Abstract
Dimeric Salmonella typhimurium orotate phosphoribosyltransferase (OMP synthase, E.C. 2.4.2.10), a key enzyme in de novo pyrimidine nucleotide synthesis, has been co-crystallized in a complete substrate complex of E•MgPRPP•orotate, and the structure solved to 2.2 Å resolution. This structure resembles that for Saccharomyces cerevisiae OMP synthase in showing a dramatic and asymmetric reorganization around the active site-bound ligands, but shares the same basic topology previously observed in complexes of OMP synthase from S. typhimurium and Escherichia coli. The catalytic loop (residues 99–109) contributed by subunit A is reorganized to close the active site situated in subunit B and to sequester it from solvent. Furthermore, the overall structure of subunit B is more compact, due to movements of the amino-terminal hood and elements of the core domain. The catalytic loop of subunit B remains open and disordered, and subunit A retains the more relaxed conformation observed in loop-open S. typhimurium OMP synthase structures. A non-proline cis-peptide formed between Ala71 and Tyr72 is seen in both subunits. The loop-closed catalytic site of subunit B reveals that both the loop and the hood interact directly with the bound pyrophosphate group of PRPP. In contrast to dimagnesium hypoxanthine-guanine phosphoribosyltransferases, OMP synthase contains a single catalytic Mg2+ in the closed active site. The remaining pyrophosphate charges of PRPP are neutralized by interactions with Arg99A, Lys100B, Lys103A, and His105A. The new structure confirms the importance of loop movement in catalysis by OMP synthase, and identifies several additional movements that must be accomplished in each catalytic cycle. A catalytic mechanism based on enzymic and substratea-ssisted stabilization of the previously documented oxocarbenium transition state structure is proposed.
INTRODUCTION
Orotate phosphoribosyltransferase (OMP synthase E.C. 2.4.2.10) catalyzes the formation of the nucleosidic bond in de novo pyrimidine nucleotide synthesis. Orotate and the ribosyl 5-phosphate donor PRPP react with the loss of pyrophosphate to form OMP, the precursor to UTP. OMP synthase is a discrete protein in bacteria and fungi. In higher eukaryotes, this activity is part of the bifunctional UMP synthase (OMP synthase, OMP decarboxylase) (1), the locus for hereditary orotic aciduria (2). The enzyme is one of 10 phosphoribosyltransferases (PRTases) characterized by their base specificity (3). The PRTase group is now known to contain several distinct evolutionary families, based on different architectures. The Rossmann-fold-based Type I PRTases constitute the enzymes of de novo and salvage purine and pyrimidine biosynthesis, including hypoxanthine-guanine PRTase (HGPRTase), GPAT, adenine PRTase, OMP synthase and uracil PRTase (4). The Type I PRTases show minimal sequence conservation among different substrate specificity classes, mainly centered on a short PRPP binding motif (5). More widespread conservation of sequence is observed among enzymes for each substrate class. PRPP synthase, in which the β-γ pyrophosphoryl group of ATP reacts with O1 of ribose 5-phosphate, also exhibits the Type I fold (6). The Type II enzymes, of TIM barrel architecture, are represented by pyridine PRTase, including quinolinate PRTase (7), nicotinate PRTase (8), and nicotinamide PRTase (9).
The paradigm for the Type I enzymes has been HGPRTase, exemplified by the human (10) and bacterial (11) enzymes as well as enzymes from the eukaryotic parasites Tritrichomonas foetus (12) Plasmodium falciparum (13), and Toxoplasma gondii (14), solved in complexes including base, PRPP (15), and nucleotide (16). A complex with a transition state analog has also been solved (17) and provides a view along the reaction coordinate. A major finding from the HGPRTase structures is that the enzyme can exist in a relatively open form, whose active site, including bound substrate, is exposed to bulk solvent (10), which is not compatible with the development of high-energy transition states for group transfer. Active site closure is provided by a disordered loop of peptide, formed by an extension of the nucleotide fold, which projects into solvent adjacent to the active site. This “flexible loop”, sometimes referred to as Loop II (16), adopts a folded and ordered β-structure in transition state analog complexes, and several residues from the loop project into the active site. In the loop-closed complexes, active site water molecules are highly ordered, with low B-factors, and are positioned away from the developing oxocarbenium ion. In each catalytic cycle, the flexible loop needs to move between its open position to a closed to interact with bound ligands (17, 18). Other Type I PRTases contain a homolog of the flexible loop. Motional studies have been done on the GPAT loop (19), and in that case X-ray diffraction studies have captured the loop in open and closed positions.
As seen with HGPRTases, the catalytic loop of OMP synthase (residues from 99–109 in the S. typhimurium enzyme) adopts an open and disordered conformation in X-ray structures of E•OMP and E•MgPRPP•orotate complexes (20, 21). Notably, this dynamic loop exhibits the highest sequence conservation of any contiguous region in the protein, indicating significant functional importance. Chemical modification and mutagenesis studies (22) have shown that loop residues Lys103, and Lys100, are important for activity. Complementation experiments have demonstrated that Lys103 exercises its catalytic function in the active site formed by the adjacent subunit, while Lys100 acts in the active site formed by its own subunit (23). A three-dimensional structure of the highly homologous E. coli enzyme, in complex with an inhibitory sulfate anion, captured one of the two loops in a more structured "loop-closed" conformation atop the active site of the adjacent subunit (24). In this case Lys103 formed a hydrogen bond with the sulfate anion, which occupied the position of the β-phosphate of bound PRPP in the earlier structure. A structure of the yeast enzyme has captured a symmetric ternary complex of enzyme, MgPRPP and orotate in which each loop is closed over the adjacent active site of the dimer (25). The groups of Witte and McClard have also shown that the yeast enzyme demonstrates half-of-the-sites binding, leading to a proposal that the enzyme follows a complex “double Theorell-Chance” mechanism (26). Because the yeast and bacterial enzymes are structurally similar, it quite likely that findings for each have relevance to a general mechanism.
The emerging dynamic picture is that the loop from one subunit moves from a solvent-extended and disordered to a highly structured one that occludes solvent from the active site of the adjacent subunit. It has been shown that as for HGPRTase, chemistry of group transfer is rapid, whereas a subsequent step, proposed to be related to loop opening, is rate determining (27). NMR has shown that the catalytic loop of OMP synthase moves between the flexible solvent-extended open position and a closed position. Overall PRPP binding and release from binary complexes is a two-step partitioning process in which the loop moves between open and closed positions and PRPP dissociates from the loop-open complex (28).
We have crystallized a complete substrates complex of enzyme, orotate, and MgPRPP. The structure shows one catalytic loop in a stabilized conformation positioned over the ligands in the adjacent subunit, with several residues of the loop forming new contacts to bound substrate. The major part of the loop from the other subunit extends into solution and remains disordered.
METHODS
Enzyme
Recombinant S. typhimurium OMP synthase was expressed in E. coli and purified to homogeneity (22). For crystallization, an ammonium sulfate suspension of the enzyme was centrifuged and the precipitate dissolved in 20 mM Na-HEPES, pH 7.2. The solution was then repeatedly concentrated on a Centricon-10 (Amicon) and rediluted with the same buffer.
Crystallization
S. typhimurium OMP synthase (15 mg/ml, 0.65 mM) was co-crystallized with a 1:0.8 molar stoichiometry of orotate, with excess PRPP and MgCl2 by the hanging drop vapor diffusion method at 18°C. A 10 µl drop of 0.1 mM orotate was pipetted onto a siliconized cover slide and allowed to dry completely, 2 µl of protein solution containing 3 mM PRPP and 6 mM MgCl2 was placed on top of the dried substrate and mixed with 2 µl of the reservoir solution. This solution was equilibrated against 700 µl of reservoir solution composed of 1.4 M trisodium citrate brought to pH 6.5 with HCl. The dimensions of the rectangular crystals were approximately 0.4 × 0.3 × 0.2 mm. Prior to data collection, the crystal was placed in reservoir buffer supplemented with 20% glycerol, 0.006 mM orotate, 3 mM PRPP and 6 mM MgCl2.
Data Collection and Reduction
X-ray diffraction data were recorded from crystals flash-frozen in a stream of nitrogen at −178°C, on an image plate detector (Rigaku R-AXIS IV) coupled to a rotating-anode X-ray generator (Rigaku RU-H3R) using CuKα radiation (50 kV and 100 mA). The data were processed using the programs DENZO and SCALEPACK (29). Diffraction from these crystals is consistent with the orthorhombic space group C222 with cell parameters a = 105.57, b = 154.23 and c = 52.60 Å. On the basis of the space group symmetry, the unit-cell volume and the molecular weight of 23,546 Da, a single dimeric protein molecule per asymmetric unit was proposed (Vm = 2.27 Å3 Da−1 and solvent content = 45.47%). The Rsym on intensities, for 29410 reflections to 2.2 Å resolution (99.9% complete) was 4.7%. 91.3% of reflections had I/σ (I) > 1.0.
Structure Solution and Refinement
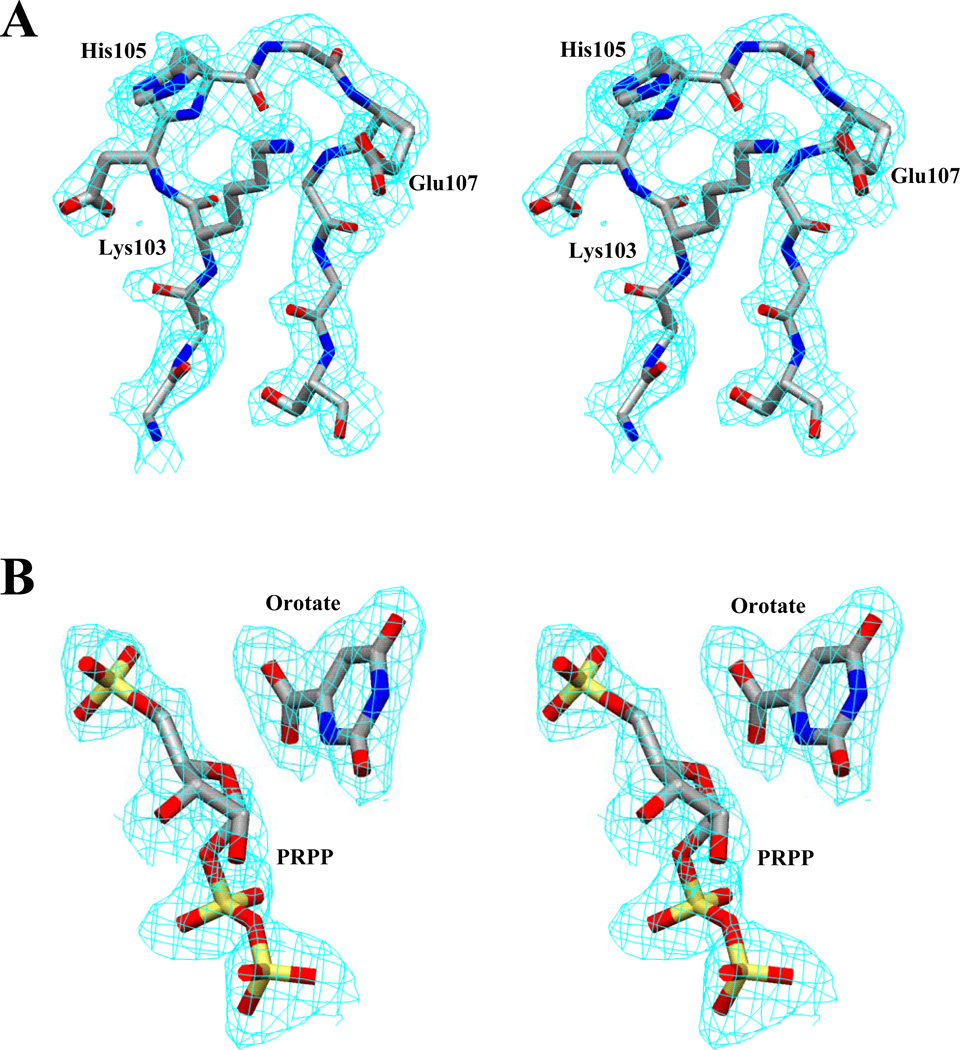
The structure was solved by molecular replacement using the AmoRe software package implemented in CCP4 (30) with the partially closed E. coli OMP synthase dimer (97 % sequence identity) as the search model (24). The best solution after rigid body refinement gave an R-factor of 46.6% and a correlation coefficient of 38.3% using 8.0 - 4.0 Å data. The OMP synthase complex structure was further refined using Xplor (31), without inclusion of the catalytic loop residues (102–108). Subsequently, the catalytic loop for subunit A and other smaller loops were built into the electron density map using the program O (32). The orotate, PRPP, Mg2+ and water molecules were included in the model, when structural refinement had reached Rfree of 33.0%. PRPP and orotate coordinates were taken from the Hetero-Compound Information Center Database (33), based on a non-productive base-analog complex with Toxoplasma gondii HGPRTase (34). The ligands were readily identified from significant and well-resolved peaks in the Fo-Fc map. 2Fo-Fc and Fo-Fc maps showed clear electron density for a cis peptide bond connecting Ala71 and Tyr72 in both subunits. Water molecules were built into 3σ density in Fo-Fc maps and retained in the model if the B-factors refined to below 60 Å2. The final model contains 419 residues (102B – 108B are not included), two orotate molecules, one PRPP molecule, one Mg2+ (no PRPP or Mg2+ was observed in the A subunit), and 384 water molecules with Rwork and Rfree of 22.5% and 28.9% respectively. The final model was analyzed with PROCHECK (35), with 91.2% of residues in the most favored regions, 8.5% in additional allowed regions and only Glu163 (A subunit) in disallowed regions, although this residue fits well in the electron density map. Details of the data collection and refinement parameters are given in Table 1. The catalytic loop of subunit A and the PRPP•orotate of subunit B fit well in the 1σ level 2Fo-Fc electron density map as shown in Figures 1A and 1B, respectively.
Table 1.
Data collection and refinement statistics.
| Data collection: | |
| Data range (Å) | 20.0 - 2.0 (2.07-2.00) |
| Completeness (%) | 99.9 (99.1) |
| Rsym (%) | 4.7 (37.5) |
| I/σ(I) | 20.35 (4.23) |
| Mosaicity | 0.47 |
| Observation (no) | 419629 |
| Unique reflections (no) | 29410 |
| Space group | C222 |
| a (Å) | 105.57 |
| b (Å) | 154.23 |
| c (Å) | 52.60 |
| Refinement: | |
| Missing residues 102 – 108 in the B monomer | |
| Resolution range (Å) | 8.0–2.2 |
| σ cutoff applied | 2σ(F) |
| No. of unique reflections | 24601 |
| Rwork (90% data) (%) | 22.5 |
| Rfree (10% data) (%) | 28.9 |
| Total no. of non-H atoms | 3725 |
| No. of protein atoms | 3274 |
| No. other atoms | 66 |
| No. of metal atoms | 1 |
| No. of water molecules | 384 |
| RMS deviations from ideal geometry: | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.944 |
| Torsion angles (°) | 13.786 |
| Improper angles (°) | 1.544 |
| Average B values (Å2) | |
| All atoms | 29.21 |
| All protein atoms | 27.05 |
| Main chain atoms | 25.31 |
| Side chain atoms | 28.87 |
| Water molecules | 44.80 |
| Phi-Psi map statistics | |
| Most favored regions | 91.2% |
| Additional allowed regions | 8.5% |
| Disallowed regions | 0.3% |
Figure 1.
Stereo view of the 2Fo-Fc electron density map at 1σ for the structure model. (A) Catalytic loop of subunit A. (B) Bound PRPP and orotate in subunit B.
Figures were made using PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC.
RESULTS
General Description of the Structure
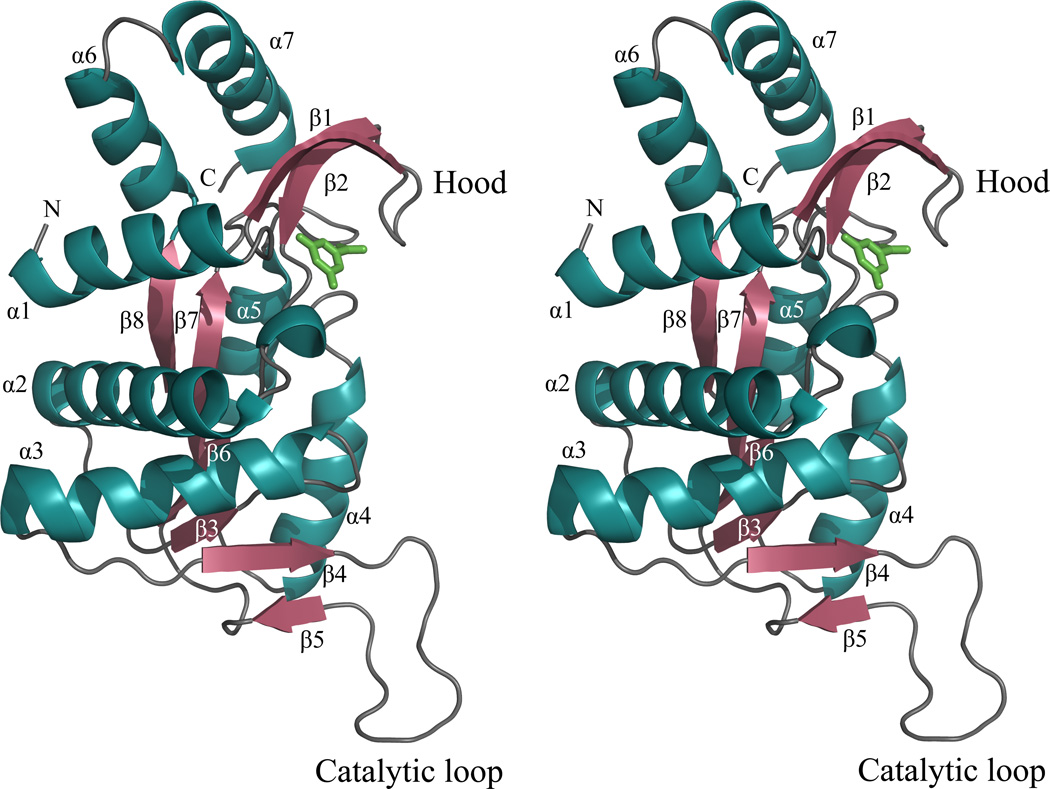
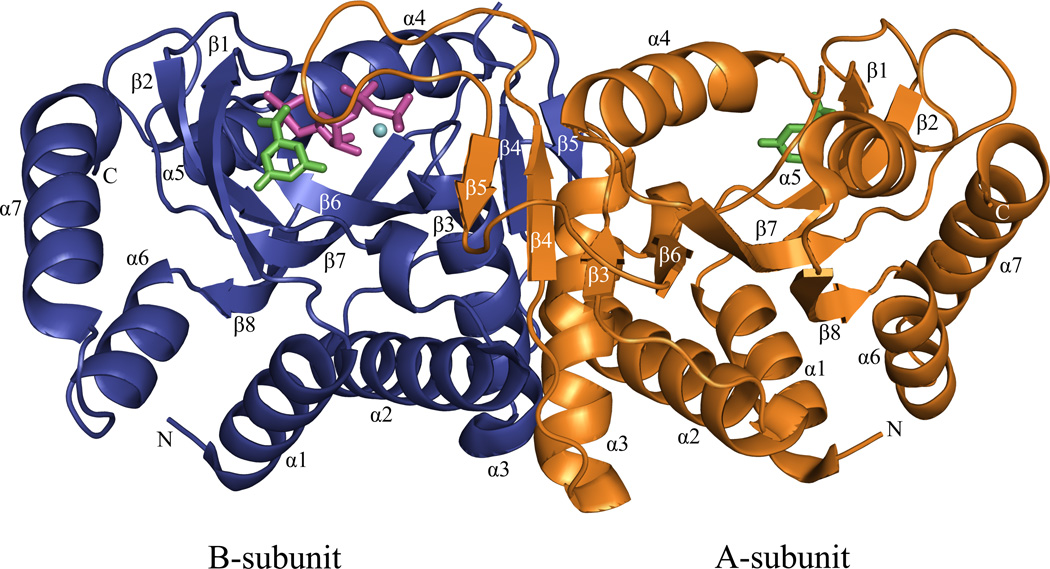
The E•MgPRPP•orotate complex is an asymmetric dimer similar to that observed in previous OMP synthase structures and other members of the PRTase family. The structure of the subunit is shown as a stereo ribbon diagram in Figure 2, and the dimer is shown in Figure 3. The Type I PRTase fold observed in Salmonella typhimurium and E. coli OMP synthases in previous complexes is followed by the complete substrates complex here. The structure contains an amino terminal β-hairpin "hood" anchored by an α-helix, a Rossmann fold core containing the prominent catalytic loop, and a pair of carboxyl terminal α-helices. These structural elements were renumbered here from previous publications (21) to include the β-strands of the hood. Strikingly, the catalytic loop of subunit A is seen in a previously undescribed conformation atop the active site of subunit B, where it forms a compact β-structure. The dimer interface is very similar to that described previously (20, 21), and when interface areas of the two subunits are superpositioned, they appear nearly identical. The exception to this occurs in the main chains and side chains of Arg99A and Lys100B, which have moved as discussed later. The asymmetry of the catalytic loop and at Arg99 and Lys100 is important for function.
Figure 2.
A stereo ribbon diagram of the A subunit of the OMP synthase complex. Orotate (green).
Figure 3.
A ribbon diagram of the dimeric OMP synthase complex. Orotate (green), PRPP (magenta) and Mg2+ (cyan) are drawn with ball and stick model. No PRPP or Mg2+ was observed in the A subunit. Subunit A (orange), subunit B (blue)
Asymmetry of the Dimer
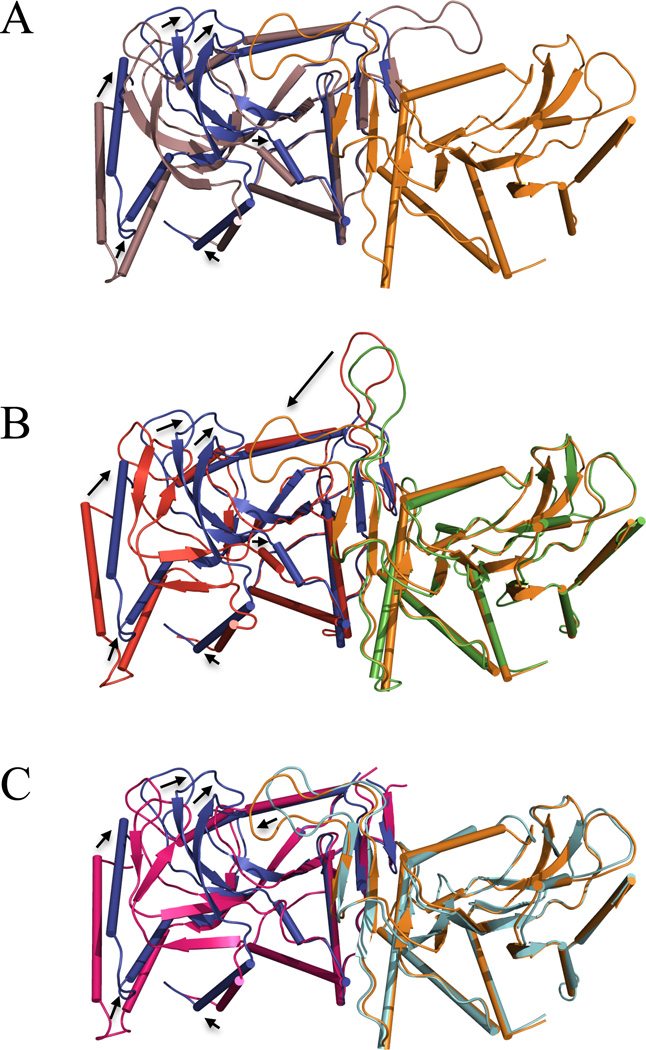
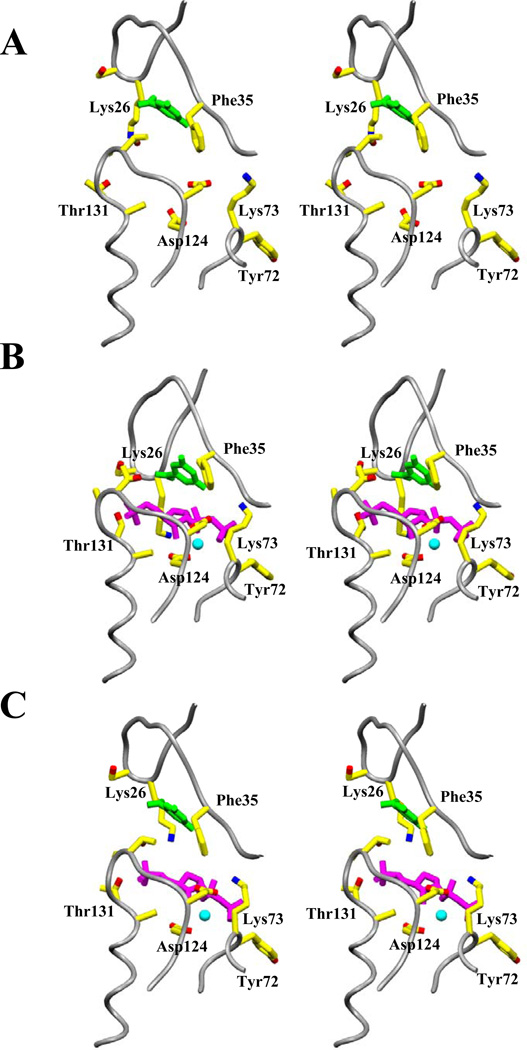
Previously reported structures of S. typhimurium E•OMP (1STO) and E•MgPRPP•orotate (1OPR) complexes were fully symmetric (20, 21). The SO42− bound complex of the E. coli enzyme (1ORO) was asymmetric with one of the catalytic loops disposed towards the catalytic site of the adjacent subunit, while the other loop was disordered (24). In the current complex (1LH0), the dimer is highly asymmetric, with movement of the catalytic loop from subunit A and dramatic structural rearrangements of subunit B, yielding a more buried active site than the previous sulfate complex. Figure 4 shows overlays of subunit A and B (panel A), the current structure and the more loop-open E•MgPRPP•orotate structure of Scapin et al. (21, Panel B), and the current structure and the asymmetric sulfate complex of Henriksen et al. (24, Panel C). A few key movements are described here. The hood region (Cα of Lys26B) has moved 5.4 Å toward the subunit interface (Cα of Tyr95B), together with α6 and α7. The segment composed of residues 124–130, and its extension α4 (130–142), shows a 1.6 Å rms deviation between the subunit A and B structures. The difference is mainly the result of an upward motion of the backbone at 5-phosphate binding residues 127B–130B, where the side chain of Thr128 forms a new hydrogen bond with the orotate carboxylate. In subunit A, the phosphate-binding residues are comparatively disordered (with poor electron density). Most visibly, the catalytic loop of subunit A has moved 22 Å (Cα of Gly106A 1LH0 vs. 1OPR, 21) and reordered to the top of subunit B active site. Lys100 of subunit B extends into the subunit B active site to interact with PRPP. As part of the loop movement, Arg99A makes strong interactions across the subunit interface with PRPP in subunit B that pull the A loop toward subunit B. The loop movement pulls Lys100A away from the PRPP binding site of subunit A. Competition between the two subunits for preferential hydrogen bonding by Arg99 and Lys100 thus may drive the observed asymmetry, as discussed below. It should be noted that in the C222 space group, subunit A and subunit B experience different packing interactions, including close packing around the carboxyl terminal of subunit B. It is not possible a priori to determine whether these crystal packing interactions result from, or help induce, the observed asymmetry.
Figure 4.
Superposition of OMP synthase structures. (A) Subunit A of the current structure (1LH0, violet) is overlaid on subunit B of 1LH0 (blue) of the dimer on the left side and subunit A is shown on the right side in orange. (B) The two subunits (red and green) of the previous loop-open E•MgPRPP•orotate dimer (1OPR, 21) are overlaid on the current complex (1LH0, blue and orange). (C) The two subunits of the sulfate complex in pink and cyan (1ORO, 23) are overlaid on the current structure (1LH0) in blue and orange. In each panel, residues of the interface were used for superpositioning. Black arrows show major movements. Ligands are not shown.
Structure of the Active Site
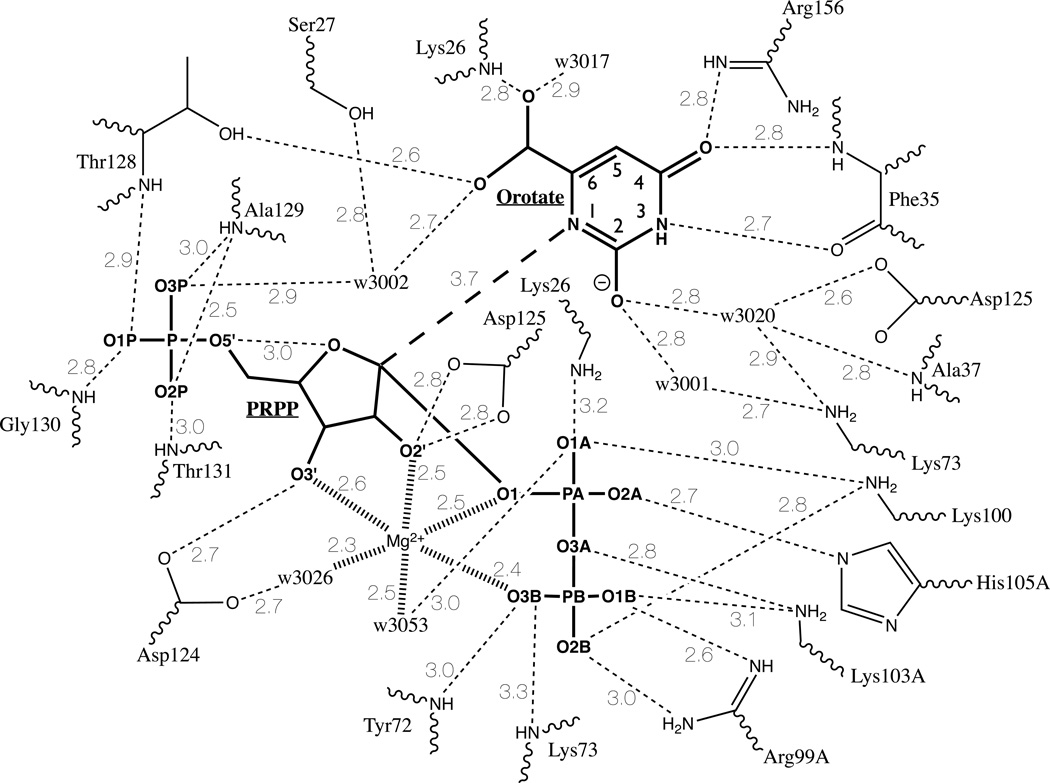
The catalytic site of loop-closed subunit B is formed by contributions from the hood, residues at the carboxyl ends of β3, β4, β6, β7, and residues from the catalytic loop of subunit A. The hood forms a cover to the base-binding site in all complexes solved to date (20, 21, 24). The ε-amino group of Lys26B interacts with the pyrophosphate oxygen O1A of PRPP (3.2 Å) and with the carboxylate of Glu135B in α4 (2.8 Å; this distance is 9.0 Å in subunit A) (Figure 5). The main chain oxygen and nitrogen of Phe35B interact with bound orotate, which also stacks against the side chain of Phe34B in the hood domain. The guanidino group of Arg156B from the end of β7 interacts with orotate O4 (2.8 Å). The conserved PRPP binding motif (5) extends from 120–132, which encompasses β6 and its extension into a tight loop. The top of β6 provides Asp124B, whose carboxyl interacts with O3' of PRPP (2.7 Å), and Asp125B whose carboxyl is near O2' of PRPP and O2 of orotate (Figure 5). The highly conserved residues Thr128B, Ala129B, Gly130B, Thr131B, and Ala132B provide interactions via their amide nitrogens and the side chain hydroxyls with the 5-phosphate of PRPP. The main chain nitrogens of Tyr72B and Lys73B in β3 project into the active site where they interact with O3B of the pyrophosphate of PRPP. Residues from β4 and the catalytic loop provide interactions to the bound pyrophosphate group of PRPP through the guanidino nitrogens of Arg99A, the ε-amino of Lys100B, ε-amino of Lys103A, and the ε-imino nitrogen of His105A.
Figure 5.
Cartoon of binding interactions in the active site of Subunit B. Substrate covalent bonds are shown in bold. Hydrogen bonds (< 3.5 Å) are shown as dashed lines. Bold dashed line indicates N1 (orotate) to C1 (PRPP) reaction coordinate. Mg2+ coordination bonds are shown as hashed lines. Solvent water molecules are indicated by “w” with the four digits of the pdb designation shown. Note that residues Lys26. Lys73, and Asp125 are each shown twice in the diagram.
The non-proline cis-peptide bond between Ala71 and Tyr72 is unusual. In OMP synthase, the previous PRPP and orotate complex contained a trans-peptide bond at this position, although the sulfate complex showed the cis-peptide bond. HGPRTase has a non-proline cis-peptide at the homologous position (14).
Structure of the Catalytic Loop
The catalytic loop of subunit A lies tightly against the active site formed by subunit B. The loop in this conformation exhibits β-strand-like geometry over much of its length with 104A–108A forming a hydrogen-bonded turn (Figure 1A). There are 3 backbone hydrogen bonds of about 3 Å that are internal to the loop, N Lys103A - O Gly109A, N His105A - O Lys103A, and N Gly108A - O His105A. Two additional intra-loop bonds involve side chain nitrogens of Lys103A and Arg99A to Oε of Glu107A. The loop also interacts with the hood through N Gly106A forming a 2.8 Å hydrogen bond to the main chain carbonyl oxygen of Thr24B. In addition, hydrogen bonds form between the loop and bound ligands: the ε-amino group of Lys103A forms hydrogen bonds to the α-β bridging oxygen O3A (2.8 Å) and β-phosphate oxygen O1B (3.1 Å), the Nε nitrogen of His105A forms a hydrogen bond (2.7 Å) to the α-phosphate oxygen O2A. All these interactions contribute to stabilizing the closed conformation.
Orotate Binding
Orotate binding is similar in the two subunits of the 1LH0 complex, but somewhat different in detail from that reported earlier (21). In the closed subunit B, orotate is bound by residues Lys26B and Phe35B contributed by the hood, Arg156B from β7, and Thr128B from the phosphate-binding loop formed by the extension of β6. Bound orotate forms a stacking interaction with the side chain of Phe34B (see Figure 5). Orotate N1 is 3.7 Å from C1' of PRPP, its site for nucleophilic attack, and is close (3.9 Å) to O4' of PRPP. For bond formation between N1 of orotate and C1' of PRPP to occur, N1 must be in a deprotonated state. Figure 5 shows bound orotate in its N1 lactim, with O2 as the deprotonated enolate. This dianionic form of the base has been documented through transition state analysis of the S. typhimurium, human and Plasmodium OMP synthases (36, 37). The N1-O2 lactam could also be accommodated by the X-ray data. Two bound water molecules, w3020 and w3001, form interactions with O2 and may be involved in its deprotonation. The yeast OMP synthase shows two similarly positioned water molecules, and the authors propose their catalytic role in deprotonation from the lactim (25). In 1LH0, orotate N3 interacts with the main chain carboxyl oxygen of Phe35B (2.7 Å), as observed previously (21). Orotate O4 forms H-bonds from the main chain nitrogen of Phe35B and the guanidino nitrogen of Arg156B. The carboxylate of orotate is known to be a major contributor to base specificity (22, 39). One of the carboxylate oxygens is close (3.5 Å) to O4' of PRPP, which may enhance oxocarbenium ion formation. The same orotate carboxylate atom forms a hydrogen bond (2.8 Å) to the main chain nitrogen of Lys26B, and to one water molecule (w3017; 2.9 Å). The other carboxylate oxygen interacts with the side chain oxygen of Thr128B (2.6 Å) and a water molecule (w3002; 2.7 Å).
In subunit A, the active site is more loop-open and the bound orotate molecule makes similar but fewer interactions. The major difference in orotate binding between the two subunits is the hydrogen bond to Thr128 present in the B subunit. The latter hydrogen bond may be a critical determinant of the compact nature of B subunit, pulling the phosphate binding loop, and PRPP, towards the hood.
MgPRPP Binding
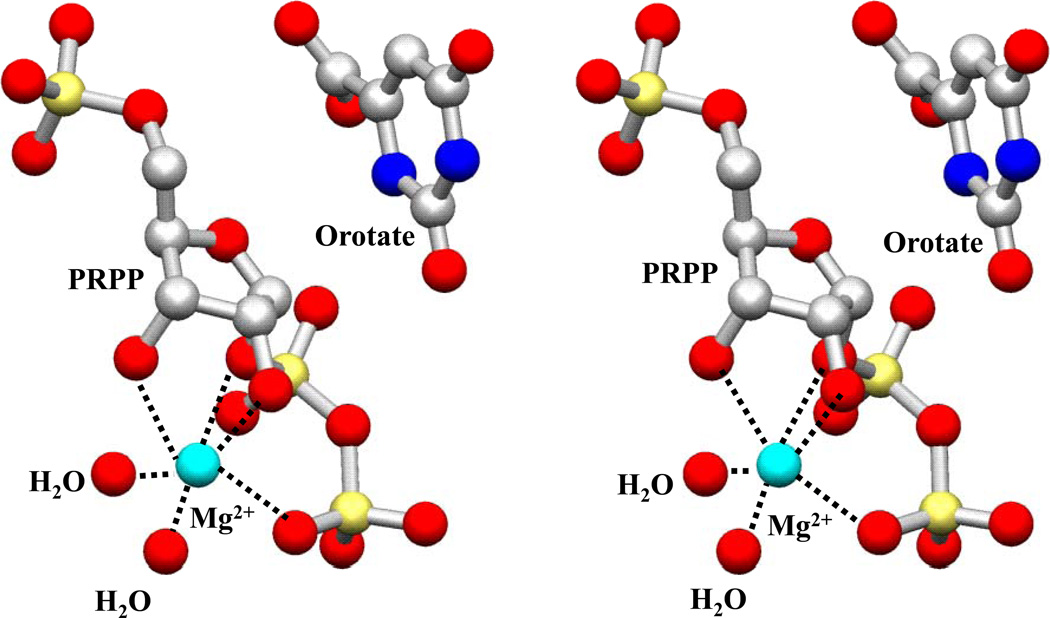
OMP synthase and GPAT (40) are distinct among Type I PRTases in having a single Mg2+ ion in the catalytic complex. The OMP synthase Mg2+ ion is six-coordinate (Figure 6) with a somewhat distorted octahedral geometry. The Mg2+ interacts with O2' (2.5 Å) and O3' (2.6 Å) of the ribosyl group, pyrophosphate oxygen O3B (2.4 Å) and C1’-PP-bridging O1 (2.5 Å) atoms of the pyrophosphoryl group and two water molecules (w3053; 2.5 Å, and w3026; 2.3 Å). The Mg2+ ion makes no direct interactions with the enzyme. This pattern is similar to that the first Mg2+ in the HGPRTase transition state analog structure (17). Disruption of the C1'-OPPi bond may relieve the distortion of coordination seen here. Neutralization of the pyrophosphate charge is accomplished by the positive charges from the catalytic loop (Arg99A, Lys100B, Lys103A) and the hood (Lys26B). The sugar conformation is difficult to discern unambiguously at 2.2 Å resolution; however, the refined PRPP is consistent with the furanose conformation reported in the Toxoplasma gondii HGPRTase•MgPRPP structure (34) that served as the starting point for the refinement.
Figure 6.
Stereo view diagram of the coordination of Mg2+ ion in subunit B.
The 5-phosphate group of PRPP is remarkable in that every oxygen participates in multiple hydrogen-bonds that firmly anchor the group to the main chain N atoms of residues of the 5-phosphate-binding loop (Thr128B-Gly132B; Figure 5). The hydrogen bond network serves to position O5' within 3.0 Å of the ribosyl ring oxygen. The sugar atoms O2' and O3' interact with the residues Asp125B (O2'; 2 bonds of 2.8 Å). Asp124B (O3'; 2.7 Å) and one water molecule (w3001, O3'; 3.0 Å, this interaction is not shown in Figure 5).
In subunit B, the pyrophosphoryl group of PRPP interacts to form a strikingly extensive and tight hydrogen bond network with residues from both catalytic loops, the hood, the β3-α3 turn, bound Mg2+ and several waters. The catalytic loop of subunit B contributes Lys100B, whose the ε-amino group forms hydrogen bonds to α- and β-phosphate. Subunit A provides Arg99A, which forms two bonds with β-phosphate. The catalytic loop of subunit A also provides Lys103A, which forms hydrogen bonds to α- and β-phosphate, and His105A, whose ε-imino nitrogen interacts with α-phosphate. The ε-amino group of hood residue Lys26B extends across orotate to form a hydrogen bond to the α-phosphate. The backbone nitrogens of Tyr72B and Lys73B interact with β-phosphate. All of the interactions of the pyrophosphoryl group are with residues that have moved from their positions in other structures.
The PRPP binding seen in subunit B of 1LH0 is strikingly different from that in the previous substrates complex, 1OPR, which is symmetric (Figure 7). In 1OPR the 5-phosphate of PRPP does not interact closely with the phosphate binding pocket in subunit A (3.0–4.1 Å NH—O distances), nor is it so close to O4' (4.9 Å). Residues 128–132 have higher (28–57 vs. an average 30 Å2) B-factors in this subunit. The side chain of Thr128 makes a 2.6 Å hydrogen bond with a 5-phosphate oxygen replacing one to the orotate carboxylate in subunit B and no hydrogen bonding interaction with Asp125A (4.7 Å) is present in subunit A. The pyrophosphoryl group makes remarkably different contacts in the open subunit. Hydrogen bonds to Lys100A are absent, only a single hydrogen bond is formed to Arg99B, and hydrogen bonds to catalytic loop residues Lys103B and His105B are not present. The interactions of main chain nitrogens of Tyr72A (2.7 Å) and Lys73A (2.3 Å) with the β-phosphate are preserved.
Figure 7.
Stereo views comparing the substrate binding area of PRPP complexes. The backbone is shown in worm format with gray color, and selected side chains (yellow), orotate (green), PRPP (red) and Mg2+ (light blue) are shown in ball and stick. (A) Subunit A, containing only orotate, of the current complex (lLH0). (B) Subunit B of the current loop-closed E•MgPRPP•orotate complex (lLH0). (C) Corresponding area of the previous loop-open E•MgPRPP•orotate complex (1OPR, 21), resembling the more open subunit A of lLH0.
Active Site Water Molecules
A major function of loop closure in Type I PRTases is to exclude solvent from the developing transition state (10, 21, 23). However, the active site of subunit B contains a total of 10 water molecules, with B-factors ranging from 15.5 to 51.2 Å2, compared to the overall B-factor of 29 Å2 for atoms of the protein. Since OMP synthase is very effective in avoiding alternative hydrolysis reactions of PRPP and OMP (CG, unpublished), these water molecules are clearly restrained from reacting with the developing transition state.
DISCUSSION
The key finding of our work is that S. typhimurium OMP synthase can exist in a conformer with its catalytic loop in a highly structured β-form that sequesters the active site from bulk solvent and contributes key residues to the active site. Unlike HGPRTase, the OMP synthase loop moves across the subunit interface to affect catalysis in the adjacent site. Although this loop-down conformer had been anticipated (22–24, 26, 41), its direct observation here permits much fuller interpretation of the mechanistic roles of specific residues. Remarkably, loop movement from subunit A is accompanied by a dramatic tightening of the structure of subunit B, with the formation of many new hydrogen bonds.
The current structure results from co-crystallization. The complex, like that published previously (1OPR, 21), contains orotate, PRPP, and Mg2+ with no observational OMP or PPi. Figure 7 presents a comparison of the loop-open, orotate-containing active site of subunit A of 1LH0 (Figure 7A) versus the loop-closed orotate- and MgPRPP-containing active site of subunit B (Figure 7B). Figure 7C shows the structure of 1OPR, with an active site structure resembling that of subunit A of 1LH0. The previous 1OPR complex was the result of a crystal soaking procedure, and crystal lattice constraints or crystallization components may have prevented the achievement of the loop-closed conformation. In solution, the on-enzyme equilibrium between E•MgPRPP•orotate and E•OMP•PPi Michaelis complexes (Kint) is 1:2, with rapid (300 s−1) interconversion (27). The selective stabilization of one form may also be linked to barriers imposed by the crystal lattice. An alternative is that crystallization has captured a non-productive complex. However, the nearly ideal positions of catalytic residues and substrates suggests a productive form is being viewed. PRPP is an unstable molecule, susceptible to base catalyzed elimination of β-phosphate following intramolecular attack of O2' on the α-phosphate (42). The intact PRPP in the complex may originate in part from the large molar excess present during crystal growth and because cryoprotectant solution contained fresh PRPP. However, unpublished studies (CG laboratory) have shown that OMP synthase in fact protects bound PRPP in binary complexes from base-catalyzed degradation, and this protection may be enhanced in a completely closed complex.
The rearrangement observed in subunit B of 1LH0 versus 1OPR, and the striking asymmetry of the dimer in 1LH0, can be rationalized on the basis of interactions in the individual subunits. The presence of PRPP sets up a competition for hydrogen bonds donated by Lys100 and Arg99. As subunit B captures the catalytic loop of subunit A, Lys100A is pulled away from its position in the PRPP binding site of subunit A. In subunit B, the correct positioning of Lys100B and Arg99A allows PRPP movement and permits the catalytic loop of subunit A to deploy over subunit B, promoting the interaction of Lys103A and His105A with bound PRPP. The position of PRPP in 1OPR remains closer to the subunit interface, and both catalytic loops, which are being pulled by the interaction of Lys100 with PRPP, remain open and disordered. The failure of the hood of subunit A of 1LH0 to move toward the phosphate binding loop appears to result from the lack of stabilizing interactions with the catalytic loop of subunit B, rather than any obvious steric conflict. It should be highlighted that the position of catalytic loop B is not represented in the electron density and thus it may not be in the fully solvent-extended conformation seen earlier (21). The structural asymmetry seen here does raise the very interesting possibility that enzyme function is also asymmetric. Related observations by the groups of McClard and Witte on the kinetics, binding and structure of the yeast enzyme, and of the bacterial complex which we presented in 1LH0, have led to a detailed mechanism for alternating sites cooperativity (25, 26).
The functional correlates of the structural asymmetry seen here remain to be established. Previous chemical modification kinetics and stoichiometries (41), steady-state (43) and pre-steady-state (27) kinetic studies have provided no evidence for functional kinetic cooperativity by the S. typhimurium enzyme, although the McClard group has observed such behavior for the yeast enzyme (26). NMR and partial proteolysis studies of binary complexes identified forms of the S. typhimurium enzyme with both catalytic loops in a closed position and although the structure of the loop was not characterized in that work, heterogeneity might have been observed through chemical shift exchange. In addition, binding studies with binary complexes have shown that each of the four substrates for the enzyme can bind to two sites per dimer with a single KD (27). Net reverse catalysis by OMP synthase involves rapid chemistry (100 s−1) and slower PRPP release, which requires catalytic loop opening at 400 s−1, and partitioning of the open complex between loop closure (12,000 s−1) and PRPP release (2200 s−1, 28). This indicates a delicate poise between forces serving to stabilize the loop-closed structure (including the many bonds described here) and those tending to promote its return to a solvent-extended position. These may include hydration of the loop and the active site itself. It is quite possible that interactions between the two catalytic sites may help subunit B release product as subunit A fills and recaptures the loop of subunit B. This could produce the flattening of the free-energy profile that enhances enzyme efficiency.
Comparisons to other Ribosyltransferases
Growing knowledge of protein and transition state structures of ribosyltransferases has resulted in a general proposal for their action (44) in which oxocarbenium-like transition states are stabilized by the 5’ oxygen and a second oxygen, here the carboxyl of orotate, and that reaction requires migration of the developing oxocarbenium entity (C1'-O4') in the reaction trajectory. Here, that movement appears to be about 1.8 Å (the 4.8 Å N-C-O axis, minus 3.0 Å for the N1-C1' and C1'-OPP covalent bonds), although a Michaelis-like E•OMP•MgPPi complex will help define this motion better. OMP synthase provides a paradigm in this group because its reaction is freely reversible and a transition state structure is known (36, 37).
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health (GM48623 to C.G. and S.C.A.).
We thank Bradley Bizzle for preparation of the enzyme, and Eric Barr for assistance with ChemDraw. CG wishes to thank Drs. Richard Furneaux and Peter Tyler (Industrial Research Ltd, New Zealand) for pointing out the potential role of orotate in stabilizing the oxocarbenium character of the transition state.
Abbreviations
- OMP synthase
orotate phosphoribosyltransferase
- PRTase
phosphoribosyltransferase
- HGPRTase
hypoxanthine-guanine PRTase
- OMP
orotidine 5'-phosphate
- PRPP
D-5-phosphoribosyl-α-1-pyrophosphate
- GPAT
-
glutamine PRPP amidotransferase
We use the ' symbol to denote atoms of the ribosyl ring of PRPP or OMP.
Footnotes
Coordinates for this structure have been deposited in the Protein Data Bank (PDB) and are assigned accession number 1LH0.
REFERENCES
- 1.Jones ME. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Ann. Rev. Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 2.Suttle DP, Becroft DMO, Webster DR. Disorders of Pyrimidine Metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic Basis of Inherited Disease. 6th ed. 1989. pp. 1095–1126. [Google Scholar]
- 3.Musick WD. Structural features of the phosphoribosyltransferases and their relationship to the human deficiency disorders of purine and pyrimidine metabolism. CRC Crit. Rev. Biochem. 1981;11:1–34. doi: 10.3109/10409238109108698. [DOI] [PubMed] [Google Scholar]
- 4.Smith JL. Forming and inhibiting PRT active sites. Nat. Struct. Biol. 1999;6:502–504. doi: 10.1038/9266. [DOI] [PubMed] [Google Scholar]
- 5.Hove-Jensen B, Harlow KW, King CJ, Switzer RL. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J. Biol. Chem. 1986;261:6765–6771. [PubMed] [Google Scholar]
- 6.Eriksen TA, Kadziola A, Bentsen AK, Harlow KW, Larsen S. Structural basis for the function of Bacillus subtilis phosphoribosylpyrophosphate synthetase. Nat. Struct. Biol. 2000;7:303–308. doi: 10.1038/74069. [DOI] [PubMed] [Google Scholar]
- 7.Eads JC, Ozturk D, Wexler TB, Grubmeyer C, Sacchettini JC. A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase. Structure. 1997;5:47–58. doi: 10.1016/s0969-2126(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 8.Shin DH, Oganesyan N, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum. J. Biol. Chem. 2005;280:18326–18335. doi: 10.1074/jbc.M501622200. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Zhang X, Bheda P, Revollo JR, Imai S, Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat. Struct. Mol. Biol. 2006;13:661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- 10.Eads JC, Scapin G, Xu Y, Grubmeyer C, Sacchettini JC. The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase with bound GMP. Cell. 1994;78:944–948. doi: 10.1016/0092-8674(94)90301-8. [DOI] [PubMed] [Google Scholar]
- 11.Vos S, Parry RJ, Burns MR, de Jersey J, Martin JL. Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase. J. Mol. Biol. 1998;282:875–889. doi: 10.1006/jmbi.1998.2051. [DOI] [PubMed] [Google Scholar]
- 12.Munagala N, Basus VJ, Wang CC. Role of the flexible loop of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus in enzyme catalysis. Biochemistry. 2001;40:4303–4011. doi: 10.1021/bi0026932. [DOI] [PubMed] [Google Scholar]
- 13.Shi W, Li CM, Tyler PC, Furneaux RH, Cahill SM, Girvin ME, Grubmeyer C, Schramm VL, Almo SC. The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor. Biochemistry. 1999;38:9872–9880. doi: 10.1021/bi990664p. [DOI] [PubMed] [Google Scholar]
- 14.Heroux A, White EL, Ross LJ, Davis RL, Borhani DW. Crystal structure of Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase with XMP, pyrophosphate, and two Mg2+ ions bound: insights into the catalytic mechanism. Biochemistry. 1999;38:14495–14506. doi: 10.1021/bi990508i. [DOI] [PubMed] [Google Scholar]
- 15.Balendiran GK, Molina JA, Xu Y, Torres-Martinez J, Stevens R, Focia PJ, Eakin AE, Sacchettini JC, Craig SP. Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding. Protein Sci. 1999;8:1023–1031. doi: 10.1110/ps.8.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heroux A, White EL, Ross LJ, Borhani DW. Crystal structures of the Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase-GMP and -IMP complexes: comparison of purine binding interactions with the XMP complex. Biochemistry. 1999;38:14485–14494. doi: 10.1021/bi990507q. [DOI] [PubMed] [Google Scholar]
- 17.Shi W, Li CM, Tyler PC, Furneaux RH, Grubmeyer C, Schramm VL, Almo SC. The 2.0 Å structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor. Nat. Struct. Biol. 1999;6:588–596. doi: 10.1038/9376. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Eads JC, Sacchettini JC, Grubmeyer C. Kinetic mechanism of human hypoxanthine-guanine phosphoribosyltransferase: rapid phosphoribosyl transfer chemistry. Biochemistry. 1997;36:3700–3712. doi: 10.1021/bi9616007. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Burgner JW, Krahn JM, Smith JL, Zalkin H. Tryptophan fluorescence monitors multiple conformational changes required for glutamine phosphoribosylpyrophosphate amidotransferase interdomain signaling and catalysis. Biochemistry. 1999;39:11659–11669. doi: 10.1021/bi991060o. [DOI] [PubMed] [Google Scholar]
- 20.Scapin G, Grubmeyer C, Sacchettini JC. Crystal structure of orotate phosphoribosyltransferase. Biochemistry. 1994;33:1287–1294. doi: 10.1021/bi00172a001. [DOI] [PubMed] [Google Scholar]
- 21.Scapin G, Ozturk DH, Grubmeyer C, Sacchettini JC. The crystal structure of the orotate phosphoribosyltransferase complexed with orotate and alpha-D-5-phosphoribosyl-1- pyrophosphate. Biochemistry. 1995;34:10744–10754. doi: 10.1021/bi00034a006. [DOI] [PubMed] [Google Scholar]
- 22.Ozturk DH, Dorfman RH, Scapin G, Sacchettini JC, Grubmeyer C. Locations and functional roles of conserved lysine residues in Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry. 1995;34:10755–10763. doi: 10.1021/bi00034a007. [DOI] [PubMed] [Google Scholar]
- 23.Ozturk DH, Dorfman RH, Scapin G, Sacchettini JC, Grubmeyer C. Structure and function of Salmonella typhimurium orotate phosphoribosyltransferase: protein complementation reveals shared active sites. Biochemistry. 1995;34:10764–10770. doi: 10.1021/bi00034a008. [DOI] [PubMed] [Google Scholar]
- 24.Henriksen A, Aghajari N, Jensen KF, Gajhede M. A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase. Biochemistry. 1996;35:3803–3809. doi: 10.1021/bi952226y. [DOI] [PubMed] [Google Scholar]
- 25.González-Segura L, Witte JF, McClard RW, Hurley TD. Ternary complex formation and induced asymmetry in orotate phosphoribosyltransferase. Biochemistry. 2007;46:14075–14086. doi: 10.1021/bi701023z. [DOI] [PubMed] [Google Scholar]
- 26.McClard RW, Holets EA, MacKinnon AL, Witte J. Half-of-sites binding of Orotidine 5’-Phosphate and α-D-5-Phosphorylribose 1-Diphosphate to orotate phosphoribosyltransferase from Saccharomyces cerevisiae supports a novel variant of the Theorell-Chance mechanism with alternating site catalysis. Biochemistry. 2006;45:5330–5342. doi: 10.1021/bi051650o. [DOI] [PubMed] [Google Scholar]
- 27.Wang GP, Lundegaard C, Jensen KF, Grubmeyer C. Kinetic mechanism of OMP synthase: a slow physical step following group transfer limits catalytic rate. Biochemistry. 1999;38:275–283. doi: 10.1021/bi9820560. [DOI] [PubMed] [Google Scholar]
- 28.Wang GP, Cahill SM, Liu X, Girvin ME, Grubmeyer C. Motional dynamics of the catalytic loop in OMP synthase. Biochemistry. 1999;38:284–295. doi: 10.1021/bi982057s. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- 31.Brunger AT. X-PLOR Version 3.1. New Haven, CT, USA: Yale University Press; 1992. [Google Scholar]
- 32.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Kleywegt GJ, Jones TA. Databases in Protein Crystallography. Acta Cryst. 1998;D54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 34.Heroux A, White EL, Ross LJ, Kuzin AP, Borhani DW. Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis. Structure. 2000;8:1309–1318. doi: 10.1016/s0969-2126(00)00546-3. [DOI] [PubMed] [Google Scholar]
- 35.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 36.Tao W, Grubmeyer C, Blanchard JS. Transition state structure of Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry. 1996;35:14–21. doi: 10.1021/bi951898l. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Luo M, Schramm VL. Transition states of Plasmodium falciparum and human orotate phosphoribosyltransferases. J. Am. Chem. Soc. 2009;131:4685–4694. doi: 10.1021/ja808346y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data or Biochemical Research. 3rd ed. Oxford: Oxford Science Publications; 1986. [Google Scholar]
- 39.Niedzwicki JG, Iltzsch MH, el Kouni MH, Cha S. Structure-activity relationship of pyrimidine base analogs as ligands of orotate phosphoribosyltransferase. Biochem. Pharmacol. 1984;33:2383–2395. doi: 10.1016/0006-2952(84)90710-x. [DOI] [PubMed] [Google Scholar]
- 40.Krahn JM, Kim JH, Burns MR, Parry RJ, Zalkin H, Smith JH. Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry. 1997;36:11061–11068. doi: 10.1021/bi9714114. [DOI] [PubMed] [Google Scholar]
- 41.Grubmeyer C, Segura E, Dorfman R. Active site lysines in orotate phosphoribosyltransferase. J. Biol. Chem. 1993;268:20299–20304. [PubMed] [Google Scholar]
- 42.Khorana HG, Fernandes JF, Kornberg A. Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J. Biol. Chem. 1958;230:941–948. [PubMed] [Google Scholar]
- 43.Bhatia M, Vinitsky A, Grubmeyer C. Kinetic mechanism of orotate phosphoribosyltransferase from Salmonella typhimurium. Biochemistry. 1990;29:10480–10487. doi: 10.1021/bi00498a009. [DOI] [PubMed] [Google Scholar]
- 44.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]