Abstract
The extracellular matrix fibrils of Myxococcus xanthus are essential for the social lifestyle of this unusual bacterium. These fibrils form networks linking or encasing cells and are tightly correlated with cellular cohesion, development, and social (S) gliding motility. Previous studies identified a set of bacterial chemotaxis homologs encoded by the dif locus. It was determined that difA, difC, and difE, encoding respective homologs of a methyl-accepting chemotaxis protein, CheW, and CheA, are required for fibril production and therefore S motility and development. Here we report the studies of three additional genes residing at the dif locus, difB, difD, and difG. difD and difG encode homologs of chemotaxis proteins CheY and CheC, respectively. difB encodes a positively charged protein with limited homology at its N terminus to conserved bacterial proteins with unknown functions. Unlike the previously characterized dif genes, none of these three newly studied dif genes are essential for fibril production, S motility, or development. The difB mutant showed no obvious defects in any of the processes examined. In contrast, the difD and the difG mutants were observed to overproduce fibril polysaccharides in comparison with production by the wild type. The observation that DifD and DifG negatively regulate fibril polysaccharide production strengthens our hypothesis that the M. xanthus dif genes define a chemotaxis-like signal transduction pathway which regulates fibril biogenesis. To our knowledge, this is the first report of functional studies of a CheC homolog in proteobacteria. In addition, during this study, we slightly modified previously developed assays to easily quantify fibril polysaccharide production in M. xanthus.
Myxococcus xanthus is a gram-negative bacterium with a lifestyle that is unusual for a prokaryote. Like other bacteria, M. xanthus cells grow and divide as vegetative cells when nutrients are abundant. During vegetative growth, cells utilize their gliding motility to move over surfaces in search of more favorable or less adverse environments (41, 49). The gliding movement of M. xanthus cells often occurs in large cell groups in a coordinated manner (17, 47). Since M. xanthus preys on other organisms, translocation in large groups is advantageous because more antimicrobial compounds and hydrolytic enzymes can be released collectively, allowing more effective killing and feeding (12). In addition to the vegetative cell cycle, M. xanthus cells can undergo a developmental cycle upon nutrient limitation (13). During the developmental cycle, hundreds of thousands of preexisting cells coordinate their movement to allow the orderly and timely aggregation necessary for fruiting body formation. M. xanthus cells eventually differentiate into environmentally resistant myxospores within mature fruiting bodies. As a unicellular organism, M. xanthus provides one of the simplest systems for studying social behaviors and cell-cell interactions during both its vegetative and developmental cycles.
The gliding motility of M. xanthus is controlled by two distinct motility systems, the adventurous (A) and the social (S) gliding motility systems (18, 19). Genetic analysis indicates that these two systems function independently in the sense that cells with a mutation in one system are still motile due to the activities of the remaining system; however, cells with mutations in both systems are rendered nonmotile. S motility is manifested by the movement of cell groups or rafts of cells, whereas A motility enables the movement of well-isolated cells. The gliding motility of M. xanthus is essential for both the wolf-pack-like feeding during vegetative growth and the organization of aggregation during the developmental cycle (12, 23). Experimental evidence reveals a tight correlation between S motility and fruiting body formation, as most mutants of the S motility system are defective in development to various extents (19, 31).
Due to its importance for the lifestyle of M. xanthus, S motility has been the focus of extensive scientific research. It has become evident that two cell surface components, the polarly localized type IV pili (22, 51) and the peritrichous extracellular matrix fibrils (2, 6, 43, 50, 56), are critical for functional S motility. Recent advances suggest that M. xanthus S motility resembles twitching motility mechanistically in that both forms of motility require the presence of type IV pili and close cell proximity (32, 37). It is believed that both twitching and S motilities are powered by the retraction of type IV pili (21, 33, 44, 45). The peritrichous extracellular matrix fibrils, the other structure crucial for M. xanthus S motility, constitute the extracellular matrix that connects adjacent cells (2, 5). The fibrils are composed of polysaccharides with roughly equivalent amounts of associated proteins; the polysaccharides appear to form the backbone of the structure (4). Although the role of fibrils in S motility is not fully understood, a recent study proposed that the fibril polysaccharide may provide the anchor for retracting pili (30). Lipopolysaccharides have also been implicated in M. xanthus gliding motility and development (9, 55), but the function of lipopolysaccharides in these processes remains to be elucidated.
In M. xanthus, both pili and fibrils appear to be controlled by two separate chemotaxis-like pathways. Strains with mutations in frz (“frizzy”) chemotaxis gene have defects in cell reversal rates (7), and it has been suggested that the Frz pathway may partially control the type IV-pilus-mediated S motility (41, 45). On the other hand, the production of fibrils is controlled, at least in part, by the dif (defective in fruiting and fibrils) chemotaxis pathway (6, 54, 56). It was previously shown that three genes at the dif locus, difA, difC, and difE, are required for S motility, fruiting body formation, and fibril biogenesis (6, 54, 56). difA encodes a methyl-accepting chemoreceptor protein (MCP), difC encodes a CheW homolog, and difE encodes a CheA homolog (6, 54). Based on these results, the dif genes were hypothesized to define a chemotaxis-like signal transduction pathway that regulates the biogenesis of fibrils. The defects of the dif mutants in development and S motility may be attributed to their defects in fibril production.
Here we report studies of three additional genes at the M. xanthus dif locus, difB, difD, and difG. Sequence analysis indicates that DifD is homologous to CheY and DifG is homologous to CheC, a chemotaxis protein present in Bacillus subtilis and some other bacteria but absent in the enteric bacteria (27). DifB shows homology to a conserved family of hypothetical bacterial proteins with unknown function. We constructed and studied strains with in-frame deletions in each of these three genes. Unlike difA, difC, and difE, the three remaining genes at the dif locus, difB, difD, and difG, are not absolutely required for M. xanthus S motility, fruiting body formation, or fibril biogenesis. Surprisingly, we discovered that both difD and difG mutants overproduce fibril polysaccharides in comparison with production by the wild type. Therefore, rather than being positive regulators like DifA, DifC, and DifE, DifD and DifG negatively regulate the production of fibril polysaccharides in M. xanthus. In addition, during the course of this study, we slightly modified previously described dye binding assays to easily quantify the production of fibril polysaccharides in M. xanthus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The M. xanthus strains and plasmid constructs used in this study are listed in Table 1. M. xanthus was grown at 32°C on Casitone-yeast extract (CYE) agar plates or in CYE liquid medium (10). Colony-forming (CF) agar plates were used as the development-inducing medium for M. xanthus (16). XL1-Blue (Stratagene), the Escherichia coli strain used for plasmid construction, was grown and maintained at 37°C on Luria-Bertani agar plates or in Luria-Bertani liquid medium (34). Unless noted otherwise, agar plates contained 1.5% agar. Kanamycin was added to media at 100 μg/ml for selection purposes when appropriate.
TABLE 1.
M. xanthus strains and plasmids
| M. xanthus strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| DK1622 | Wild type | 22 |
| YZ602 | ΔdifB | This study |
| YZ603 | ΔdifE | This study |
| YZ604 | ΔdifG | This study |
| YZ613 | ΔdifD | This study |
| Plasmids | ||
| pBJ113 | Gene replacement vector with KG cassette; Kanr | 20 |
| pWB117 | difB in-frame deletion in pBJ113 | This study |
| pWB118 | difE in-frame deletion in pBJ113 | This study |
| pWB119 | difG in-frame deletion in pBJ113 | This study |
| pWB120 | difD in-frame deletion in pBJ113 | This study |
Construction of dif locus mutants.
Mutants with in-frame deletions at the dif locus were constructed by a two-step homologous recombination gene replacement protocol by using a modified positive-negative kanamycin/galactose (KG) cassette selection method (46). DNA fragments with an internal in-frame deletion were generated by a two-step, overlap PCR procedure (39) by using PfuTurbo DNA polymerase (Stratagene). The fragments with appropriate deletions were blunt-end ligated into the SmaI site of pBJ113 (20) to create plasmids pWB117 through pWB120 (Table 1). The deletion plasmid constructs were electroporated (24) into DK1622, a wild-type M. xanthus strain (22). Kanamycin-resistant transformants were subsequently plated on CYE agar plates supplemented with 1% galactose. Deletion mutants were identified by a galactose-resistant and kanamycin-sensitive phenotype and by PCR and Southern analyses of chromosomal DNA (39, 46). Double mutants were constructed by Mx4-mediated generalized transduction (35).
Motility assays.
Motility on hard-agar plates was examined by spotting 5 μl of a cell suspension of approximately 5 × 109 cells/ml onto the center of a standard CYE agar plate. After 2 days of incubation at 32°C, overall colony morphology, colony expansion, and colony edge morphology were examined and documented macroscopically and microscopically. Motility on low-percentage- or soft-agar surfaces was examined as previously described (42). Briefly, 5 μl of cells at the concentration described above was spotted onto the center of a CYE plate containing 0.4% agar and incubated at 32°C for 5 days before documentation.
Assessment of fruiting body development.
To examine fruiting body formation, exponentially growing cells from overnight cultures were harvested and resuspended in MOPS (morpholinepropanesulfonic acid) buffer (10 mM MOPS [pH 7.6], 2 mM MgSO4) at approximately 5 × 109 cells/ml. Five microliters of this cell suspension was spotted in triplicate onto the surface of CF agar plates. Development was examined and documented after 5 days of incubation at 32°C.
Analysis of cellular cohesion.
The agglutination assay described by Wu et al. (53) was used to determine the cellular cohesion of various M. xanthus strains. Exponentially growing overnight cultures of M. xanthus were harvested and resuspended to approximately 2.5 × 108 cells/ml in CYE medium, and the optical density (OD) at 600 nm was recorded every 10 min for a total of 2 h. Agglutination is expressed as relative absorbance for each time point, which was calculated by dividing the OD at each time point by the initial OD for each strain.
Examination of fibril production.
To detect fibril-specific protein antigens, whole-cell lysates were prepared from 5 × 107 cells. Cell lysates were then separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and analyzed by immunoblot analysis by using standard protocols (39). Monoclonal antibody (MAb) 2105, a MAb against the fibril protein FibA, was used as the primary antibody (15, 25).
To examine the polysaccharide portion of the fibrils, two different assays were performed. The first was a plate assay to determine the binding of the fluorescent dye calcofluor white, as described previously (11, 36). Briefly, cells from overnight cultures were pelleted and resuspended in MOPS buffer at approximately 5 × 109 cells/ml, and 5-μl volumes of these suspensions were spotted on the surface of CYE agar plates impregnated with 50 μg of calcofluor white/ml. The plates were incubated at 32°C for 6 days before they were examined and photographed under the illumination of a handheld long-wavelength (365-nm) UV light source.
The second assay was a liquid colorimetric assay to measure the binding of Congo red and trypan blue. The liquid assay, adapted from that of Arnold and Shimkets (3), was used to quantitatively determine the relative level of fibril polysaccharide production. All strains tested were harvested at near-identical culture densities (approximately 3.5 × 108 cells/ml), washed, and resuspended to approximately 2.8 × 108 cells/ml in MOPS buffer. Stock solutions of the dyes were prepared in deionized distilled water at 150 μg/ml for Congo red and 100 μg/ml for trypan blue. A total of 900 μl of the cell suspension was mixed with 100 μl of a dye stock solution to give final concentrations of 2.5 × 108 cells/ml and either 15 μg of Congo red/ml or 10 μg of trypan blue/ml. Control samples containing each dye in MOPS buffer only were included, and triplicate assays were performed for all samples. All samples were vortexed briefly and incubated undisturbed in the dark at room temperature for 30 min. The cell suspensions were then pelleted at 16,000 × g in a benchtop centrifuge for 5 min, and the absorbances of the supernatants were measured at 490 and 585 nm for Congo red and trypan blue, respectively. The reported percentage of dye bound by each sample was calculated from the quotient obtained by dividing the absorbance of each sample by the absorbance of the control.
Nucleotide sequence accession numbers.
The nucleotide sequences of the genes studied here have been deposited in GenBank under the accession numbers AF076485 and AY327119.
RESULTS
Sequence analysis of difB, difD, and difG.
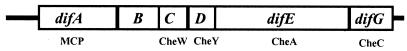
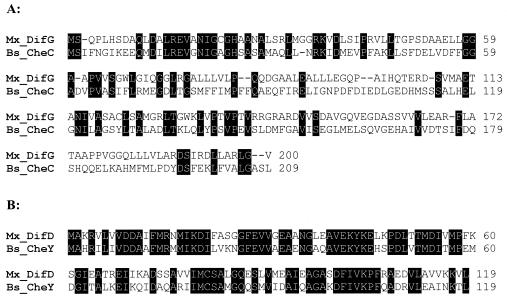
It was previously reported that five open reading frames existed at the dif locus (54). We recognized later that an additional gene, difG, lies immediately downstream of difE (Fig. 1). The last two nucleotides of the predicted start codon for difG, a GTG rather than an ATG, overlap with the first two nucleotides of the stop codon of difE, a TGA. difG is predicted to encode a protein of 200 amino acids with 27% identity to the CheC chemotaxis protein of B. subtilis (Fig. 2A). The homology is extended over the entire length of both proteins, and a conserved-domain search identified DifG as a CheC homolog with high confidence (1). DifD shows a very high degree of homology to several CheY proteins (54); the highest is 62% identity to CheY of B. subtilis (Fig. 2B). As previously reported, difA, difC, and difE encode homologs of the chemotaxis proteins MCP, CheW, and CheA, respectively (54). DifB encodes a positively charged protein of 222 amino acids with 27 lysine (K) and 14 arginine (R) residues and a total of 31 net positive charges (data not shown). The N terminus of DifB shows limited similarity to a conserved but uncharacterized family of hypothetical bacterial proteins (data not shown).
FIG. 1.
M. xanthus dif locus and homology. All of the dif genes read from left to right. The homology of the encoded polypeptides to chemotaxis proteins is indicated below the relevant genes.
FIG. 2.
Homology of M. xanthus DifG (Mx_DifG) to B. subtilis CheC (Bs_CheC) (A) and DifD (Mx_DifD) to B. subtilis CheY (Bs_CheY) (B). DifG shares 27% identity with B. subtilis CheC. DifD shares 62% identity with B. subtilis CheY. Identical residues are shaded in black, and the sequences shown comprise the entire proteins.
Construction of mutants with in-frame deletions in difB, difD, difE, and difG.
In previous studies, mutants with in-frame deletions in difA and difC were constructed and examined, but the difE mutations had been insertions only (6, 29, 54, 56). Since difG is immediately downstream of difE, it was not clear whether the defects of the difE insertion mutant were the result of difE disruption or polar effects on difG or other genes downstream. Similarly, a difB insertion mutant was reported to have a dsp (dispersed growth) phenotype, but the defects could be the result of a polar effect by the insertion (29). To examine the functions of difB, difD, difE, and difG, we constructed mutants with in-frame deletions of these four genes as described in Materials and Methods. In strain YZ602, amino acid residues 4 through 219 were deleted from DifB; in YZ613, amino acids 26 through 99 of DifD were deleted; YZ603 contains a deletion of amino acid residues 4 to 840 of DifE; and YZ604 contains a deletion of amino acids 4 to 199 of DifG.
Examination of motility of the new dif mutants.
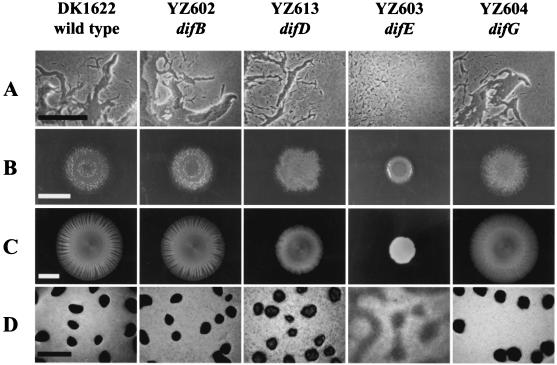
It was shown previously that DifA and DifC are essential for S motility (6, 54). To determine if any of the new dif deletion mutants were defective in S motility, these mutants were examined on both hard (1.5%)- and soft (0.4%)-agar plates as described in Materials and Methods. Microscopically, the colony edges of all mutants on hard agar, except for YZ603 (difE), consisted of both single cells and cell groups, suggesting that both motility systems were present (Fig. 3A). Among these mutants, the colony morphology of YZ602 (difB) appeared the most similar to that of the wild type on hard and soft agar. The colonies of both the wild type and YZ602 displayed slightly glossy centers with rough and dry-looking edges on hard agar (Fig. 3B); on soft agar, they exhibited comparable colony patterns and levels of spreading (Fig. 3C). The colony morphologies of YZ613 (difD) and YZ604 (difG) appeared somewhat similar to each other on hard agar, since both had an extremely dry and rough appearance over the entire colony. Of these two, YZ613 (difD) deviated most significantly in colony morphology from that of the wild type on both surfaces: it displayed a considerable reduction in spreading on soft agar and irregular colony edges on both agar surfaces. YZ604 (difG) appeared similar to the wild type with respect to the degree or level of colony expansion, indicating little if any defects in motility. YZ603 (difE) appeared glossy over the entire colony and spread substantially less than did the wild-type strain on both hard- and soft-agar surfaces; however, it must have retained A motility due to the many single cells observed at its advancing colony edges on hard agar.
FIG. 3.
Colony edge morphology, colony spreading, and fruiting body formation. The strains and their genotypes are indicated above each column. (A) Colony edge morphology on hard agar; scale bar, 100 μm. (B) Colony expansion on hard (1.5%) agar; scale bar, 1 cm. (C) Colony expansion on soft (0.4%) agar; scale bar, 1 cm. (D) Development on CF media; scale bar, 500 μm.
The above results suggested that the difE mutant lacked S motility but that the mutants of difB, difD, and difG possessed both A and S motilities. To confirm the motility status for each strain, secondary mutations in each motility system were introduced into each of the dif deletion mutants. Colony expansion of the double mutants was examined on hard-agar surfaces. Except for YZ603 (difE), which lost motility in a background lacking A motility, all mutants retained motility regardless of the mutant background (data not shown). These results indicated that the difB, difD, and difG genes are not absolutely required for either motility system. Since YZ603 (difE) exhibited the same defects as the previously described difE insertion mutant (54, 56; see also sections below), it may therefore be considered an internal negative control for this study.
Characterization of development.
Since the difA and the difC mutants were previously shown to be defective in fruiting body formation (6, 54), the new dif deletion mutants were examined for development as described in Materials and Methods. On CF media, all mutants with the exception of YZ603 (difE) formed obvious fruiting bodies or aggregates (Fig. 3D). However, obvious developmental defects were observed for YZ613 (difD), which formed aggregates that were translucent and irregularly shaped. The fruiting bodies of YZ604 (difG) appeared slightly bigger or less compact than those of the wild type. Similar results were obtained when development was examined on TPM (10 mM Tris, pH 7.6; 8mM MgSO4; 1 mM KH2PO4) medium (28), and the defects observed for YZ613 (difD) were even more pronounced than those on CF (data not shown). In the process of this study, we noticed that both YZ613 and YZ604 consistently formed fruiting bodies and aggregates that were more resistant to dispersion by sonication, suggesting alterations in cell-cell interaction and/or the architecture of the aggregates.
Assessment of cellular cohesion.
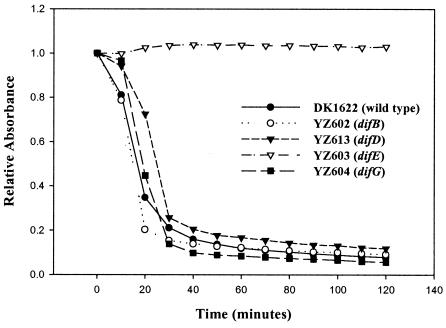
Cellular cohesion or agglutination is closely associated with S motility and requires the presence of both extracellular fibrils and pili (2, 6, 22, 43, 50, 51, 56). Since some of the new mutants displayed defects or abnormalities in both colony expansion and development, and because the previously characterized dif mutants were defective in agglutination (6, 56), the cellular cohesion of these new mutants was examined by the agglutination assay described in Materials and Methods. The agglutination assay is based on the ability of cells to clump and sediment out of suspension, which in time drastically reduces the apparent OD. As shown in Fig. 4, all of the new dif mutants, except YZ603 (difE), have agglutination patterns comparable to that of the wild-type strain, suggesting that the difB, difD, and difG mutants produce both fibrils and pili. To confirm that these strains were not defective in the production of type IV pili, the mutants were subjected to immunoblot analysis by using polyclonal antibodies against M. xanthus PilA (52). Both surface pili and pili from whole-cell lysates were examined as described previously (48, 52). The results confirmed that all of the dif mutants were capable of producing surface pili (data not shown).
FIG. 4.
Agglutination and cellular cohesion assays. Cells were grown overnight in CYE medium, and the OD at 600 nm was adjusted to approximately 0.5 with CYE medium. Absorbance was measured every 10 min for 2 h. Relative absorbance was obtained by dividing the absorbance at each time point by the initial absorbance for each strain. The graph represents the data from one of many experiments with similar results.
Examination of extracellular fibril production.
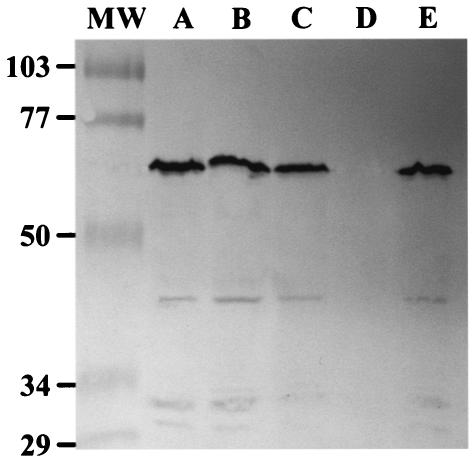
Previous studies demonstrated that both difA and difC mutants are defective in fibril production (6, 56). One of the fibril proteins, FibA, a metalloprotease (25), was found missing from the cell surfaces of difA and difC mutants (6, 56). Even though the fibA mutant could agglutinate properly, it formed less-organized fruiting bodies (25), somewhat reminiscent of the phenotypes displayed by the difD and difG mutants. Furthermore, the difficulty of disrupting or dispersing the aggregates formed by the difD and difG mutants might indicate changes in cell surface properties. The new dif mutants were therefore examined for fibril biogenesis. First, the presence of FibA was examined by immunoblot analysis using MAb 2105, as described in Materials and Methods. All of the new dif mutants, with the exception of YZ603 (difE), showed levels of reactivity with MAb 2105 comparable to those of the wild type (Fig. 5), suggesting that the difB, difD, and difG mutants are not defective in the production of FibA, which is associated with the extracellular fibrils.
FIG. 5.
Immunoblot analysis of the fibril protein FibA. Whole-cell lysates were prepared from 5 × 108 cells, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and probed with anti-FibA MAb 2105. Lanes: MW, molecular weight standards in thousands; A, DK1622 (wild type); B, YZ602 (difB); C, YZ613 (difD); D, YZ603 (difE); E, YZ604 (difG). The most predominate band is at a molecular weight of ca. 66,000. The multiple banding patterns are consistent with previous reports and likely reflect self-processing of FibA, an M4 metalloprotease (25).
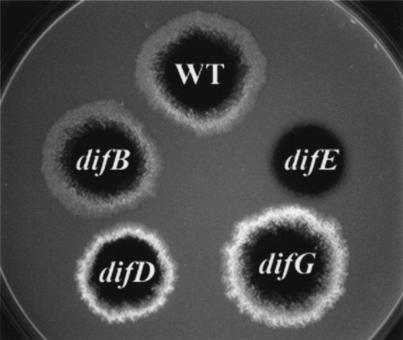
Next, the production of fibril polysaccharide was examined by the binding of the fluorescent dye calcofluor white (Fig. 6). The binding of the dye, visualized by fluorescence under the illumination of UV light, demonstrates the production of fibril polysaccharides (11, 36, 56). The wild-type strain fluoresced when illuminated by UV light, whereas the difE deletion mutant exhibited no fluorescence, indicating undetectable levels of binding to the fluorescent dye. The difB mutant fluoresced at a level comparable to that of the wild type. Surprisingly, YZ613 (difD) and YZ604 (difG) showed substantially increased intensities of fluorescence, suggesting an overproduction of fibril polysaccharides.
FIG. 6.
Binding of calcofluor white. Five microliters of cells at approximately 5 × 109 cells/ml was spotted onto CYE plates containing 50 μg of calcofluor white/ml. After incubation for 6 days at 32°C, the plates were photographed under the illumination of long-wavelength (365-nm) UV light. The diameter of the plate shown is 9 cm. WT, wild type.
Fibril polysaccharide production was analyzed more quantitatively by the binding of two separate dyes in colorimetric assays. Previous studies identified Congo red and trypan blue as dyes capable of binding to cells with fibrils (3, 11). By using a slightly modified version of the procedure of Arnold and Shimkets (3) as described in Materials and Methods, the relative levels of fibril polysaccharide production by each new dif mutant were determined by the amount of dye bound by the cells (Table 2). Under our assay conditions, the wild type bound 40.0 and 14.8% of Congo red and trypan blue, respectively. Consistent with the results from the calcofluor white binding assay, YZ602 (difB) exhibited levels of binding of Congo red and trypan blue comparable to those of the wild-type strain. In contrast, the difD and difG mutants bound significantly more of both dyes. YZ613 (difD) bound 87.1 and 51.5% and YZ604 (difG) bound 69.7 and 36.4% of Congo red and trypan blue, respectively. These results further demonstrate that both the difD and difG mutants overproduce M. xanthus fibril polysaccharides.
TABLE 2.
Binding of Congo red and trypan bluea
| Strain | % of dye bound
|
|
|---|---|---|
| Congo red | Trypan blue | |
| DK1622 (wild type) | 40 ± 0.8 | 14.8 ± 0.2 |
| YZ602 (difB) | 37.4 ± 0.4 | 15.7 ± 0.8 |
| YZ613 (difD) | 87.1 ± 0.2 | 51.5 ± 0.4 |
| YZ603 (difE) | 13.3 ± 0.6 | 1.4 ± 1.9 |
| YZ604 (difG) | 69.7 ± 0.6 | 36.4 ± 0.7 |
All values reflect a percentage of dye bound in a suspension of 2.5 × 108 cells/ml with either 15 μg of Congo red/ml or 10 μg of trypan blue/ml. Details of the experiment are described in Materials and Methods.
DISCUSSION
We report here studies of three additional genes at the dif locus in M. xanthus (Fig. 1). Further sequence analysis indicated that difG, which is immediately downstream of difE, encodes a homolog of the CheC chemotaxis protein from B. subtilis (Fig. 2A). difD, which is immediately upstream of difE, encodes a CheY homolog (Fig. 2B). difB, which is further upstream of difD (Fig. 1), encodes a positively charged protein with limited homology at its N terminus to a conserved but uncharacterized family of hypothetical bacterial proteins. Due to complications of possible polar effects from insertion mutations, we constructed and analyzed in-frame deletions of difB, difD, difE, and difG. The difE deletion resulted in defects similar to those of a previously described difE insertion (54, 56), confirming the requirement of difE for the biogenesis of fibrils, and thus S motility and development, in M. xanthus. On the other hand, none of the difB, difD, or difG mutants exhibited phenotypes similar to those of the difA, difC, and difE mutants. We observed no obvious defects in the difB mutant under our study conditions. difD and difG mutants showed slight defects in development and altered colony morphology or colony expansion patterns. However, through qualitative and quantitative analyses, we showed that both difD and difG mutants overproduced fibril polysaccharides (Fig. 6 and Table 2). By using a colorimetric assay which measured the binding of trypan blue, the difD and difG mutants were found to produce as much as three and two times the amount of fibril polysaccharides produced by the wild type, respectively (Table 2). Considering that fibril-associated proteins do not appear to significantly impact the structure of fibrils and that the fibril polysaccharides may form the backbone of fibrils (4, 25, 30), the overproduction of fibril polysaccharides in difD and difG mutants may possibly represent an overproduction of fibrils. However, because no obvious FibA overproduction was detected in our experiments, the possibility remains that it is simply the fibril polysaccharide that is overproduced in these two mutants and not the fibrils themselves. Although there is a distinction between the fibril and the fibril polysaccharide, these two terms are used interchangeably in the rest of the discussion for convenience.
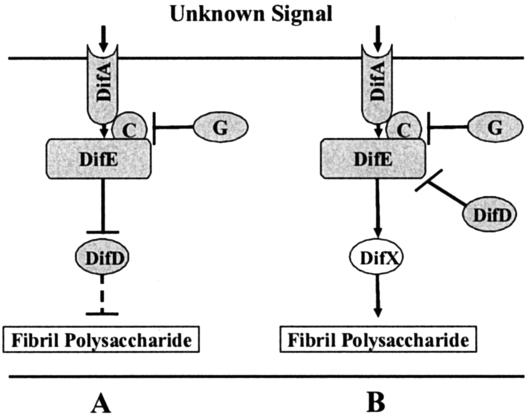
The finding that the difD deletion mutant overproduced fibril polysaccharides was unexpected and intriguing. In enteric bacteria, four major players, MCPs, CheW, CheA, and CheY, constitute the central chemotaxis pathway (8, 14). Mutations in a particular MCP gene in enteric bacteria generally cause defects in chemotaxis to the specific attractants or repellents sensed by the mutated chemoreceptor. Mutations in the genes for the other three components, CheW, CheA, and CheY, result in the same chemotaxis defects: the complete elimination of a chemotaxis response. Although it is not known what signals regulate the Dif pathway in M. xanthus, we had expected a difD mutant to have the same defects as those previously described for the difA, difC, and difE mutants. The present data may be explained by two working models (Fig. 7). The first model, depicted in Fig. 7A, proposes that, instead of the usual stimulatory or positive effects of CheA on CheY as seen in bacterial chemotaxis, DifE (CheA-like) inhibits the activity of DifD (CheY-like), which in turn negatively regulates fibril biogenesis. If this model is correct and since DifD is predicted to be a single-domain protein like CheY, it is reasonable to assume that there are additional components between DifD and the direct regulation of fibril polysaccharides. In the second model (Fig. 7B), DifX, a hypothetical interacting partner or substrate for the putative DifE kinase, is proposed. DifX could be a response regulator capable of positively regulating the expression of genes involved in fibril biogenesis, or it could be an enzyme or a group of enzymes catalyzing the synthesis of fibril polysaccharides. In this model, we propose that DifD inhibits the activity of DifE to stimulate DifX, perhaps by diverting the phosphate flow from DifX to itself, as does CheY1 in Sinorhizobium meliloti chemotaxis (40). In this context, it is perhaps relevant that preliminary results with the yeast two-hybrid system indicated direct interactions between DifE and DifD (Z. Li and Z. Yang, unpublished results). In both models, DifG is proposed to have an inhibitory effect on fibril biogenesis based on the observation that the difG mutant overproduced fibril polysaccharides. We tentatively placed DifG on the pathway as interacting with the putative complexes formed by DifA, DifC, and DifE. The formation of complexes by DifA, DifC, and DifE is extrapolated from the knowledge that the chemoreceptors, CheW, and CheA exist as multimolecular ternary complexes in enteric bacteria (14). The speculation that DifG interacts with the presumed complexes of DifA, DifC, and DifE is based primarily on the studies of CheC in B. subtilis (27). It was proposed that B. subtilis CheC, which shares homology with flagellar switch proteins, may interact with the flagellar switch and the chemoreceptor-CheW-CheA complex to bring about the adaptation in chemotactic responses (27). cheC mutants of B. subtilis have increased levels of MCP methylation and appear to promote the activity of the CheA kinase (38). Our findings are consistent with the model proposed for the function of CheC in B. subtilis.
FIG. 7.
Two working models for the regulation of fibril polysaccharides by the Dif pathway. Arrows indicate positive or stimulatory effects, and bars represent negative or inhibitory effects. (A) A model proposing that DifE negatively effects DifD and that DifD in turn has an inhibitory effect on the production of fibril polysaccharides. The dashed line indicates the possible involvement of multiple components downstream of DifD. (B) A model proposing that an additional response regulator, DifX, positively regulates fibril polysaccharide production. It is further proposed that DifE stimulates the function of DifX and DifD inhibits the stimulatory effects of DifE on DifX. In both models, DifG is proposed to have an inhibitory effect on fibril polysaccharide production by interacting with the signaling complex of DifA, DifC, and DifE.
As far as we are aware, DifG is the first CheC homolog to be identified and studied in proteobacteria. It was reported previously that the B. subtilis CheC protein interacted the strongest with CheD in the yeast two-hybrid analysis and that CheD homologs existed in all prokaryotic species in which CheC was present (27). In most of the bacterial and archaeal species with both CheC and CheD homologs, the genes that encode these homologs are located in the same operon. M. xanthus appears to be the exception in that there are no CheD homologs in the vicinity of difG. In fact, BLAST searches revealed no CheD homologs in the nearly completed M. xanthus genome sequence (Z. Yang, unpublished results). This finding is consistent with the conclusion that CheC in B. subtilis plays a role in chemotaxis adaptation that is independent from CheD (27). We prefer the model shown in Fig. 7B for two reasons. First, in most bacterial signal transduction pathways, kinases usually stimulate the functions or activities of their cognate response regulators that are responsible for regulating the downstream processes, whether transcriptional or otherwise. The model proposed in Fig. 7A goes against this well-known dogma. Second, preliminary genetic analysis indicated that a difD difE double mutant displayed a phenotype similar to the parental difE mutant (our unpublished results). This suggests that difE is epistatic to difD and that DifD is therefore unlikely to function downstream of DifE in the pathway.
While the working models are consistent with our data, we recognize that they are speculative and not comprehensive with regard to the regulation of fibril biogenesis and that the interactions among the Dif proteins predicted by the models remain to be examined. In addition to the dif locus, there are two other loci with apparent regulatory functions in fibril biogenesis. At the sglK locus, fibR and sglK were found to have somewhat opposite effects (50). Mutations in sglK, like mutations in difA, difC, and difE, lead to defects in fibril biogenesis, S motility, and development (50, 56). SglK is therefore a positive regulator of fibril production. On the other hand, mutations in fibR were found to produce increased amounts of the fibril protein FibA (25, 50). FibR therefore appears to be a negative regulator of FibA and possibly other fibril proteins. The stk gene was identified by transposon insertion mutations that resulted in a more cohesive or stickier phenotype (11). stk mutations were found to restore group motility to certain S motility mutants and led to increased production of fibrils. Stk therefore appears to be a negative regulator of fibril production. Interestingly, both Stk and SglK were found to be homologs of DnaK, a chaperone in the HSP70 family (50). How the Dif pathway may interact with SglK, FibR, and Stk is yet to be investigated. The difference between the fibR mutant and the difD and difG mutants should be noted. Using immunoblotting, we did not observe an obvious overproduction of FibA (Fig. 5). The overproduction of fibrils in stk mutants was observed by using scanning electron microscopy (11). Since polysaccharides form the backbones of fibrils, it is probably safe to assume that stk mutants overproduce fibril polysaccharides. The level of FibA production in stk mutants remains to be examined. In addition, our current models have not yet taken into account that difA, difE, and fibA are all implicated in the chemotactic response of M. xanthus cells to phosphatidylethanolamine attractants (25, 26).
During this study, we made slight modifications to previously described assays for quantifying the production of fibril polysaccharides in M. xanthus. A dye binding assay was reported previously for the determination of the kinetic parameters of Congo red binding to M. xanthus cells (3). In addition, it was reported that the binding of both Congo red and trypan blue, analyzed by plate assays, correlated with the amount of fibrils on the cell surface as examined by scanning electron microscopy (11). Using the absorbance at the wavelengths with peak absorbance (490 nm for Congo red and 585 nm for trypan blue), we determined that there is a linear relationship between the absorbance value and the amount of dye in a solution over a wide range (data not shown). It is therefore feasible to determine the amount of dye bound by M. xanthus cells by measuring the amount of unbound dye in a colorimetric assay, as noted previously (3). The wide range of possible working concentrations of the dye affords the flexibility to allow the measurements of various levels of fibril polysaccharide production. Our results indicate that compared to Congo red, trypan blue has a substantially reduced background level of binding to fibril-deficient strains. In general, we observed little if any binding of trypan blue to cells defective in fibril production. On the other hand, fibril-defective mutants still showed significant binding to Congo red in our assay. This is consistent with previous findings that, in addition to fibrils, there is an additional receptor on M. xanthus cells for Congo red (3, 50). Based on these observations, we suggest that trypan blue provides the more sensitive and effective measurement for the relative amount of fibril polysaccharide production in M. xanthus. The ability to quantify fibril polysaccharide production in various mutants should greatly facilitate the studies of the regulation of fibrils in M. xanthus.
Acknowledgments
We are grateful to Jill Sible for granting access to her microscope facilities. We thank Larry Shimkets and John Kirby for helpful discussions. We thank the laboratories of Heidi Kaplan and Dale Kaiser for generously providing antibodies, plasmids, and/or protocols. We thank Wenyuan Shi for his support and encouragement.
This work was supported by grant MCB-0135434 from the National Science Foundation to Z. Yang.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. W., and L. J. Shimkets. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, J. W., and L. J. Shimkets. 1988. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J. Bacteriol. 170:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behmlander, R. M., and M. Dworkin. 1994. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behmlander, R. M., and M. Dworkin. 1991. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J. Bacteriol. 173:7810-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellenger, K., X. Ma, W. Shi, and Z. Yang. 2002. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J. Bacteriol. 184:5654-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackhart, B. D., and D. R. Zusman. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA 82:8767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 9.Bowden, M. G., and H. B. Kaplan. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30:275-284. [DOI] [PubMed] [Google Scholar]
- 10.Campos, J. M., and D. R. Zusman. 1975. Regulation of development in Myxococcus xanthus: effect of 3′:5′-cyclic AMP, ADP, and nutrition. Proc. Natl. Acad. Sci. USA 72:518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dana, J. R., and L. J. Shimkets. 1993. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J. Bacteriol. 175:3636-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dworkin, M. 1973. Cell-cell interactions in the myxobacteria. Symp. Soc. Gen. Microbiol. 23:125-142. [Google Scholar]
- 13.Dworkin, M. 2000. Introduction to myxobacteria, p. 221-242. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 14.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13:457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill, J., E. Stellwag, and M. Dworkin. 1985. Monoclonal antibodies against cell-surface antigens of developing cells of Myxococcus xanthus. Ann. Inst. Pasteur Microbiol. 136A:11-18. [DOI] [PubMed] [Google Scholar]
- 16.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 17.Hartzell, P. L., and P. Youderian. 1995. Genetics of gliding motility and development in Myxococcus xanthus. Arch. Microbiol. 164:309-323. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 19.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 20.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser, D., C. Manoil, and M. Dworkin. 1979. Myxobacteria: cell interactions, genetics, and development. Annu. Rev. Microbiol. 33:595-639. [DOI] [PubMed] [Google Scholar]
- 24.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 25.Kearns, D. B., P. J. Bonner, D. R. Smith, and L. J. Shimkets. 2002. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J. Bacteriol. 184:1678-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearns, D. B., B. D. Campbell, and L. J. Shimkets. 2000. Myxococcus xanthus fibril appendages are essential for excitation by a phospholipid attractant. Proc. Natl. Acad. Sci. USA 97:11505-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirby, J. R., C. J. Kristich, M. M. Saulmon, M. A. Zimmer, L. F. Garrity, I. B. Zhulin, and G. W. Ordal. 2001. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol. Microbiol. 42:573-585. [DOI] [PubMed] [Google Scholar]
- 28.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 29.Lancero, H., J. E. Brofft, J. Downard, B. W. Birren, C. Nusbaum, J. Naylor, W. Shi, and L. J. Shimkets. 2002. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J. Bacteriol. 184:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y., H. Sun, X. Ma, A. Lu, R. Lux, D. Zusman, and W. Shi. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacNeil, S. D., A. Mouzeyan, and P. L. Hartzell. 1994. Genes required for both gliding motility and development in Myxococcus xanthus. Mol. Microbiol. 14:785-795. [DOI] [PubMed] [Google Scholar]
- 32.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 33.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.O'Connor, K. A., and D. R. Zusman. 1986. Genetic analysis of Myxococcus xanthus and isolation of gene replacements after transduction under conditions of limited homology. J. Bacteriol. 167:744-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramaswamy, S., M. Dworkin, and J. Downard. 1997. Identification and characterization of Myxococcus xanthus mutants deficient in calcofluor white binding. J. Bacteriol. 179:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez, A. M., and A. M. Spormann. 1999. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 181:4381-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosario, M. M., J. R. Kirby, D. A. Bochar, and G. W. Ordal. 1995. Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry 34:3823-3831. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schmitt, R. 2002. Sinorhizobial chemotaxis: a departure from the enterobacterial paradigm. Microbiology 148:627-631. [DOI] [PubMed] [Google Scholar]
- 41.Shi, W., T. Kohler, and D. R. Zusman. 1993. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol. Microbiol. 9:601-611. [DOI] [PubMed] [Google Scholar]
- 42.Shi, W., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimkets, L. J. 1986. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J. Bacteriol. 166:837-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 46.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 47.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 48.Wall, D., S. S. Wu, and D. Kaiser. 1998. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J. Bacteriol. 180:759-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward, M. J., and D. R. Zusman. 1997. Regulation of directed motility in Myxococcus xanthus. Mol. Microbiol. 24:885-893. [DOI] [PubMed] [Google Scholar]
- 50.Weimer, R. M., C. Creighton, A. Stassinopoulos, P. Youderian, and P. L. Hartzell. 1998. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J. Bacteriol. 180:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 52.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z., Y. Geng, D. Xu, H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123-1130. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Z., D. Guo, M. G. Bowden, H. Sun, L. Tong, Z. Li, A. E. Brown, H. B. Kaplan, and W. Shi. 2000. The Myxococcus xanthus wbgB gene encodes a glycosyltransferase homologue required for lipopolysaccharide O-antigen biosynthesis. Arch. Microbiol. 174:399-405. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]