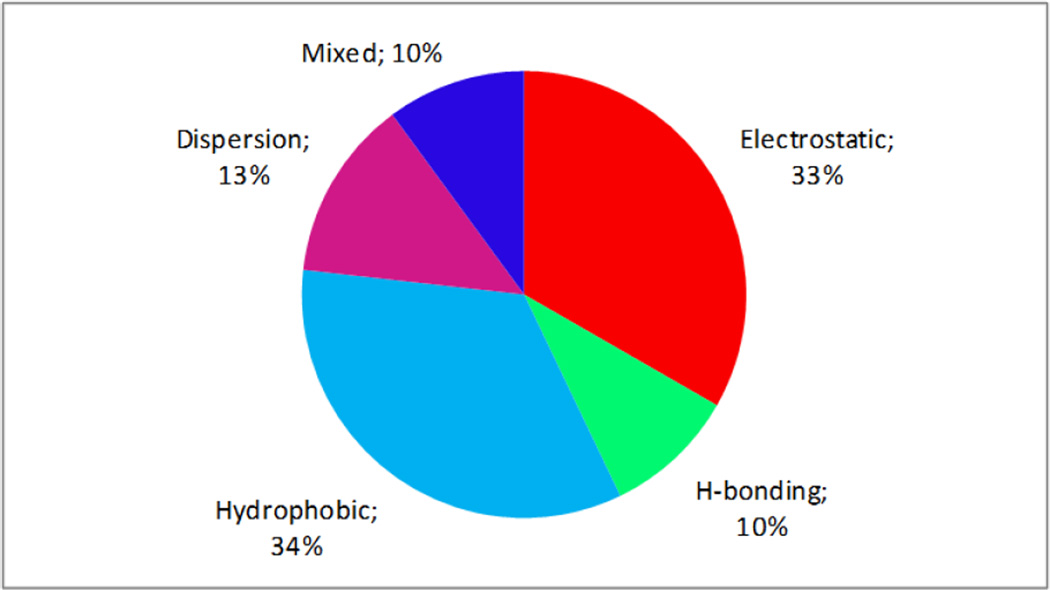
One of the aims of this study was to analyze the role of physico-chemical factors in toxicity variation. Within the framework of SiRMS (like in CoMFA approach [61]) it is possible to define the relative influence of the different physical and chemical factors on the character of the molecules interaction with the biological target [25]. For this purpose it is necessary to summarize and compare the normalized contributions of simplexes in the obtained model separately for every differentiation group. Thus, the relative contribution of simplexes, where differentiation of vertexes corresponds to the partial charges on atoms reflects the role of electrostatic factors; the relative contribution of simplexes, where atoms are differentiated by lipophilicity reflects the role of hydrophobic factors, etc. The results of such analysis show that hydrophobic and electrostatic interactions of toxicants with their biological target are the most important (one third per each) (see Fig. 5). Hence, one can deduce an assumption that compound transport to the site of action (which depends on its lipophilicity) and interaction of nitroaromatic compounds with a biological target (which depends strongly on electrostatic factors and including the reactivity of nitroaromatics) have a big influence on the level of toxicity of investigated compounds. Interestingly that no relationship between log(IGC50) and corresponding Hammett constants (σ, σn, σ0, σ+, σ−) was found for monosubstituted nitroaromatic compounds (R < 0.25).
Figure 5.
Relative influence of some physicochemical factors on variation of toxicity estimated on the basis of consensus model 16.
The contributions of different substituents to toxicity variation of investigated compounds were also estimated using the capabilities of HiT QSAR (Fig. 6). Such average* contributions of substituents to toxicity were estimated for different sets corresponding to mechanisms A (models 1–3) and B (models 4–6) as well as a combined set (models 7–13). As expected (see above), the behavior of all three curves is quite similar on a qualitative level. Generally, one can see that the insertion of the following substituents increases toxicity: Halogen (F<Cl<Br<I), O-Alk (O-CH3<O-C2H5<O-C4H9), methyl, ethyl, chloromethyl, nitro group, and, especially, phenyl and aminophenyl. OH, CHO and COOC2H5 substituents do not show clear pattern as to their influence on toxicity and COOH, CONH2 and CH2OH groups decrease it. As expected, there is no correlation between the substituents’ contributions to toxicity and their Hammett constants (R = −0.24 – 0.08). However, the correlation between toxicity and the substituents’ lipophilicity values is relatively high (R2 = 0.81). These observations may indicate that the toxicity of nitroaromatic compounds mostly depends on their transport properties (defined by their lipophilicity) and their reactivity is less important.
Figure 6.
Contributions of different substituents in benzene ring to nitroaromatics toxicity change.
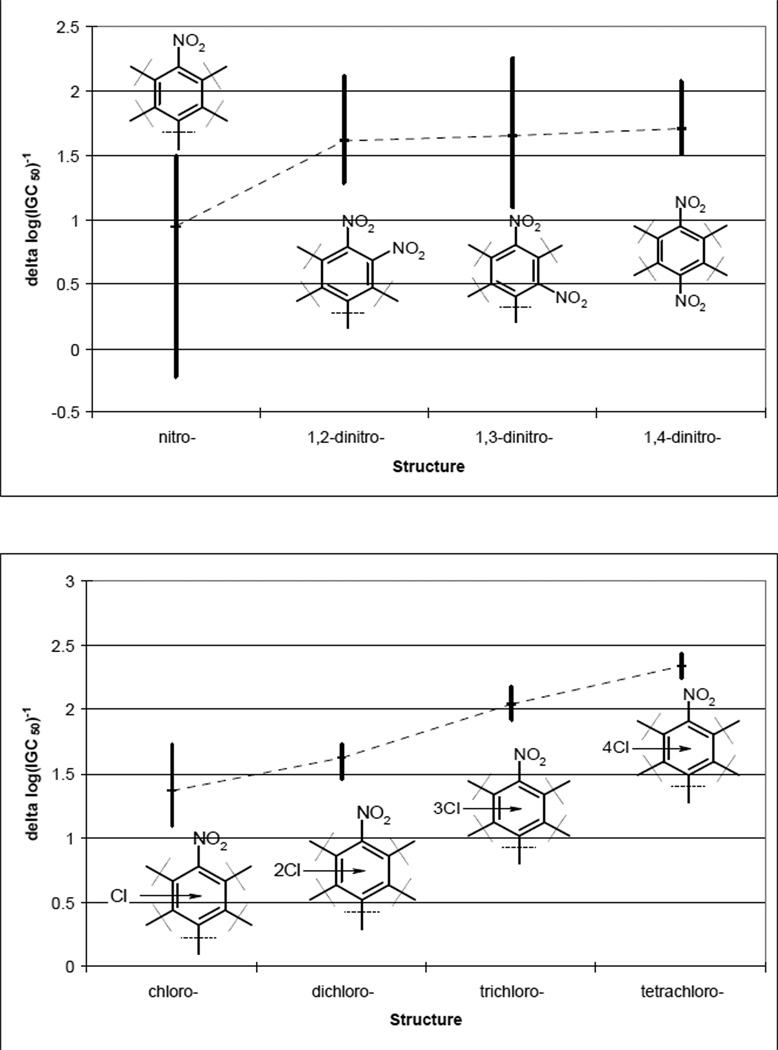
It is important to analyze the contribution of individual chemical functional groups on toxicity. As expected [10], the insertion of the second nitro-group makes compounds more toxic (Fig. 7). The trend of toxicity increase is as follows: ortho-dinitro < meta-dinitro < para-dinitro (Fig. 7). The insertion of chlorine atoms into benzene ring leads to an increased toxicity (Fig. 7). Thus, the most toxic compound amongst those considered in this study is 2,3,5,6-tetrachloro-1,4-dinitrobenzene (Table 2). Relative positions of chlorine atoms are not important – the differences between isomers’ toxicity are minor (Table 2). This situation is significantly different from the trends observed when studying chemical toxicity in rats [10]. In that case, 2,5-dichloronitrobenzene is one of the least toxic compounds, but 2,6-dichloronitrobenzene is highly toxic. The difference between their toxicity is about 2 logarithmic units. The behavior of carboxyl group in the cases mentioned above is also substantially different. COOH group is believed to increase the toxicity in rats [10] but has an opposite effect on toxicity in Tetrahymena pyriformis (Fig. 6). Most probably, features of nitroaromatic compound metabolism are significantly different for these two various biological species. However, it is unlikely to be related to the peculiarities of nitroaromatic compounds concerning nucleophilic substitution or radical reduction. As it was shown before in rats [10] as well as in the present study, polar effects of substituents in benzene ring do not effect toxicity significantly.
Figure 7.
Contributions of insertion of nitro- (a) and chlorine- (b) groups to nitroaromatics toxicity change.
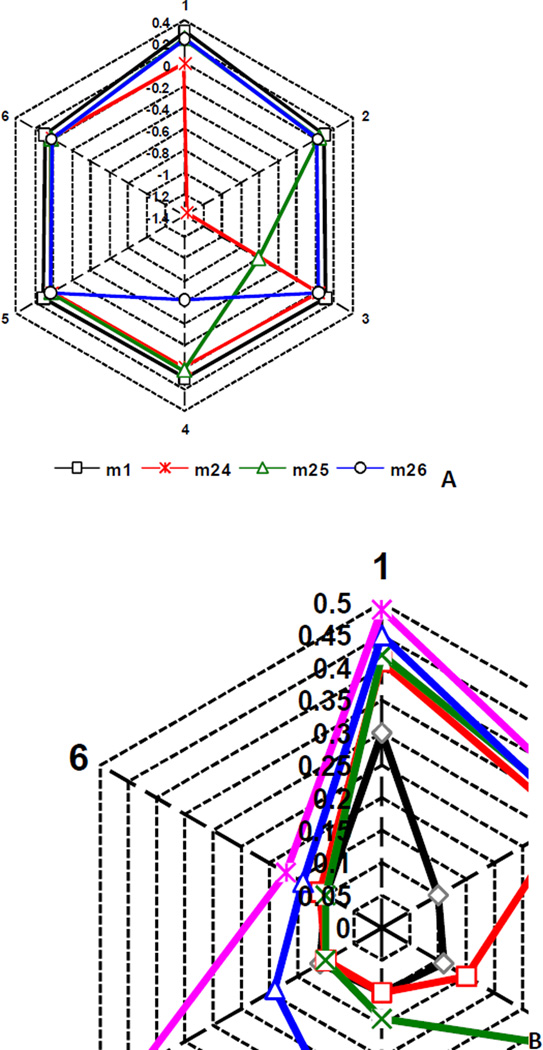
As mentioned previously [10], HiT QSAR approach allows one to estimate the influence of different fragments not only on the investigated property, but also on each other. In Fig. 8, the toxicity of each molecule is represented as six separate contributions (peaks on the hexagon),  where Ci – i-th (i = 1–6) C atom on benzene ring and X – substituent (H, Cl, NO2, COOH). In other words, relative contributions of each carbon of aromatic ring and its substituents were analyzed separately. Peaks corresponding to certain fragments are increasing according to their contribution to toxicity. Unsubstituted nitrobenzene 1 (black) was used as a starting point in all cases. Carbon with nitro group is always in position 1. The strongest mutual influence of substituents on toxicity was observed for isomers of nitrobenzoic acid (compounds 24–26). The results of analyzing this influence are shown in Fig. 8A. As obvious from the Fig. 8A, insertion of carboxyl group in ortho-position to nitro group (compound 24, red) has the most negative influence on toxicity (1.4 logarithmic units). Nitro group toxicity was also decreased in this case (0.3 LU). The situation for two other isomers (compounds 25, green and 26, blue) is nearly the same. Negative contribution of carboxyl group to toxicity is substantially lower (0.6 LU) and positive contribution of nitro group is almost the same as in nitrobenzene.
where Ci – i-th (i = 1–6) C atom on benzene ring and X – substituent (H, Cl, NO2, COOH). In other words, relative contributions of each carbon of aromatic ring and its substituents were analyzed separately. Peaks corresponding to certain fragments are increasing according to their contribution to toxicity. Unsubstituted nitrobenzene 1 (black) was used as a starting point in all cases. Carbon with nitro group is always in position 1. The strongest mutual influence of substituents on toxicity was observed for isomers of nitrobenzoic acid (compounds 24–26). The results of analyzing this influence are shown in Fig. 8A. As obvious from the Fig. 8A, insertion of carboxyl group in ortho-position to nitro group (compound 24, red) has the most negative influence on toxicity (1.4 logarithmic units). Nitro group toxicity was also decreased in this case (0.3 LU). The situation for two other isomers (compounds 25, green and 26, blue) is nearly the same. Negative contribution of carboxyl group to toxicity is substantially lower (0.6 LU) and positive contribution of nitro group is almost the same as in nitrobenzene.
Figure 8.
The analysis of structural fragment influence on toxicity for some substituted nitrobenzenes.
Similar analysis was carried out for chlorine-substituted nitrobenzenes 4 (red), 56 (green), 73 (blue) and 83 (purple) (Fig. 8B). The influence of an increase in the number of chlorine atoms in benzene ring was traced. The effect of each additional chlorine atom on toxicity is bigger than that of the previous chlorine atom. If for 2-chloronitrobenzene 4 the incremental contribution of the chlorine substituent is equal to 0.33 LU, then for the fourth chlorine atom in 2,3,4,5-tetrachloronitrobenzene 83 it increased to 0.43 LU. The relative contribution to toxicity of the nitro group increases in proportion to the number of chlorine substituents (0.30 LU for nitrobenzene 1 and 0.49 LU for 2,3,4,5-tetrachloronitrobenzene 83). The addition of a new chlorine atom also leads to an appreciable increase of contribution to toxicity of the aromatic C-H group nearest to chlorine. Thus, for 2-chloronitrobenzene 4 the contribution of 3-CH group is increased by 0.05 LU in comparison with nitrobenzene 1, and for 2,3,4,5-tetrachloronitrobenzene 83 the contribution of 5-CH is increased by 0.32 LU.
Insertion of the second nitro-group into nitrobenzene effects the contributions to toxicity of not only the initial nitro group but also C-H fragments (Fig. 8C). Contributions of nitro groups are increased in comparison with nitrobenzene 1: for ortho-isomer 66 (red) by 0.13 LU; for meta-isomer 14 (green) by 0.08 LU and for para-isomer 67 (blue) by 0.14 LU. For every C-H fragment, the value of the contribution to toxicity is maximally increased in comparison with nitrobenzene 1 in case of para-isomer 67 on 0.07 LU. Thus, the substituents in the benzene ring affect not only their own contributions to toxicity, but also the effect on toxicity of their neighboring groups and can even increase the contribution to toxicity of C-H groups.
4. Conclusions
Robust 2D QSTR models were obtained for 95 nitroaromatic compounds tested against Tetrahymena pyriformis. Their predictivity was successfully validated on an external test set consisting of 63 nitroaromatic compounds. We have established that the consideration of the possible chemical mechanism of the toxicity of nitroaromatic compounds is not obligatory but is desirable for the development of predictive QSTR models. Toxicities of 48 novel explosives such as RDX, HMX, CL-20, FOX-7, HBT and other compounds of military interest were predicted with the consensus model. The results suggest that most polychloronitrobenzenes are moderately toxic but the toxicity of the newest explosives such as FOX-7 and HBT is expected to be relatively high.
We have examined the influence of structural features of nitroaromatic compound on their toxicity. We found that parameters of substituents in the aromatic ring that characterize their hydrophobicity and ability to be involved in electrostatic interactions are the most significant underlying factors with respect to the compound toxicity. Furthermore, the mutual influence of substituents in the aromatic ring plays an important role in toxicity of nitroaromatic compounds. Contributions of substituents to toxicity are certainly non-additive. Mutual influence of substituents effects activation of aromatic C-H fragments (increasing theirs toxicity) to a considerable degree.
We have succeeded to obtain robust and predictive (R2ext=0.64) mechanism-based (local) QSAR model with limited AD. At the same time mechanism-free consensus model has better coverage but was less predictive. Finally, we have shown that the best results (R2ext=0.65 and 100% coverage) could be obtained by using global consensus model combining predictions made with both local mechanism-based (local) and mechanism-free QSAR models. Thus, approach combining local mechanism-based and mechanism-free QSAR models in one global consensus model allow one to achieve the highest external predictive accuracy and the largest coverage could find general application in chemical toxicity prediction for diverse chemicals.
Acknowledgements
The use of trade, product, or firm names in this report is for descriptive purposes only and does not imply endorsement by the U.S. Government. Results in this study were funded and obtained from research conducted under the Environmental Quality Technology Program of the United States Army Corps of Engineers by the USAERDC. Permission was granted by the Chief of Engineers to publish this information. The findings of this report are not to be construed as an official Department of the Army position unless so designated by other authorized documents. AT acknowledges the support from the NIH grants R21GM076059 and GM66940
Footnotes
Abbreviations: HiT QSAR – Hierarchical Technology for Quantitative Structure - Activity Relationships; IGC50 – 50% inhibition growth concentration; CART – Classification and Regression Trees; QSAR/QSTR – Quantitative Structure Activity/Toxicity Relationship; SiRMS – Simplex representation of molecular structure; PLS – Partial Least Squares or Projections to Latent Structures; AD –applicability domain;
The contributions have been averaged over all corresponding models for all compounds containing substituents in different positions in aromatic ring.
References
- 1.Patai S. The Chemistry of Amino, Nitroso, and Nitro Compounds and Their Derivatives. New York, USA: John Wiley & Sons Inc.; 1982. [Google Scholar]
- 2.Feuer H, Nielsen AT. Nitro Compounds: Recent Advances in Synthesis and Chemistry. New York: VCH Publishing; 1990. [Google Scholar]
- 3.Neilson AH, Allard A-S. Environmental Degradation and Transformation of Organic Chemicals. Boca Raton, Florida: CRC Press; 2008. [Google Scholar]
- 4.Talmage SS, Opresko DM, Maxwell CJ, Welsh CJ, Cretella FM, Reno PH, Daniel FB. Nitroaromatic munition compounds: environmental effects and screening values. Rev. Environ. Contam. Toxicol. 1999;161:1–156. doi: 10.1007/978-1-4757-6427-7_1. [DOI] [PubMed] [Google Scholar]
- 5.Rickert DE. Toxicity of Nitroaromatic Compounds. Bristol, Pennsylvania: Hemisphere Publishing Corp.; 1984. [Google Scholar]
- 6.Robidoux PY, Svendsen C, Caumartin J, Hawari J, Ampleman G, Thiboutot S, Weeks JM, Sunahara GI. Chronic toxicity of energetic compounds in soil determined using the earthworm (Eisenia andrei) reproduction test. Environ. Toxicol. Chem. 2000;19:1764–1773. [Google Scholar]
- 7.Donlon BA, Razo-Flores E, Field JA, Lettinga G. Toxicity of N-substituted aromatics to acetoclastic methanogenic activity in granular sludge. Appl. Environ. Microbiol. 1995;61:3889–3893. doi: 10.1128/aem.61.11.3889-3893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cronin MTD, Schultz TW. Development of Quantitative Structure-Activity Relationships for the Toxicity of Aromatic Compounds to Tetrahymena pyriformis: Comparative Assessment of the Methodologies. Chem. Res. Toxicol. 2001;14:1284–1295. doi: 10.1021/tx0155202. [DOI] [PubMed] [Google Scholar]
- 9.Katritzky AR, Oliferenko P, Oliferenko A, Lomaka A, Karelson M. Nitrobenzene toxicity: QSAR correlations and mechanistic interpretations. J. Phys. Org. Chem. 2003;16:811–817. [Google Scholar]
- 10.Kuz’min VE, Muratov EN, Artemenko AG, Gorb LG, Qasim M, Leszczynski J. The effects of characteristics of substituents on toxicity of the nitroaromatics: HiT QSAR study. J. Comp. Aid. Mol. Des. 2008;22:747–759. doi: 10.1007/s10822-008-9211-x. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal WK, Khadikar PV. QSAR prediction of toxicity of nitrobenzenes. Bioorg. Med. Chem. 2001;9:3035–3040. doi: 10.1016/s0968-0896(01)00211-5. [DOI] [PubMed] [Google Scholar]
- 12.Cronin MTD, Gregory BW, Schultz TW. Quantitative structure-activity analyses of nitrobenzene toxicity to Tetrahumena pyriformis. Chem. Res. Toxicol. 1998;11:902–908. doi: 10.1021/tx970166m. [DOI] [PubMed] [Google Scholar]
- 13.Bearden AP, Schultz TW. Comparison of Tetrahymena and Pimephales Toxicity Based on Mechanism of Action. SAR & QSAR in Env. Res. 1998;9:127–153. doi: 10.1080/10629369808039153. [DOI] [PubMed] [Google Scholar]
- 14.Schultz TV, Lin DT, Arnold LM. QSARs for monosubstituted anilines eliciting the polar narcosis mechanism of action. Sci. Total Environ. 1991;109:569–580. doi: 10.1016/0048-9697(91)90210-6. [DOI] [PubMed] [Google Scholar]
- 15.Schultz TW, Lin DT, Wesley SK. QSARs for monosubstitutued phenols and and the polar narcosis mechanism of toxicity. Qual. Assur. Good Pract. Regul. Law. 1992;1:132–143. [PubMed] [Google Scholar]
- 16.Vaes WHJ, Ramos EU, Verhaar HJM, Hermens JLM. Acute toxicity of nonpolar versus polar narcosis: Is there a difference? Environmental Toxicology and Chemistry. 1998;17:1380–1384. [Google Scholar]
- 17.Estrada E, Uriarte E. Quantitative Structure-Toxicity Relationships Using Tops-Mode. 1. Nitrobenzene Toxicity to Tetrahymena Pyriformis. SAR & QSAR in Env. Res. 2001;12:309–324. doi: 10.1080/10629360108032919. [DOI] [PubMed] [Google Scholar]
- 18.Dearden JC, Cronin MTD, Schultz TW, Lin DT. QSAR study of the toxicity of nitrobenzenes to Tetrahymena pyriformis. QSAR. 1995;14:427–432. [Google Scholar]
- 19.Schmitt H, Altenburger R, Jastorff B, Schuurmann G. Quantitative structure-activity analysis of the algae toxicity of nitroaromatic compounds. Chemical Research in Toxicology. 2000;13:441–450. doi: 10.1021/tx9901635. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko IV. Combinatorial QSAR Modeling of Chemical Toxicants Tested against Tetrahymena pyriformis. J. Chem. Inf. Model. 2008;48:766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
- 21.Cronin MTD, Bryant SE, Dearden JC, Schultz T. Quantitative structure-activity study of the toxicity of benzonitriles to the ciliate Tetrahymena pyriformis. SAR QSAR Environ. Res. 1995;3:1–13. doi: 10.1080/10629369508233989. [DOI] [PubMed] [Google Scholar]
- 22.Cronin MTD, Schultz TW. Structure-toxicity relationships for phenols to Tetrahymena pyriformis. Chemosphere. 1996;32:1453–1468. doi: 10.1016/0045-6535(96)00054-9. [DOI] [PubMed] [Google Scholar]
- 23.Mekenyan O, Roberts DW, Karcher W. Molecular orbital parameters as predictors of skin sensitization potential of halo- and pseudohalobenzenes acting as SNAr electrophiles. Chemical Research in Toxicology. 1997;10:994–1000. doi: 10.1021/tx960104g. [DOI] [PubMed] [Google Scholar]
- 24.Fourches D, Muratov EN, Tropsha A. Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuz’min VE, Artemenko AG, Muratov EN. Hierarchical QSAR technology on the base of Simplex representation of molecular structure. J. Comp. Aid. Mol. Des. 2008;22:403–421. doi: 10.1007/s10822-008-9179-6. [DOI] [PubMed] [Google Scholar]
- 26.Artemenko AG, Muratov EN, Kuz'min VE, Kovdienko NA, Hromov AI, Makarov VA, Riabova OB, Wutzler P, Schmidtke M. Identification of individual structural fragments of N,N'-(bis-5-nitropyrimidyl)dispirotripiperazine derivatives for cytotoxicity and antiherpetic activity allows the prediction of new highly active compounds. J. Antimicrob. Chemother. 2007;60:68–77. doi: 10.1093/jac/dkm172. [DOI] [PubMed] [Google Scholar]
- 27.Kuz’min VE, Artemenko AG, Lozitska RN, Fedtchouk AS, Lozitsky VP, Muratov EN, Mescheriakov AK. Investigation of anticancer activity of macrocyclic Schiff bases by means of 4D-QSAR based on simplex representation of molecular structure. SAR QSAR Environ. Res. 2005;16:219–230. doi: 10.1080/10659360500037206. [DOI] [PubMed] [Google Scholar]
- 28.Kuz’min VE, Muratov EN, Artemenko AG, Gorb LG, Qasim M, Leszczynski J. The effect of nitroaromatics’ composition on their toxicity in vivo: Novel, efficient non-additive 1D QSAR analysis. Chemosphere. 2008;72:1373–1380. doi: 10.1016/j.chemosphere.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 29.Muratov EN, Artemenko AG, Kuz'min VE, Lozitsky VP, Fedchuk AS, Lozitska RN, Boschenko YA, Gridina TL. Investigation of Anti-influenza Activity Using Hierarchic QSAR Technology on the Base of Simplex Representation of Molecular Structure. Antivir. Res. 2005;65:A62–A63. [Google Scholar]
- 30.Muratov EN, Varlamova EV, Artemenko AG, Kuz'min VE, Makarov VA, Riabova OB, Wutzler P, Schmidtke M. QSAR analysis of [(biphenyloxy)propyl]isoxazoles – agents against coxsackievirus B3. Future Med. Chem. 2011;3:31–43. doi: 10.4155/fmc.10.278. [DOI] [PubMed] [Google Scholar]
- 31.Jolly WL, Perry WB. Estimation of atomic charges by an electronegativity equalization procedure calibration with core binding energies. J. Am. Chem. Soc. 1973;95:5442–5450. [Google Scholar]
- 32.Wang R, Fu Y, Lai L. A new atom–additive method for calculating partition coefficients. J. Chem. Inf. Comput. Sci. 1997;37:615–621. [Google Scholar]
- 33.Ioffe BV. Chemistry Refractometric Methods. Himiya, Leningrad: 1983. [Google Scholar]
- 34.Kuz’min VE, Artemenko AG, Lozitsky VP, Muratov EN, Fedtchouk AS, Dyachenko NS, Nosach LN, Gridina TL, Shitikova LI, Mudrik LM, Mescheriakov AK, Chelombitko VA, Zheltvay AI, Vanden Eynde J-J. The analysis of structure- anticancer and antiviral activity relationships for macrocyclic pyridinophanes and their analogues on the basis of 4D QSAR models (simplex representation of molecular structure) Acta Biochim. Polon. 2002;49:157–168. [PubMed] [Google Scholar]
- 35.Kuz’min VE, Artemenko AG, Muratov EN, Volineckaya IL, Makarov VA, Riabova OB, Wutzler P, Schmidtke M. Quantitative Structure-Activity Relationship studies of [(biphenyloxy)propyl]isoxazole derivatives – human rhinovirus 2 replication inhibitors. J. Med. Chem. 2007;50:4205–4213. doi: 10.1021/jm0704806. [DOI] [PubMed] [Google Scholar]
- 36.Lindgren F, Geladi P, Rannar S, Wold S. Interactive variable selection (IVS) for PLS. Part 1: Theory and Algorithms. J. Chemometr. 1994;8:349–363. [Google Scholar]
- 37.Rannar S, Lindgren F, Geladi P, Wold S. A PLS kernel algorithm for data sets with many variables and fewer objects. Part 1: Theory and algorithm. J. Chemometr. 1994;8:111–125. [Google Scholar]
- 38.Hasegawa K, Miyashita Y, Funatsu K. GA strategy for variable selection in QSAR studies: GA-based PLS analysis of calcium channel anatgonists. J Chem Inf Comput Sci. 1997;37:306–310. doi: 10.1021/ci960047x. [DOI] [PubMed] [Google Scholar]
- 39.Carhart RE, Smith DH, Venkataraghavan R. Atom pairs as molecular features in structure - activity studies. Definition and application. J. Chem. Inf. Comput. Sci. 1985;25:64–73. [Google Scholar]
- 40.Kuz'min VE, Artemenko AG, Kovdienko NA, Tetko IV, Livingstone DJ. Lattice model for QSAR studies. J Mol Model. 2000;6:517–526. [Google Scholar]
- 41.Vitiuk NV, Kuz’min VE. Mechanistic models in chemometrics for the analysis of multidimensional data of researches. Analogue of dipole-moments method in the structure (composition)-property relationships analysis. Zh. Anal. Khimii. 1994;49:165–167. [Google Scholar]
- 42.Lindgren F, Geladi P, Rannar S, Wold S. Interactive variable selection (IVS) for PLS. Part 1: Theory and algorithms. J Chemometrics. 1996;8:349–363. [Google Scholar]
- 43.Kubinyi H. Evolutionary variable selection in regression and PLS analyses. J Chemometrics. 1996;10:119–133. [Google Scholar]
- 44.Kuz’min VE, Muratov EN, Artemenko AG, Varlamova EV, Gorb LG, Wang J, Leszczynski J. Consensus QSAR modeling of phosphor-containing chiral AChE inhibitors. QSAR Comb. Sci. 2009;28:664–677. [Google Scholar]
- 45.Breiman L, Friedman JH, Olshen RA, CJ S. Classification and Regression Trees. Wadsworth, Belmont: 1984. [Google Scholar]
- 46.Tropsha A, Gramatica P, Gombar V. The Importance of Being Earnest: Validation is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003;22:69–77. [Google Scholar]
- 47.Muratov EN, Artemenko AG, Varlamova EV, Polischuk PG, Lozitsky VP, Fedtchuk AS, Lozitska RN, Gridina TL, Koroleva LS, Sil'nikov VN, Galabov AS, Makarov VA, Riabova OB, Wutzler P, Schmidtke M, Kuz'min VE. Per aspera ad astra: application of Simplex QSAR approach in antiviral research. Future Med. Chem. 2010;2:1205–1226. doi: 10.4155/fmc.10.194. [DOI] [PubMed] [Google Scholar]
- 48.Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol. Inf. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 49.Tropsha A, Golbraikh A. Predictive QSAR Modeling Workflow Model Applicability Domains and Virtual Screening. Curr. Pharm. Des. 2007;13:3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- 50.Todeschini R, Ballabio D, Consonni V, Manganaro A, Mauri A. Canonical Measure of Correlation (CMC) and Canonical Measure of Distance (CMD) between sets of data. Part 1. Theory and simple chemometric applications. Analytica Chimica Acta. 2009;548:45–51. doi: 10.1016/j.aca.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 51.Todeschini R, Consonni V, Pavan M. A distance measure between models: a tool for similarity/diversity analysis of model populations. Chemometr. Int. Lab. Sys. 2004;70:55–61. [Google Scholar]
- 52.Davison ML. Introduction to Multidimensional Scaling and Its Applications. Applied Psychological Measurement. 1983;7:373–379. [Google Scholar]
- 53.QSAR, Expert, and Group. The report from the expert group on (Quantitative) Structure-Activity Relationships [(Q)SARs] on the principles for the validation of (Q)SARs. Journal. 2004:206. [Google Scholar]
- 54.Muratov EN, Kuz'min VE, Artemenko AG. Domain applicability: How far are ideal and reality? Journal. 2008 Available in web: http://oasys2.confex.com/acs/235nm/techprogram/P1145213.HTM. [Google Scholar]
- 55.Minieka E. Optimization Algorithms for Networks and Graphs. New York: Marcel Dekker; 1978. [Google Scholar]
- 56.Jaworska J, Nikolova-Jeliazkova N, Aldenberg T. QSAR Applicability Domain Estimation by Projection of the Training Set in Descriptor Space: A Review. Altern. Lab. Anim. 2005;33:445–459. doi: 10.1177/026119290503300508. [DOI] [PubMed] [Google Scholar]
- 57.Kubinyi H. Quantitative structure-activity relationships (QSAR) and molecular modeling in cancer research. J. Cancer Res. Clin. Oncol. 1990;116:529–537. doi: 10.1007/BF01637071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gramatica P. Principles of QSAR models validation: internal and external. QSAR Comb. Sci. 2007;26:694–701. [Google Scholar]
- 59.Golbraikh A, Tropsha A. Beware of Q2. J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 60.Tetko IV, Sushko I, Pandey AK, Tropsha A, Zhu H, Papa E, Öberg T, Todeschini R, Fourches D, Varnek A. Critical assessment of QSAR Models to predict environmental toxicity against Tetrahymena pyriformis: Focusing on applicability domain and overfitting by variable selection. J. Chem. Inf. Model. 2008;48:1733–1746. doi: 10.1021/ci800151m. [DOI] [PubMed] [Google Scholar]
- 61.Cramer RD, Patterson DI, Bunce JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape binding to carrier proteins. J. Am. Chem. Soc. 1988;110:5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 62.Dearden JC, Cronin MTD, Schultz TW, Lin DT. QSAR Study of the Toxicity of Nitrobenzenes to Tetrahymena pyriformis. Quant. Struct.-Act. Relat. 1995;14:427–432. [Google Scholar]
- 63.Schmitt H, Altenburger R, Jastorff B, Schuurmann G. Quantitative Structure-Activity Analysis of the Algae Toxicity of Nitroaromatic Compounds. Chem. Res. Toxicol. 2000;13:441–450. doi: 10.1021/tx9901635. [DOI] [PubMed] [Google Scholar]