1 Introduction
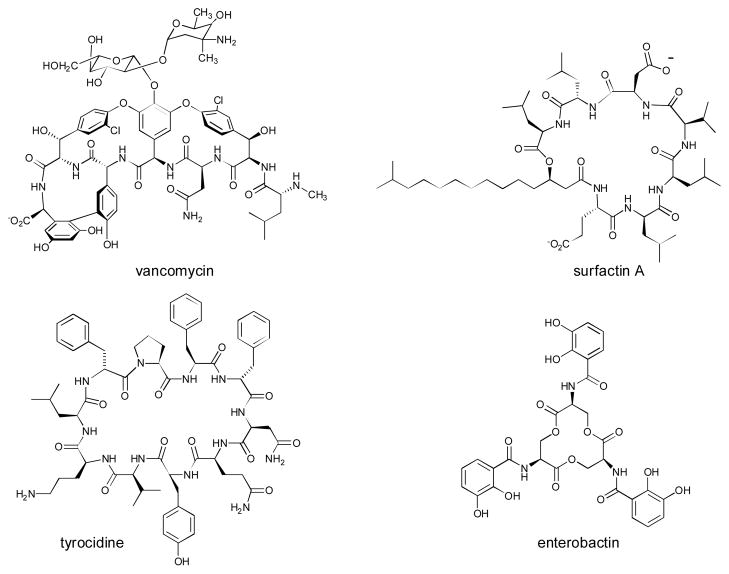
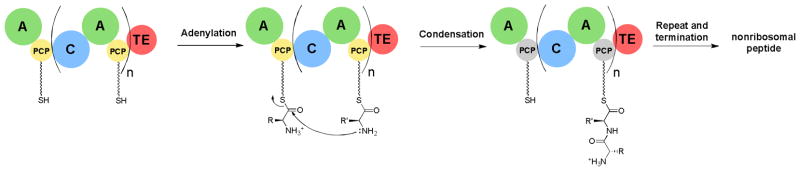
Nonribosomal peptide natural products constitute a large, structurally diverse family of biologically active compounds (Fig. 1).1 The secondary metabolites are synthesized on large multidomain, modular enzyme assemblies (nonribosomal peptide synthetases, NRPSs). NRPSs contain three core domains: adenylation (A), condensation (C) and peptidyl carrier protein (PCP). Together, these domains constitute the repeating module responsible for activation and incorporation of a single amino acid into the growing peptide chain (Fig. 2). The basic chemical logic and structure of NRPSs has been reviewed in recent literature.2–5 This review will focus on the structure and unusual chemistry of NRPSs published in recent years.
Fig. 1.
Example nonribosomal peptide (NRP) natural products.
Fig. 2.
Schematic of an nonribosomal peptide synthetase (NRPS) assembly line. The peptidyl carrier domain (PCP) has a phosphopantethiene prosthetic group terminated with a thiol (wavy lines). The adenylation (A) domain activates amino acids and the condensation (C) domain makes the amide linkages. Termination is frequently catalyzed by a thioesterase (TE) domain.
NRPSs utilize a thiol-template mechanism that is functionally similar to the enzymology used by fatty acid and polyketide synthases.6,7 A central feature is the phosphopantetheine prosthetic group, providing a flexible arm, about 20 Å in length, terminating with a thiol.8 This prosthetic group, which is derived from coenzyme A, is added to small carrier domains of NRPSs by a phosphopantetheinyl transferase (PPTase).9 The first step in an NRPS assembly line is the selection of a specific amino acid by the A domain. Using a chemical activation pathway that is shared with the aminoacyl-tRNA synthetases of ribosomal enzymology, A domains use ATP to make an amino acid mixed anhydride-AMP ester intermediate.10 The A domain then loads the activated amino acid to the phosphopantetheinyl (holo-)PCP domain resulting in a covalent thioester adduct. After amino acids are loaded onto each PCP domain, the C domain of the downstream module catalyzes the coupling of the amino group of the donor substrate with the upstream thioester, forming an amide bond and releasing the phosphopantetheine arm. The growing peptide is further lengthened according to a synthetic order that is co-linear with the modules in the synthetase. At the conclusion of the peptide elongation process, the mature peptide remains tethered to the final module as a thioester. Release from the PCP domain is frequently catalyzed by a C-terminal thioesterase (TE) domain, hydrolyzing the bound intermediate to a carboxylate and regenerating the phosphopantetheine-functionalized synthetase. As a result of the modular, co-linear organization of the synthesis, the overall process is frequently compared to an assembly line. Based on the wide range of chemical structures seen in NRP natural products, molecular diversity is obtained by combining a large precursor pool of building blocks with alternate linkages and enzymatic tailoring.
2 Structures of NRPS domains
X-ray crystal and NMR structures have been determined for both individual NRPS domains and multidomain constructs. Atomic resolution structures provide details of active site chemistry and the organization of the macromolecular assembly.
2.1 Peptidyl carrier protein domains
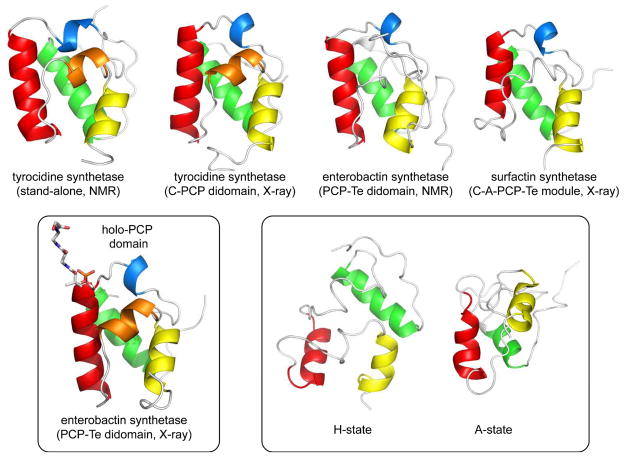
The peptidyl carrier protein (PCP) domain is a small (~9kDa) subunit that carries the phosphopantetheinyl prosthetic group and is thus the site of covalent linkage for amino acids and the growing peptide chain.6,7 The domain is functionally homologous to acyl carrier protein (ACP) domains integral to fatty acid and polyketide biosynthesis. The first structure determined of a PCP domain was the NMR structure of an excised domain from the tyrocidine NRPS of B. brevis published in 2000.11 The small domain is a distorted four-helix bundle and subsequent NMR and X-ray structures of PCP domains from other synthetases revealed a common motif (Fig. 3A).12–15 The functional serine residue is at the end of one of the helices on a disordered loop region. A conserved serine residue in the motif (I/L)GG(D/H)SL is the site of attachment for the pantetheine prosthetic group as a seryl phosphodiester.
Fig. 3.
Representative structures of PCP domains from NRPS assembly line constructs. (a) Structures of apo-PCP domains, PDB codes: 2GDW, 2JGP, 2ROQ, 2VSQ (left to right). (b) Structure of a holo-PCP domain showing site of phosphopantethiene attachment, PDB code, 3TEJ. (c) Alternate conformations of a tyrocidine PCP domain observed in the context of binding partners, PDB codes, 2GDX, 2GDY. The N- and C-termini of the domains are indicated.
The carrier domains deliver substrates to multiple protein partners presumably through formation of specific protein/protein interactions. The small size of the PCP domain and phosphopantetheinyl arm in relation to that of the other enzyme players in the pathway suggests that a significant amount of domain reorganization is necessary for the NRPS assembly line to function. Distinct conformations of PCP domains have been observed by changing the state of the PCP (holo vs. apo) or by adding protein binding partners.16 As illustrated in Figure 3B, re-examination of the tyrocidine PCP NMR spectra revealed two conformations that were in slow exchange on the time-scale of the experiment. Both the apo and holo (with phosphopantetheine loaded) forms of the PCP domain each adopt two conformations. The A/H state (Fig. 3A) is shared between the two, and a distinct state (A-state or H-state) is unique to each form of the carrier domain. Each of the structural conformations represents a significant reorientation of the helical bundle and the position of the prosthetic group. Importantly, these states showed differential interaction with partner enzymes. In addition to NMR studies of PCP domains, structures of carrier domains in the context of multidomain NRPS fragments have been determined. The didomains PCP-C12 and PCP-TE13 along with the tetra-domain C-A-PCP-TE14 all show the carrier domain in an A/H conformation. In addition, the recent structure of a phosphopantetheine loaded PCP-TE didomain provides structural insight into prosthetic group attachment of a holo-PCP domain (Fig. 3C).15
2.2 Adenylation domains
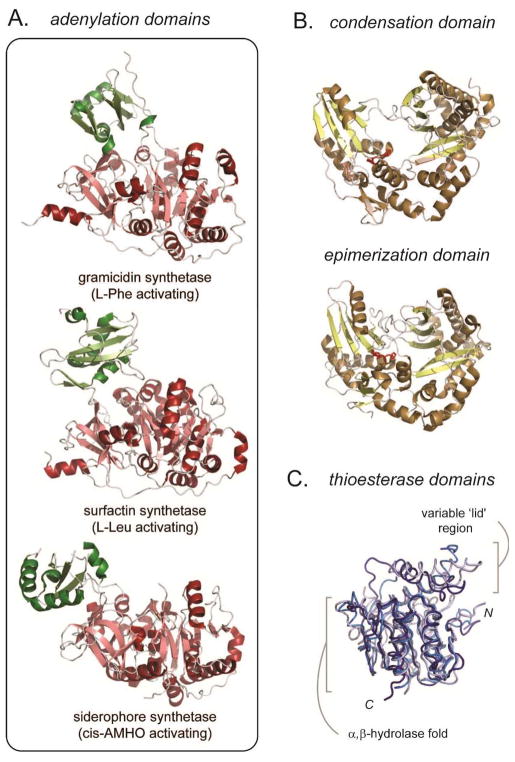
Adenylation (A) domains are responsible for both activating the amino acid and loading the building block onto the adjacent PCP domain. An adenylation domain from the gramicidin S synthetase provided the first atomic resolution structure for an NRPS domain (Fig. 4A).12 The structure demonstrated that A domains share a common fold with known adenylation enzymes such as firefly luciferase and acyl-CoA ligases.13 A domains contain a large N-terminal region with a majority of the catalytic functionality and a smaller C-terminal subdomain. The chemistry of A domains involves two steps: ATP-dependent activation of the amino acid by adenylation of the carboxylic acid forming an aminoacyl adenylate and reaction of the activated substrate with the phosphopantetheine arm of a PCP to generate an aminoacyl thioester. It is believed that the interplay between the two mobile subdomains orchestrate the two distinct chemistries. Examination of the structure of the gramicidin A domain allowed identification of core residues responsible for substrate recognition, and led to the formulation of a code for the assignment of amino acid specificity based on primary sequence.19–21 In addition to the A domain from gramicidin S synthetase, the X-ray structures of aryl acid activating adenylation domains from the bacillibactin22 and acinetobactin23 NRPS biosynthetic pathways have been determined showing a similar subdomain architecture. More recently, the structure of an A domain from the eukaryote N. lolii was determined.24 The fungal enzyme activates the large amino acid cis-AMHO (Nδ-cis-anhydromevalonyl-Nδ-hydroxy-L-ornithine) as part of the biosynthesis of a siderophore natural product. The work provides a structural basis for amino acid binding in high organisms and confirms the inherent mobility of the two subdomains (Fig. 4A).
Fig. 4.
Representative structures of catalytic domains in NRPS assembly lines. (a) X-ray structures of three adenylation domains, PDB codes 1AMU, 2VSQ, 1ITE (top to bottom), illustrating the flexibility of the two subdomains relative to one another. (b) Representative structure of a condensation and epimerization domain from tyrocidine synthetase. The condensation domain is from a didomain PCP-C construct, PDB code: 2JGP, and epimerization domain is a stand-alone construct, PDB code: 2XHG. Two histidine residues implicated in catalysis are shown in red. (c) Overall of three NRPS thioesterase domains, PDB codes: 1JMK, dark blue; 2CB9, blue; 3TEJ, light blue. The flexible, substrate specific ‘lid’ region is illustrated.
2.3 Condensation domains
NRPS condensation (C) domains are responsible for the formation of amide bonds between the amino acid building blocks. The domain couples the amine of a PCP-bound phosphopantetheinyl amino acid to the upstream peptidyl thioester, transferring the growing chain to the downstream PCP domain (see Fig. 2). NRPS C domains share sequence homology with the diverse family of acyltransferases, for example the widely studied chloramphenicol acetyltransferase.25 Common among the family is a conserved HHXXXDG motif, which is believed to play a role in the chemistry of amide bond formation. The first X-ray structure determined for an NRPS C domain was VibH, a free-standing domain in the vibriobactin biosynthetic pathway in V. cholera.26 The structure shows a pseudodimeric fold with a deep active site cleft formed between the two distinct subdomains with the catalytic histidines located at the base of the cleft (Fig. 4B). The X-ray crystal structure of a PCP-C didomain construct from the tyrocidine synthetase provided the structural information for a C domain that acts in cis within an NRPS assembly line.12 Overall the structure is similar to the stand-alone VibH21. Using this structure as a guide, a mechanism of catalysis based on electrostatic stabilization of reaction intermediates, as opposed to the previously suggested acid/base catalysis by the histidines of the HHXXXD motif, has been proposed.27–29
One hallmark of NRPS pathways is the ability to incorporate D-amino acids into peptide natural products through α-carbon epimerization in the context of the assembly line. Epimerization (E) domains are homologous to C domains by sequence with similar active site motifs. The structure of an E domain from tyrocidine synthetase has been deposited into the PDB database (code 2XHG) and supports the prediction of structural homology between C and E domains (Fig. 4B).
2.4 Thioesterase domains
Thioesterase (TE) domains are often found at the C-terminus of an NRPS and catalyze release of the peptide from the assembly line through hydrolysis of the phosphopantetheinyl thioester to form a free acid or by cyclization using an internal nucleophile.30 TE domains are members of the large family of α,β-hydrolases containing an active site catalytic triad with serine acting as a nucleophile to cleave the thioester. The X-ray structure of the TE domain from the last module of surfactin synthetase confirmed an α,β-hydrolase fold with a catalytic triad consisting of Ser-His-Asp.31 The active site forms a large hydrophobic cleft to accommodate the lipopeptide substrate in a conformation that promotes head-to-tail cyclization. The structure also reveals a flexible ‘lid’ region covering the active site reminiscent of lipase structures.32 The structure of the TE domain from the fengycin NRPS has also been determined.33 This TE domain shares overall structural features with the surfactin thioesterase including a large active site cleft and a ‘lid’ region with conformational flexibility (Fig. 4C). In addition, as discussed in the next sections, thioesterase domain structures have been determined in the context of the termination domain of surfactin synthetase14 and a didomain fragment of enterobactin synthetase.15–16 Taken together, structural and biochemical studies suggest that the large, protected active site plays a key role in sequestering the reactive intermediate and controlling the timing and regiospecifcity of hydrolytic bond cleavage.
2.5 C-A-PCP-TE termination module
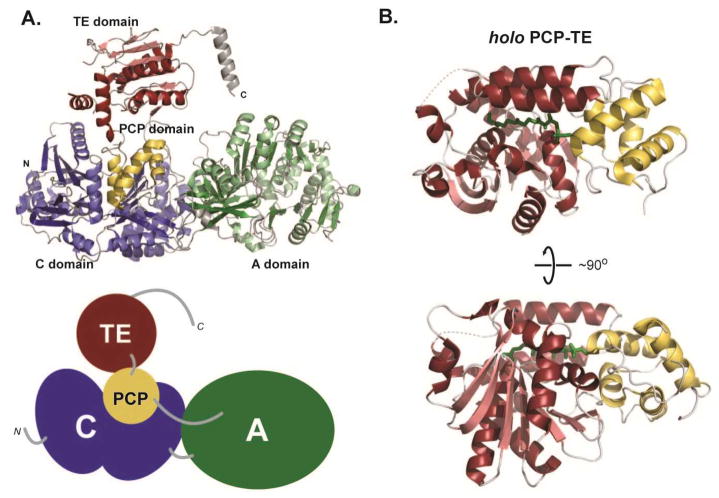
The termination module of surfactin synthetase is a 144 kDa four-domain protein responsible for the incorporation of the final amino acid (L-Leu) into the surfactin peptide and macrocyclization. In the 2.6 Å X-ray structure of the C-A-PCP-TE construct, the entire protein chain is evident in the electron density maps.14 The structural folds of the individual domains in this module are similar to structures of monomeric domains (Fig. 5A). The deviations observed in this multidomain structure include slight differences in the subdomains of both the C and A domains. The A domain contains bound L-Leu and is proposed to occupy a confirmation that occurs before ATP binding. The TE domain is very similar to the previously described ‘open’ state observed in the structure of this individual domain.31 The overall module architecture is dominated by the C domain and the N-terminal subdomain of the A domain. These two domains form an extensive interface that is likely invariant during the catalytic process. Both the PCP domain and the C-terminal subdomain of the A domain are adjacent to the C-A platform and appear to be more flexible. The large distance between the C and A domain active sites necessitates mobility of the smaller domains during the multiple steps of catalysis. The orientation of the PCP domain with respect to other domains suggests that the termination module crystallized in an orientation where the PCP domain interacts with the acceptor site of the C domain. The linker regions between domains also suggest that the C/A interaction is conformationally stable based on the well-ordered intervening segment. In contrast, the linkers between the A/PCP and PCP/TE domains, though shorter, appear less structured, suggesting flexibility. The X-ray structure also contains a serendipitous observation relevant to NRPS module/module interactions. A C-terminal helix that was not part of the synthetase, but that originated from the cloning vector, forms an interaction with the C-domain of an adjacent synthetase in the crystal lattice. Though not biologically relevant, this helix is homologous to predicted linkers present in non-terminal modules. Thus, the helical interaction could mimic the natural structural elements that link NRPS modules in trans.
Fig. 5.
Multidomain structures of NRPSs. (a) Structure of the four-domain termination module from surfactin synthetase, PDB code: 1VSQ, in ribbon representation along with a cartoon of the domain organization. (b) Two orientations of a didomain structure of PCP-TE with a phosphopantethiene analog loaded on to the PCP domain, PDB code: 1TEJ.
2.6 PCP-TE didomain structures
The NMR structure of the 37 kDa PCP-TE didomain construct from the E. coli enterobactin NRPS synthetase provides a detailed picture of the functional intra- and intermolecular interactions between NRPS domains.16 NMR analysis shows that the two domains form a compact structure with an extensive hydrophobic interface. Based on the orientation between the active site serine of each domain, the observed conformation appears to be catalytically relevant for PCP delivery of the substrate into the active site of the TE. As with the previously described structures of TEs,31,33 the enterobactin TE contains a dynamic ‘lid’ region consisting of two α-helices. Overall, the structure displays plasticity that allows the PCP domain to interact with protein partners in addition to the TE. Based on NMR measurements, a second conformation of the didomain construct was proposed with a more open structure. In addition, NMR titrations with the interacting partners (PPtase and the upstream C domain) showed that the presence of these domains modulate the PCP-TE didomain interactions. Recently, the X-ray structure of a similar enterobactin PCP-TE construct was solved by conjugating a phosphopantetheine analog designed to fix the mobility of the two domain fragment (Fig. 5B).15 The structure provides the first high resolution picture of an NRPS holo-carrier domain interacting with a partner enzyme.
3 Noncanonical chemistry in NRP pathways
The continued isolation of natural products and the identification of associated gene clusters have revealed novel structures and enzymology.34 In NRP pathways, unique building blocks, non-traditional synthetase chemistries, and elaborate tailoring strategies come together to create an ever-expanding array of complex metabolites with varied biological activity. The progress in this area exposes the remarkable diversity of biosynthetic machinery and highlights the potential to exploit these systems for bioengineering endeavors.1,2,35 The following highlights recent examples of noncanonical chemistry described in NRPS pathways.
3.1 Building block biosynthesis
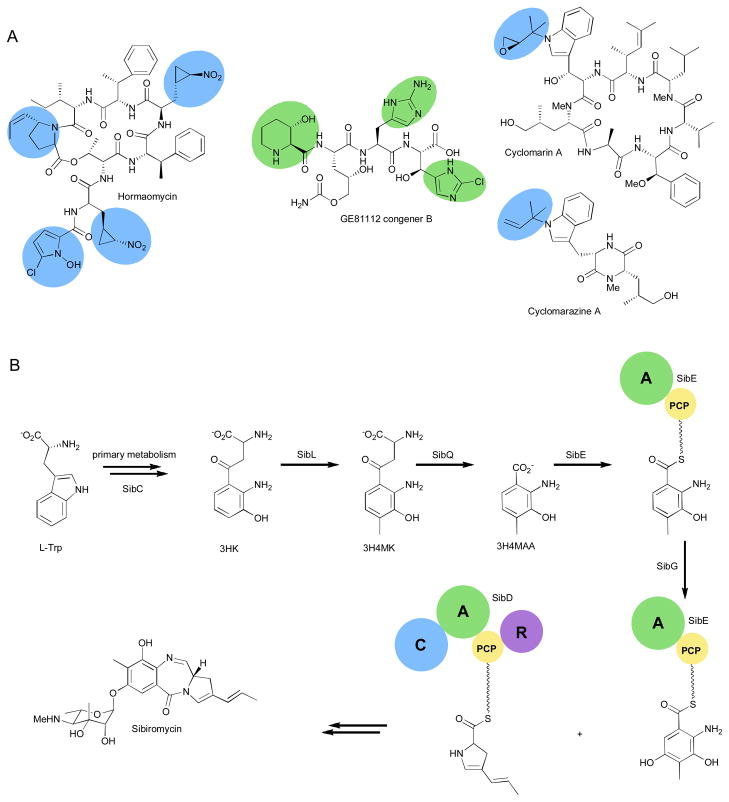
One hallmark of NRPS machinery is the utilization of diverse building blocks beyond the twenty common proteinogenic amino acids.36 The bacterial signaling metabolite, hormaomycin, is a prime example of the wide array of amino acids that can be incorporated into NRPs (Fig. 6A).37 Isolation and structure elucidation revealed several unique residues are incorporated into the depsipeptide.38–40 Precursor feeding studies, isolation of the gene cluster, and in vivo and in vitro investigations have uncovered the pathways to these unusual molecules.37 At least five enzymes are required to convert L-Pro to N-hydroxy-5-chloro-2-pyrrole carboxylic acid using a PCP tethered approach. The propenyl-pyrrolidine carboxylic acid is made from L-Tyr through a series of transformations homologous to enzymes in the pathways to tomaymycin, sibiromycin, and anthramycin.37 The particularly unique amino acid 4-nitrocyclopropyl alanine appears twice in the natural product. Feeding studies established that L-Lys is the biosynthetic precursor,41 but no genes could be attributed to the biosynthesis and further study of unassigned enzymes from the cluster may shed some light on the assembly of this building block.
Fig. 6.
Unusual building block incorporation. (a) Representative nonribosomal peptides with unusual building blocks. (b) Details of the biosynthesis of sibiromycin.
The pyrrolo[1,4]benzodiazepine, sibiromycin, is an antitumor antibiotic which contains a propenyl-pyrrole similar to hormaomycin.42 It also contains a highly substituted anthranilic acid moiety for which the biosynthesis has recently been elucidated (Fig. 6B).43,44 Feeding studies showed that L-Trp is the precursor45 which is first converted to L-kynurenine through primary metabolism and then to L-3-hydroxykynurenine (L-3HK) by SibC. SibL was identified as a SAM-dependent C-methyltranferase that accepts either D- or L-3HK and installs a methyl at C4 to form 3-hydroxy-4-methylkynurenine (3H4MK). Next, SibQ is a PLP-dependent kynureninase that converts 3H4MK to the corresponding anthranilic acid (3H4MAA) and alanine. The NRPS didomain SibE activates 3H4MAA forming the adenylate which is then loaded onto the SibE thiolation domain. The final step to the fully decorated anthranilic acid is accomplished by the FAD-dependent hydroxylase SibG that acts only on the PCP tethered 3H4MAA. The second NRPS enzyme, SibD, then selects, activates, and incorporates the propenyl-pyrrole that is then eliminated from the assembly line through reductive elimination, undergoes spontaneous ring closure, and is decorated with a unique sugar moiety by SibH to form the mature natural product.43 With just two NRPS enzymes and one tailoring enzyme the producing organism is able to create a highly active compound thanks to the complexity established by the unique building blocks.
Modifications to PCP-bound amino acid building blocks are a common occurrence among NRPS assemblies.46,47 The gene cluster of the GE81112 series of tetrapeptide natural products revealed more modules than amino acids present in the structure. In vivo studies showed that all the synthetases were active and the authors postulate that the seemingly extra A-PCP didomains are used to regulate building block biosynthesis, specifically 3-hydroxy-6-chlorohistidine biosynthesis. This natural product also contains 5-hydroxy-2-aminopentanoic acid for which the biosynthetic genes have not been identified and a hydroxyl-pipecolic acid derived from L-Lys.48 The timing of hydroxylation of these building blocks is not currently known and may happen as the building blocks are synthesized or after the peptide is released.49–52 Further studies of this pathway should help to assign roles to each of the genes in the cluster.
Although on assembly line modification are becoming common, there are still a fair amount of solution modifications being identified. For example, a prenyltransferase, CymD, from the cyclomarin/cyclomarazine pathways was recently characterized and shown to prenylate free tryptophan in solution. This feature allowed the group to create a CymD knockout mutant and conduct feeding studies with other modified residues to create cyclomarin analogues.53 On assembly versus solution modifications can be hard to predict. A fungal prenyltransferase was found to act mostly to remove prenyl groups from preformed peptides as apposed to those free in solution.54 Though these modifications can complicate biosynthetic studies, this variety in substrate recognition can aid in compiling a library of biosynthetic machinery for use in future engineering efforts.
3.2 Unusual assembly-line chemistry
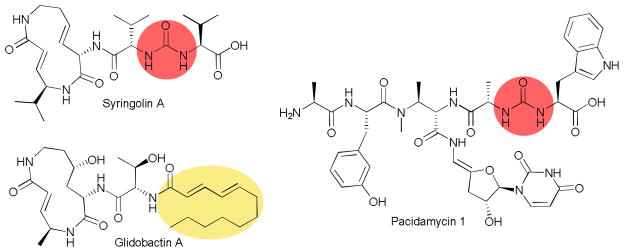
In most linear NRPS pathways, chain elongation is achieved through attack of the carbonyl of the tethered amino acid of one module by the free amine of the tethered amino acid on the next module. This assembly line chemistry results in a polarized peptide backbone. A few NRP natural products have been isolated and characterized that reverse the polarity by employing an ureido group.55–59 Biosynthetic studies of these unique compounds have revealed that the NRPS assembly line is responsible for creating the ureido linkage.60 In the case of the syrbactin proteasome inhibitor syringolin A, a single NRPS C-A-PCP module, SylC, is responsible for using two molecules of valine, ATP, and bicarbonate/CO2 to create the unique linkage (Fig. 7). From this data, two mechanisms were proposed. The adenylation mechanism has the first valine activated and tethered to the PCP domain, then the amine is carboxylated and the carboxyl group adenylated. Then the second valine can attack the activated carboxyl to form the ureido group and release AMP. The second mechanism is a cyclization mechanism where activation of the carboxylate is not required. On isolation of the gene cluster of a second NRP containing an ureido group, pacidamycin, two NRPS were found to be necessary to select two different amino acids to tether via the ureido linkage.61 This provides further evidence for the adenylation mechanism for ureido bond formation and exposes additional reactions that should be catalyzed within the C domain.
Fig. 7.
Selected nonribosomal peptides, highlighting unusual assembly-line chemistry.
Another syrbactin family member, glidobactin, has a similar structure and function, but lacks the ureido functionality. Instead, the N-terminus is acylated by a fatty acid moiety. In this case a C-A-PCP initiation module was described that is responsible for incorporating the fatty acid chain onto the amino termini of the first amino acid in the pathway. The C domain recognizes a variety of acyl-CoA donors and loads them to the free amine of the PCP domain tethered threonine residue. The acyl-threonine is then passed down the assembly line to complete the biosynthesis of the natural product.62 The work illustrates the potential of this system and related N-acyl initiation pathways to provide analogs for improved target specificity and activity.
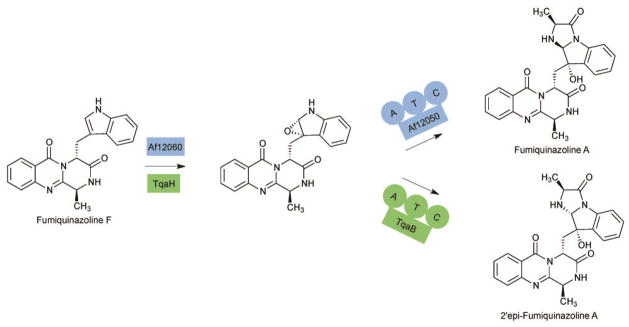
Several complex and unusual NRPS derived natural products have been isolated from fungi.63–69 The biosynthetic organization of fungal systems frequently differs from prokaryotic NRPSs in the chain termination strategies. Often the canonical TE domain is replaced with a terminal condensation, reduction, or thiolation domain.70,71 Though terminal condensation domains are present in many gene clusters, the mechanism for these enzymes hadn’t been established. Recently, two A-PCP-C modules have been described in the pathways to fumaquinazoline and tryptoquialanine that are responsible for the biosynthesis of a shared pathway intermediate that differs only in the stereochemistry at one carbon (Fig. 8). Fumiquinazoline F is first oxidized across the C2′-C3′ bond and then alkylated by the A-PCP-C module. Stereochemical control was at first postulated to be a result of the oxidation step, but swapping of the oxidases between pathways had no effect on the stereochemical outcome. However, by creating chimeric A-PCP-C modules, it has been shown that the C domains determine the final configuration at C2′. The C domain therefore must couple the loaded L-ala to the free oxidized Fumiquinazoline F and create a chiral center by amine addition to the C2′ position.72 Further biochemical and structural characterization of these two C domains could shed light onto how these enzymes can afford the two diastereomers.
Fig. 8.
Conversion of Fumiquinazoline F to Fumiquinazoline A and 2′-epi-Fumiquinazoline A by homologous oxidases and an A-T-C modules.
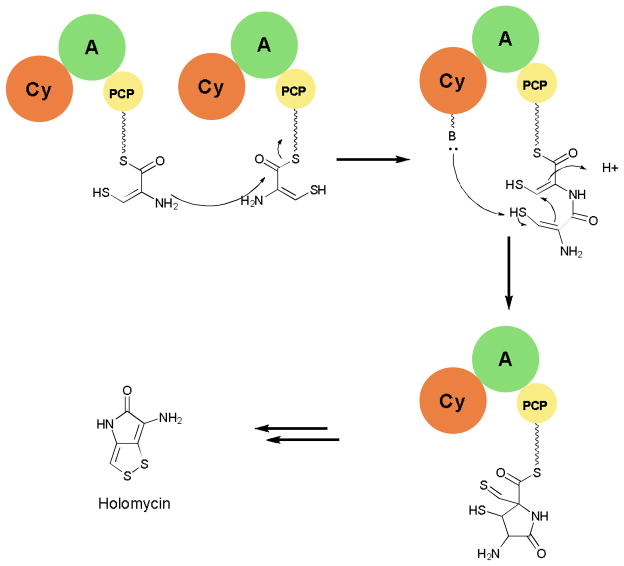
Another example of unusual C domain activity can be seen in the pathway of holomycin and related natural products. Here a tridomain module with the organization Cy-A-PCP plus an additional stand-alone C domain and a cysteine oxidase are proposed to be responsible for selecting, activating, oxidizing, coupling, and cyclizing two cysteine residues to yield a cyclodithiol-PCP-domain tethered intermediate (Fig. 9).73,74 Further TE catalyzed hydrolysis, decarboxylation, acylation, and cyclodisulfide bond formation reactions afford the active natural product.75 Further investigations of this pathway will give more evidence for the proposed roles of each domain during the biosynthesis and allow for better understanding of the biosynthesis of related natural products.
Fig. 9.
Reactions catalyzed by Cy-A-PCP module from the holomycin biosynthesis.
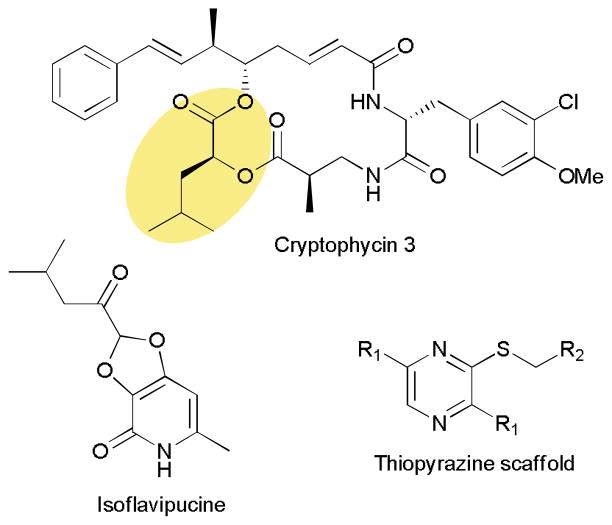
Modification of building blocks for NRPSs frequently occurs prior to selection and loading onto the synthetase. Epimerization domains are the main exception and are responsible for switching the stereochemistry at a single position.1,2,76 A wide variety of adenylation domains have been characterized, even those that activate D-amino acids so it is intriguing to find cases where on-assembly tailoring of the selected amino acid occurs.77–79 Biosynthetic studies of the anticancer agents cryptophycins revealed a dimodular NRPS, which in the second module contains a ketoreductase (KR) domain (Fig. 10).80 This module selects and loads 2-ketoisocaproic acid (2-KIC) onto the PCP domain, then the KR reduces the α-ketone to a hydroxyl forming 2-hydroxyisocaproic acid-PCP (2-HIC-PCP). This module in turn creates an ester bond while adding this last building block onto the growing chain. In vitro adenylation assays found that both 2-KIC and 2-HIC are activated and loaded equally well, but the biological availability of 2-HIC must be very low necessitating a dedicated reductase to perform this essential transformation.76 It is interesting to note that A domains that select for 2-hydroxy acids directly can be found in other pathways together with NADPH-dependent reductases to convert the 2-keto acid to the 2-hydroxy acid.81–84 Not only is this mechanism of on-assembly modification of note, but the modular organization allowed the group to use a chemoenzymatic approach to create a library of analogs exploiting the promiscuity of this module.80
Fig. 10.
Example peptidic natural products derived from noncanonical NRPS assembly lines.
Chemoenzymatic approaches are an attractive method to prepare target compounds that are otherwise difficult to synthesize using traditional synthetic chemistry.85 Module and domain swapping techniques have proven useful in creating several natural product analogs with potentially improved biological properties.72,86 During such an experiment, an NRP synthetase module was swapped from a hybrid PKS-NRPS assembly with unexpected results.87 Switching the NRPS module from preaspyridone synthetase with that of the cyclopiazonic acid pathway led to the creation of an analog with altered amino acid composition.87,88 When the NRPS module from the isoflavipucine pathway was used a thiopyrazine containing compound was detected (Fig. 10). Experiments revealed that this pair of NRPS and PKS enzymes did not interact and the NRPS alone was responsible for converting two equivalents of L-Leu into the thiopyrazine. Altering the amino acid and thiol donor allowed the group to create a library of 63 thiopyrazines in significant (up to 30 mg/L) quantities.89 Not only is the novel assembly line chemistry unusual, but it also differed from the in vivo function highlighting the potential of biosynthetic enzymes to be applied toward entering new chemical space.
3.3 Unusual Termination Strategies
Frequently, termination in NRP biosynthesis is carried out by a final C-A-PCP-TE module that loads the final amino acid onto the chain and then releases the chain from the assembly via hydrolysis or macrocyclization utilizing hydroxyl, amine, or thiol nucleopiles.1 Examples of alternative mechanisms of release are becoming more and more common with reductive elimination and macrocyclization using a carbon based nucleophile among the most prevalent.70,71 There have been extensive reviews related to this topic recently,90,91 so here we will only report the most recent and unique among the myriad of termination strategies employed in NRPS assembly lines.
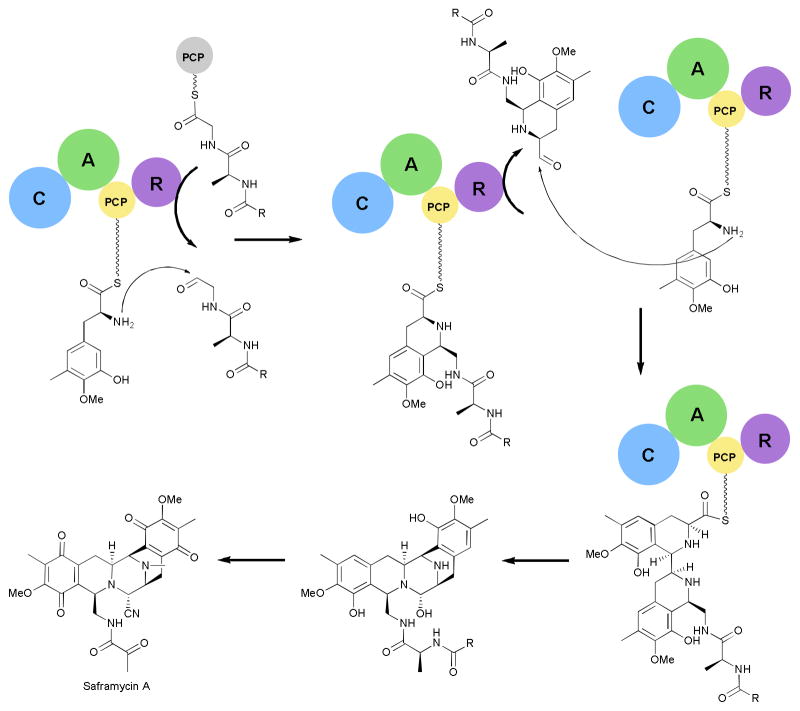
In vitro characterization of the termination module from the saframycin pathway revealed seven transformations that are accomplished by the single enzyme (Fig. 11).92 The C-A-PCP-R module selects and loads a tyrosine derivative onto the PCP domain.93 Then instead of the traditional peptide bond formation, the downstream peptide is reductively eliminated from the corresponding PCP domain to yield an aldehyde that is then coupled to the tyrosyl amine through a Pictet-Spengler reaction.92 Then the PCP-bound intermediate undergoes the second reductive elimination followed by a second Pictet-Spengler reaction with another tyrosine derivative. Finally, the C-A-PCP-R module catalyzes a final reductive elimination and intramolecular cyclization. Tailoring enzymes complete the transformation of the intermediate into the final natural product.94 This termination module not only contains a multifunctional reductase, but also a condensation domain capable of performing two Pictet-Spengler reactions. Further investigations into the biosynthesis of the saframycins may aid in the production of a related compound, ecteinascidin 743, which is a clinically approved anticancer compound that is currently produced by a semi-synthetic route.95
Fig. 11.
Saframycin biosynthetic pathway. The reductase (R) domain catalyzes three reductive eliminations while the condensation domain performs two Pictet-Spengler reactions. R=C13H27.
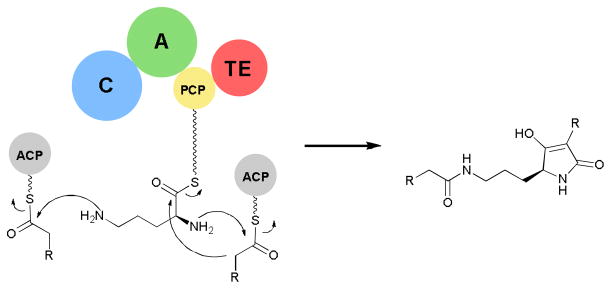
Although C-A-PCP-TE termination modules are common, new chain termination chemistries beside hydrolysis and macrocyclization are possible. Gene cluster analysis of the tetramic acid containing macrolactam, HSAF, revealed a single hybrid PKS/NRPS gene.96 In vitro analysis of the excised NRPS enzyme revealed an ornithine specific A domain. Incubations of the ornithine loaded NRPS with steroyl-ACP and subsequent MS analysis revealed the NRPS module alone was sufficient to acylate both amines of ornithine and form the tetramic acid core scaffold via a Dieckmann-type cyclization (Fig. 12). Further studies to probe the mechanism of this reaction sequence may provide a useful tetramic acid producing enzyme scaffold to use for further bioengineering of tetramic acid products.
Fig. 12.
Reactions catalyzed by single C-A-PCP-TE module during biosynthesis of the tetramic acid, HSAF. R=long chain alkyl.
3.4 Post-assembly line tailoring chemistry
Tailoring of NRPs can occur before, during, and after assembly into the complete peptide chain.97 The section on starter unit biosynthesis highlights some recent examples of interesting tailoring enzymology that happens before the assembly line. Some transformations towards a mature natural product occur while the growing chain is attached to the assembly line.80 The reasoning for this timing isn’t always clear and determining the exact order of reactions can often be challenging. During dapdiamide E antibiotic biosynthesis, an epoxide is introduced partway through the biosynthesis.98 Given the structure of these compounds and the reactivity of epoxides in general, it makes sense that epoxidation would happen later on in the biosynthesis to avoid any interactions with nucleophilic groups that may cause ring opening. Biosynthetic studies showed that first fumarate and 2,3-diaminopropanoate are coupled by the stand-alone C domain, DdaG. Then the peptide is loaded onto the A-PCP didomain, DdaD, which tethers the molecule for epoxidation by the Fe(II)/α-ketogluterate dependent dioxygenase, DdaC. Removal by a TE domain and coupling to valine by a second stand-alone C domain complete the biosynthesis. Given that this system consists more of solution phase reactions than assembly line steps it is odd that DdaC only acts on a carrier protein tethered intermediate. It is possible that this configuration is due to the evolutionary origin of these enzymes perhaps gathered from other pathways over time.
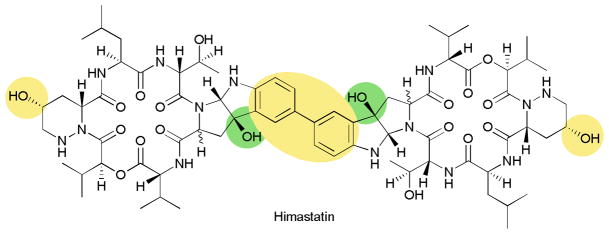
NRPS derived peptides are often subject to a wide range of ‘tailoring’ transformations including oxidations, methylations, acylations and glycosylations.97,99,100 As one recent example, after the release of the peptide during himastatin biosynthesis, three P450s are responsible for creating the mature natural product.101 First, a Trp is oxidized to a pyrroloindole moiety, then a second P450 hydroxylates the D-Pip, finally the third P450 creates the aryl-aryl linkage that gives the natural product its potency (Fig. 13). Tailoring can be very important for selectivity and activity of natural products and deserves further investigation. Though the biochemical studies of such enzymes can often be daunting, a greater understanding of these systems can be invaluable towards the future of bioengineering.
Fig. 13.
Structure of Himastatin, highlighting the oxidative tailoring.
4 Conclusions
The macromolecular machinery responsible for the biosynthesis of nonribosomal peptide natural products performs complex and often unique chemistry. NMR and X-ray crystallographic structural analyses of NRPS domains and multidomain fragments have provided insight into the details of macromolecular organization and chemistry. The diversity of chemistry found in the biosynthetic pathways to nonribosomal peptides continues to expand. Spurred by widespread genomic sequencing efforts, novel and unique biosynthetic gene clusters are continually being described and characterized. A notable area of expansion in recent years is in the areas of fungal and eukaryotic pathways. Further investigations into the structure and chemistry of NRPS systems will expand the knowledge of complex enzyme catalyzed chemistry and aid efforts to exploit biosynthetic machinery toward the production of desired small molecules.
Acknowledgments
Work in the S.D.B. laboratory on NRPS biosynthesis is supported by the NIH (GM086570) and NSF (CAREER-0645653).
References
- 1.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 2.Koglin A, Walsh CT. Nat Prod Rep. 2009;26:987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finking R, Marahiel MA. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CT. Acc Chem Res. 2007;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 6.Hertweck C. Angew Chem Int Ed Engl. 2009;48:4688–716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 7.Leibundgut M, Maier T, Jenni S, Ban N. Curr Opin Struct Biol. 2008;18:714–725. doi: 10.1016/j.sbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Walsh CT, Gehring AM, Weinreb PH, Quadri LE, Flugel RS. Curr Opin Chem Biol. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 9.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 10.Freist W, Cramer F. Angew Chem Int Ed Engl. 2003;32:190–200. [Google Scholar]
- 11.Weber T, Baumgartner R, Renner C, Marahiel MA, Holak TA. Structure. 2000;8:407–418. doi: 10.1016/s0969-2126(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 12.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanovic A, Samel SA, Essen LO, Marahiel MA. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Zheng T, Bruner SD. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dotsch V. Science. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 17.Conti E, Stachelhaus T, Marahiel MA, Brick P. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti E, Franks NP, Brick P. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 19.Stachelhaus T, Mootz HD, Marahiel MA. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 20.Challis GL, Ravel J, Townsend CA. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 21.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May JJ, Kessler N, Marahiel MA, Stubbs MT. Proc Natl Acad Sci USA. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drake EJ, Duckworth BP, Neres J, Aldrich CC, Gulick AM. Biochemistry. 2010;49:9292–9305. doi: 10.1021/bi101226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TV, Johnson LJ, Johnson RD, Koulman A, Lane GA, Lott JS, Arcus VL. J Biol Chem. 2010;285:2415–2427. doi: 10.1074/jbc.M109.071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie AG. J Mol Biol. 1990;213:167–186. doi: 10.1016/S0022-2836(05)80129-9. [DOI] [PubMed] [Google Scholar]
- 26.Keating TA, Marshall CG, Walsh CT, Keating AE. Nat Struct Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 27.Lewendon A, Murray IA, Kleanthous C, Cullis PM, Shaw WV. Biochemistry. 1988;27:7385–7390. doi: 10.1021/bi00419a032. [DOI] [PubMed] [Google Scholar]
- 28.Stachelhaus T, Walsh CT. Biochemistry. 2000;39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 29.Marshall CG, Hillson NJ, Walsh CT. Biochemistry. 2002;41:244–250. doi: 10.1021/bi011852u. [DOI] [PubMed] [Google Scholar]
- 30.Kohli RM, Walsh CT. Chem Comm. 2003;7:297–307. doi: 10.1039/b208333g. [DOI] [PubMed] [Google Scholar]
- 31.Bruner SD, Weber T, Kohli RM, Schwarzer D, Marahiel MA, Walsh CT, Stubbs MT. Structure. 2002;10:301–310. doi: 10.1016/s0969-2126(02)00716-5. [DOI] [PubMed] [Google Scholar]
- 32.Cygler M, Schrag JD, Sussman JL, Harel M, Silman I, Gentry MK, Doctor BP. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samel SA, Wagner B, Marahiel MA, Essen LO. J Mol Biol. 2006;359:876–889. doi: 10.1016/j.jmb.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 34.Sieber SA, Marahiel MA. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 35.Pickens LB, Tang Y, Chooi YH. Annu Rev Chem Biomol Eng. 2011;2:211–236. doi: 10.1146/annurev-chembioeng-061010-114209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh CT, Fischbach MA. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofer I, Crusemann M, Radzom M, Geers B, Flachshaar D, Cai X, Zeeck A, Piel J. Chem Biol. 2011;18:381–391. doi: 10.1016/j.chembiol.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Omura S, Mamada H, Wang NJ, Imamura N, Oiwa R, Iwai Y, Muto N. J Antibiot (Tokyo) 1984;37:700–705. doi: 10.7164/antibiotics.37.700. [DOI] [PubMed] [Google Scholar]
- 39.Rössner E, Zeeck A, König WA. Angew Chem Int Ed Eng. 1990;29:64–65. [Google Scholar]
- 40.Zlatopolskiy BD, Radzom M, Zeeck A, de Meijere A. Eur J Org Chem. 2006;2006:1525–1534. [Google Scholar]
- 41.Brandl M, Sergei Kozhushkov I, Boris Zlatopolskiy D, Alvermann P, Geers B, Zeeck A, de Meijere A. Eur J Org Chem. 2005;2005:123–135. [Google Scholar]
- 42.Parker KA, Babine RE. Tet Lett. 1982;23:1763–1766. [Google Scholar]
- 43.Giessen TW, Kraas FI, Marahiel MA. Biochemistry. 2011;50:5680–5692. doi: 10.1021/bi2006114. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Khullar A, Chou S, Sacramo A, Gerratana B. Appl Environ Microbiol. 2009;75:2869–2878. doi: 10.1128/AEM.02326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurley LH. Acc Chem Res. 1980;13:263–269. [Google Scholar]
- 46.Galm U, Wendt-Pienkowski E, Wang L, George NP, Oh TJ, Yi F, Tao M, Coughlin JM, Shen B. Mol Biosyst. 2009;5:77–90. doi: 10.1039/b814075h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaillancourt FH, Vosburg DA, Walsh CT. ChemBioChem. 2006;7:748–752. doi: 10.1002/cbic.200500480. [DOI] [PubMed] [Google Scholar]
- 48.Binz TM, Maffioli SI, Sosio M, Donadio S, Muller R. J Biol Chem. 2010;285:32710–32719. doi: 10.1074/jbc.M110.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin X, Zabriskie TM. ChemBioChem. 2004;5:1274–1277. doi: 10.1002/cbic.200400082. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Thomas MG, O’Connor SE, Hubbard BK, Burkart MD, Walsh CT. Biochemistry. 2001;40:11651–11659. doi: 10.1021/bi0115434. [DOI] [PubMed] [Google Scholar]
- 51.Buntin K, Rachid S, Scharfe M, Blöcker H, Weissman KJ, Müller R. Angew Chem Int Ed Engl. 47:4595–4599. doi: 10.1002/anie.200705569. [DOI] [PubMed] [Google Scholar]
- 52.Samel SA, Marahiel MA, Essen LO. Mol Biosyst. 2008;4:387–393. doi: 10.1039/b717538h. [DOI] [PubMed] [Google Scholar]
- 53.Schultz AW, Lewis CA, Luzung MR, Baran PS, Moore BS. J Nat Prod. 2010;73:373–377. doi: 10.1021/np9006876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou H, Zheng X, Li SM. J Nat Prod. 2009;72:44–52. doi: 10.1021/np800501m. [DOI] [PubMed] [Google Scholar]
- 55.Harada K-I, Mayumi T, Shimada T, Suzuki M, Kondo F, Watanabe MF. Tet Lett. 1993;34:6091–6094. [Google Scholar]
- 56.Walther T, Renner S, Waldmann H, Arndt H-D. ChemBioChem. 2009;10:1153–1162. doi: 10.1002/cbic.200900035. [DOI] [PubMed] [Google Scholar]
- 57.Chem RH, Buko AM, Whittern DN, McAlpine JB. J Antibiot. 1989;42:512–520. doi: 10.7164/antibiotics.42.512. [DOI] [PubMed] [Google Scholar]
- 58.Isomo F, Sakaida Y, Takahashi S, Kinoshita T, Nakamura T, Inukai M. J Antibiot. 1993;46:1203–1207. doi: 10.7164/antibiotics.46.1203. [DOI] [PubMed] [Google Scholar]
- 59.Chatterjee S, Nadkarni SR, Vijaykumar EK, Patel MV, Ganguli BN, Fehlhaber HW, Vertesy L. J Antibiot. 1994;47:595–598. doi: 10.7164/antibiotics.47.595. [DOI] [PubMed] [Google Scholar]
- 60.Imker HJ, Walsh CT, Wuest WM. J Am Chem Soc. 2009;131:18263–18265. doi: 10.1021/ja909170u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang W, Ostash B, Walsh CT. Proc Natl Acad Sci USA. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imker HJ, Krahn D, Clerc J, Kaiser M, Walsh CT. Chem Biol. 2010;17:1077–1083. doi: 10.1016/j.chembiol.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daniel JFdS, Rodrigues Filho E. Nat Prod Rep. 2007;24:1128–1141. doi: 10.1039/b618086h. [DOI] [PubMed] [Google Scholar]
- 64.Pruksakorn P, Arai M, Kotoku N, Vilchèze C, Baughn AD, Moodley P, Jacobs WR, Jr, Kobayashi M. Bioorg Med Chem Lett. 2010;20:3658–3663. doi: 10.1016/j.bmcl.2010.04.100. [DOI] [PubMed] [Google Scholar]
- 65.Peltola J, Ritieni A, Mikkola R, Grigoriev PA, Pócsfalvi G, Andersson MA, Salkinoja-Salonen MS. Appl Environ Microbiol. 2004;70:4996–5004. doi: 10.1128/AEM.70.8.4996-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao S, Smith KS, Deveau AM, Dieckhaus CM, Johnson MA, Macdonald TL, Cook JM. J Med Chem. 2002;45:1559–1562. doi: 10.1021/jm0155953. [DOI] [PubMed] [Google Scholar]
- 67.Rabindran SK, Ross DD, Doyle LA, Yang W, Greenberger LM. Cancer Res. 2000;60:47–50. [PubMed] [Google Scholar]
- 68.Cole RJ. J Food Prot. 1981;44:715–722. doi: 10.4315/0362-028X-44.9.715. [DOI] [PubMed] [Google Scholar]
- 69.Valdes JJ, Cameron JE, Cole RJ. Environ Health Perspect. 1985;62:459–463. doi: 10.1289/ehp.8562459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eisfeld K. Physiology and Genetics. Vol. 15. Springer; Berlin, Heidelberg: 2009. pp. 305–330. [Google Scholar]
- 71.Cramer RA, Jr, Stajich JE, Yamanaka Y, Dietrich FS, Steinbach WJ, Perfect JR. Gene. 2006;383:24–32. doi: 10.1016/j.gene.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 72.Haynes SW, Ames BD, Gao X, Tang Y, Walsh CT. Biochemistry. 2011;50:5668–5679. doi: 10.1021/bi2004922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang S, Zhao Y, Qin Z, Wang X, Onega M, Chen L, He J, Yu Y, Deng H. Process Biochem. 2011;46:811–816. [Google Scholar]
- 74.Li B, Walsh CT. Proc Natl Acad Sci U S A. 2010;107:19731–19735. doi: 10.1073/pnas.1014140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B, Walsh CT. Biochemistry. 2011;50:4615–4622. doi: 10.1021/bi200321c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stein DB, Linne U, Hahn M, Marahiel MA. ChemBioChem. 2006;7:1807–1814. doi: 10.1002/cbic.200600192. [DOI] [PubMed] [Google Scholar]
- 77.Tang G-L, Cheng Y-Q, Shen B. J Biol Chem. 2007;282:20273–20282. doi: 10.1074/jbc.M702814200. [DOI] [PubMed] [Google Scholar]
- 78.Scott-Craig JS, Panaccione DG, Pocard JA, Walton JD. J Biol Chem. 1992;267:26044–26049. [PubMed] [Google Scholar]
- 79.Weber G, Schörgendorfer K, Schneider-Scherzer E, Leitner E. Curr Genet. 1994;26:120–125. doi: 10.1007/BF00313798. [DOI] [PubMed] [Google Scholar]
- 80.Ding Y, Rath CM, Bolduc KL, Hakansson K, Sherman DH. J Am Chem Soc. 2011;133:14492–14495. doi: 10.1021/ja204716f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Y, Orozco R, Kithsiri Wijeratne EM, Espinosa-Artiles P, Leslie Gunatilaka AA, Patricia Stock S, Molnár I. Fungal Genet Biol. 2009;46:353–364. doi: 10.1016/j.fgb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 82.Xu Y, Orozco R, Wijeratne EMK, Gunatilaka AAL, Stock SP, Molnár I. Chem Biol. 2008;15:898–907. doi: 10.1016/j.chembiol.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 83.Haese A, Schubert M, Herrmann M, Zocher R. Mol Microbiol. 1993;7:905–914. doi: 10.1111/j.1365-2958.1993.tb01181.x. [DOI] [PubMed] [Google Scholar]
- 84.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. Proc Natl Acad Sci USA. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giessen TW, Marahiel MA. FEBS Lett. 2012 doi: 10.1016/j.febslet.2012.01.017. (In Press http://dx.doi.org/10.1016/j.febslet.2012.01.017) [DOI] [PubMed]
- 86.Stachelhaus T, Schneider A, Marahiel M. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 87.Xu W, Cai X, Jung ME, Tang Y. J Am Chem Soc. 2010;132:13604–13607. doi: 10.1021/ja107084d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Walsh CT. Biochemistry. 2009;48:8746–8757. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qiao K, Zhou H, Xu W, Zhang W, Garg N, Tang Y. Org Lett. 2011;13:1758–1761. doi: 10.1021/ol200288w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kopp F, Marahiel MA. Nat Prod Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- 91.Du L, Lou L. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 92.Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Nat Chem Biol. 2010;6:408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- 93.Mikami Y, Takahashi K, Yazawa K, Arai T, Namikoshi M, Iwasaki S, Okuda S. J Biol Chem. 1985;260:344–348. [PubMed] [Google Scholar]
- 94.Li L, Deng W, Song J, Ding W, Zhao Q-F, Peng C, Song W-W, Tang G-L, Liu W. J Bacteriol. 2008;190:251–263. doi: 10.1128/JB.00826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cuevas C, Francesch A. Nat Prod Rep. 2009;26:322–337. doi: 10.1039/b808331m. [DOI] [PubMed] [Google Scholar]
- 96.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 98.Hollenhorst MA, Bumpus SB, Matthews ML, Bollinger JM, Jr, Kelleher NL, Walsh CT. J Am Chem Soc. 2010;132:15773–15781. doi: 10.1021/ja1072367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dimise EJ, Widboom PF, Bruner SD. Proc Natl Acad Sci USA. 2008;105:15311–15316. doi: 10.1073/pnas.0805451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Proc Natl Acad Sci USA. 2009;106:92549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma J, Wang Z, Huang H, Luo M, Zuo D, Wang B, Sun A, Cheng YQ, Zhang C, Ju J. Angew Chem Int Ed Engl. 2011;50:7797–7802. doi: 10.1002/anie.201102305. [DOI] [PubMed] [Google Scholar]